Abstract
NAD(P)H:rubredoxin oxidoreductase (NROR) has been purified from the hyperthermophilic archaeon Pyrococcus furiosus. The enzyme is exceedingly active in catalyzing the NADPH-dependent reduction of rubredoxin, a small (5.3-kDa) iron-containing redox protein that had previously been purified from this organism. The apparent Vmax at 80°C is 20,000 μmol/min/mg, which corresponds to a kcat/Km value of 300,000 mM−1 s−1. The apparent Km values measured at 80°C and pH 8.0 for rubredoxin, NADPH, and NADH were 50, 5, and 34 μM, respectively. The enzyme did not reduce P. furiosus ferredoxin. NROR is a monomer with a molecular mass of 45 kDa and contains one flavin adenine dinucleotide molecule per mole but lacks metals and inorganic sulfide. The possible physiological role of this hyperactive enzyme is discussed.
Pyrococcus furiosus is a member of the hyperthermophilic archaea, microorganisms that thrive at extreme temperatures and inhabit shallow and deep-sea volcanic environments (23, 24). It is an obligate anaerobe and grows optimally at 100°C by the fermentation of carbohydrates and peptides to organic acids, CO2 and H2. If elemental sulfur (S0) is present in the medium, it is reduced to H2S. The primary electron acceptor for these oxidative fermentative pathways is the small (7.5-kDa), iron-sulfur-containing redox protein ferredoxin (1). The oxidation of reduced ferredoxin is thought to be coupled to H2 production and S0 reduction via ferredoxin:NADP oxidoreductase and sulfhydrogenase (12, 15). Sulfhydrogenase is a bifunctional enzyme which catalyzes the oxidation of NADPH and the reduction of protons and S0 to H2 and H2S, respectively (15).
A second small, iron-containing redox protein termed rubredoxin (∼5.3 kDa) has also been purified from P. furiosus (3). Rubredoxin from several species of anaerobic bacteria has been characterized (11), but its physiological function is not clear. Neither sulfhydrogenase or ferredoxin:NADP oxidoreductase efficiently reduces rubredoxin with NADH or NADPH as the electron donor (12, 15). An intriguing question, therefore, is whether P. furiosus contains an NAD(P)H-dependent enzyme which specifically uses rubredoxin as an electron acceptor. Herein the purification and properties of such an enzyme, which is termed NAD(P)H:rubredoxin oxidoreductase (NROR), are described.
Purification of NROR.
NROR activity was determined anaerobically by the NADPH-dependent reduction of P. furiosus rubredoxin at 494 nm (molar absorptivity of 9,220 M−1 cm−1 [3]) at 80°C. The reaction mixture (2.0 ml) contained 100 mM EPPS [N-(2-hydroxyethyl)-piperazine-N′-(3-propanesulfonic acid)] buffer (pH 8.0), NADPH (0.3 mM), and rubredoxin (10 μM). As determined by this assay, the specific activity of cell extracts from six different batches of P. furiosus cells was 2.2 ± 0.8 μmol of rubredoxin reduced/min/mg. The enzyme appeared to be located in the cytoplasm, since after ultracentrifugation (110,000 × g for 2 h) of the cell extract, more than 90% of the activity was in the supernatant fraction. This also contained 83% of the cellular glutamate dehydrogenase activity, a known cytoplasmic enzyme (14, 21). From a small-scale purification of NROR by using the NADPH-dependent reduction of rubredoxin to measure its activity, it was found that the pure enzyme also catalyzed the NADH-dependent reduction of benzyl viologen (BV). In this case the assay mixture (2.0 ml) contained 50 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] buffer (pH 10.2), NADH (0.3 mM), and BV (1 mM). A molar absorptivity of 7,800 M−1 cm−1 was used for reduced BV. This NADH:BV oxidoreductase assay was used for the large-scale purification of NROR, where 1 U of activity equals 1 μmol of NADH oxidized/min/mg.
P. furiosus (DSM 3638) was routinely grown at 90°C in a 600-liter fermentor with maltose as the carbon source as described previously (4). NROR was purified at 23°C under strictly anaerobic conditions (4). Frozen cells (400 g [wet weight]) were thawed in 1.5 liters of buffer A (50 mM Tris-HCl [pH 8.0] containing 10% [vol/vol] glycerol, 2 mM dithiothreitol, and 2 mM sodium dithionite) containing DNase I (10 μg/ml) and were lysed by incubation at 35°C for 2 h. A cell extract was obtained by centrifugation at 50,000 × g for 80 min. The supernatant (1.3 liters) was loaded onto a column (8 by 21 cm) of DEAE-Sepharose Fast Flow (Pharmacia LKB, Piscataway, N.J.) equilibrated with buffer A. The column was eluted with a linear gradient (9.0 liters) from 0 to 0.5 M NaCl in buffer A, and 90-ml fractions were collected. Elution of NROR activity started as 0.2 M NaCl was applied to the column. Those fractions containing NROR activity were combined (810 ml), concentrated by ultrafiltration (type PM-30 membrane; Amicon, Beverly, Mass.), and washed with buffer B. Buffer B was the same as buffer A except that sodium dithionite was omitted. The concentrated sample (150 ml) was applied to a column (5 by 12 cm) of blue Sepharose (Pharmacia LKB) equilibrated with buffer B. The column was eluted with a linear gradient (1.4 liters) of 0 to 2.0 M NaCl in buffer B, and 50-ml fractions were collected. Elution of NROR activity started as 1.6 M NaCl was applied. Those fractions containing NROR activity were combined (600 ml), concentrated to 30 ml by ultrafiltration (PM-30 membrane), and applied to a column (6 by 60 cm) of Superdex 200 (Pharmacia LKB) equilibrated with buffer B containing 50 mM KCl. Fractions of 25 ml were collected. Those containing NROR activity were combined (50 ml) and applied to a column (2.6 by 10 cm) of Q-Sepharose (high performance; Pharmacia LKB) equilibrated with buffer B. The column was eluted with a gradient (0.5 liter) of 0 to 1.0 M KCl in buffer B, and 20-ml fractions were collected. Elution of NROR activity started as 0.3 M KCl was applied. Those fractions judged to be homogeneous on the basis of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (10) were combined (80 ml), concentrated by ultrafiltration (PM-30 membrane), and stored as pellets in liquid N2.
The results of a typical purification are given in Table 1. Three peaks of NADH-dependent BV reduction activity were separated after chromatography on the blue Sepharose column (data not shown), but only data for the peak corresponding to NROR, which accounted for ∼40% of the total BV reduction activity, are shown in Table 1. One of the other two peaks from this column contained ferredoxin:NADP oxidoreductase, which also catalyzes the NADH-dependent reduction of BV (12), but the nature of the enzyme responsible for the third peak of NADH:BV oxidoreductase activity is not known. The specific activities of pure NROR were 490 μmol of NADH oxidized/min/mg in the NADH:BV oxidoreductase assay (Table 1) and 2,750 μmol of NADPH oxidized/min/mg in the NADPH:rubredoxin assay. In the latter assay, the enzyme was purified 1,830-fold, with a recovery of activity (NROR) of about 47% relative to that of the cell extract (1.5 U/mg).
TABLE 1.
Purification of NROR from P. furiosus
| Step | Amt of protein (mg) | Activity (U) | Sp act (U/mg) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 38,300 | 28,700 | 0.7 | 100 | 1 |
| DEAE-Sepharose Fast Flow | 7,400 | 26,200 | 3.5 | 92 | 5 |
| Blue Sepharose | 145 | 15,700 | 110 | 55 | 140 |
| Superdex 200 | 16 | 6,630 | 420 | 23 | 560 |
| Q-Sepharose (high performance) | 9.8 | 4,800 | 490 | 17 | 650 |
Biophysical properties of NROR.
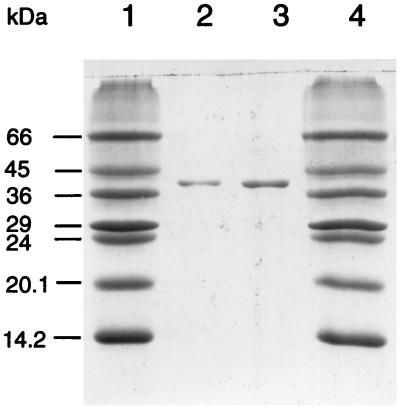
SDS-polyacrylamide gel electrophoresis (10) of purified NROR showed only one protein band, and this had a mass of 45,000 Da (Fig. 1). By using a calibrated column (1.6 by 60 cm) of Superdex 200 (Pharmacia LKB) equilibrated with 50 mM Tris-HCl buffer (pH 8.0) containing KCl (200 mM), NROR was eluted with a mass of 51,000 ± 5,000 Da. Therefore, the protein appears to be monomeric. The N-terminal amino acid sequence, MKVVIVGNGPGGFELAKQLSQTYEV, was determined after electroblotting the enzyme from an SDS gel onto a polyvinylidene difluoride protein sequencing membrane (see, for example, reference 13). Database searches indicated that this sequence in NROR bound flavin since it showed similarity to the sequences of such domains present in NADH dehydrogenase of Escherichia coli (25) and NADH oxidase (nox-1 and nox-2) of Archaeoglobus fulgidus (9). The presence of a flavin chromaphore was also suggested by the UV-visible absorption spectra of the oxidized (as purified) NROR. This exhibited peaks near 375 and 450 nm which decreased in intensity upon the addition of sodium dithionite or NADPH (data not shown). As determined by using a molar absorptivity of 11,300 M−1 cm−1 at 450 nm (2), the holoenzyme contains one flavin moiety per mole, while plasma emission spectroscopy (13) showed the presence of 1.8 g-atoms of P per mol of enzyme. These data suggest that NROR contains one flavin adenine dinucleotide (FAD) molecule per mole. This was confirmed by thin-layer chromatography analysis of the acid-extracted cofactor which migrated at the same position as that of FAD (data not shown). Mass spectroscopy analyses, carried out by the Mass Spectrometry Facility at the University of Georgia, revealed that the flavin cofactor had a mass of 786 Da, which corresponds to that of FAD. No metals could be detected NROR (>0.05 atoms/mol) as measured by plasma emission spectroscopy, while chemical analysis (20) showed that it contained two cysteinyl residues. The enzyme was thermostable, with times required for a 50% loss in catalytic activity (with 0.4 mg/ml in 50 mM EPPS, pH 8.0) at 80 and 95°C of about 12 and 2 h, respectively.
FIG. 1.
SDS–12.5% polyacrylamide gel electrophoresis of P. furiosus NROR. Lanes 1 and 4 contain molecular mass markers. Lanes 2 and 3 contain 1 and 2 μg of NROR, respectively.
Catalytic properties of NROR.
Purified NROR catalyzed the NADPH-dependent reduction of various electron carriers. These included (where 100% activity equals 51 μmol reduced/min/mg at 50°C with the indicated concentration of electron carrier) methyl viologen (6%, 1 mM), benzyl viologen (100%, 1 mM), FAD (42%, 150 μM), flavin mononucleotide (44%, 150 μM), menadione (72%, 200 μM), 2,6-dichlorophenol indophenol (365%, 100 μM), cytochrome c (8%, 50 μM), and iron (III) citrate (2%, 100 μM). P. furiosus ferredoxin (40 μM) was not utilized as an electron acceptor by NROR with either NADPH or NADH as the electron donor. The most efficient electron acceptor was rubredoxin (1,160%, 9.5 μM). The kinetics obtained with this protein are summarized in Table 2. The apparent Vmax at 80°C was approximately 20,000 μmol/min/mg, which corresponds to a kcat/Km value of 300,000 mM−1 s−1. These data suggest that rubredoxin is the physiological substrate of NROR, while NADPH (Km, 5 μM) rather than NADH (Km, 34 μM) is the preferred electron donor (Table 2). Surprisingly, NROR exhibited high catalytic activity even at a low temperature. For example, the apparent Vmax at 25°C was approximately 300 μmol/min/mg. However, as shown in Table 2, upon an increase in the temperature to 80°C, the apparent Km increased fivefold and the apparent Vmax increased almost 2 orders of magnitude. Table 2 also shows that NROR catalyzed the NADPH-dependent reduction of DTNB (5,5′-dithiobis[2-nitrobenzoic acid]) (45%, 2.5 mM) at 80°C, indicating that this enzyme can function as a disulfide reductase (8). The pH optima for the reduction of DTNB and rubredoxin were similar (7.6 and 7.0, respectively). NROR also functioned as an NADPH peroxidase and catalyzed the NADPH-dependent reduction of peroxide (Table 2).
TABLE 2.
Kinetics of NROR from P. furiosus
| Substrate (concn range, mMa) | Cosubstrate (concn, mM) | Apparent Km (mM) | Apparent Vmax (U/mg) | kcat (s−1) | kcat/Km(mM−1 s−1) |
|---|---|---|---|---|---|
| NADH (0.03–0.2) | BV (1.0) | 0.034 | 472 | 354 | 10,400 |
| NADPH (0.01–0.2) | BV (1.0) | 0.005 | 455 | 341 | 68,300 |
| BV (0.4–2.0) | NADH (0.3) | 1.25 | 1,000 | 750 | 600 |
| DTNB (0.05–2.5) | NADPH (0.3) | 0.25 | 25 | 19 | 75 |
| H2O2 (5–55) | NADPH (0.3) | 9.5 | 12 | 9 | 1 |
| Rubredoxinb (0.0013–0.0075) | NADPH (0.3) | 0.01 | 294 | 221 | 22,050 |
| Rubredoxin (0.0013–0.0075) | NADPH (0.3) | 0.05 | 20,000 | 15,000 | 300,000 |
Concentration range used to determine kinetic values.
The activity was determined at 25°C. All other assays were performed at 80°C.
Physiological role of NROR.
While the reduction by NROR of both a disulfide (DTNB) and a peroxide occurred at significant rates at 80°C (>10 μmol reduced/min/mg), these rates are about 3 orders of magnitude less than the rate of rubredoxin reduction under the same conditions (Table 2). Therefore, it seems unlikely that the primary role of NROR in vivo is either as an NADPH peroxidase or as a disulfide reductase. The measured NADPH peroxidase activity probably occurred by a nonspecific reaction, perhaps with the flavin group, but DTNB-dependent disulfide reductase activity of NROR was inhibited by 55% when the enzyme (0.6 mg/ml in 50 mM Tris, pH 7.8) was treated with iodoacetate (9 mM) at 25°C for 10 h. This result indicates that two cysteinyl residues of NROR may be the catalytic site for the disulfide reductase activity. Disulfides, such as thioredoxin (7) and glutaredoxin (19), have been identified in P. furiosus, but whether these can be reduced by NROR remains unknown.
On the other hand, the fact that NROR is such a remarkably efficient enzyme for reducing rubredoxin (kcat/Km = 3 × 105 mM−1 s−1 at 80°C) suggests that this is its sole physiological function and that reduced rubredoxin plays an important role in vivo. The question is, what is the purpose of the rapid reduction of rubredoxin? Unfortunately, the function of this protein in P. furiosus or in any other of the anaerobic organisms from which it has been purified is not known. Rubredoxin has been proposed to play a role in nitrate reduction (22), methanogenesis from H2 and CO2 (16), O2 reduction (5, 6), and CO oxidation (18), but the nature of the enzymes involved in oxidizing and reducing the protein either is not specific or is not consistent among the various organisms.
Two enzymes that catalyze the NADH-dependent reduction of rubredoxin have been characterized for mesophilic anaerobes. That from Clostridia acetobutylicum (17), which is a FAD-containing monomer with a molecular mass of 41 kDa, closely resembles P. furiosus NROR in its molecular properties. In contrast, NROR from Desulfovibrio gigas consists of two different subunits with molecular masses of 27 and 32 kDa, and it contains both FAD and flavin mononuclotide (5). Interestingly, the specific activities of the two mesophilic enzymes in the NADH-dependent reduction of rubredoxin were 46 and 12 U/mg, respectively (5, 17). A value of 294 U/mg was obtained for the P. furiosus enzyme when assayed under comparable conditions (at 25°C), which approach 80° below the optimum growth temperature of the organism. At present it is not clear why P. furiosus NROR is so active at such a “low” temperature. Obviously, much more is to be learned about the role of this enzyme and of rubredoxin in P. furiosus and other microorganisms.
Acknowledgments
We thank H.-S. Huang and Z.-H. Zhou for providing P. furiosus rubredoxin.
This research was supported by a grant (BCS-9632657) from the National Science Foundation.
REFERENCES
- 1.Aono S, Bryant F O, Adams M W W. A novel and remarkably thermostable ferredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Bacteriol. 1989;171:3433–3439. doi: 10.1128/jb.171.6.3433-3439.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaucamp K, Bergmeyer H U, Beutler H-O. Coenzymes, metabolites, and other biochemical reagents. In: Bergmeyer H U, editor. Methods of enzymatic analysis. I. New York, N.Y: Verlag Chemie, Academic Press, Inc.; 1974. p. 532. [Google Scholar]
- 3.Blake P R, Park J-B, Bryant F O, Aono S, Magnuson J K, Eccleston E, Howard J B, Summers M F, Adams M W W. Determinants of protein hyperthermostability: purification and amino acid sequence of rubredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochemistry. 1991;30:10885–10895. doi: 10.1021/bi00109a012. [DOI] [PubMed] [Google Scholar]
- 4.Bryant F O, Adams M W W. Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J Biol Chem. 1989;264:5070–5079. [PubMed] [Google Scholar]
- 5.Chen L, Liu M-Y, LeGall J, Fareleira P, Santos H, Xavier A V. Purification and characterization of an NADH-rubredoxin oxidoreductase involved in the utilization of oxygen by Desulfovibrio gigas. Eur J Biochem. 1993;216:443–448. doi: 10.1111/j.1432-1033.1993.tb18162.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Liu M-Y, LeGall J, Fareleira P, Santos H, Xavier A V. Rubredoxin oxidase, a new flavo-heme-protein, is the site of oxygen reduction to water by the “strict anaerobe” Desulfovibrio gigas. Biochem Biophys Res Commun. 1993;193:100–105. doi: 10.1006/bbrc.1993.1595. [DOI] [PubMed] [Google Scholar]
- 7.Guagliardi A, de Pascale D, Cannio R, Nobile V, Bartolucci S, Rossi M. The purification, cloning, and high level expression of a glutaredoxin-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:5748–5755. doi: 10.1074/jbc.270.11.5748. . (Erratum, 272:20961, 1997.) [DOI] [PubMed] [Google Scholar]
- 8.Holmgren A, Björnstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–207. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 9.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.LeGall J, Moura J J G, Peck H D, Jr, Xavier A V. Hydrogenase and other iron-sulfur proteins from sulfate-reducing and methane-forming bacteria. In: Spiro T G, editor. Iron-sulfur proteins. New York, N.Y: John Wiley & Sons; 1982. pp. 207–214. [Google Scholar]
- 12.Ma K, Adams M W W. Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus: a new multifunctional enzyme involved in the reduction of elemental sulfur. J Bacteriol. 1994;176:6509–6517. doi: 10.1128/jb.176.21.6509-6517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma K, Adams M W W. An unusual oxygen-sensitive, iron- and zinc-containing alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1999;181:1163–1170. doi: 10.1128/jb.181.4.1163-1170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma K, Robb F T, Adams M W W. Purification and characterization of NADP-specific alcohol dehydrogenase and glutamate dehydrogenase from the hyperthermophilic archaeon Thermococcus litoralis. Appl Environ Microbiol. 1994;60:562–568. doi: 10.1128/aem.60.2.562-568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma K, Schicho R N, Kelly R M, Adams M W W. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc Natl Acad Sci USA. 1993;90:5341–5344. doi: 10.1073/pnas.90.11.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nolling J, Ishii M, Koch J, Pihl T D, Reeve J N, Thauer R K, Hedderich R. Characterization of a 45-kDa flavoprotein and evidence for a rubredoxin, two proteins that could participate in electron transport from H2 to CO2 in methanogenesis in Methanobacterium thermoautotrophicum. Eur J Biochem. 1995;231:628–638. doi: 10.1111/j.1432-1033.1995.0628d.x. [DOI] [PubMed] [Google Scholar]
- 17.Petitdemange H, Marczak R, Blusson H, Gay R. Isolation and properties of reduced nicotinamide adenine dinucleotide-rubredoxin oxidoreductase of Clostridium acetobutylicum. Biochem Biophys Res Commun. 1979;91:1258–1265. doi: 10.1016/0006-291x(79)91202-6. [DOI] [PubMed] [Google Scholar]
- 18.Ragsdale S W, Ljungdahl L G, DerVartanian D V. Isolation of carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum enzyme. J Bacteriol. 1983;155:1224–1237. doi: 10.1128/jb.155.3.1224-1237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raia C A, D’Auria S, Guagliardi A, Bartolucci S, De Rosa M, Rossi M. Characterization of redox proteins from extreme thermophilic archaebacteria—studies on alcohol dehydrogenase and thioredoxins. Biosens Bioelectron. 1995;10:135–140. [Google Scholar]
- 20.Riddles P W, Blakeley R L, Zerner B. Reassessment of Ellman’s reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 21.Robb F T, Park J-B, Adams M W W. Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism of the hyperthermophilic archaebacterium, Pyrococcus furiosus. Biochim Biophys Acta. 1992;1120:267–272. doi: 10.1016/0167-4838(92)90247-b. [DOI] [PubMed] [Google Scholar]
- 22.Seki Y, Seki S, Satoh M, Ikeda A, Ishimoto M. Rubredoxin from Clostridium perfringens: complete amino acid sequence and participation in nitrate reduction. J Biochem. 1989;106:336–341. doi: 10.1093/oxfordjournals.jbchem.a122854. [DOI] [PubMed] [Google Scholar]
- 23.Stetter K O. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 1996;18:149–158. [Google Scholar]
- 24.Stetter K O, Fiala G, Huber G, Huber R, Segerer G. Hyperthermophilic microorganisms. FEMS Microbiol Rev. 1990;75:117–124. [Google Scholar]
- 25.Young I G, Rogers B L, Campbell H D, Jaworowski A, Shaw D C. Nucleotide sequence coding for the respiratory NADH dehydrogenase of Escherichia coli. UUG initiation codon. Eur J Biochem. 1981;116:165–170. doi: 10.1111/j.1432-1033.1981.tb05314.x. [DOI] [PubMed] [Google Scholar]