Abstract
To determine whether N-terminal sequences are involved in the transmembrane signaling mechanism of EnvZ, the nucleotide sequences of envZ genes from several enteric bacteria were determined. Comparative analysis revealed that the amino acid sequence between Pro41 and Glu53 was highly conserved. To further analyze the role of the conserved sequence, envZ of Escherichia coli was subjected to random PCR mutagenesis and mutant alleles that produced a high-osmolarity phenotype, in which ompF was repressed, were isolated. The mutations identified clustered within, as well as adjacent to, the Pro41-to-Glu53 sequence. These findings suggest that the conserved Pro41-to-Glu53 sequence is involved in the signal transduction mechanism of EnvZ.
In Escherichia coli, EnvZ is involved in sensing changes in the osmolarity of the external environment (2, 6, 8, 15). During adaptation to osmolarity stress, E. coli differentially regulates the genes encoding the outer membrane porin proteins, OmpF and OmpC. The response regulator OmpR controls the expression of the ompF and ompC genes. When cells are grown either under high-osmolarity conditions or in the presence of membrane-perturbing agents such as procaine, the level of OmpR-phosphate in the cell increases, which stimulates the expression of ompC and the repression of ompF (1, 4, 5, 9, 11, 16, 18). Modulation of the intracellular levels of OmpR-phosphate thereby controls the relative expression of the ompF and ompC genes (18, 24).
EnvZ functions as a dimer (17, 26) and undergoes transautophosphorylation on His243, using ATP as the phosphate donor (7, 19, 22, 27). The phosphate group is subsequently transferred to Asp55 of OmpR. EnvZ also possesses a phosphatase activity that stimulates the dephosphorylation of OmpR-phosphate. The sum of the kinase and phosphatase activities controls the level of OmpR-phosphate in the cell (5, 8, 15, 18). Hsing et al. recently presented a model in which the positioning of His243 relative to the ATP-binding domain determines whether EnvZ functions as a kinase or a phosphatase (8).
While the biochemical and structural properties of the cytoplasmic signalling domain have been extensively studied, the domains involved in the sensing function of EnvZ have not been elucidated. The periplasmic domain of EnvZ encompasses the region from Pro41 to Arg162 (3). It is flanked by two transmembrane sequences, TM1 (Leu16 to Leu40) and TM2 (Tyr163 to Ile179). It was found that replacement of the periplasmic region from Arg55 to Arg146 did not affect EnvZ function (12). Furthermore, EnvZ of Xenorhabdus nematophilus, which possesses a small periplasmic loop rather than the large domain found in the EnvZ proteins of most enteric bacteria, was able to complement an envZ-null strain of E. coli (20). These results raise the question of which regions of EnvZ are essential for sensing osmolarity signals. In the present study, we have taken both a comparative and a genetic approach to address this question.
Bacterial strains and plasmids.
The bacterial strains and plasmids utilized in this study are described in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| S. flexneri | Wild type | ATCC 120022 |
| Enterobacter cloacae | Wild type | ATCC 23355 |
| Y. enterocolitica | Wild type | Lab strain |
| P. vulgaris | Wild type | ATCC 13315 |
| E. coli | ||
| AT142 | F−envZ::Kan lacU169 araD139 rpsL relA thiA flbB | 12 |
| AT142pcnB | pcnB::Tn10 of AT142 | 12 |
| WH57 | ompF-lacZ of AT142 | 7 |
| LEO544 | pcnB::Tn10 of WH57 | 12 |
| Plasmids | ||
| pMRL25 | ompR-envZ in pBR322 | 12 |
| pKS2 | pMRL25 with envZ H243N | 19 |
| pJW49 | pMRL25 with a SalI site at bp 5–10 of envZ | This study |
| pJW32P | pJW49 with envZ L32P | This study |
| pJW35A | pMRL25 with envZ L35A | This study |
| pJW35P | pJW49 with envZ L35P | This study |
| pJW43A | pMRL25 with envZ L43A | This study |
| pJW43P | pJW49 with envZ L43P | This study |
| pJW48E | pJW49 with envZ K48E | This study |
Identification of a conserved N-terminal sequence.
In an attempt to find conserved sequences in the N-terminal region that may be involved in the sensing function of EnvZ, a comparative approach was taken in which EnvZ proteins from several genera within the Enterobacteriaceae family were analyzed. To this aim, the nucleotide sequences of the envZ genes of Shigella flexneri, Enterobacter cloacae, Yersinia enterocolitica, and Proteus vulgaris were determined. To obtain the nucleotide sequences of the various envZ genes, DNA fragments were PCR amplified directly from single bacterial colonies, using the following degenerate primer set: 5′GC(A/T)AA(C/T)GC(A/C/T)GA(A/G)CAGATG (Ala35 to Met40 of OmpR) and 5′CGG(C/G)GT(A/G)CG(C/T)AA(A/G)TC(A/G)TG (Pro248 to His243 of EnvZ). The nucleotide sequences of the different envZ genes were determined by a combination of direct sequence analysis of the PCR products and subcloning into M13.
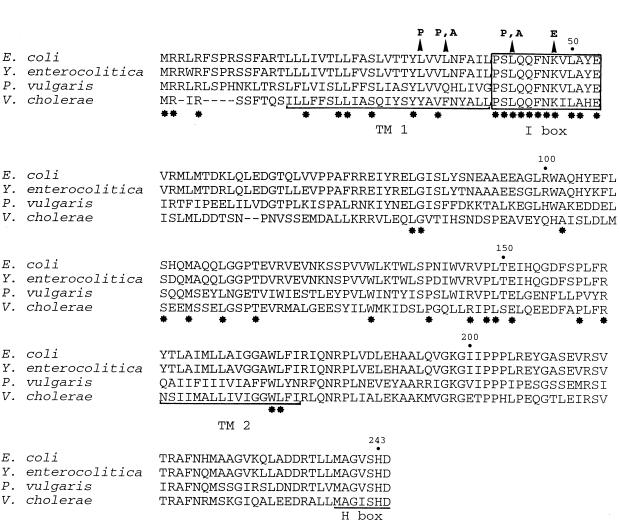
Figure 1 shows the sequence alignment of amino acids Met1 to Asp244 of various EnvZ proteins. The recently determined sequence of EnvZ from Vibrio cholerae (23) is also shown in Fig. 1. The amino acid sequences of the EnvZ proteins of S. flexneri and Enterobacter cloacae were nearly identical to that of the E. coli protein (Table 2), so they were not included in the sequence comparison. This comparison revealed that a 13-residue sequence encompassing Pro41 to Glu53 of E. coli EnvZ was 100% identical to the corresponding Proteus sequence and 85% identical to that in V. cholerae (Table 2). We refer to this highly conserved sequence as the identity box or I box. The I box is located at the junction between TM1 and the periplasmic domain. In contrast to the highly conserved nature of the I box, the overall degree of identity in the transmembrane and periplasmic domains was low. In the TM1 domains of the various EnvZ proteins, 6 of the 25 residues were identical (24% identity), while the periplasmic and TM2 domains exhibited only 14 and 12% amino acid sequence identity, respectively.
FIG. 1.
Comparison of amino acid sequences of the Met1-to-Asp244 regions of various EnvZ proteins. Invariant residues in the N-terminal region (Met1 to Ile179) are indicated by stars beneath the sequence. The I box is enclosed in a box. The TM1 and TM2 domains and the sequence containing the H243 site of phosphorylation (H box) are underlined. Amino acid substitutions of mutants isolated in this study are indicated by arrowheads above the sequence.
TABLE 2.
Amino acid sequence identity relative to the E. coli protein
| Bacterium | % Identity for:
|
|
|---|---|---|
| EnvZa | I box | |
| S. flexneri | 99.6 | 100 |
| Enterobacter cloacae | 98.4 | 100 |
| Y. enterocolitica | 92.2 | 100 |
| P. vulgaris | 45.8 | 100 |
| V. choleraeb | 35.7 | 85 |
Met1 to Ile179.
Taken from reference 23.
The distinctive characteristics of the I box include the presence of a proline residue (Pro41), four polar residues (Ser42, Gln44, Gln45, and Asn47), and two charged residues (Lys48 and Glu53). Furthermore, the nonpolar residues Leu43, Phe46, and Leu50 are conserved in the EnvZ proteins. Based on secondary-structure predictions, the I-box sequence exists as an amphiphilic alpha helix in which Leu43 and Leu50 are positioned on the nonpolar face of the helix (26). The biochemical properties of this region of EnvZ of X. nematophilus were also conserved (20). EnvZ of X. nematophilus contains a proline residue (Pro50), three polar residues (Thr42, Ser44, and Ser47), and two charged residues (Glu41 and Asp46), as well as the invariant nonpolar residues mentioned above.
Finally, OmpR was found to be highly conserved. The OmpR proteins in the E. coli-Proteus group exhibited >89% amino acid identity while those from V. cholerae and X. nematophilus exhibited 82% (26) and 74% (20) identity, respectively, to OmpR of E. coli.
I-box and TM1 mutations.
The highly conserved nature of the I-box sequence suggested that it might be involved in perceiving osmolarity signals. High-osmolarity conditions stimulate an increased kinase-to-phosphatase ratio in EnvZ, which results in elevated OmpR-phosphate levels and the repression of ompF (5, 8, 26). Therefore, mutations that either enhance the kinase activity or decrease the phosphatase activity of EnvZ would generate a high-osmolarity-type signal. If the I-box region was involved in modulating the kinase-to-phosphatase ratio of EnvZ, mutations that stimulate elevated OmpR-phosphate levels, and the concomitant repression of ompF, would be predicted to occur in this region of the molecule. To test this prediction, a genetic screen was designed to isolate strains with mutations in the N-terminal region of EnvZ that cause repression of ompF. A PCR approach was used in which the DNA fragment encoding the N-terminal region of EnvZ (Met1 to Glu106) was amplified under mutagenic conditions (10). The resultant PCR fragments were ligated into a plasmid encoding OmpR and the C-terminal region of EnvZ (Phe107 to Gly450), and the recombinant plasmids were transformed into the envZ-null strain LEO544, which contains both an ompF-lacZ reporter gene fusion and the pcnB80 allele, which is used to maintain plasmids at low copy numbers (7, 13). Since OmpR is phosphorylated by acetyl phosphate in envZ-null strains (7, 12), ompF is expressed at low levels in LEO544, and hence this strain forms red colonies on MacConkey-lactose agar. LEO544 transformed with envZ alleles that cause ompF to be completely repressed form white colonies (LacZ−) on MacConkey-lactose agar. The formation of white colonies could result either from envZ mutations that stimulate higher OmpR-phosphate levels (i.e., kinase positive and phosphatase negative), resulting in the repression of ompF, or from mutations that reduce OmpR-phosphate to levels that are insufficient to activate ompF expression (i.e., kinase negative and phosphatase positive; see reference 18). These possibilities can be distinguished since in the former case, OmpC is produced, while in the latter instance it is not produced (see Fig. 2). In this screen, envZ-null alleles are not recovered since they produce red colonies. To ensure that this was the case, an ompR-envZ-containing plasmid carrying the envZ-null allele, H243N (19), was transformed into LEO544. As expected, the resultant strain formed red colonies.
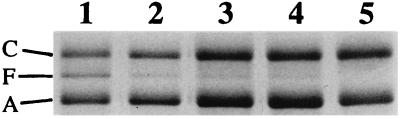
FIG. 2.
Outer membrane protein analysis. AT142pcnB cells containing various envZ alleles were grown on MacConkey medium, and outer membrane proteins were prepared and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (5). Lane 1, pJW49 (wild type); lane 2, pJW32P; lane 3, pJW35P; lane 4, pJW43P; lane 5, pJW48E. C, OmpC; F, OmpF; A, OmpA.
Using this screen, seven white colonies containing single missense mutations in envZ were obtained. The following single-amino-acid substitutions were identified: Leu32 to Pro (L32P), Leu35 to Pro (L35P), Leu43 to Pro (L43P), and Lys48 to Glu (K48E). Mutants with the L35P allele were isolated three times, and mutants with the L43P allele were isolated twice. Leu32 and Leu35 are located in the TM1 domain adjacent to the I box, while Leu43 and Lys48 are located within the I-box sequence (Fig. 1). Thus, the mutations isolated clustered in the C-terminal end of TM1 and in the I-box sequence.
The effect that the mutations had on the kinase-to-phosphatase ratio was assessed by analyzing the production of OmpC in the mutant strains. If the kinase-to-phosphatase ratio was elevated, OmpC would be produced at increased levels. On the other hand, OmpC would not be produced in strains in which the kinase-to-phosphatase ratio was low (18). To distinguish between these possibilities, the mutant strains were grown in MacConkey medium and the outer membrane proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2). The relative amounts of OmpF and OmpC in the outer membrane were then analyzed by densitometric scanning (Table 3). Figure 2 shows that OmpC was produced by all of the mutant strains. Densitometric scanning revealed that the levels of OmpC were elevated relative to that of the wild-type strain (Table 3). Thus, the mutations in TM1 and the I-box sequence caused an elevation of the intracellular levels of OmpR-phosphate, indicating that the kinase-to-phosphatase ratio in the mutant EnvZ molecules was increased.
TABLE 3.
Porin protein production on MacConkey medium
| Strain | Relative % of porin protein produceda
|
|
|---|---|---|
| OmpF | OmpC | |
| Wild type | 14 | 34 |
| L32P | 4 | 40 |
| L35P | 2 | 43 |
| L43P | 2 | 40 |
| K48E | 1 | 51 |
The values represent the relative amounts of OmpF and OmpC, expressed as a percentage of the total absorbance of OmpA, OmpF, and OmpC, as determined by scanning densitometry of the outer membrane proteins.
To ensure that the mutant EnvZ proteins had properly assembled into the cytoplasmic membrane, vesicles were prepared (12) and EnvZ was detected by Western blot analysis, using enhanced chemiluminescence (Sigma Co.). Membrane vesicles prepared from the envZ-null strain harboring plasmids encoding either the wild-type or a mutant EnvZ protein contained a protein with a molecular weight of 50,000, representing full-length EnvZ (data not shown). These findings indicated that the mutant EnvZ proteins were incorporated into the membrane.
Growth of mutant strains in nutrient broth.
To further evaluate the effect that the TM1 and I-box mutations had on EnvZ function, we grew cells in nutrient broth. OmpR-phosphate is maintained at moderate levels, and OmpF is produced at high levels, in cells grown under nutrient broth conditions (5, 18). By growing the mutant strains under these conditions, we could determine the extent to which the kinase-to-phosphatase ratio had been reset in the mutant EnvZ proteins. Strains with the TM1 mutation L35P or the I-box mutation L43P were selected for this analysis. Table 4 shows that in wild-type cells grown in nutrient broth, OmpF was produced at approximately threefold-higher levels than OmpC. In contrast, OmpF production was markedly decreased, but not fully repressed, and OmpC production was increased in the mutant strains. These results indicated that the L35P and L43P mutations had elevated the kinase-to-phosphatase ratio of EnvZ sufficiently to trigger a switch in the relative amounts of OmpF and OmpC produced by the cell. However, the amount of OmpR-phosphate present in the mutant cells was apparently not large enough to reduce OmpF to low levels. To address the question of whether the mutant EnvZ proteins were able to sense high-osmolarity signals and further increase the levels of OmpR-phosphate, the mutant cells were grown in nutrient broth containing 20% sucrose. Table 4 shows that OmpF production was further reduced in the mutant strains grown under high-osmolarity conditions. This result suggested that the mutant proteins were still able to perceive high-osmolarity stimuli and set the kinase-to-phosphatase ratio to higher levels, resulting in an increase in OmpR-phosphate levels and a concomitant decrease in OmpF production.
TABLE 4.
Porin protein production in nutrient broth medium
| Strain | Relative % of porin protein produced ina:
|
|||
|---|---|---|---|---|
| NB
|
NB + sucrose
|
|||
| OmpF | OmpC | OmpF | OmpC | |
| Wild type | 51 | 18 | 7 | 53 |
| L35P | 19 | 48 | 8 | 50 |
| L43P | 17 | 50 | 6 | 55 |
| L35A | 54 | 20 | 3 | 60 |
| L43A | 18 | 58 | 2 | 66 |
The values represent the relative amounts of OmpF and OmpC as described in the footnote in Table 3. NB, nutrient broth.
Alanine substitutions at Leu35 and Leu43.
The mutations of strains isolated in this study consisted of either proline substitutions or a charge reversal. Proline substitutions could indirectly affect EnvZ function by inducing conformational alterations. For example, proline replacement in transmembrane domains has been shown to disrupt alpha-helical structure (14). To introduce conservative substitutions in TM1 and the I-box sequence, alanine residues were substituted for Leu35 and Leu43 by site-directed mutagenesis (Sculptor in vitro system; Amersham). The resulting L35A and L43A envZ alleles were transformed into the envZ-null strain AT142pcnB, and porin production in cells grown in nutrient broth was analyzed. Table 4 shows that OmpF was repressed and OmpC was stimulated in cells containing the L43A form of EnvZ. In addition, OmpF production was further reduced in cells grown under high-osmolarity conditions. These results further support the idea that Leu43 is involved in the signal transduction mechanism of EnvZ. In contrast, cells containing the L35A form of EnvZ produced OmpF and OmpC at levels similar to those found in the wild-type strain (Table 4). Thus, the OmpF repression observed when Leu35 was replaced by a proline residue appeared to be due to an induced secondary-structure alteration in TM1.
Summary.
The Pro41-to-Glu53 sequence of EnvZ, referred to as the I box, was found to be highly conserved in enteric bacteria and V. cholerae. The I box is located in the periplasmic domain of EnvZ, in close proximity to the cytoplasmic membrane. We showed that replacement of Leu43 with either a proline or an alanine residue stimulated a reduction in OmpF production under conditions in which OmpF is normally produced at high levels. Mutations in EnvZ that cause ompF to be repressed had been previously determined to occur at Pro41, Leu43, Gln44, and Leu50 (8, 22, 26). It was proposed recently that Leu43, Leu50, and Leu57 are involved in a dimeric leucine zipper-like structure and that this structure may play a role in osmotic signal transduction (26). Based on the present and previous information, we propose that the I box is directly involved in sensing osmolarity signals. Leu43 appears to be particularly critical in this function. We envision that the I box undergoes conformational alteration by sensing changes in the physical and/or chemical properties of the cytoplasmic membrane that are induced by osmolarity stress. Alternatively, the I box may directly sense changes in extracellular water activity (25). A conformational change in the I box would in turn affect the secondary structure of TM1, which has been proposed to be critical in maintaining the proper balance between the kinase and phosphatase activities of EnvZ (8, 21).
Nucleotide sequence accession numbers.
The partial sequences reported herein have been deposited in the GenBank DNA database under the following accession numbers: AF030314 (ompR) and AF030415 (envZ) for S. flexneri; AF030315 (ompR) and AF030416 (envZ) for Enterobacter cloacae; AF030316 (ompR) and AF030417 (envZ) for Y. enterocolitica; and AF030317 (ompR) and AF030418 (envZ) for P. vulgaris.
Acknowledgments
We are grateful to M. Krebs, M. Leonardo, and M. Majors for critical reading of the manuscript. We thank K. Skarphol for providing pKS2.
This study was supported by Public Health Service grant GM44671.
REFERENCES
- 1.Aiba H, Nakasai F, Mizushima S, Mizuno T. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J Biol Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- 2.Egger L A, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phospho-relay. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 3.Forst S, Comeau D, Norioka S, Inouye M. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J Biol Chem. 1987;262:16433–16438. [PubMed] [Google Scholar]
- 4.Forst S, Delgado J, Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forst S, Delgado J, Rampersaud A, Inouye M. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J Bacteriol. 1990;172:3473–3477. doi: 10.1128/jb.172.6.3473-3477.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forst S, Roberts D. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res Microbiol. 1994;145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 7.Hsing W, Silhavy T J. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsing W, Russo F D, Bernd K K, Silhavy T J. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 10.Kenney T J, Churchward G. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J Bacteriol. 1996;178:3564–3571. doi: 10.1128/jb.178.12.3564-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan C-Y, Igo M M. Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J Bacteriol. 1998;180:171–174. doi: 10.1128/jb.180.1.171-174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonardo M R, Forst S. Re-examination of the role of the periplasmic domain of EnvZ in sensing of osmolarity signals in Escherichia coli. Mol Microbiol. 1996;22:405–413. doi: 10.1046/j.1365-2958.1996.1271487.x. [DOI] [PubMed] [Google Scholar]
- 13.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson I, Saaf A, Whitley P, Gafvelin G, Waller C, von Heijne G. Proline-induced disruption of a transmembrane alpha-helix in its natural environment. J Mol Biol. 1998;284:1165–1175. doi: 10.1006/jmbi.1998.2217. [DOI] [PubMed] [Google Scholar]
- 15.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 105–127. [Google Scholar]
- 16.Rampersaud A, Inouye M. Procaine, a local anesthetic, signals through the EnvZ receptor to change the DNA binding affinity of the transcriptional activator protein OmpR. J Bacteriol. 1991;173:6882–6888. doi: 10.1128/jb.173.21.6882-6888.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts D L, Bennett D W, Forst S A. Identification of the site of phosphorylation on the osmosensor, EnvZ, of Escherichia coli. J Biol Chem. 1994;269:8728–8733. [PubMed] [Google Scholar]
- 18.Russo F D, Silhavy T J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 19.Skarphol K, Waukau J, Forst S A. Role of His243 in the phosphatase activity of EnvZ in Escherichia coli. J Bacteriol. 1997;179:1413–1416. doi: 10.1128/jb.179.4.1413-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabatabai N, Forst S. Molecular analysis of the two-component genes, ompR and envZ, in the symbiotic bacterium Xenorhabdus nematophilus. Mol Microbiol. 1995;17:643–652. doi: 10.1111/j.1365-2958.1995.mmi_17040643.x. [DOI] [PubMed] [Google Scholar]
- 21.Tokishita S, Kojima A, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J Biochem. 1992;111:703–713. doi: 10.1093/oxfordjournals.jbchem.a123823. [DOI] [PubMed] [Google Scholar]
- 22.Tokishita S, Mizuno T. Transmembrane signal transduction by the Escherichia coli osmotic sensor, EnvZ: intermolecular completion of transmembrane signalling. Mol Microbiol. 1994;13:435–444. doi: 10.1111/j.1365-2958.1994.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 23.Tow A L, Coyne V E. Cloning and characterisation of a novel ompB operon from Vibrio cholerae 569B. Biochim Biophys Acta. 1999;1444:269–275. doi: 10.1016/s0167-4781(98)00277-2. [DOI] [PubMed] [Google Scholar]
- 24.Waukau J, Forst S. Molecular analysis of the signaling pathway between EnvZ and OmpR in Escherichia coli. J Bacteriol. 1992;174:1522–1527. doi: 10.1128/jb.174.5.1522-1527.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood J M. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev. 1999;63:230–262. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaku H, Mizuno T. The membrane-located osmosensory kinase, EnvZ, that contains a leucine zipper-like motif functions as a dimer in Escherichia coli. FEBS Lett. 1997;417:409–413. doi: 10.1016/s0014-5793(97)01329-x. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]