Homologous recombination is a fundamental cellular process that rearranges genes within and between chromosomes, promotes DNA repair, and guides segregation of chromosomes at division. It provides, therefore, a potent evolutionary force that serves both to promote genetic diversity and to conserve genetic identity. Failure to repair damaged DNA is associated at a cellular level with sensitivity to radiation, altered mutation rates, DNA and chromosomal aberrations, and reduced viability.
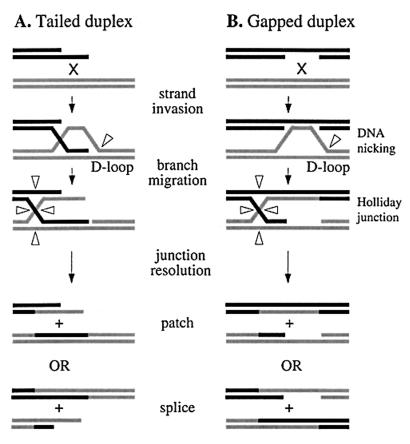
Genetic recombination occurs via breakage and reunion of DNA chains and generally conserves sequence information. Precision is achieved through the simple expedient of pairing complementary single strands from each molecule to form a heteroduplex intermediate that registers homology. Illegitimate exchange can be aborted at this stage by enzymes that recognize base pair mismatches (57). More than 30 years ago, Holliday (19) proposed a model of meiotic recombination in which homologous chromatids exchange single DNA strands to form a joint molecule with a four-way junction at the point of exchange. Resolution of the junction by symmetrical strand cleavage, coupled with repair of mismatches, provided plausible explanations for the formation of recombinants and for the patterns of marker segregation. Many features of the model have since been found wanting. In particular, recombination is now known to initiate at a double-strand break or single-strand gap in one DNA molecule (Fig. 1) and is often asymmetric or nonconservative in outcome (Fig. 1A) (46). However, the general idea that recombination entails a sequence of reactions that form and then resolve heteroduplex intermediates has withstood the test of time.
FIG. 1.
Schematic diagram showing molecular pathways for homologous recombination by single-strand invasion at ends (A) and gaps (B). Arrowheads indicate sites of DNA strand cutting.
The molecular pathways of homologous recombination have been dissected in detail in Escherichia coli (23, 26, 49). Heteroduplex intermediates are usually formed by the RecA filament, a structure that overcomes the natural tendency for Watson-Crick strands to remain paired and at the same time makes use of this property to exchange single strands between duplex molecules in a way that secures homologous alignment (49). The solution is so elegant that it has been retained throughout evolution (45). RecA polymerizes on regions of single-stranded DNA generated at the ends of (broken) duplex DNA by nucleases (e.g., RecBCD) that preferentially degrade one strand (Fig. 1A) or at single-strand gaps (Fig. 1B). Polymerization proceeds in the 5′-to-3′ direction to form a helical nucleoprotein filament that can extend to the adjacent duplex. Within the filament, the DNA is extended, and if the DNA is duplex, it is underwound. Extending the DNA is critical for DNA pairing, in which a homologous duplex is sought, brought into alignment within the filament, and driven rapidly to exchange strands with the resident molecule (49). Strand exchange links the two molecules together and creates a heteroduplex joint. If pairing is limited to the single-stranded region bound by RecA, the exchange forms a D-loop (Fig. 1A). However, if it extends into the duplex, strand exchange is reciprocal and generates a Holliday junction. In both cases, strand exchange is unidirectional and proceeds with a 5′-to-3′ polarity. The joint molecules formed by RecA are then eliminated by other enzymes. In E. coli, these enzymes include RuvABC, RecG, and RusA.
The availability of partial and complete genome sequences from diverse species has facilitated a study of the conservation of these enzymes during evolution. This minireview summarizes our findings in the context of the enzymology of Holliday junction processing.
HOLLIDAY JUNCTION PROCESSING BY RuvABC
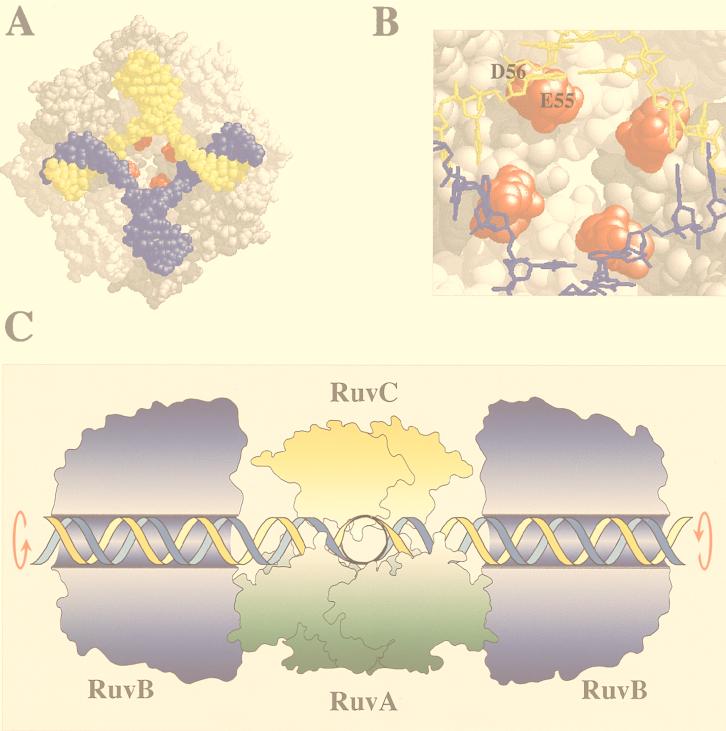
The RuvAB and RuvC proteins have been shown to catalyze the branch migration and resolution of Holliday junction recombination intermediates. There is genetic and biochemical evidence to suggest that in E. coli at least, these two activities can be coordinated in a single tripartite complex (Fig. 2C). The availability of crystal structures for RuvA and RuvC and electron microscope images of RuvB have greatly advanced our understanding of the structure of this complex and how it operates.
FIG. 2.
Holliday junction processing by RuvABC. (A) Structure of the RuvA tetramer complexed with a Holliday junction (produced with RasMol) (18). The acidic pins are shown in red. (B) Locations of the acidic pin residues (Glu55 and Asp56; colored red) of RuvA, showing their proximity to the phosphate backbone at the junction core. (C) Model of branch migration and resolution by the RuvABC resolvasome. A tetramer of RuvA holds the Holliday junction in a square planar conformation (side view). RuvB hexamer rings encircle duplexes on each side of the RuvA-junction complex. Branch migration is achieved by drawing the duplexes through these rings, generating heteroduplex DNA. A RuvC dimer bound to the upper face of the junction scans for specific target sequences as the DNA passes across RuvA. Symmetrically related incisions are introduced to yield nicked duplexes which can be sealed by DNA ligase. Outlines of RuvA and RuvC were taken from the crystal structures of these proteins (3, 38). The RuvB structure is based on electron micrographs of RuvAB-junction complexes (36). An animated model of branch migration by RuvAB can be viewed at the Krebs Institute web site (23a).
A tetramer of the 22-kDa RuvA protein forms a grooved platform on which the Holliday junction is held in a square planar configuration (Fig. 2A) (18, 33, 38). A pair of helix-hairpin-helix motifs from each RuvA subunit position lysine residues for contact with the phosphate backbone of the DNA (18, 37). In the center of the RuvA tetramer are four protrusions, each consisting of two acidic residues from each monomer (Fig. 2A and B). These acidic “pins” project towards the crossover point of the duplex arms and may be involved in strand separation (38) or in structure selectivity (20a). RuvB is a hexameric ring helicase that assembles on opposing arms of the junction via contacts with RuvA. Branch migration of the Holliday junction is achieved by the RuvB helicase motor drawing the DNA through the complex, with each duplex arm rotating within the channels on the surface of RuvA (36, 38). The crystal structure of Mycobacterium leprae RuvA has revealed that two tetramers of RuvA can bind the junction, creating a stable protein-junction sandwich through which the DNA is guided (39). The octameric version of RuvA may represent a specialized form of RuvAB specifically active in branch migration and distinct from the RuvABC resolution complex.
The dimeric RuvC endonuclease resolves the Holliday junction into duplex products by the introduction of symmetrically related nicks in two of the four DNA strands (13, 21). Upon binding a junction, RuvC imposes an unfolded twofold symmetric structure on the DNA which may position the scissile bonds for cleavage in the active site (5). It shows a preference for cleavage between the third and fourth positions of a tetranucleotide sequence with the consensus 5′-(A/T)TT(G/C)-3′ (4).
There is good evidence to suggest that RuvC is accommodated on the RuvAB-junction complex to form a branch migration-resolvase complex known as the resolvasome (Fig. 2C). The original notion of a relationship between branch migration and resolution came from genetic studies. Mutations in the three ruv genes confer identical phenotypes and can be suppressed by mutations that switch on expression of an alternative resolvase called RusA (31). Strains deficient in RuvAB are therefore defective in junction resolution even when RuvC is present. Subsequent biochemical analysis has demonstrated interactions between RuvAC, RuvBC, and RuvABC on synthetic Holliday junctions (11, 47, 50). In addition, the resolution activity of RuvC is stimulated by RuvAB on large Holliday junction substrates known as χ-structures (58). The resolvasome model predicts that a RuvABC-junction complex tracks along DNA, with RuvC able to scan for cleavable sequences as the DNA passes through.
Two questions arise from these studies. (i) Does the Ruv ABC resolvasome model based on the analysis of E. coli proteins represent a universal paradigm? (ii) Does the RuvAB branch migration motor have any function other than to locate junctions at cleavable sites? To help answer these questions, we investigated the conservation of RuvAB and RuvC in the available bacterial genome sequences.
CONSERVATION OF RUVABC IN BACTERIA
Genes encoding homologs of RuvA and RuvB are widespread in bacteria, whereas homologs of RuvC are less common (Fig. 3). The ruvA and ruvB genes are often found together in an operon, sometimes including ruvC upstream, although there are cases, such as in Synechocystis, where the three ruv genes are scattered around the genome in separate operons. RuvC is absent from the completed genomes of Mycoplasma genitalium, Mycoplasma pneumoniae, Bacillus subtilis, and Borrelia burgdorferi and has yet to be found in Enterococcus, Streptococcus, and Clostridium spp. (Fig. 3). A phylogenetic tree based on 16S rRNA sequences shows that Mycoplasma, Streptococcus, Enterococcus, Clostridium, and Bacillus spp. define a branch of the tree that seems to lack RuvC (Fig. 4). The absence of RuvC from B. burgdorferi is puzzling, especially as it is present in the related spirochete Treponema pallidum. As some of these genomes contain the Holliday junction resolvase RusA (Fig. 3), they could be using this enzyme instead. However, several organisms have neither RuvC nor RusA, suggesting the existence of an alternative Holliday junction resolvase whose nature has yet to be described. It would seem incongruous to have the enzymes to move Holliday junctions (RuvAB) but lack the means to resolve them.
FIG. 3.
Occurrence of Holliday junction-processing enzymes in eubacteria. Searches were performed with gapped BLAST and PSI-BLAST programs (2) and databases provided by the National Center for Biotechnology Information (32a), The Institute for Genomic Research (46a), and the Sanger Centre (40a). The definition of a homolog (or ortholog) of each gene was based on the following four criteria: (i) sequence similarity searches using the E. coli proteins and subsequent searches with the identified homologs, (ii) analysis based on the known crystal structures of RuvA and RuvC and information derived from mutant versions of the five enzymes site-directed, (iii) experimental evidence confirming the appropriate activities of predicted homologs, and (iv) conservation of distinctive motifs belonging to the family of proteins. The random expectation values (2) for the E. coli protein against the most dissimilar homolog were as follows: Mycoplasma genitalium RuvA (6 × 10−4), Mycoplasma pneumoniae RuvB (2 × 10−49), Synechocystis sp. strain RuvC (5 × 10−6), and Campylobacter jejuni RecG (2 × 10−48). Specific features used to identify each homolog included the following: the pair of helix-hairpin-helix motifs and the conserved acidic pin residues of RuvA (37), high conservation of the unusual helicase domain of RuvB, four essential acidic residues required for catalysis and conserved residues around helices 3 and 4 of RuvC (17, 40), and similarities in the helicase and N-terminal domains of RecG and the unique motif it shares with TRCF. TRCF and RecG have different N- and C-terminal domains, sharing considerable homology (38% identity between the E. coli proteins) only in the central helicase motifs (42). RusA homologs are highly divergent and were identified by searching with each member of the family. With the exception of Clostridium acetobutylicium, all contained three aspartic acid residues required for Holliday junction resolution in a conserved C-terminal domain of RusA (7). Several eukaryotic proteins that have been designated RuvB-like (TIP49, RUVBL1, and RUVBL2) were excluded due to the absence of several motifs characteristic of the bacterial RuvB family and the lack of evidence supporting their involvement in recombination. Details of alignments and the accession numbers of identified homologs are available on request. Species are placed in order relative to the phylogenetic tree shown in Fig. 4. Small black circles identify the organisms for which the genome sequence has been completed. The presence (black squares) and absence (white squares) of RuvA, RuvB, RuvC, RecG, and RusA homologs are indicated; a grey square indicates that these homologs were not found in that species, but the genome sequence is incomplete. Abbreviations: Synechocystis, Synechocystis sp. strain PCC6803; Actinobacillus actinomyc., Actinobacillus actinomycetemcomitans.
FIG. 4.
Phylogenetic tree of eubacteria based on 16s rRNA sequences. The tree contains all the organisms listed in Fig. 3 and was obtained from the Ribosomal Database Project II (30) at its website (38a). The Aquifex sequence comes from Aquifex pyrophilus and not Aquifex aeolicus, because the latter was unavailable. Horizontal branch lengths are drawn to scale; the bar is 0.1 nucleotide replacement per site in length. Species for which homologs of RuvC are absent or yet to be detected are shown in bold type. Abbreviations: Porphyromonas, Porphyromonas gingivalis; Actinobacillus actinomyc., Actinobacillus actinomycetemcomitans.
However, the possibility that RuvAB are involved in restoring stalled replication forks independently of the RuvC resolvase (41) may explain why RuvAB are so highly conserved in bacteria. It has been proposed that a Holliday junction can arise at a blocked replication fork by annealing the newly synthesized strands. Resolution of a junction at this stage would create a double-strand break. Chromosome breakage could be avoided by protecting the junction from endonuclease cleavage with RuvAB. Subsequent degradation of the accessible duplex end of the junction by RecBCD exonuclease is predicted to reestablish the replication fork. Reversing the movement of the junction by branch migration could also restore the fork. The formation of a RuvAB complex in which a RuvA octamer sandwiches the junction may provide the means to protect it from resolution by RuvC.
Sequence comparisons of the Ruv protein homologs can also provide valuable information about critical functional domains. The two helix-hairpin-helix motifs of RuvA involved in Holliday junction binding (18, 37) are conserved in all instances. The acidic pin region (Glu55 and Asp56 [Fig. 2B]) is also present with the exception of the RuvA proteins from the two Mycoplasma species. These proteins have an additional four residues at this position with two glutamic acids located nearby. Junction recognition is not impaired as a result of these modifications because purified M. pneumoniae RuvA retains specificity for Holliday junction substrates (20a). The presence of both Glu55 and Asp56 is not obligatory as 10 species contain only one of these acidic residues. Four hydrophobic residues (Leu167, -170, and -199 and Tyr172) that are likely to form an interface with RuvB (33) are also highly conserved. Motifs consistent with helicase activity are conserved in all examples of RuvB. The four acidic residues of RuvC (Asp7, -138, and -141 and Glu66) that are required for endonuclease activity (3, 40) are present in all the RuvC-like proteins, although the homolog from T. pallidum has aspartic acid-138 replaced with histidine.
PHAGE HOMOLOGS OF RUVC
Homologs of RuvC have recently been identified in a number of bacteriophages from L. lactis (6). These phage enzymes possess all four acidic residues required for Holliday junction resolution by E. coli RuvC (Fig. 5) (40). The L. lactis phage bIL66 RuvC homolog is lethal in recA and recBC mutants of E. coli, probably by nicking at replication forks, resulting in double-strand DNA breaks (6). It is likely that phage bIL66 RuvC is more like the general branch-cutting endonucleases from T4 and T7 (54). In fact, T7 endonuclease I also has a lethal effect in E. coli recA cells (35).
FIG. 5.
Alignment of L. lactis and S. pyogenes phage RuvC homologs. Protein sequences from L. lactis phages bIL66 and bIL67 were chosen as representatives of the larger family of phage proteins. The bIL66 RuvC has percent identity values of 98, 96, and 93 to homologs from sk1, 712, and bIL170, respectively. bIL67 RuvC protein is 93% identical to those from c2 and vML3. Residues in boldtype in the E. coli RuvC sequence (Eco) represent those conserved among the eubacterial RuvC family (the structure of E. coli RuvC is represented under the sequence). Residues in boldtype in the bIL66, bIL67, and Spy (S. pyogenes) RuvC homologs represent matches with the E. coli RuvC sequence. Circles indicate the four acidic residues required for catalysis by E. coli RuvC; triangles mark residues thought to be involved in sequence specificity of Holliday junction resolution (3, 17). Residues considered similar are as follows: A and G; D and E; F, I, L, M, V, W, and Y; K and R; N and Q; and S and T.
The structure of the L. lactis phage RuvC proteins is interesting in that the central portion of the protein, encompassing α-helices 2 and 4, differs significantly from that of E. coli RuvC (Fig. 5). In E. coli RuvC, this region is thought to be involved in sequence specificity of Holliday junction resolution. Mutations in the conserved loop between helices 3 and 4 results in proteins with altered preferences for cleavage at resolution hot spots. In particular, the orientation of two conserved lysine residues present in these helices appears to position the junction for cleavage (17). The replacement of this region in the phage RuvC enzymes may enable them to cleave Holliday junctions irrespective of the sequences at the crossover point. This would convert them into more general branch-cutting enzymes, perhaps more suited to the requirements of phage replication and recombination. On a more-general point, the sequence specificity of RuvC may ensure that a four-way (Holliday) intermediate is the only possible target, as the symmetrically arranged consensus sequence would be missing from other branched DNA structures.
Two proteins similar to the phage RuvC family were also found on the chromosome of Streptococcus pyogenes (Fig. 5). The copies, which are 93% identical, appear to be part of cryptic prophages. One is associated with a RecT homolog 1,505 bp upstream and a phage r1t ORF22 homolog 359 bp downstream. The other contains homologs of ORF19 and ORF24 from Streptococcus thermophilus phage φ01205 located just downstream.
BRANCH MIGRATION BY RECG
The RuvABC proteins appear to constitute a simple system for the processing of Holliday junctions. However, the situation is complicated by a functional overlap between ruv and recG. Strains with mutations in the ruvABC genes, while exhibiting a significant defect in DNA repair, show only a slight reduction in genetic recombination. This relative proficiency in recombination is eliminated by mutation of recG (25). The product of recG is a 76-kDa structure-specific DNA helicase which, like RuvAB, can catalyze the branch migration of Holliday junctions (27). It can recognize and unwind a variety of branched structures, including four- and three-strand junctions, and appears to counter the strand exchange reaction mediated by RecA in vitro. These findings have led to a model whereby RecG targets the three-way junction at a D-loop created by 3′-end invasion (Fig. 1A) and drives the junction into duplex-duplex regions to form a Holliday junction (32, 52, 53). This junction can then be resolved by the action of RuvABC.
RecG also shares a functional relationship with the replication protein PriA, a DNA helicase required for primosome assembly at D-loops. Mutations in priA that probably affect this helicase activity have been isolated as suppressors of the DNA repair and recombination defects associated with recG mutations (1). The functions of these two branch-specific helicases in replication and recombination at D-loops have yet to be clearly defined. In keeping with its activity on D-loops, RecG also unwinds R-loops, formed by the assimilation of homologous RNA into duplex DNA (15, 48). An unrelated helicase (UvsW) with properties similar to RecG, including the ability to dissociate R-loops, functions in phage T4 recombination and repair and the regulation of replication origins (8).
CONSERVATION OF RECG IN BACTERIA
RecG homologs are widespread in bacteria with the exception of M. genitalium, M. pneumoniae, and Chlamydia trachomatis, all of which have relatively small genomes of 0.58, 0.81, and 1.05 Mb, respectively (Fig. 3). It could be argued that RecG is a luxury activity for more-complex organisms. However, it is present in other bacteria with only slightly larger genomes, such as T. pallidum (1.14 Mb) and Rickettsia prowazekii (1.10 Mb). The evolutionary conservation of RecG, together with the profound recombination and repair defect found in a recG ruv double mutant (25), implies that RecG plays an important and distinct role in itself and is not simply an alternative to RuvAB.
The helicase domain of RecG is highly conserved in all instances, with the N-terminal 250 amino acids, implicated in junction DNA specificity (28), showing more limited similarity. The product of the mfd gene, TRCF (transcription-repair coupling factor), is highly homologous to RecG in the region encompassing the seven helicase motifs (42). The similarity is consistent with a common evolutionary origin and suggests a shared enzymatic function. TRCF targets the UvrABC excision repair system to lesions in the transcribed strand. It recognizes the RNA polymerase stalled at a damaged site, releases the polymerase and the transcript (possibly through helicase activity), and directs UvrAB to the lesion through TRCF-UvrA interactions. The occurrence of TRCF in bacteria generally mirrors that of RecG. Exceptions are Chlamydia trachomatis where TRCF is present and RecG is absent, and Aquifex aeolicus where the opposite situation takes place. Both enzymes are absent from the two Mycoplasma species. RecG and TRCF share an additional conserved motif of 37 to 40 residues located adjacent to helicase motif VI (residues 606 to 642 of E. coli RecG and 926 to 965 of E. coli TRCF). Mutations in the conserved residues in this region eliminate the ability of RecG to complement the DNA repair defect associated with recG mutation (28a).
HOLLIDAY JUNCTION RESOLUTION BY RUSA
The recombination and DNA repair defects associated with mutations in ruv can be suppressed by insertion of IS2 or IS10 elements upstream of the rusA coding region, which activate transcription of the normally silent gene on the cryptic prophage DLP12 (29, 31). The 14-kDa RusA protein is a homodimeric Holliday junction-specific endonuclease (43). It is sequence specific like the RuvC resolvase, although it prefers to cut 5′ of a CC dinucleotide (9, 16). RusA displays less structure specificity than RuvC and can bind a variety of branched DNA structures (10). Complete suppression of the ruv defect is dependent on the activity of RecG (31). RecG may be required by RusA to branch migrate Holliday junctions to cleavable sequences or simply to generate four-way junctions from D-loops.
With the exception of A. aeolicus, RusA homologs are found in phage genomes (HK022 and 97, mycobacteriophage TM4, L. lactis r1t, and Staphylococcus aureus φPVL) or associated with phage sequences on the chromosomes of a number of bacteria (Fig. 3) (7). The gene is frequently located downstream of replication-associated proteins (including homologs of E. coli dnaC and λ O and P) and upstream of lysis genes. The molecular organization of the sequences from E. coli K-12, Neisseria gonorrhoeae, and phages 82, HK022, and 97 are similar and generally correspond to the nin region of phage λ. An unrelated structure-specific endonuclease, called Rap (NinG), has been isolated from this region of λ. Rap is capable of nicking a wide range of branched DNA molecules including D-loops (44, 44a). It can also resolve large Holliday junction substrates known as χ-structures, apparently by the introduction of symmetrically related nicks (44a). RusA homologs show the highest conservation in the C-terminal half of the protein where the three catalytic aspartic acid residues (positions 70, 72, and 91) reside (7). The RusA homolog from L. lactis phage r1t has been partially purified and resolves Holliday junctions in a manner similar to that of E. coli RusA (44a).
A. aeolicus is the only member of the eubacteria to lack the RuvABC system (Fig. 3). Phylogenetic trees based on 16S rRNA have shown that this bacterium is deeply rooted (Fig. 4), branching at the base of the eubacterial domain close to Archaea and Eucarya (12). RuvABC may therefore be a more recent acquisition by eubacteria, although the possibility that Aquifex lost this system cannot be discounted. A. aeolicus does have RecG and RusA which in E. coli are known to comprise a proficient system for resolving Holliday junctions (31). The A. aeolicus version of rusA (Aq1953) is located beside genes that are not normally associated with phages, namely, rnhB (RNase HII), rplS (ribosomal protein L19), and lplA (lipoate protein ligase). The lack of associated phage-like sequences suggests that A. aeolicus rusA could be a genuine host gene. Since Aquifex is an ancient bacterium, the uncoupled branch migration and resolution system provided by RecG and RusA may have been the original paradigm for recombination in the ancestor of Bacteria and perhaps even Archaea and Eucarya. If this scenario were correct, bacteriophages would have acquired RusA as a beneficial factor for removing recombination intermediates at some later date. A cellular origin for RusA accords with its structure selectivity, which is more like RuvC than the less discriminating branch-cutting enzymes from phages T4 and T7.
HOLLIDAY JUNCTION PROCESSING IN ARCHAEA AND EUCARYA
Homologs of RuvABC, RecG, and RusA are chiefly absent from the currently available archaeal and eukaryotic sequences. As the Archaea have DNA and RNA metabolism systems that are more like eukaryotes, one might expect them to resemble each other rather than eubacteria. However, a homolog of RecG was found on chromosome II of Arabidopsis thaliana (GenBank accession no. AC005560). This gene is remarkably similar to E. coli RecG (35% identity; 50% similarity), with most of the residues present in the bacterial RecG family conserved. It has an additional 132 residues at the N terminus which may carry localization signals or a domain for interaction with other proteins. The Arabidopsis RecG protein resembles most closely the RecG homologs from Synechocystis and Aquifex. Such an obvious RecG-like protein is absent from the completed Archaea, Saccharomyces cerevisiae, and Caenorhabditis elegans genome sequences. It is possible that Arabidopsis picked up RecG from bacteria or from mitochondrional or chloroplast progenotes.
S. cerevisiae contains a Holliday junction resolvase unrelated to RuvC or RusA known as CCE1 (22, 54). An equivalent enzyme from Schizosaccharomyces pombe has also been found (34, 51, 56). Although encoded by a nuclear gene, it is targeted to the mitochondrion. S. cerevisiae CCE1 behaves very like the eubacterial resolvases and shows a preference for cleaving 5′ of a CT dinucleotide (55). Surprisingly, CCE1 shows 43% similarity to MRS1, a mitochondrial RNA splicing enzyme (24). This raises questions regarding the origin of enzymes that recognize junction-like structures in DNA and RNA.
For the most part, enzymes involved in branch migration and resolution of Holliday junctions in the Archaea and Eucarya have yet to be characterized, although junction-resolving activities have been detected in extracts from mammalian cells (14, 20). The identification and characterization of such activities from these two domains of life remain an obstacle to overcome. It will be fascinating to see how much they resemble and operate like their eubacterial counterparts.
ACKNOWLEDGMENTS
We thank those organizations who provided access to their unpublished sequence data including The Institute for Genomic Research, the Sanger Centre, and the University of Oklahoma.
This work was funded in part by the Royal Society and Medical Research Council.
REFERENCES
- 1.Al-Deib A A, Mahdi A A, Lloyd R G. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 4.Bennett R J, Dunderdale H J, West S C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- 5.Bennett R J, West S C. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol. 1995;252:213–226. doi: 10.1006/jmbi.1995.0489. [DOI] [PubMed] [Google Scholar]
- 6.Bidnenko E, Ehrlich S D, Chopin M-C. Lactococcus lactis phage operon coding for an endonuclease homologous to RuvC. Mol Microbiol. 1998;28:823–834. doi: 10.1046/j.1365-2958.1998.00845.x. [DOI] [PubMed] [Google Scholar]
- 7.Bolt E L, Sharples G J, Lloyd R G. Identification of three aspartic acid residues essential for catalysis by the RusA Holliday junction resolvase. J Mol Biol. 1999;286:403–415. doi: 10.1006/jmbi.1998.2499. [DOI] [PubMed] [Google Scholar]
- 8.Carles-Kinch K, George J W, Kreuzer K N. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan S N, Harris L, Bolt E L, Whitby M C, Lloyd R G. Sequence-specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J Biol Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- 10.Chan S N, Vincent S D, Lloyd R G. Recognition and manipulation of branched DNA by the RusA Holliday junction resolvase of Escherichia coli. Nucleic Acids Res. 1998;26:1560–1566. doi: 10.1093/nar/26.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies A A, West S C. Formation of RuvABC-Holliday junction complexes in vitro. Curr Biol. 1998;8:725–727. doi: 10.1016/s0960-9822(98)70282-9. [DOI] [PubMed] [Google Scholar]
- 12.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.Dunderdale H J, Benson F E, Parsons C A, Sharples G J, Lloyd R G, West S C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 14.Elborough K M, West S C. Resolution of synthetic Holliday junctions in DNA by an endonuclease from calf thymus. EMBO J. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 1997;16:203–209. doi: 10.1093/emboj/16.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraud-Panis M J, Lilley D M. Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J Mol Biol. 1998;278:117–133. doi: 10.1006/jmbi.1998.1681. [DOI] [PubMed] [Google Scholar]
- 17.Hagan N F P, Vincent S D, Ingleston S M, Sharples G J, Bennett R J, West S C, Lloyd R G. Sequence-specificity of Holliday junction resolution: identification of RuvC mutants defective in metal binding and target site recognition. J Mol Biol. 1998;281:17–29. doi: 10.1006/jmbi.1998.1934. [DOI] [PubMed] [Google Scholar]
- 18.Hargreaves D, Rice D W, Sedelnikova S E, Artymiuk P J, Lloyd R G, Rafferty J B. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6Å resolution. Nat Struct Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- 19.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 20.Hyde H, Davies A A, Benson F E, West S C. Resolution of recombination intermediates by a mammalian activity functionally analogous to Escherichia coli RuvC resolvase. J Biol Chem. 1994;269:5202–5209. [PubMed] [Google Scholar]
- 20a.Ingleston, S. M., G. J. Sharples, and R. G. Lloyd. Unpublished data.
- 21.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. E. coli RuvC protein is an endonuclease that resolves the Holliday structure, an intermediate of homologous recombination. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleff S, Kemper B, Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Krebs Institute, University of Sheffield. 6 September 1996, revision date. Animated model. [Online.[ http://www.shef.ac.uk/uni/academic/I-M/mbb/ruva/ruva.html. [22 July 1999, last date accessed.]
- 24.Kreike J, Schulze M, Ahne F, Lang B F. A yeast nuclear gene, MRS1, involved in mitochondrial RNA splicing: nucleotide sequence and mutational analysis of two overlapping open reading frames on opposite strands. EMBO J. 1987;6:2123–2129. doi: 10.1002/j.1460-2075.1987.tb02479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd R G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 2236–2255. [Google Scholar]
- 27.Lloyd R G, Sharples G J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahdi A A, McGlynn P, Levett S D, Lloyd R G. DNA binding and helicase domains of the Escherichia coli recombination protein RecG. Nucleic Acids Res. 1997;25:3875–3880. doi: 10.1093/nar/25.19.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Mahdi, A. A., and P. McGlynn. Unpublished results.
- 29.Mahdi A A, Sharples G J, Mandal T N, Lloyd R G. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- 30.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 32a.National Center for Biotechnology Information. 21 July 1999 revision sequence date. Information. database. [Online.] National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov. [22 July 1999, last date accessed.]
- 33.Nishino T, Ariyoshi M, Iwasaki H, Shinagawa H, Morikawa K. Functional analyses of the domain structure in the Holliday junction binding protein RuvA. Structure. 1998;6:11–21. doi: 10.1016/s0969-2126(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 34.Oram M, Keeley A, Tsaneva I. Holliday junction resolvase in Schizosaccharomyces pombe has identical endonuclease activity to the CCE1 homologue YDC2. Nucleic Acids Res. 1997;26:594–601. doi: 10.1093/nar/26.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panayotatos N, Fontaine A. An endonuclease specific for single-stranded DNA selectively damages the genomic DNA and induces the SOS response. J Biol Chem. 1985;260:3173–3177. [PubMed] [Google Scholar]
- 36.Parsons C A, Stasiak A, Bennett R J, West S C. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 37.Rafferty J B, Ingleston S M, Hargreaves D, Artymiuk P J, Sharples G J, Lloyd R G, Rice D W. Structural similarities between Escherichia coli RuvA and other DNA-binding proteins and a mutational analysis of its binding to the Holliday junction. J Mol Biol. 1998;278:105–116. doi: 10.1006/jmbi.1998.1697. [DOI] [PubMed] [Google Scholar]
- 38.Rafferty J B, Sedelnikova S E, Hargreaves D, Artymiuk P J, Baker P J, Sharples G J, Mahdi A A, Lloyd R G, Rice D W. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science. 1996;274:415–421. doi: 10.1126/science.274.5286.415. [DOI] [PubMed] [Google Scholar]
- 38a.Ribosomal Database Project II. 31 July 1995, release date. Sequences. [Online.] Ribosomal Database Project II, Michigan State University. http://www.cme.msu.edu/RDP. [22 July 1999, last date accessed.]
- 39.Roe S M, Barlow T, Brown T, Oram M, Keeley A, Tsaneva I R, Pearl L H. Crystal structure of an octameric RuvA-Holliday junction complex. Mol Cell. 1998;2:361–372. doi: 10.1016/s1097-2765(00)80280-4. [DOI] [PubMed] [Google Scholar]
- 40.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc Natl Acad Sci USA. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Sanger Centre. 21 July 1999, revision date. Sequence database. [Online.] Sanger Centre. http://www.sanger.ac.uk. 22 July 1999, last date accessed.]
- 41.Seigneur M, Bidneko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 42.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 43.Sharples G J, Chan S C, Mahdi A A, Whitby M C, Lloyd R G. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharples G J, Corbett L M, Graham I R. λ Rap protein is a structure-specific endonuclease involved in phage recombination. Proc Natl Acad Sci USA. 1998;95:13507–13512. doi: 10.1073/pnas.95.23.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Sharples, G. J. Unpublished results.
- 45.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi N K, Yamamoto K, Kitamura Y, Luo S, Yoshikura H, Kobayashi I. Non-conservative recombination in E. coli. Proc Natl Acad Sci USA. 1992;89:5912–5916. doi: 10.1073/pnas.89.13.5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.The Institute for Genomic Research. 7 April 1999, revision dates. Sequence database. [Online.] The Institute for Genomic Research. http://www.tigr.org. [22 July 1999, last date accessed.]
- 47.van Gool A J, Shah R, Mézard C, West S C. Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 1998;17:1838–1845. doi: 10.1093/emboj/17.6.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vincent S D, Mahdi A A, Lloyd R G. The RecG helicase of Escherichia coli dissociates R-loops. J Mol Biol. 1996;264:713–721. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- 49.West S C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- 50.Whitby M C, Bolt E L, Chan S N, Lloyd R G. Interactions between RuvA and RuvC at Holliday junctions: inhibition of junction cleavage and formation of a RuvA-RuvC-DNA complex. J Mol Biol. 1996;264:878–890. doi: 10.1006/jmbi.1996.0684. [DOI] [PubMed] [Google Scholar]
- 51.Whitby M C, Dixon J. A new Holliday junction resolving enzyme from Schizosaccharomyces pombe that is homologous to CCE1 from Saccharomyces cerevisiae. J Mol Biol. 1997;272:509–522. doi: 10.1006/jmbi.1997.1286. [DOI] [PubMed] [Google Scholar]
- 52.Whitby M C, Lloyd R G. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 1995;14:3302–3310. doi: 10.1002/j.1460-2075.1995.tb07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitby M C, Ryder L, Lloyd R G. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993;75:341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]
- 54.White M F, Giraud-Panis M-J E, Pöhler J R, Lilley D M J. Recognition and manipulation of branched DNA structure by junction-resolving enzymes. J Mol Biol. 1997;269:647–664. doi: 10.1006/jmbi.1997.1097. [DOI] [PubMed] [Google Scholar]
- 55.White M F, Lilley D M J. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J Mol Biol. 1996;257:330–341. doi: 10.1006/jmbi.1996.0166. [DOI] [PubMed] [Google Scholar]
- 56.White M F, Lilley D M J. Characterization of a Holliday junction-resolving enzyme from Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:6465–6471. doi: 10.1128/mcb.17.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Worth L, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalysed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zerbib D, Mézard C, George H, West S C. Co-ordinated actions of RuvABC in Holliday junction processing. J Mol Biol. 1998;281:621–630. doi: 10.1006/jmbi.1998.1959. [DOI] [PubMed] [Google Scholar]