Significance
Nucleotide excision repair (NER) clears genomes of DNA adducts formed by UV light, environmental agents, and antitumor drugs. Defects in NER cause the skin cancer-prone disease xeroderma pigmentosum, yet NER also counteracts the efficacy of antitumor agents. NER operates through the stepwise assembly of the multiprotein complex and culminates in the excision of the DNA damage. We show that two interaction sites between the XPA and RPA proteins are critical for NER activity by structurally organizing the NER complex and licensing the DNA incision reactions. Our studies furthermore suggest that XPA and RPA constrict the DNA to assume a U-shape and place the two incision sites on the damaged DNA in close proximity to coordinate the two incision reactions.
Keywords: DNA repair, nucleotide excision repair, xeroderma pigmentosum protein A, replication protein A
Abstract
The xeroderma pigmentosum protein A (XPA) and replication protein A (RPA) proteins fulfill essential roles in the assembly of the preincision complex in the nucleotide excision repair (NER) pathway. We have previously characterized the two interaction sites, one between the XPA N-terminal (XPA-N) disordered domain and the RPA32 C-terminal domain (RPA32C), and the other with the XPA DNA binding domain (DBD) and the RPA70AB DBDs. Here, we show that XPA mutations that inhibit the physical interaction in either site reduce NER activity in biochemical and cellular systems. Combining mutations in the two sites leads to an additive inhibition of NER, implying that they fulfill distinct roles. Our data suggest a model in which the interaction between XPA-N and RPA32C is important for the initial association of XPA with NER complexes, while the interaction between XPA DBD and RPA70AB is needed for structural organization of the complex to license the dual incision reaction. Integrative structural models of complexes of XPA and RPA bound to single-stranded/double-stranded DNA (ss/dsDNA) junction substrates that mimic the NER bubble reveal key features of the architecture of XPA and RPA in the preincision complex. Most critical among these is that the shape of the NER bubble is far from colinear as depicted in current models, but rather the two strands of unwound DNA must assume a U-shape with the two ss/dsDNA junctions localized in close proximity. Our data suggest that the interaction between XPA and RPA70 is key for the organization of the NER preincision complex.
Nucleotide excision repair (NER) is the primary repair pathway for the removal of bulky DNA adducts induced by UV irradiation, environmental mutagens, and anticancer agents from our genomes (1). Individuals with mutations in NER genes suffer from xeroderma pigmentosum, a disease characterized by extreme sensitivity to UV irradiation and a highly increased incidence of skin cancer (2). NER is multistep pathway involving more than 30 proteins and is initiated by XPC–RAD23B, which recognizes aberrations in duplex DNA (3). For some lesions, such as cyclopyrimidine dimers (CPDs) in chromatin, this recognition additionally requires UV-DDB and its associated ubiquitin ligase complex (4). XPC–RAD23B recruits the 10-subunit complex TFIIH to lesions (5), which, using the XPB translocase and XPD helicase, opens the DNA and verifies the lesion (6, 7). Xeroderma pigmentosum protein A (XPA) and replication protein A (RPA) then join the complex to stabilize the open DNA (5, 8) and help to position the ERCC1–XPF and XPG endonucleases (9). ERCC1–XPF first incises the DNA 5′ to the lesion, replication proteins initiate gap filling, and following 3′ incision by XPG, gap filling is completed and the resulting nick sealed (10–12). Progression through the NER pathway is driven by multiple protein–protein and protein–DNA interactions among the factors and substrates (13, 14).
The focus of this work is the critical role of XPA and RPA in organizing the preincision complex and licensing the dual incision reaction. XPA is a small protein consisting of only 273 residues that acts as a scaffold by interacting with several NER proteins and the DNA substrate (15). XPA has a central globular core and disordered N and C termini. It binds to single-stranded/double-stranded DNA (ss/dsDNA) junctions with its globular core right at the junction and a long C-terminal α-helix extending away and interacting with the duplex arm (16–19). The binding of XPA to DNA stimulates interactions with NER proteins, including RPA (20–24), and ERCC1–XPF (25, 26).
RPA, the ubiquitous eukaryotic ssDNA binding protein, is a heterotrimer consisting of the RPA70, RPA32, and RPA14 subunits (27). The RPA ssDNA binding apparatus consists of four OB-fold domains (RPA70A, RPA70B, RPA70C, and RPA32D) and two recruitment domains on long flexible linkers: the OB-fold domain RPA70N and the winged helix domain RPA32C. XPA binds to both RPA32C and RPA70AB (20, 24, 28). Using pull-down assays, it was reported that residues 4 to 29 of XPA bind to RPA32 (21). However, NMR analysis revealed the RPA32C-binding motif as XPA29–46, a region that also binds other DNA repair proteins, including UNG2 and RAD52 (20). XPA has a second weaker interaction with RPA70AB through its DNA binding domain (DBD). This interaction was originally attributed to XPA residues 153 to 176 (23, 29), and certain mutations in that region result in reduced NER activity. However, none of the methods used in those studies directly tested whether these residues bind to RPA70AB. Recently, we used NMR chemical-shift perturbation analysis to map the RPA70AB interaction and found that XPA residues 98 to 129 were involved and that there is no role for 153 to 176 (24). We reported a structural model in which residues 101 to 114 in and around the XPA DBD Zn binding motif interact with RPA70AB. This model was confirmed by observing that mutations in several of these residues resulted in reduction in binding affinity and in vitro NER activity (24). Thus, the XPA DBD engages both RPA70AB and the ss/dsDNA junction in the NER bubble.
Here, we report investigations of how the two contacts between XPA and RPA contribute to the NER reaction using a structural approach and mutations in XPA that disrupt the binding interface with both RPA32C and RPA70AB. Biochemical and cellular assays show synergistic contributions to the physical interaction between RPA and XPA and to overall NER activity. Using integrated structural modeling, we show that the two contacts between XPA and RPA can be engaged simultaneously and generate models of how they shape the NER preincision complex.
Results
XPA Mutations Inhibit Binding to RPA.
XPA has been shown to interact with RPA32C via disordered residues 29 to 46 and with RPA70AB through the zinc binding motif in the DBD (20, 24). We set out to inhibit the interaction between XPA and RPA and assess the importance of these interactions for NER using site-specific mutagenesis to generate a series of XPA mutations in these two regions (Fig. 1A and Table 1). We first assessed the two interaction interfaces individually and then in combination.
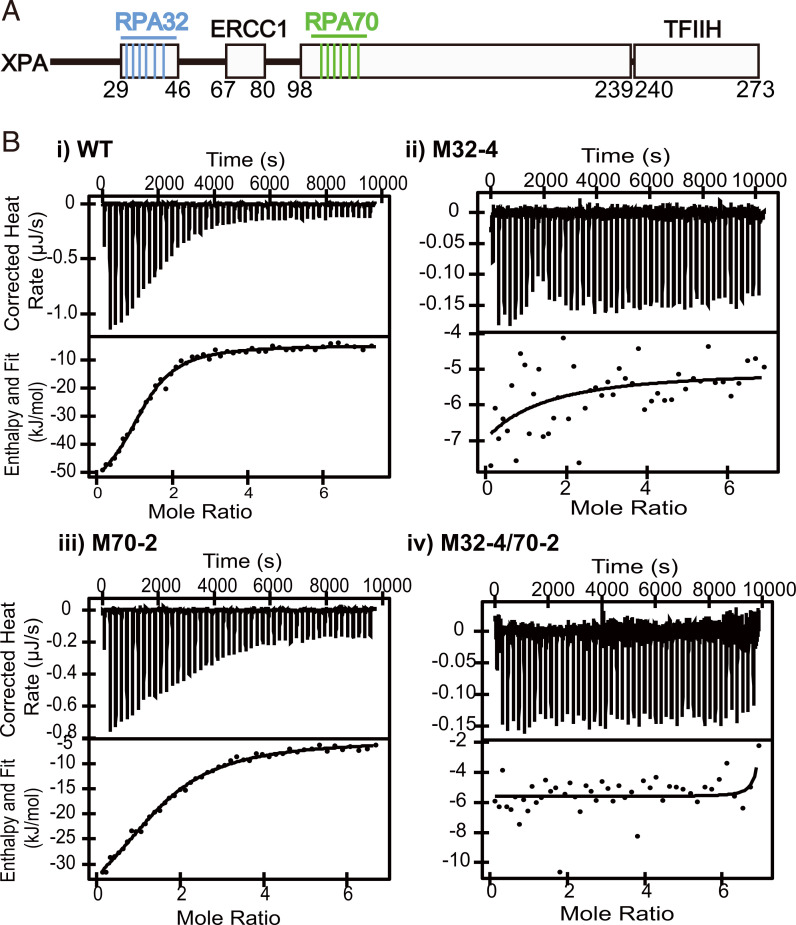
Fig. 1.
Interaction of WT, M32-4, M70-2, and M32-4/70-2 with RPA and DNA. (A) Domain map of XPA. Interaction sites with other protein and mutants introduced in this study are highlighted. (B) Isothermal titration calorimetry thermograms of XPA mutants and full-length RPA showing the raw heat release (Upper) and integrated heat release (Lower). XPA proteins used are indicated. For the titrations in i–iv, the first injection of 1 µL was removed for analysis. The thermograms in B are representative of one of three or more replicates.
Table 1.
Mutant XPA proteins used in this study
| Name | XPA mutation | RPA binding site |
|---|---|---|
| M32-1 | R34E | 32C |
| M32-3 | R34E/R39E/R42E | 32C |
| M32-4 | R30E/K31E/R32E/R34E | 32C |
| M32-6 | R30E/K31E/R32E/R34E/R39E/R42E | 32C |
| M70-1K | E106K | 70AB |
| M70-1A | F112A | 70AB |
| M70-2 | E106K/F112A | 70AB |
| M70-4 | D101N/E106K/K110E/F112A | 70AB |
| M70-6 | D101N/E106K/K110E/E111K/F112A/D114N | 70AB |
| M32-1/70-1 | R34E/F112A | 32C and 70AB |
| M32-1/70-2 | R34E/E106K/F112A | 32C and 70AB |
| M32-3/70-2 | R34E/R39E/R42E/E106K/F112A | 32C and 70AB |
| M32-4/70-2 | R30E/K31E/R32E/R34E/E106K/F112A | 32C and 70AB |
All XPA mutants in this study are named by labeling the binding site in RPA to which they are directed (RPA32C or RPA70AB) and the number of mutations made.
We have previously mapped the interaction of XPA29–46 to an acidic binding surface on RPA32C (20). There is a strong electrostatic component to this interaction, mediated by six basic XPA residues: Arg30, Lys31, Arg32, Arg34, Arg39, and Arg42. To test their role in mediating the interaction with RPA32C, a series of charge reversal mutations of these residues to Glu were made to obtain the strongest possible inhibitory effect (Table 1). We have previously identified residues Asp101, Glu106, Lys110, Glu111, Phe112, and Asp114 of XPA as important in mediating the interaction with RPA70AB and have shown that mutations in these residues diminish the interaction between XPA and RPA70 and inhibit the NER reaction in vitro (24). These same mutations were used here for the cell-based assays, both alone and in combination with the RPA32C mutations. All XPA mutants used in this study were named by labeling the binding site to which they are directed (RPA32C or RPA70AB) and the number of mutations made (Table 1). We made two distinct single mutations in the RPA70AB interaction domain of XPA, E106K, and F112A, and these are denoted as M70-1K and M70-1A, respectively.
To validate the mutant design, we tested the ability of the mutant XPA proteins to interact with RPA and DNA. After purifying the recombinant proteins (SI Appendix, Fig. S1A), the affinity of RPA for XPA WT and variants with mutations in the RPA32C and RPA70AB binding motifs was measured using isothermal titration calorimetry. A dissociation constant (Kd) of 3.6 ± 1.2 µM was determined for the WT protein. Whereas mutations in the RPA32C binding motif had a dramatic effect on the binding of XPA to RPA, mutations in the RPA70AB binding motif had a much more limited effect, no greater than a threefold reduction in affinity compared to the WT protein (Fig. 1B and Table 2). The loss of binding between RPA and the M32-4 mutant, independent of a mutation in the RPA70AB binding motif, supports the previous finding that the RPA32C–XPA29–46 interaction is stronger than the interaction between RPA70AB and XPA98–239 (20, 24).
Table 2.
Thermodynamic parameters for the binding of XPA to RPA
| XPA protein | Kd (µM) | n | ΔH (kJ/mol) | -TΔS (kJ/mol) |
|---|---|---|---|---|
| WT | 3.6 ± 1.2 | 1.3 ± 0.1 | −46 ± 5.4 | 15 ± 5.4 |
| M32-4 | n/o | n/o | n/o | n/o |
| M70-2 | 11 ± 0.5 | 2.0 ± 0.3 | −38 ± 2.9 | 10 ± 2.8 |
| M32-4/70-2 | n/o | n/o | n/o | n/o |
n/o, no observable binding (SI Appendix, Fig. S1).
We next tested the ability of the XPA mutants to bind DNA substrates mimicking NER intermediates using electric mobility shift assays (EMSA)(SI Appendix, Fig. S1C). We have previously shown that mutations in the RPA70AB binding interface do not significantly affect protein folding or DNA binding ability (24). A similar observation was made for the XPA proteins with mutations in the RPA32C binding motif, as EMSA measurements showed their DNA binding affinity was comparable to WT–XPA (SI Appendix, Fig. S1C). We note that the shape of the DNA-bound bands was slightly different, possibly due to the presence of additional negatively charged residues in the protein. Proteins with mutations in both the RPA32C and RPA70AB interaction interfaces also showed robust DNA binding activity, albeit with about twofold lower binding affinity than WT protein (SI Appendix, Fig. S1C).
To further evaluate any perturbations of XPA binding to the DNA substrate, we quantified DNA binding affinity by microscale thermophoresis using the fluorescently tagged NER junction mimic reported previously (30). This approach was used to measure the affinity of the M70-2 mutant in the XPA DBD construct (24) and it had no discernable effect. A similar observation was made here for the full-length mutant protein. In addition, as anticipated based on their location in the XPA N-terminal region distant from the DBD, very little effect on DNA binding affinity was observed for the M32-4 mutation (Kd = 8.4 ± 2.3 µM vs. 5.4 ± 1.1 µM for the WT, respectively) (SI Appendix, Fig. S1B). In the construct combining the M32-4 and M70-2 mutations, there was a slightly larger effect (Kd = 10 ± 3 µM). We are unsure of the origin of these very modest effects on binding of DNA, but they are clearly much smaller than the many orders of magnitude of the effect on XPA–RPA interaction caused by mutations in the RPA32C binding motif.
Inhibition of XPA Interaction with RPA70AB Reduces UV-Induced Damage Repair.
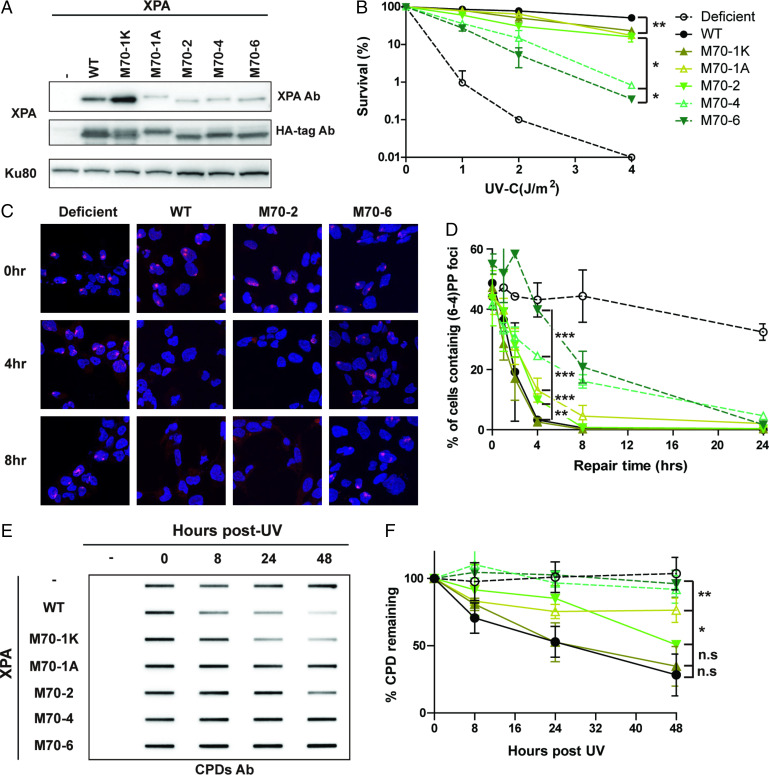
We have previously shown that the mutations in the RPA70AB binding motif of XPA inhibit the physical interaction and reduce the biochemical NER activity on a lesion-containing plasmid (24). To investigate the effect in cells, we generated stable XP2OS (XPA mutant) cell lines expressing XPA with mutations in the RPA70AB interaction domain by lentiviral transfections. We verified that these cells expressed WT and mutant XPA proteins at comparable levels (Fig. 2A). The cells were first tested for hypersensitivity to UV irradiation. Compared to WT, M70-1K, M70-1A, and M70-2 showed a slight but statistically significant increase in UV sensitivity (Fig. 2B). Cells expressing M70-4 and M70-6 with a larger number of mutations exhibited a further increase in UV sensitivity (Fig. 2B).
Fig. 2.
Mutations in the RPA70 interaction domain of XPA cause defects in the repair of UV lesions. (A) Expression level of WT and RPA70-interaction mutant XPA in XP2OS cells transduced with HA-tagged XPA. Proteins were detected with anti-XPA and anti-HA antibodies, using Ku80 as a loading control. (B) Clonogenic survival assays. Cells were treated with the indicated UV dose, grown for 10 d and stained with methylene blue. Survival rates were normalized to nontreated cells. The P value was compared to XPA WT. (C) Representative figures of cells irradiated through a 5-µM micropore filter with UV (100 J/m2) and stained with a (6, 4) PP antibody after the indicated times of repair. (6, 4) PP foci are red and cell nuclei are stained blue with DAPI. For microscope analysis, 10X magnification was used with Axio observer 7 (D) Quantification of C: 100 cells were counted for each sample and the data represent at least two independent experiments. The P value was measured compared to XPA WT. (E) Determination of CPD repair kinetics using slot-blot assays. Cells were irradiated with 5 J/m2 genomic DNA isolated at the indicated time points and the levels determined using an anti-CPD antibody CAC-NM-DND-002. (F) Quantification of E, normalized to WT at 0 h. The P value was compared to XPA WT. *P < 0.05, **P < 0.01, ***P < 0.001.
To obtain deeper insights, we examined the repair kinetics of the UV lesions 6–4 photoproducts [(6, 4) PPs] and CPDs in these cells by: 1) measuring the adduct levels at sites of local UV irradiation in cell nuclei assay and 2) measuring the level of adducts in globally irradiated cells using slot-blot assays. Following irradiation through micropore filters, 40∼60% of cells contained sites of (6, 4) PP damage. In XPA-deficient cells, the (6, 4) PPs persisted for 24 h post-UV irradiation, whereas these adducts were repaired within 4 h in WT–XPA cells (Fig. 2 C and D). The kinetics of (6, 4) PP repair corresponded well to the results of survival assays: M70-1K exhibited WT kinetics of repair, M70-1A and M70-2 had slightly slower repair kinetics, and M70-4 and M70-6 had the slowest repair kinetics, although the (6, 4) PPs were still repaired at much higher levels than in XPA-deficient cells (Fig. 2 C and D). The determination of repair rates using slot blot assays with (6, 4) PP antibodies showed similar results while allowing for better quantification (SI Appendix, Fig. S2 A and B). The repair of (6, 4) PPs was near WT levels in XPA 70-M1K, reduced by about twofold in M70-1A and M70-2, and by about fivefold in M70-4 and M70-6 cells. The effects of the mutations were also measured for CPD repair kinetics, which are known to be repaired significantly more slowly than (6, 4) PPs. The differences in CPD repair rates paralleled those observed for (6, 4) PPs. The results from the repair of the CPDs at sites of local UV damage were more difficult to quantify, due to the slower repair rates leading to foci with gradually decreasing intensity (SI Appendix, Fig. S2 E and F). However, data from the slot blots of the globally irradiated cells revealed an approximately twofold decrease in CPD repair for M70-1A and M70-2 and an approximately fivefold decrease for M70-4 and M70-6 (Fig. 2 E and F and SI Appendix, Fig. S2 C–F). The CPD repair rates for M70-4 and M70-6 were statistically different from WT cells (Fig. 2 E and F and SI Appendix, Fig. S2 C and D). Together, our data indicate that progressive weakening of the XPA–RPA70AB interaction leads to hypersensitivity to UV irradiation due to reduced rates of NER.
Interaction between XPA and RPA32C Promotes NER and Cellular Resistance to UV Irradiation.
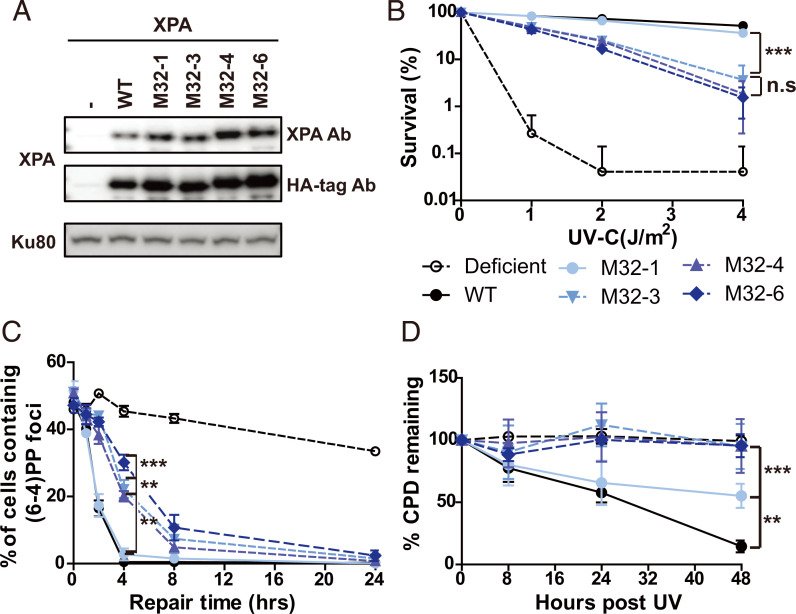
We took an equivalent cell-based approach to assess the importance of the interaction of XPA with RPA32C for the repair of UV lesions by NER. We generated cells with mutations in the RPA32C-interaction motif of XPA (Table 1) and picked clones with comparable XPA expression levels (Fig. 3A). We then tested their sensitivity to UV irradiation and lesion repair kinetics. Following UV treatment, M32-1 cells showed similar sensitivity as WTXPA cells (Fig. 3B), while the M32-3, M32-4, and M32-6 cells showed greater, statistically significant hypersensitivity to UV, although they were less sensitive than XP-A–deficient cells (Fig. 3B). The survival rates of M32-3, M32-4, and M32-6 were not statistically different from each other. We next investigated the repair of UV-induced (6, 4) PPs and CPDs of the mutant cells by local UV irradiation assays and slot-blot assays. The results were consistent between the two assays, with the M32-1 single mutant showing similar levels of (6, 4) PP repair as XPA-WT, while M32-3, M32-4, and M32-6 showed a three- to sixfold decrease in the rate of repair (Fig. 3C and SI Appendix, Fig. S3 A and B). Similarly, there was no difference in the repair of CPDs between WT and M32-1, whereas CPDs persisted in M32-3, M32-4, and M32-6 up to 48 h at similar levels as in XPA-deficient XP2OS cells (Fig. 3D and SI Appendix, Fig. S3 C–F). In conclusion, a progression in the degree of defect in the interaction between XPA and RPA32C or RPA70AB led to increasingly reduced, but still significant NER activity and ability to repair UV-induced damage.
Fig. 3.
Mutations in the RPA32 interacting domain of XPA lead to a cellular defect in the repair of UV lesions. (A) Expression level of WT and RPA32-interaction mutant XPA in XP2OS cells transduced with HA-tagged XPA. Proteins were detected anti-XPA and anti-HA antibodies, using Ku80 as a loading control. (B) Clonogenic survival assays. Cells were treated with the indicated UV dose, grown for 10 d, and stained with methylene blue. Survival rates were normalized to nontreated cells. (C) Quantification of C: 100 cells were counted for each sample and the data represent at least two independent experiments. The P value was compared to XPA WT. (D) Determination of CPD repair kinetics using slot-blot assays. Cells were irradiated with 5 J/m2 genomic DNA isolated, and adduct levels determined with an anti-CPD antibody. Data were normalized to WT at 0 h. The P value was measured compared to XPA WT. **P < 0.01, ***P < 0.001.
Interactions of XPA with RPA32C and RPA70AB Contribute Synergistically to NER.
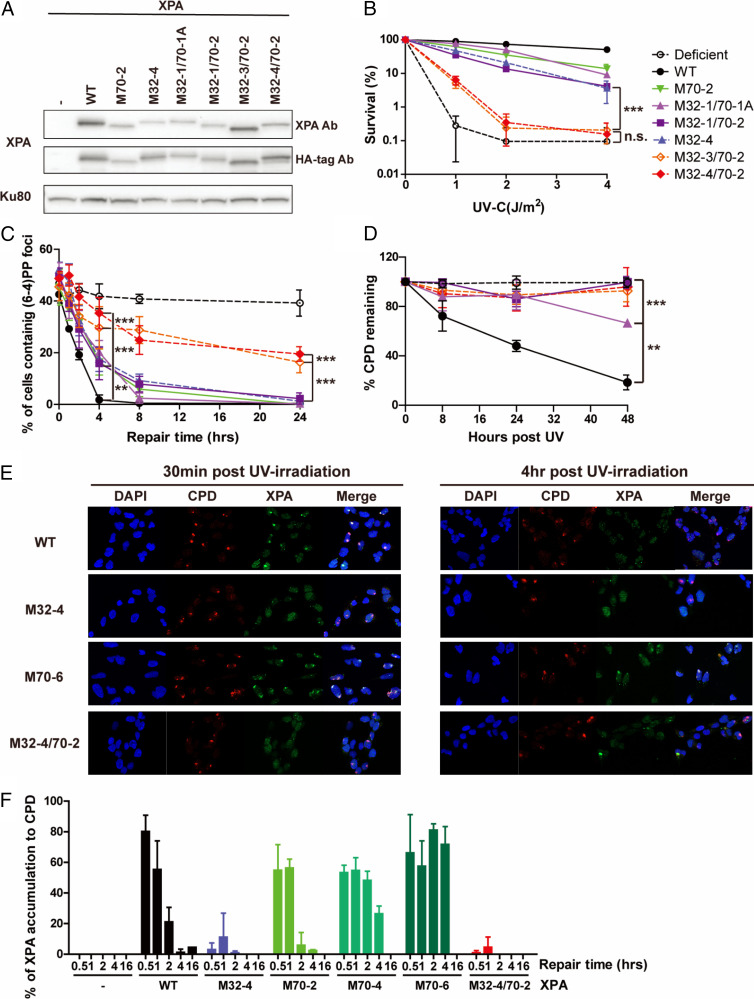
Following the analysis of the individual contacts with RPA, we combined mutations in the RPA32C and RPA70AB binding interfaces of XPA to determine if loss of these physical interactions have independent or synergistic effects on NER activity and sensitivity to UV irradiation. To this end, we generated cells with mutations in both the RPA32C and RPA70AB interaction interfaces and compared their activities with the corresponding mutants in just one or the other interface (Fig. 4A and Table 1). We first tested the sensitivity of those cells to different doses of UV-C (Fig. 4B). Combining a limited number of mutations, such as M32-1 and M70-2, led to twofold reduction in sensitivity compared to individual M70-2, clearly an additive effect (Fig. 4B). The effect was even more dramatic when combining M70-2 and M32-3 or M32-4 mutations. The difference in UV sensitivity of the M32-3/70-2 and M32-4/70-2 was no longer statistically significantly different from the XP2OS patients at a dose UV of 4 J/m2, while the corresponding individual M32-3, M32-4, or M70-2 mutations individually only showed moderate UV sensitivity (Fig. 4B).
Fig. 4.
The RPA32 and 70 binding domains of XPA synergistically contribute to NER activity in cells. (A) XPA expression levels in XP2OS cells transduced with WT and RPA32/70-interaction mutant XPA detected anti-XPA and anti-HA antibodies, using Ku80 as a loading control. (B) Clonogenic survival assays. Cells were treated with the indicated UV dose, grown for 10 d and stained with methylene blue. Survival rates were normalized to nontreated cells. The P value was compared to M32-4. (C) Quantification of (6, 4) PPs repair at sites of local UV damage. Cells were irradiated through a 5-µM micropore filter with UV (100 J/m2) and stained for (6, 4) PP. One-hundred cells were counted for each sample and the data represent at least two independent experiments. The P value was compared to XPA WT. (D) Determination of CPD repair kinetics using slot-blot assays. Cells were irradiated with 5 J/m2 genomic DNA isolated at the indicated time points and adduct levels determined with an anti-CPD antibody. Band intensities were normalized to WT at 0 h. The P value was compared to XPA WT. (E) Representative figure of colocalization assay with CPD and XPA. Cells were irradiated through a 5-µM micropore filter with UV (100 J/m2) and stained for CPD and XPA to measure the accumulation of XPA to CPD. XPA was stained by HA-tag antibody. For microscope analysis, 10X magnification was used with Axio observer 7(F) Quantification of E: 100 cells were counted for each sample and data represent two independent experiments. Percent of colocalization was calculated by dividing number of cells containing colocalization by number of cells containing CPD foci. **P < 0.01, ***P < 0.001.
The progressive effect of cumulative mutations was also reflected in the repair kinetics of UV lesions. M32-1/70-1A and M32-1/70-2 cells exhibited slower repair of (6, 4) PPs at the 8-h time point in both locally damaged sites and globally irradiated cells. A more dramatic loss in the ability to repair UV damage was observed for M32-3/70-2 and M32-4/70-2 (Fig. 4C and SI Appendix, Fig. S4 A, B, and E). In these cells, up to 50% of (6, 4) PPs remained at 24 h post-UV, while (6, 4) PPs were never observed at these time points in any of the individual RPA-binding domain mutants. Indeed, in the slot blot assays, the repair of (6, 4) PPs in these cells was not statistically different from the XP2OS cells. The defects in repair kinetics were equally significant for CPDs. Whereas WT–XPA removed 80% of the CPDs within 48 h, these lesions persisted in cells expressing XPA with mutations in both RPA interaction domains to the same degree as in the XPA patient cells (Fig. 4D and SI Appendix, Fig. S4 C, D, F, and G). Together, these data clearly show that combining mutations in two XPA–RPA interaction interfaces are synergistic, leading to greater NER deficiency than mutations in either of the individual contacts alone.
XPA Interaction with RPA32C Stimulates Recruitment to NER Complexes, and Interaction with RPA70 Is Needed for Completion of NER.
Having shown that both the RPA32C and RPA70AB interaction domains of XPA are important for NER, we aimed to determine what their respective roles might be. We therefore tested the kinetics of the arrival and departure of XPA with the XPA–RPA32C and XPA–RPA70AB interaction mutations at UV-induced DNA damage. In WT cells, the XPA protein arrived at UV damaged sites within 30 min and was released within 4 h (Fig. 4 E and F). In the RPA32C interaction, M32-4–expressing cells, XPA localization to DNA damage was dramatically reduced (3% of UV-induced DNA damage at 0.5 h and 10% of UV-induced DNA damage at 1 h), suggesting that recruitment and association with the NER complex was diminished. In contrast, in cells expressing the RPA70AB interaction mutants, XPA associated with UV damage like the WT cells at 0.5 h and 1 h, but remained bound at damaged sites for extended periods of time (80% for M70-6 and 50% for M70-4 vs. 20% for WT at 2 h, 70% for M70-6 and 25% for M70-4 vs. 1% of WT at 4 h) (Fig. 4F). The M32-4/70-2 double-interaction domain mutant barely localized to UV damage, as expected from the M32-4 mutant (Fig. 4 E and F). These results indicate that RPA32C and RPA70AB have distinct roles for XPA in NER: the RPA32C interaction with XPA is required for recruitment of XPA to UV-induced damage, while RPA70AB interaction to XPA is important for positioning of XPA for completion of NER, likely to license full complex assembly and 5′ incision by ERCC1–XPF.
Mutations in XPA–RPA Interaction Domains Inhibit NER In Vitro Activity.
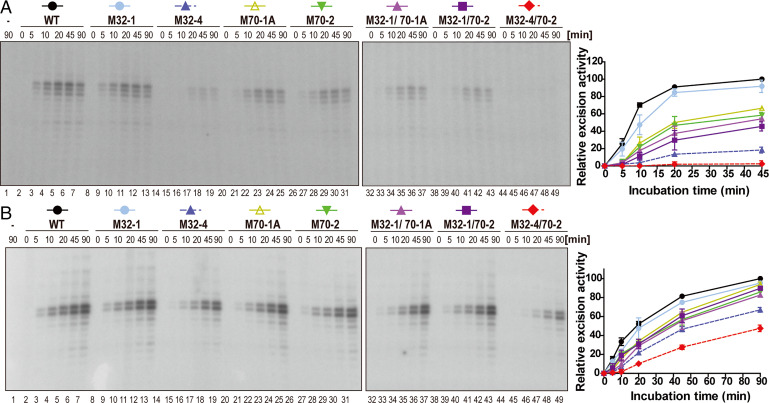
We next tested the intrinsic ability of the mutant XPA proteins to mediate the NER reaction using biochemical experiments by monitoring the excision of a damage-containing oligonucleotide from a plasmid containing a site-specific dG-acetylaminofluorene (AAF) lesion using NER proficient cell extracts or the purified core NER proteins (XPC–RAD23B, TFIIH, XPA, RPA, ERCC1–XPF, and XPG) in the presence of WT and mutant XPA proteins. We first carried out the reaction using extracts from XPA-deficient XP2OS cell extracts supplemented with purified XPA WT and mutant proteins. While incubation with extract alone did not yield a signal, we observed robust, time-dependent accumulation of the excision product in the presence of WT and M32-1 proteins (Fig. 5A, lanes 1 to 13). In contrast, complementation with the M32-4 protein led to an about fivefold reduction in excision activity. (Fig. 5A, lanes 14 to 19). The activity of M70-1A and M70-2 was decreased by about twofold (Fig. 5A, lanes 20 to 31). Combining these last two mutations with M32-1 lead to a further reduction in activity (Fig. 5A, lanes 32 to 43). No excision activity was detectable in the presence of the M32-4/70-2 protein, consistent with the dramatic reduction observed in cellular activity (Fig. 5A, lanes 44 to 49).
Fig. 5.
In vitro NER activity is diminished by mutations in the RPA32 and RPA70 interaction domains of XPA. (A) NER activity of XPA-deficient cell extracts with complemented with WT and mutant XPA. A plasmid containing a site-specific AAF lesion was incubated with XP2OS cell extract and the purified XPA (50 nM) proteins for 0 to 90 min. The excision products were detected by annealing to a complementary oligonucleotide with a 4-dG overhang, which was used as a template for a fill-in reaction with [α-32P] dCTP. Quantification of the data are from two independent experiments. (B) In vitro NER activity of WT and mutant XPA using purified NER proteins. Assays were conducted as in A, except that 20 nM XPA was used with together purified XPC-RAD23B (5 nM), TFIIH (10 nM), RPA (42 nM), XPG (27 nM), and XPF-ERCC1 (13 nM) proteins.
The overall NER activity was higher when purified proteins were used for the NER reaction, but the same order of activity level was observed for the various mutant versions of XPA (Fig. 5B). The activity of the M32-4 protein was reduced by about twofold and for the M32-4/70-2 protein by about fourfold at the 45-min time point (Fig. 5B, lanes 6, 18, and 48). In conclusion, the results from the biochemical experiments are in good agreement with the cellular studies and corroborate the observation that the multiple weak interactions between XPA and both RPA32 and RPA70 are required for progression through the NER pathway.
The Two XPA–RPA Contacts Can Be Simultaneously Accommodated.
Having established that the interactions between XPA and RPA both affect NER proficiency, we considered whether they act either together or independently in promoting repair. One particularly relevant question in this context is whether both contacts can be engaged simultaneously or if they would need to be spaced in time along the NER trajectory. We therefore set out to determine the structure of the complex of XPA and RPA on model NER bubble substrates. As noted above, XPA and RPA are modular proteins with multiple globular and disordered domains and therefore are expected to retain considerable flexibility even when bound to each other. Such systems are not readily amenable to traditional high-resolution structural techniques, such as X-ray crystallography. However, in this case a lower-resolution solution technique, small-angle X-ray scattering (SAXS), is sufficient to provide the structural information of interest.
We have previously established that the XPA DBD can form stable ternary complexes with RPA70AB and model substrates mimicking the ss/dsDNA junction of the NER bubble either 3′ or 5′ to the lesion (24). At first glance, binding to the 5′ junction poses a topological problem because engaging the ssDNA with 5′-3′ polarity (70A-70B-70C-32D) would seem to place RPA70AB far from the XPA DBD at the 5′ ss/dsDNA junction. However, we found that RPA70AB formed similar complexes with the XPA DBD by inverting its orientation to retain 5′-3′ polarity on the ssDNA. Thus, we continued to test if RPA-bound XPA is able to engage both 3′ and 5′ ss/dsDNA junction substrates.
Following the strategy used for study of the complex of XPA DBD and RPA70AB bound to model substrates (24), we designed and optimized Y-shaped DNA substrates for both ss/dsDNA junctions 3′ and 5′ to the lesion. These substrates were composed of dsDNA and a short ssDNA overhang for XPA and a longer ssDNA overhang for RPA. The optimal substrates contained a 10-bp duplex with one 8-nt and one 30-nt overhang (Fig. 6). Complexes formed with these substrates remained stable for days after isolation by size-exclusion chromatography (SEC). Initial SAXS trials were performed with the full-length XPA and RPA proteins, but both had regions that retained a high degree of flexibility because they did not contact the other protein or the DNA substrate. Consequently, we prepared truncation constructs optimized for SAXS analysis: XPA1–239 and RPAΔ32NΔ70N (including deletion of the long RPA70N-RPA70A linker). High-quality SAXS data were collected for complexes of XPA1–239 and RPAΔ32NΔ70N bound to both the 3′ and 5′ junction substrate. Analysis of these data (detailed description provided in SI Appendix) revealed: 1) the solutions were free of aggregation; 2) both complexes contain globular domains with a limited number of flexible loops and linkers; and 3) the ternary complex is stable and globular, which indicates that direct analysis of the distance distribution function, P(r), in terms of a molecular shape is feasible (SI Appendix, Fig. S1 and Table S1). Ab initio shape calculations were therefore performed with density from solution scattering (DENSS) to obtain molecular envelopes for the two complexes (Fig. 6 and SI Appendix, Fig. S6).
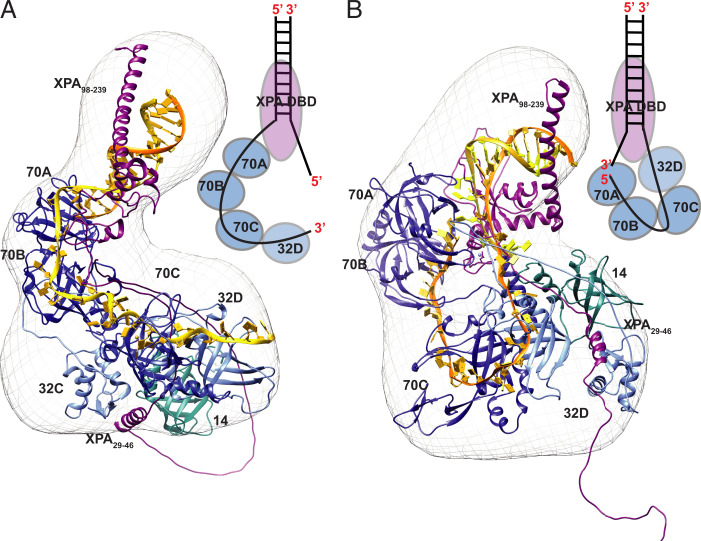
Fig. 6.
SAXS-based structural model of the complex of XPA1–239 and RPAΔ32NΔ70N complex engaged on two model NER substrates mimicking the (A) 3′ ss/dsDNA junction and (B) 5′ ss/dsDNA junction. A schematic of XPA occupying the 3′ (A) and 5′ (B) DNA junctions and RPA subunits engaging the two DNA substrates is illustrated next to the structures. Ribbon diagrams of the XPA–RPA bound to the model substrates with RPA subunits in shades of blue (RPA70ABC-dark blue, RPA32-light blue and RPA14-cyan), XPA DBD in purple and DNA in yellow for the 3′ end and orange for 5′ end. The structures with the SAXS molecular envelopes place XPA DBD and RPA70AB interaction at the top portion of the envelope, revealing the reversal in the orientation of RPA32C/XPA29–46 interaction site for the two substrates. The final representative structural model is shown fit into the ab initio SAXS molecular envelope (mesh) generated by DENSS from the SAXS data.
A variety of previously determined XPA and RPA structures and SAXS molecular envelopes were used to generate our structural models (SI Appendix includes a description). The final representative models show reasonable fits to the corresponding experimental scattering profiles (3′ junction, χ = 2.8; 5′ junction, χ = 4.75) and the corresponding molecular envelopes. The model for the 3′ junction complex has a smoothly curved and compact arrangement of domains with the trimer core (RPA70C/32D/14) and RPA32C/XPA29–46 positioned within the larger lobe of the molecular envelope, and XPA DBD/RPA70AB in the smaller extended lobe (Fig. 6A). The model for the 5′ junction complex has a similar curved arrangement of domains but is less compact (Fig. 6B). Importantly, both ss/dsDNA junction substrates are seen to be readily accommodated, consistent with our past observation that even with RPA bound to the undamaged strand there are no steric factors that inhibit XPA from binding to either junction of the NER bubble.
Despite their overall similarity, careful comparisons of the two models reveal some very clear differences, particularly in the orientation of the RPA domains. The positioning of the RPA32C–XPA29–46 contact is on opposite faces of the molecular envelope in the two complexes. Intriguingly, because RPA binds ssDNA in the 5′-3′ orientation, the path of the ssDNA in the complex with the 5′ ss/dsDNA junction substrate is very different from that with the 3′ substrate. In the complex with the 3′ junction substrate, the ssDNA overhang mimicking the undamaged strand extends smoothly from the 3′ toward the 5′ junction. In contrast, the ssDNA in the complex with the 5′ junction substrate is highly bent and U-shaped to accommodate both the 5′-3′ polarity on the ssDNA and the RPA70AB-XPA interaction. As a result of this topological challenge, the ssDNA overhang mimicking the undamaged strand ends up remarkably close to the 3′ ss/dsDNA junction, placing the two ss/dsDNA junctions in close proximity. The implications of these observations for the structure of the NER preincision complex are discussed below.
Discussion
In this study, we have analyzed the roles of the two distinct interaction sites between XPA and RPA in NER at the biochemical, structural, and cellular level. Our data, summarized in Table 3, show that both interactions are required for full NER activity and that mutations in the RPA-binding surfaces of XPA show additive defects, both physically and functionally. We propose that although similarly important for overall NER activity, the roles of these two contacts are distinctly different. Our data are consistent with a model in which the stronger XPA–RPA32C interaction is important to colocalize the two proteins, while the interaction between XPA–RPA70AB is needed for the proper positioning on the DNA bubble and licensing of the 5′ incision by ERCC1–XPF (Table 3).
Table 3.
Summary of the properties of mutant XPA cells and proteins analyzed in this study
| RPA binding | DNA binding | UV sensitivity | (6, 4) PP repair | Localization to damage | Release from damage | In vitro NER activity | |
|---|---|---|---|---|---|---|---|
| WT | +++ | +++ | — | +++ | +++ | +++ | +++ |
| M32-4 | — | ++ | ++ | + | — | n/o | + |
| M70-2 | ++ | +++ | + | ++ | +++ | ++ | ++ |
| M70-6 | +* | ++* | ++ | + | +++ | — | —* |
| M32-4/70-2 | — | + | +++ | — | — | n/o | — |
| XP-A cells | — | — | +++ | — | — | — | — |
n/o, not observed.
*For M70-6, data denoted by an asterisk are from Topolska-Woś et al. (24).
In our mechanistic models of how RPA functions, RPA32C and RPA70N serve as critical modules to recruit partner proteins to ssDNA bound by the multivalent DNA binding apparatus comprised of domains 70A, 70B, 70C, and 32D (32). XPA29–46 was one the three homologous motifs first identified as characteristic of RPA32C binding sites in DNA repair proteins (20). Subsequent studies have identified motifs with this signature sequence in a number of other RPA binding partners including UNG1, RAD52, SMARCAL1, and TIPIN (31). As we have seen for XPA, the highly modular RPA protein invariably engages its protein binding partners in a multivalent mode with one interaction involving a primary recruitment module and a second weaker interaction that involves the tandem RPA70AB high-affinity ssDNA binding domains. Despite having lower affinity, the RPA70AB interaction is critical to function because it positions the binding partner close to the DNA substrate. Hence, the multivalent mode of binding to RPA fine-tunes XPA function; the interaction with RPA32C increases the local concentration thereby increasing the effective affinity of XPA DBD for RPA70AB and the efficiency of positioning the NER nucleases ERCC1–XPF and XPG. This multivalent interaction mode provides an additional critical element to the NER machinery: weak intrinsic affinity (e.g., between XPA DBD and RPA70AB) implies a higher on-off rate associated, which is perfectly matched to the rapid structural transformations that are required for the progression through the NER trajectory (32, 33).
The functional data show that both contacts are required for NER function, although the mechanism for dysfunction when one of the two contacts are defective is different. Our studies of the kinetics of association and dissociation of XPA at sites of UV damage show that XPA mutations in the RPA32C binding motif are defective in recruitment to the NER machinery, whereas mutations in the RPA70AB binding site, leading to increased residence times at UV damage, are defective in positioning of other NER factors and maintaining the stability of the preincision and incision complexes. The loss of recruitment via interaction with RPA32C is likely compensated by the fact that strong binding of RPA to the undamaged strand will not be affected, so free RPA can still find the strand via diffusion but its arrival on site will be slower. The defect in positioning via interaction with RPA70AB is likely compensated by interaction of XPA with other NER factors. In this case, XPA will not be held in place as effectively and so the NER machinery will be less efficient overall as the incision requires having all factors in place. We propose that these compensatory mechanisms are what provides the residual activity of the various XPA mutants observed in the functional assays.
One unanswered question is whether XPA and RPA are recruited simultaneously to the damage site. In prevailing general models, XPA and RPA are recruited together or XPA is recruited only after TFIIH creates the bubble and RPA is engaged. Reports of XPA, but not RPA, contacts with XPC and TFIIH supports a sequential model in which XPA is recruited first (7, 34). RPA would then be recruited by the combination of its interaction with XPA and its strong affinity for the ssDNA generated as TFIIH unwinds the dsDNA around the lesion. However, cellular studies suggest that XPA and RPA can independently associate with sites of local UV damage (35). This leads to the question, whether RPA can only bind after the bubble is fully open, or whether RPA70AB could engage earlier as it binds tightly to as little as 8-nt of ssDNA. Interaction in this mode would fit well with a model in which RPA and XPA are recruited together to the bubble. Ongoing efforts to structurally examine the dynamic NER trajectory will reveal the intricate molecular mechanisms that govern the assembly and mechanics of the NER machinery (14, 36).
We have previously shown that the XPA DBD can interact with RPA70AB equally well bound to a model of the NER bubble with the ss/dsDNA junction 3′ or 5′ to the lesion (24). This led us to question if XPA remains engaged to a ss/dsDNA junction in the NER bubble or if it shifts from one to the other over the course of the NER trajectory. The structural analysis of the constructs containing both XPA–RPA contacts reported here revealed that the ability to engage either junction is retained. Nevertheless, studies of the isolated XPA–RPA complex are limited by the absence of the steric restrictions enforced by the other components of the NER complexes at various stages of the NER trajectory, the most significant of which is the 10-subunit TFIIH.
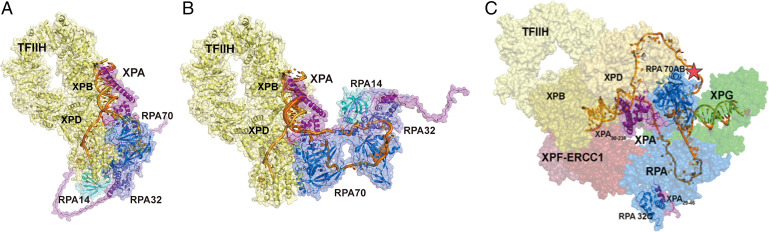
To determine if the binding of XPA and RPA at either junction would be compatible with constraints imposed by other NER factors, we generated models of the XPA–RPA–TFIIH complex bound to DNA substrates with XPA located at either junction (Fig. 7 A and B). Intriguingly, we find that both models can readily accommodate TFIIH without major structural reorganization. The close proximity of the two ss/dsDNA junctions in both models is particularly noteworthy as it is consistent with the curved nature of the RPA–DNA binding apparatus and the potential for XPA to be bound at either junction at different points of the NER trajectory (24). As a further test of XPA binding at either junction, we used the XPA–RPA–TFIIH models to build the corresponding architectural models of the full NER preincision complex (Fig. 7C). These models are based on the following observations: 1) XPD binds the lesion-containing strand and lesion and XPB binds the duplex adjacent to the 5′ junction; 2) in the cryoelectron microscopy (cryo-EM) structure, XPA interacts with TFIIH via XPD, XPB, as well as p8 and p52 (19); 3) the RPA-bound nondamaged strand has a U-shape (37, 38); 4) ERCC1–XPF is located at the 5′ junction, positioned for the 5′ incision (39); and 5) XPG, by virtue of its interaction with TFIIH via p62 and XPD, binds to the “backside” of TFIIH (13, 40) and interacts with the duplex adjacent to the 3′ junction (41, 42).
Fig. 7.
Structural models of NER complexes: (A and B) The SAXS models of XPA–RPA–DNA are aligned by superimposing XPA DBD on the XPA–TFIIH cryo-EM structure (PDB ID: 6R04). (A) TFIIH–XPA–RPA bound to a 3′ ssDNA overhang junction. (B) TFIIH–XPA–RPA bound to a 5′ ssDNA overhang junction. (C) Model of the preincision complex bound to the 5′ junction substrate. This model was prepared by aligning XPA DBD in the cryo-EM structure of TFIIH–XPA (TFIIH in yellow) with our XPA–RPA–5′ junction model (XPA in purple and RPA in blue) and placing the endonucleases XPF–ERCC1 (PDB ID: 6SXB) in red and XPG in green (PDB ID: 6Q0W) along the path of the dsDNA. The lesion is indicated as a star on the NER bubble (gold). The two interaction points of XPA and RPA are highlighted as ribbons.
Prevailing models of NER place XPA at the 5′ junction, based on the interaction of XPA with ERCC1–XPF to mediate 5′ incision and the cryo-EM structure of the XPA–TFIIH complex with TFIIH at the 5′ junction (19). In our corresponding structural model for the preincision complex, RPA70AB must be bound on the undamaged strand placing it close to the 3′ junction. Hence, to meet both of these constraints, the 5 and 3′ junctions must be positioned close together. Perhaps the most striking feature of this model is the U shape of the two strands in the bubble. This arrangement is consistent with structures of the NER nucleases bending the DNA at the ss/dsDNA junction (39, 41, 42). Our current work identifies the RPA–XPA interactions as one of the key elements stabilizing this arrangement and shows that the details of the physical interaction between XPA and RPA are critically important to efficient and effective repair of lesions by the NER pathway.
Materials and Methods
Plasmids.
HA-tagged XPA cDNA was cloned into a pWPXL vector. Mutated pWPXL-XPA and pBG100-XPA expression vectors were generated by site-directed mutagenesis from pWPXL-XPA WT and pBG100-XPA WT using the KOD-mutagenesis kit (Toyobo, TYB-SMK-101). The primers used for inverse PCR mutagenesis are shown in SI Appendix.
Antibodies.
The following antibodies were used for Western blot: anti-XPA (Santacruz, sc-853), anti-HA (Abcam, ab9110), Ku80 (Cell signaling, 2753s), and goat anti-rabbit IgG (Enzo, ADI-SAB-300); and for local UV irradiation assay and slot-blot assays: Anti- (6, 4) PP (Cosmo Bio, CAC-NM-DND-002), anti-CPD (Cosmo Bio, CAC-NM-DND-001), and Cy3 AffiniPure goat anti-mouse IgG (Jackson Immunoresearch, 11CC16C-146).
XPA Protein Expression and Purification.
WT or mutant full-length XPA or truncated XPA1–239 with an N-terminal His6 tag was expressed in Escherichia coli Rosetta pLysS cells using our previously described method (16). Details are described in SI Appendix.
EMSA.
EMSA was conducted using a three-way junction substrate as described previously (24). Details are described in SI Appendix.
Isothermal Titration Calorimetry.
Protein samples were dialyzed against 20 mM Tris (pH 8.0), 150 mM NaCl, 3% glycerol, and 0.5 mM TCEP. Experiments were performed at 25 °C with 125 rpm stirring using an Affinity isothermal titration calorimetry instrument (TA Instruments). Details are provided in SI Appendix.
Microscale Thermophoresis.
Protein samples were exchanged into 50 mM Tris⋅HCl (pH 7.8), 150 mM NaCl, 10 mM MgCl2, 0.05% Tween-20, and 1 mM DTT. Experiments were performed at room temperature using a Monolith NT.115 (NanoTemper Technologies) with standard capillaries. Details are provided in SI Appendix.
Lentiviral Cell Line Transduction.
SV40-transformed human fibroblasts XP2OS (XPA mutant) cells were transduced with lentivirus containing WT or mutant XPA proteins, as described previously (26). Details are described in SI Appendix.
Western Blot.
Protein levels in the transduced cells were assessed using standard conditions after lysing cells using RIPA buffer as described in SI Appendix.
Clonogenic Survival, Local UV Irradiation Assay, and Colocalization Assays.
Clonogenic survival, local UV irradiation, and colocalization assays were conducted as described previously (26). Details are described in SI Appendix.
Slot-Blot Assay.
Cells were irradiated with 5 J/m2 of UV-C, genomic DNA isolated after different repair times and transferred to a nitrocellulose membrane using a BioDot SF microfiltration apparatus (Bio-Rad). Lesions were detected with (6, 4) PP (1:2,000) or CPD primary antibodies (1:3,000), and anti-goat IgG mouse secondary antibody [1:2,500 for (6, 4) PPs or 1:5,000 for CPDs]. Details are described in SI Appendix.
In Vitro NER Activity Assay with Cell Extracts.
A plasmid containing a site-specific AAFlesion was incubated with XPA-deficient (XP2OS) cell extract in the absence or presence of purified WT or mutant XPA proteins as described previously (25). Details are described in SI Appendix.
In Vitro NER Activity Assay with Purified Proteins.
The same plasmid-based assay was as for the cell extract reactions. For each reaction, 5 nM of XPC-RAD23B, 10 nM of TFIIH, 20 nM of XPA, 41.6 nM of RPA, 27 nM of XPG, and 13.3 nM of XPF–ERCC1 was used. All proteins were >95% pure and produced as previously described: XPC–RAD23B (43), TFIIH (44), RPA (45), XPG (46), ERCC1–XPF (47). The reactions were conducted in repair buffer containing 45 mM Hepes-KOH pH 7.8, 5 mM MgCl2, 0.3 mM EDTA, 40 mM phosphocreatine (di-Tris salt, Sigma), 2 mM ATP, 1 mM DTT, 2.5 μg BSA, 0.5 μg creatine phosphokinase (Sigma), and NaCl (to a final concentration of 70 mM).
SAXS.
SEC-SAXS data were collected at the Advanced Light Source beamline 12.3.1 Lawrence Berkeley National Laboratory, Berkeley, California (48). Experiments were performed at 20 °C (49) and data were processed as described previously (50). Details of data acquisition and processing are provided in SI Appendix.
Computational Modeling.
The structure of the XPA1–239/DNA/RPAΔ32NΔ70N complex was generated in four steps involving a combination of homology modeling, docking, and fitting to SAXS data. The protocols utilized are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Susan Tsutakawa (Lawrence Berkeley National Laboratory), Zachary Nagel (Harvard School of Public Health), and members of the Structural Biology of DNA Repair Consortium for helpful comments and discussion. This work was supported by National Cancer Institute Grants R01 CA218315 and P01 CA092584 (to W.J.C. and O.D.S.) and Korean Institute for Basic Science Grant IBS-R022-A1 (to O.D.S.). Microscale thermophoresis instrumentation was acquired from the NIH through Grant S10 OD021483. Single-angle X-ray scattering studies were conducted at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy (DOE), Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program, supported by the DOE Office of Biological and Environmental Research. Additional support comes from the NIH project ALS-ENABLE (P30 GM124169), a High-End Instrumentation Grant S10 OD018483, and the Structural Cell Biology of DNA Repair Machines program (P01 CA092584).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207408119/-/DCSupplemental.
Data Availability
SAXS/structural model data have been deposited in SASBDB/PDB-DEV, https://pdb-dev.wwpdb.org/ (accession nos. SASDPZ3/SASDP24 (51, 52), PDBDEV_00000128 (3' junction) (53), PDBDEV_00000124 (5' junction) (54)). All other study data are included in the main text and SI Appendix.
Change History
September 8, 2022: Author Agnieszka Topolska-Woś’s name has been updated to correct a typographical error.
References
- 1.Gillet L. C., Schärer O. D., Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106, 253–276 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Cleaver J. E., Defective repair replication of DNA in xeroderma pigmentosum. Nature 218, 652–656 (1968). [DOI] [PubMed] [Google Scholar]
- 3.Sugasawa K., et al. , Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2, 223–232 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Sugasawa K., et al. , UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121, 387–400 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Volker M., et al. , Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8, 213–224 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D., Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16, 6559–6573 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riedl T., Hanaoka F., Egly J. M., The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22, 5293–5303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakasugi M., Sancar A., Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc. Natl. Acad. Sci. U.S.A. 95, 6669–6674 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagbemi A. F., Orelli B., Schärer O. D., Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst.) 10, 722–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araújo S. J., et al. , Nucleotide excision repair of DNA with recombinant human proteins: Definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 14, 349–359 (2000). [PMC free article] [PubMed] [Google Scholar]
- 11.Staresincic L., et al. , Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28, 1111–1120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mocquet V., et al. , Sequential recruitment of the repair factors during NER: The role of XPG in initiating the resynthesis step. EMBO J. 27, 155–167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araújo S. J., Nigg E. A., Wood R. D., Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 21, 2281–2291 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsutakawa S. E., et al. , Envisioning how the prototypic molecular machine TFIIH functions in transcription initiation and DNA repair. DNA Repair (Amst.) 96, 102972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugitani N., Sivley R. M., Perry K. E., Capra J. A., Chazin W. J., XPA: A key scaffold for human nucleotide excision repair. DNA Repair (Amst.) 44, 123–135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugitani N., Shell S. M., Soss S. E., Chazin W. J., Redefining the DNA-binding domain of human XPA. J. Am. Chem. Soc. 136, 10830–10833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z., et al. , Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3′- and/or 5′-ssDNA branches. Biochemistry 45, 15921–15930 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Missura M., et al. , Double-check probing of DNA bending and unwinding by XPA-RPA: An architectural function in DNA repair. EMBO J. 20, 3554–3564 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokic G., et al. , Structural basis of TFIIH activation for nucleotide excision repair. Nat. Commun. 10, 2885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mer G., et al. , Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103, 449–456 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Saijo M., Kuraoka I., Masutani C., Hanaoka F., Tanaka K., Sequential binding of DNA repair proteins RPA and ERCC1 to XPA in vitro. Nucleic Acids Res. 24, 4719–4724 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikegami T., et al. , Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nat. Struct. Biol. 5, 701–706 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Li L., Lu X., Peterson C. A., Legerski R. J., An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell. Biol. 15, 5396–5402 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topolska-Woś A. M., et al. , A key interaction with RPA orients XPA in NER complexes. Nucleic Acids Res. 48, 2173–2188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsodikov O. V., et al. , Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J. 26, 4768–4776 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orelli B., et al. , The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J. Biol. Chem. 285, 3705–3712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou Y., Liu Y., Wu X., Shell S. M., Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J. Cell. Physiol. 208, 267–273 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda T., et al. , DNA repair protein XPA binds replication protein A (RPA). J. Biol. Chem. 270, 4152–4157 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Saijo M., Takedachi A., Tanaka K., Nucleotide excision repair by mutant xeroderma pigmentosum group A (XPA) proteins with deficiency in interaction with RPA. J. Biol. Chem. 286, 5476–5483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugitani N., Voehler M. W., Roh M. S., Topolska-Woś A. M., Chazin W. J., Analysis of DNA binding by human factor xeroderma pigmentosum complementation group A (XPA) provides insight into its interactions with nucleotide excision repair substrates. J. Biol. Chem. 292, 16847–16857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldkamp M. D., Mason A. C., Eichman B. F., Chazin W. J., Structural analysis of replication protein A recruitment of the DNA damage response protein SMARCAL1. Biochemistry 53, 3052–3061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugitani N., Chazin W. J., Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog. Biophys. Mol. Biol. 117, 206–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stauffer M. E., Chazin W. J., Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 279, 30915–30918 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Li C. L., et al. , Tripartite DNA lesion recognition and verification by XPC, TFIIH, and XPA in nucleotide excision repair. Mol. Cell 59, 1025–1034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rademakers S., et al. , Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 23, 5755–5767 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan C., et al. , Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nat. Struct. Mol. Biol. 26, 397–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J., Pavletich N. P., Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brosey C. A., et al. , A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 41, 2313–2327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones M., et al. , Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation. Nat. Commun. 11, 1120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafrance-Vanasse J., et al. , Structural and functional characterization of interactions involving the Tfb1 subunit of TFIIH and the NER factor Rad2. Nucleic Acids Res. 40, 5739–5750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsutakawa S. E., et al. , Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. Proc. Natl. Acad. Sci. U.S.A. 117, 14127–14138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.González-Corrochano R., et al. , The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair. Nucleic Acids Res. 48, 9943–9958 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheon N. Y., Kim H. S., Yeo J. E., Schärer O. D., Lee J. Y., Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 47, 8337–8347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gradia S. D., et al. , MacroBac: New technologies for robust and efficient large-scale production of recombinant multiprotein complexes. Methods Enzymol. 592, 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brosey C. A., et al. , NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J. Am. Chem. Soc. 131, 6346–6347 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hohl M., Thorel F., Clarkson S. G., Schärer O. D., Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J. Biol. Chem. 278, 19500–19508 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Enzlin J. H., Schärer O. D., The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21, 2045–2053 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Classen S., et al. , Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 46, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyer K. N., et al. , High-throughput SAXS for the characterization of biomolecules in solution: A practical approach. Methods Mol. Biol. 1091, 245–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hura G. L., et al. , Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim M., et al. , Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair. SAXS/structural https://www.sasbdb.org/data/SASDPZ3/. Deposited 27 May 2022. [DOI] [PMC free article] [PubMed]
- 52.Kim M., et al. , Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair. SAXS/structural https://www.sasbdb.org/data/SASDP24/. Deposited 27 May 2022. [DOI] [PMC free article] [PubMed]
- 53.Kim M., et al. , Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair. SAXS/structural https://pdb-dev.wwpdb.org/entry.html?PDBDEV_00000128. Deposited 27 June 2022. [DOI] [PMC free article] [PubMed]
- 54.Kim M., et al. , Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair. SAXS/structural https://pdb-dev.wwpdb.org/entry.html?PDBDEV_00000124. Deposited 3 June 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SAXS/structural model data have been deposited in SASBDB/PDB-DEV, https://pdb-dev.wwpdb.org/ (accession nos. SASDPZ3/SASDP24 (51, 52), PDBDEV_00000128 (3' junction) (53), PDBDEV_00000124 (5' junction) (54)). All other study data are included in the main text and SI Appendix.