Abstract
Expression of the dnaA gene continues in the lag phase following a temperature downshift, indicating that DnaA is a cold shock protein. Steady-state DnaA protein concentration increases at low temperatures, being twofold higher at 14°C than at 37°C. DnaA protein was found to be stable at both low and high temperatures. Despite the higher DnaA concentration at low temperatures, the mass per origin, which is proportional to the initiation mass, was the same at all temperatures. Cell size and cellular DNA content decreased moderately below 30°C due to a decrease in the time from termination to division relative to generation time at the lower temperatures. Analysis of dnaA gene expression and initiation of chromosome replication in temperature shifts suggests that a fraction of newly synthesized DnaA protein at low temperatures is irreversibly inactive for initiation and for autorepression or that all DnaA protein synthesized at low temperatures has an irreversible low-activity conformation.
Escherichia coli is a mesophilic bacterium able to grow at temperatures as low as 8°C, although the optimum growth temperature is around 37°C (21). When a growing culture of E. coli is shifted to a low temperature, there is a lag (acclimation) phase where net mass increase is very low. During this lag, synthesis of most proteins is severely reduced while a number of proteins, collectively termed cold shock proteins, are synthesized at increased rates (24). After resumption of growth, many cold shock proteins are still synthesized at higher differential rates than at the optimum temperature.
Cell cycle control has been studied extensively for cells growing at or near the optimum temperature. However, very little is known about these events in cells growing at suboptimal temperatures or in cells subjected to a cold shock, i.e., a sudden shift to a low temperature. The major cell cycle event is the initiation of chromosome replication at the chromosomal origin, oriC. Initiation is normally controlled such that the mass per origin at the time of initiation, the initiation mass, is the same irrespective of growth rate, when the growth rate is varied by varying the medium composition (13, 16). The major factor in setting the initiation mass is the initiator protein DnaA (3, 19, 28), which must be accumulated to a certain level for initiation to take place (15). The initiation mass is changed when the level or activity of DnaA protein is varied (18, 35) and when extra copies of DnaA boxes are introduced into wild-type cells (10, 25). DnaA first binds to its binding sites, DnaA boxes, present in oriC and at other positions on the chromosome, including the dnaA promoter which is autorepressed (2, 7). Subsequently, an initial complex consisting of approximately 20 DnaA monomers is formed at oriC, and the double strand is melted in the AT-rich 13-mer region, allowing entry of DnaB and DnaC to generate the prepriming complex (36).
In fast-growing rich medium cultures, cells are born with four origins, which are all initiated within a very short time span, i.e., very synchronously (37). This indicates that there is an initiation cascade which might be due to release of DnaA protein from a newly replicated hemimethylated origin that is sequestered and unable to rebind DnaA (27). The released DnaA will quickly lead to initiation from remaining nonreplicated origins. This initiation synchrony is disturbed by mutations in many different genes (30), including dnaA(Ts) mutant strains grown at permissive temperatures (39).
In the present work, we have studied the initiation control in E. coli grown in rich medium at suboptimal temperatures. DnaA protein levels were monitored by immunoblot, and dnaA gene expression was assessed by a dnaA::lacZ fusion. We used flow cytometry to analyze initiation synchrony, assess DNA- and origin-per-cell distributions, and determine the initiation mass. The replication time, C, was determined by Southern blot marker frequency analysis, and the time from termination to division (D) was estimated from C and the amount of DNA per cell.
MATERIALS AND METHODS
Bacterial strain, growth media, and conditions, and enzyme measurements.
The strain used throughout this study, BBC119 (10), is a Δlac derivative of C600 that is a healthy K-12 strain and carries the λRB1 containing a dnaA::lacZ fusion (7), which allows determination of dnaA gene expression by measuring DnaA–β-galactosidase activity as described previously (3, 31). AB minimal medium (11) supplemented with 1% Casamino Acids (Difco), 0.2% glucose, and 1 μg of thiamine per ml was used for all experiments. Before the start of the experiment, cultures were kept in exponential growth at the different temperatures for more than 15 mass doublings.
Immunoblot procedure.
Sample preparation and immunoblot analysis were carried out as described previously (16). Purified DnaA protein used as standard was obtained from Ole Skovgaard, and the immunoblots were quantified with a Bio-Rad Molecular Analyst GS-700 densitometer.
Flow cytometric procedures.
Samples were prepared and flow cytometry was performed exactly as described in reference 10, based on the procedures described in references 28 and 38. The average number of origins per cell and the average light scatter per cell were determined in samples treated for 4 h at 37°C with rifampin (300 μg/ml) and cephalexin (36 μg/ml). The DNA concentration was determined by analysis of the DNA content and light scatter of samples taken directly from the exponentially growing cultures.
Determination of C by Southern blot marker frequency analysis.
Chromosomal DNA was prepared and restricted with HindIII, and Southern blot analysis was carried out as described previously (3). A hybridization probe mixture was prepared by labeling Qiaquick spin column purified PCR fragments with [35S]dATP, using DNA polymerase I Klenow fragment and random priming (Megaprime kit; Amersham). To prepare the probe mixture, we used two PCR fragments, a 1,051-bp terC and a 1,196-bp oriC fragment, which hybridize to 4.1- and 2.14-kb chromosomal HindIII fragments, respectively. The terC probe was produced by using pBD2348 (3) as the template and the primers ter1 (GTTGAAGTACTTGAGTCACC) and ter2 (CATTCAGACTTGAATGCGTG). The oriC probe was produced by using pFHC271 (17) as the template and the primers ori4 (AAGAATGGCTGGGATCGTGG) and ori6 (ATCGAGGTTACTGCGGATCA). The oriC/terC ratio was determined by measuring the intensities of hybridization to the 2.14- and the 4.1-kb fragments with the Bio-Rad Molecular Analyst GS-700 densitometer. The hybridization signals were normalized to the signals of control plasmid pTAC4511 included on the gels, where the oriC and terC bands are present in a 1:1 ratio. Plasmid pTAC4511 was constructed by inserting the 2.14-kb HindIII gidA fragment from pFHC271 into the unique HindIII site of plasmid pBD2348. Plasmid pTAC4511 was digested with HindIII and NdeI to produce oriC and terC bands of the same or similar size as those in the chromosomal DNA.
RESULTS
We have analyzed cultures grown at four different temperatures: 37°C for optimal growth; 30°C, which has been used frequently in replication studies of dna(Ts) mutants; 21°C, which is the temperature where the Arrhenius plot of growth rate versus temperature shows an abrupt bend (21); and 14°C as the low temperature.
DnaA protein concentrations and dnaA gene expression in balanced growth at different temperatures.
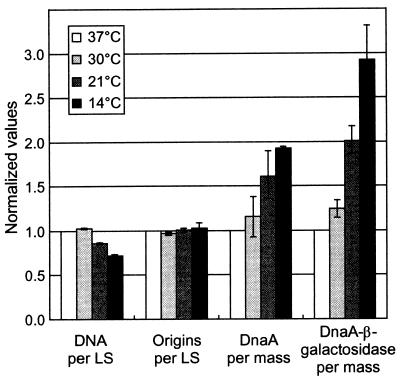
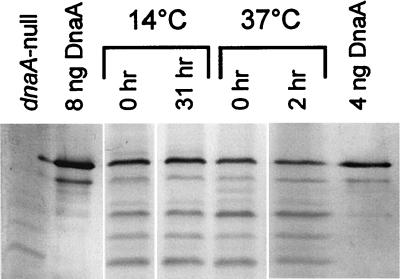
Using the dnaA::lacZ fusion, we measured dnaA gene expression at the four different temperatures and found that it increased with decreasing temperature (Fig. 1). The DnaA–β-galactosidase specific activity was threefold higher in 14°C-grown cells than in those grown at 37°C. The dnaA"lacZ fusion used is a protein fusion at codon 23 of the dnaA gene. Thus, differences in DnaA–β-galactosidase specific activity at different temperatures reflect the combined effects on transcription and translation initiation but ignore the possible effects on transcriptional polarity in dnaA and differential effects on DnaA protein stability. Therefore, we also determined the DnaA protein concentration by immunoblot analysis and found that the DnaA protein itself was also increased at lower temperatures (Fig. 1 and 2), being twofold higher in cells grown at 14°C than in those grown at 37°C. The difference between the increase in DnaA–β-galactosidase (threefold) and DnaA protein itself (twofold), is not due to proteolytic degradation of DnaA at the low temperature, since we found that DnaA is stable at both 14 and 37°C. The amount of DnaA protein stayed constant for four generations after inhibition of protein synthesis (Fig. 2). The difference might therefore be due to increased polarity in the dnaA gene at the low temperature since it has been reported that there is transcription termination in the C-terminal half of dnaA (34).
FIG. 1.
Replication control parameters at different temperatures. Cultures of strain BBC119 were grown under conditions of balanced growth in AB-thiamine-glucose-Casamino Acids supplemented medium at the indicated temperatures. Samples were taken for flow cytometric analysis of DNA, origins, and light scatter (LS), for immunoblot analysis of DnaA protein, and for determination of DnaA–β-galactosidase specific activity. The indicated values are averages of two (30 and 21°C) or three (14°C) independent experiments, and error bars indicate ranges for the different experiments. All values have been normalized to those determined in parallel for the 37°C cultures. The actual values for DNA per cell and origins per cell are shown in Table 1. We found that the cells at 37°C contained approximately 25 ng of DnaA per ml at OD450 = 1, which is in accordance with previous determinations (16), and that the specific activity of DnaA–β-galactosidase was 17 to 20 Miller units.
FIG. 2.
DnaA protein is stable at different temperatures. Cultures of strain BBC119 were grown under conditions of balanced growth in AB-glucose-Casamino-Acids medium at 14 and 37°C. Rifampin and cephalexin were added, and incubation was continued at the growth temperature. Samples were taken for immunoblot analysis at time zero and after approximately four culture generation times (corresponding to 31 h [14°C] and 2 h [37°C]).
Initiation control in balanced growth at different temperatures.
The relative origin-per-mass ratio, which is inversely proportional to initiation mass, was determined by flow cytometric analysis of cells grown at the different temperatures. Surprisingly, in view of the increasing DnaA protein concentration, we found the same origin per mass irrespective of growth temperature (Fig. 1); in other words, the initiation mass is constant. Thus, this is one of the few cases where a higher amount of wild-type DnaA protein does not lead to increased origin per mass. Another example is E. coli B/r relative to K-12 (16).
The DNA concentration at different growth temperatures was also determined by flow cytometric analysis. In view of the apparently constant origin concentration we also expected constant DNA concentration, but found it to decrease slightly but significantly below 30°C (Fig. 1). This difference is not caused by an increase in C relative to generation time (τ) at low temperature (Table 1), suggesting that the cause for the discrepancy is an overestimation of the origin concentration at low temperature.
TABLE 1.
Cell cycle parameters at different growth temperaturesa
| Growth temp (°C) | No. of origins/ cell | DNA (g.e.)/cell | τ (min) | C/τb | D/τc | C (min) | D (min) |
|---|---|---|---|---|---|---|---|
| 37 | 7.2 | 5.0 | 27.5 | 1.53 | 1.49 | 42 | 41 |
| 30 | 7.2 | 5.2 | 43 | 1.40 | 1.56 | 60 | 67 |
| 21 | 6.5 | 3.6 | 120 | 1.35 | 1.22 | 160 | 145 |
| 14 | 6.5 | 3.2 | 465 | 1.39 | 0.89 | 645 | 415 |
BBC119 was grown under conditions of balanced growth in AB-thiamine-glucose-Casamino Acids medium at the indicated temperatures. Samples were taken for flow cytometric analysis of DNA and origins.
Determined from the Southern blot analysis of origins per termini (O/T), using the formula O/T = 2C/τ.
Determined from the formula genome equivalents (g.e.)/cell = [(O/T − 1)/lnO/T] · 2D/τ (8).
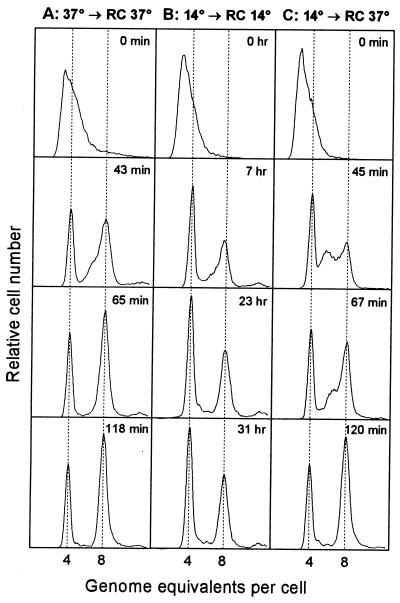
During this work it was reported that rifampin-resistant initiations are provoked by high temperature stress in E. coli (6). For the determination of origins, we had incubated samples with rifampin at 37°C irrespective of initial growth temperature. Therefore, we compared the origins per cell of a 14°C culture treated with rifampin and cephalexin at 14°C and at 37°C to those of a 37°C culture. We followed the kinetics of termination of replication in these three rifampin-treated cultures by taking samples for flow cytometry at different times after addition of the drugs (Fig. 3). When the 14°C culture completed replication at 14°C, the average cell contained 6.06 origins, whereas the average cell contained 6.80 origins after completion of replication at 37°C. The cell size was the same in cultures treated with rifampin at 14 or at 37°C (data not shown). Thus, we have overestimated the origin concentration at 14°C and probably also at 21°C.
FIG. 3.
Runout kinetics after inhibition of initiation of replication and cell division. Cultures of strain BBC119 were grown under conditions of balanced growth in AB-glucose-Casamino-Acids medium at 14 and 37°C (see Table 1 for generation times). Rifampin and cephalexin were added at time zero, and incubation was continued at the growth temperature (A and B), while half of the 14°C culture was shifted to 37°C (C). Samples were taken at the indicated times and fixed, stained, and analyzed by flow cytometry. All distributions were normalized to contain the same number of cells.
The extra replication forks initiated in rifampin at 37°C were significantly slower than normal forks at 37°C (Fig. 3C), giving rise to a peak of cells with 6 origins 45 min after drug addition. These forks had not yet terminated after 90 min (data not shown), but the last sample contained only fully replicated chromosomes. The 37°C-grown cells terminated replication within two generation times (Fig. 3A, 65 min). The 14°C-grown cells, however, terminated replication faster in rifampin at 14°C; replication was nearly completed within one generation time (Fig. 3B, 7 h).
The final origin-per-cell distribution (Fig. 3) shows that synchronous initiation occurs at multiple cellular origins at 14°C and at 37°C. Also, the extra initiations, provoked by shifting the culture to 37°C at the time of rifampin addition, are synchronous, reflecting that initiation of replication had occurred at all four origins in some cells.
Cell cycle parameters in balanced growth at different temperatures.
The major cell cycle event is initiation of replication; cell division follows C + D min later, where C is the chromosome replication time and D is the period from termination to division, during which time the daughter chromosomes are decatenated and segregated and the septum is formed. The number of origins per cell and genome equivalents of DNA per cell (Table 1) are purely a function of C and D relative to τ. The amount of DNA per cell is significantly lower in cells grown at a low temperature, and thus C or D or both relative to τ must be shorter. If the drugs used to inhibit initiation of replication and cell division work efficiently, C + D can be estimated from the number of origins per cell determined by flow cytometry (origins/cell = 2C + D/τ), and C can be estimated from the origins per DNA. But, in this case, where there are rifampin-resistant initiations at the lower temperatures, this method is not applicable.
C was instead determined by measuring the origin-to-terminus ratio by Southern blot analysis, and C/τ was then determined from the formula oriC/terC = 2C/τ. The results (Table 1) showed that C relative to τ varied little with temperature, decreasing slightly at lower temperature. Calculation of D/τ from C/τ and DNA per cell showed that D/τ decreased significantly below 30°C. The value for C of 42 min, obtained at 37°C, is in good agreement with C determined recently for another K-12 strain (5), while the D of 41 min is about twice what is normally found in fast-growing B/r cells (8). The D/τ found at 14°C would correspond to a more normal D of 24 min at 37°C.
Initiation mass and dnaA gene expression following temperature shifts.
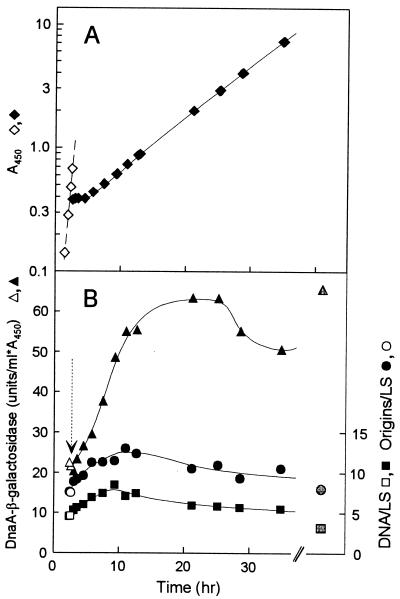
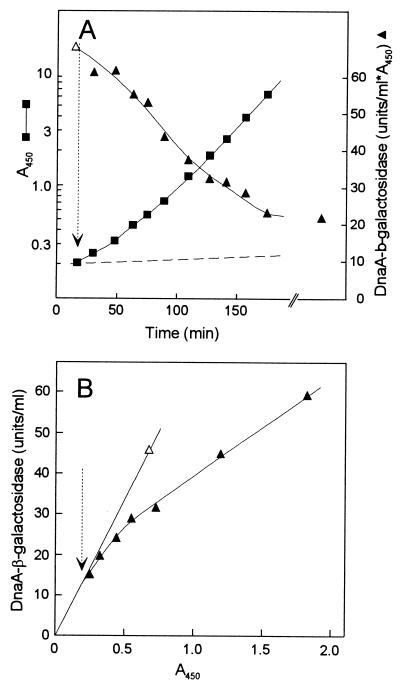
To get a clue to the cause of the higher dnaA gene expression and the reason why higher DnaA protein at a low temperature is not accompanied by higher origin per mass, we shifted cultures in balanced growth (monitored by determination of optical density at 450 nm [OD450]) from 37 to 14°C (Fig. 4), and vice versa, and measured origin per mass, DNA per mass, and DnaA–β-galactosidase activity.
FIG. 4.
Chromosome replication and dnaA gene expression following a temperature downshift. Strain BBC119 was grown under conditions of balanced growth in AB-glucose-Casamino Acids medium at 37°C, and part of the culture was shifted to 14°C at the time indicated by the arrow. (A) Growth was followed by measurement of OD450, and the culture was diluted when the OD450 reached 0.5. (B) Samples were taken for measurement of origins per light scatter (LS), DNA per light scatter, and DnaA–β-galactosidase specific activity. Origins per light scatter was determined in samples incubated with rifampin and cephalexin at 37°C. Open symbols, 37°C; black symbols, 14°C; gray symbols, steady-state 14°C values.
When the culture was shifted from high to low temperature, growth stopped for approximately 2.5 h (Fig. 4A) as previously reported for another wild type K-12 strain (24). After the lag (acclimation phase), which probably corresponds to the time required for synthesis of sufficient amounts of the cold shock proteins (23, 24), growth resumed and very quickly reached a rate identical to that in balanced growth. The dnaA gene expression, measured as DnaA–β-galactosidase activity started to increase immediately after the shift (Fig. 4B). DnaA–β-galactosidase synthesis remained high after resumption of growth, and the specific activity reached the steady-state level within two mass doublings after the shift. Also, DnaA protein concentrations increased to near steady-state levels during this time (data not shown). Initiation frequency increased immediately after the shift, resulting in a 1.7-fold increase in origins per mass within the first mass doubling. After this time, origins per mass started to decrease but did not reach the steady-state level within the time span of the shift experiment (Fig. 4B). Origins per mass measured by incubation with rifampin at 14°C followed the same kinetics (data not shown). A temperature shift from 37 to 21°C gave very similar results except that there was no growth lag, and the shift effects on dnaA gene expression, like that in steady state, and on initiation were of smaller magnitude (data not shown). The downshift experiments indicate that dnaA is one of the cold shock genes. In the first period after temperature downshift, the increase in DnaA protein synthesis results in an increase in initiation frequency. However, it seems that after some time at the low temperature the DnaA protein activity is half of the 37°C activity, leading to the same apparent initiation mass as obtained at 37°C.
When the opposite shift was carried out, growth continued without any lag (Fig. 5A), although initially with a lower rate than in steady state. The growth rate gradually increased, reaching the steady-state rate after three mass doublings, suggesting that some cold shock proteins were inhibiting growth at 37°C and had to be diluted out by growth. DnaA–β-galactosidase synthesis gradually declined after the shift, as seen most clearly from the plot in Fig. 5B, where the slope is equivalent to differential rate of synthesis. DnaA–β-galactosidase synthesis approached the steady-state rate only after more than two mass doublings. These kinetics suggest that a low temperature-stimulatory factor for dnaA gene expression had to be diluted before the normal 37°C rate could be established. It might also indicate that the excess DnaA protein from 14°C was not activated as an autorepressor upon the temperature shift. Origin per mass was virtually constant over the shift (data not shown), indicating that the excess DnaA protein was incapable of acting as an initiator.
FIG. 5.
Growth and dnaA gene expression after temperature upshift. Strain BBC119 was grown under conditions of balanced growth in AB-glucose-Casamino Acids medium at 14°C, and part of the culture was shifted to 37°C at the time indicated by the arrow. (A) Growth was followed by measurement of OD450 and the culture was diluted when the OD450 reached 0.5. Samples were taken for measurement of DnaA–β-galactosidase specific activity. (B) Differential plot of values for DnaA–β-galactosidase activity against OD450 from panel A. Open symbols: 14°C; black symbols, 37°C.
DISCUSSION
The results presented here show that the initiator protein DnaA is a cold shock protein that is synthesized at higher rates and is present at higher concentrations at low growth temperatures. In spite of a twofold-higher DnaA concentration at 14°C than at 37°C, the steady-state origin and DNA concentrations were found to be lower. This results in a more than twofold-higher DnaA-per-oriC ratio in cells grown at 14°C than in those grown at 37°C. In addition, we found that C relative to τ was very similar at all growth temperatures, whereas D relative to τ becomes significantly shorter at the lower temperatures, resulting in less DNA per cell.
Cold shock induction of the dnaA gene.
During the lag following a downshift in growth temperature, synthesis of most proteins is severely reduced while a number of proteins (cold shock proteins) are synthesized at increased rates (24). Class I cold shock proteins are those for which synthesis is increased more than 10-fold and which are expressed at very low levels at 37°C (32). Class II proteins, like GyrA, InfB, and H-NS, are synthesized at high rates at 37°C and are induced only a fewfold upon cold shock. In addition, synthesis of many ribosomal proteins is high during the lag phase (23). DnaA clearly belongs to the class II cold shock proteins: DnaA–β-galactosidase activity increases during the lag phase, and initially after resumption of growth the differential rate of synthesis is more than fourfold higher than at 37°C.
The primary cause of the growth lag is impaired initiation of translation on most mRNAs (23, 41), and this defect leads to decreased pppGpp and ppGpp levels (29). Transcription of the dnaA gene, like that of ribosomal genes, is negatively regulated by ppGpp (9). We therefore expect dnaA mRNA to increase upon the temperature downshift. mRNAs for all identified cold shock proteins, both class I and class II, carry a sequence motif, a 14-base downstream box, complementary to bases in 16S RNA around base 1475 (40). This sequence motif, which is required for translation of the cspA mRNA in the acclimation phase, has been proposed to mediate initiation of translation of all cold shock genes in the lag phase (32). The downstream box of class II genes starts at the initiation codon (32). When we inspected the dnaA mRNA sequence, we found a very nice homology to the downstream box starting at the GUG codon located 12 bp upstream of the GUG normally considered to be the start codon for dnaA (Fig. 6). Initiation of dnaA translation using the downstream box would thus produce a DnaA protein with an N-terminal extension of four amino acids. This alternative product is probably an active DnaA protein, since an N-terminal fusion with a biotinylation domain shows DnaA activity in vivo (1).
FIG. 6.
Nucleotide sequence of the dnaA mRNA translational initiation region. At the top is shown the 16S rRNA sequence which is complementary to the downstream box (40). In the dnaA mRNA sequence, the normal ribosome binding site and GUG start codon are shown in bold; at the bottom is shown the DnaA sequence translated from the first GUG start codon.
The increased DnaA concentration elicited by cold shock leads to a transient increase in origin concentration which causes a temporary increase in rrn operons and ribosomal protein operons relative to other genes due to their origin proximal localization. This increased relative ribosomal gene dosage might help to restore the translation capacity during the transition from the lag phase to renewed growth.
Rifampin-resistant initiations.
In agreement with the results of Botello and Jiménez-Sánchez (6), we observed rifampin-resistant initiations when cells grown at a low temperature were incubated with the drug at a high temperature. In our experiment, however, only a small fraction of the cells (10%) initiated after the drug addition. Botello and Jiménez-Sánchez (6) showed that the rifampin-resistant initiations were from the oriC region but were abnormal in the sense that they required RNase H and RecA. Rifampin-resistant initiations have previously been characterized when DnaA protein in some of the mutants in the P loop of the ATP binding site (e.g., dnaA601 and dnaA606) is reactivated upon downshift in temperature (14). The capacity for rifampin-resistant initiation was suggested to be dependent on DnaA since it decayed with the same half-life as the mutant DnaA proteins. Furthermore, rifampin-resistant initiation has been observed in cells overproducing wild-type DnaA protein (1). We suggest that the rifampin-resistant initiations elicited by shift to high temperature might be provoked by the higher DnaA concentrations in cells grown at low temperature.
Inactivation of DnaA protein activity at low temperature.
In contrast to the effects of increased DnaA protein levels at normal growth temperatures (3, 4), we found that the higher DnaA protein concentrations at low temperature than at 37°C did not lead to higher origin concentration. Actually, origins per mass decreased with decreasing growth temperature. Thus, initiation of chromosome replication clearly requires more DnaA protein per oriC at the lower temperatures. The downshift experiment showed that the DnaA protein synthesized upon the cold shock increased the initiation capacity and that the activity decreased slowly over several generations of growth. The temperature upshift experiment indicated that the surplus DnaA protein synthesized at the low temperature was inactive both for initiation of chromosome replication and for dnaA autoregulation.
The low activity of DnaA at the low temperature might be caused by folding problems due to the lower level of the chaperonins DnaK, GroES, and GroEL (23). These chaperonins have been shown to reactivate mutant DnaA proteins (20, 22). However, we consider this unlikely in view of the results of the temperature upshift, where levels of heat shock proteins should increase rapidly, and thus at least the DnaA synthesized at a higher rate shortly after the shift should have normal activity.
Therefore, we favor alternative hypotheses. The low DnaA activity could be due to increased synthesis of an inhibitor at low temperatures. The level of unsaturated fatty acids increases at low temperatures to maintain membrane fluidity (12). The inhibitory factor is, however, probably not the unsaturated acid phospholipids, since an increased level of unsaturation has been shown to stimulate the ATP-ADP exchange reaction that in vitro activates DnaA for initiation (43). Among the proteins, SeqA is the prime candidate for an inhibitory factor, since it seems to be an inhibitor of DnaA for both initiation and autoregulation (26, 42). Another candidate is IdiA, which inhibits DnaA initiation activity in vitro by stimulation of ATP hydrolysis (33).
Alternatively, DnaA protein, expressed in cells growing at low temperatures, may be synthesized in a form (maybe with four extra N-terminal amino acids) which shows altered binding characteristics toward the different DnaA boxes present on the chromosome. In such a model, the low-temperature DnaA protein is postulated to have lower affinity for the dnaA promoter and oriC, leading to the observed twofold-higher DnaA protein concentration at 14°C, which is needed to give a normal initiation mass at this temperature. In the temperature downshift experiment, initially there will be sufficient 37°C DnaA protein available which, together with the newly synthesized protein, acts to temporarily decrease the initiation mass. However, after prolonged growth at 14°C, the 37°C DnaA protein will be diluted out and the initiation mass will be set exclusively by the 14°C DnaA protein. In the reciprocal shift similar considerations can be applied.
We are presently investigating whether low-temperature DnaA protein is predominantly the four-amino-acid-extended form and whether artificially produced DnaA protein of this variety has lower activity at 37°C.
ACKNOWLEDGMENTS
We thank Kirsten Olesen for technical assistance and Anders Løbner-Olesen, Ulrik von Freiesleben, and Ole Skovgaard for discussions and editorial advice on the manuscript.
This work was supported by grants from the Danish Natural Science Research Council.
REFERENCES
- 1.Atlung, T. 1999. Unpublished work.
- 2.Atlung T, Clausen E, Hansen F G. Autoregulation of the dnaA gene of Escherichia coli. Mol Gen Genet. 1985;200:442–450. doi: 10.1007/BF00425729. [DOI] [PubMed] [Google Scholar]
- 3.Atlung T, Hansen F G. Three distinct chromosome replication states are induced by increasing concentrations of DnaA protein in Escherichia coli. J Bacteriol. 1993;175:6537–6545. doi: 10.1128/jb.175.20.6537-6545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlung T, Løbner-Olesen A, Hansen F G. Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in E. coli. Mol Gen Genet. 1987;206:51–59. doi: 10.1007/BF00326535. [DOI] [PubMed] [Google Scholar]
- 5.Bipatnath M, Dennis P P, Bremer H. Initiation and velocity of chromosome replication in Escherichia coli B/r and K-12. J Bacteriol. 1998;180:265–273. doi: 10.1128/jb.180.2.265-273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botello E, Jiménez-Sánchez A. A temperature upshift induces initiation of replication at oriC on the Escherichia coli chromosome. Mol Microbiol. 1997;26:133–144. doi: 10.1046/j.1365-2958.1997.5621924.x. [DOI] [PubMed] [Google Scholar]
- 7.Braun R E, O’Day K, Wright A. Autoregulation of the DNA replication gene dnaA in E. coli. Cell. 1985;40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 8.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 9.Chiaramello A E, Zyskind J W. Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J Bacteriol. 1990;172:2013–2019. doi: 10.1128/jb.172.4.2013-2019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen B B, Atlung T, Hansen F G. DnaA boxes are important elements in setting the initiation mass of Escherichia coli. J Bacteriol. 1999;181:2683–2688. doi: 10.1128/jb.181.9.2683-2688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark D J, Maaløe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 12.Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 13.Donachie W D. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 14.Hansen F G. Reinitiation kinetics in eight dnaA(Ts) mutants of Escherichia coli: rifampicin resistant initiation of chromosome replication. Mol Microbiol. 1995;15:133–140. doi: 10.1111/j.1365-2958.1995.tb02227.x. [DOI] [PubMed] [Google Scholar]
- 15.Hansen F G, Atlung T. Initiation of chromosome replication after induction of DnaA protein synthesis in a dnaA(null) rnh mutant of Escherichia coli. Mol Microbiol. 1995;15:149–154. doi: 10.1111/j.1365-2958.1995.tb02229.x. [DOI] [PubMed] [Google Scholar]
- 16.Hansen F G, Atlung T, Braun R E, Wright A, Hughes P, Kohiyama M. Initiator (DnaA) protein concentration as a function of growth rate in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1991;173:5194–5199. doi: 10.1128/jb.173.16.5194-5199.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen F G, Koefoed S, von Meyenburg K, Atlung T. Transcription and translation events in the oriC region of the E. coli chromosome. ICN-UCLA Symp Mol Cell Biol. 1981;22:37–55. [Google Scholar]
- 18.Hansen F G, Rasmussen K V. Regulation of the dnaA product in E. coli. Mol Gen Genet. 1977;155:219–225. doi: 10.1007/BF00393163. [DOI] [PubMed] [Google Scholar]
- 19.Herrick J, Kohiyama M, Atlung T, Hansen F G. The initiation mess? Mol Microbiol. 1996;19:659–666. doi: 10.1046/j.1365-2958.1996.432956.x. [DOI] [PubMed] [Google Scholar]
- 20.Hupp T R, Kaguni J M. Activation of mutant forms of DnaA protein of Escherichia coli by DnaK and GrpE proteins occurs prior to DNA replication. J Biol Chem. 1993;268:13143–13150. [PubMed] [Google Scholar]
- 21.Ingraham J L, Marr A G. Effect of temperature, pressure, pH, and osmotic stress on growth. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1570–1577. [Google Scholar]
- 22.Jenkins A J, March J B, Oliver I R, Masters M. A DNA fragment containing the groE genes can suppress mutations in the E. coli dnaA gene. Mol Gen Genet. 1986;202:446–454. doi: 10.1007/BF00333275. [DOI] [PubMed] [Google Scholar]
- 23.Jones P G, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 24.Jones P G, VanBogelen R A, Neidhardt F C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu M, Campbell J L, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 27.Løbner-Olesen A, Hansen F G, Rasmussen K V, Martin B, Kuempel P L. The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J. 1994;13:1856–1862. doi: 10.1002/j.1460-2075.1994.tb06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Løbner-Olesen A, Skarstad K, Hansen F G, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57:881–889. doi: 10.1016/0092-8674(89)90802-7. [DOI] [PubMed] [Google Scholar]
- 29.Mackow E R, Chang F N. Correlation between RNA synthesis and ppGpp content in Escherichia coli during temperature shifts. Mol Gen Genet. 1983;192:5–9. doi: 10.1007/BF00327639. [DOI] [PubMed] [Google Scholar]
- 30.Messer W, Weigel C. Initiation of chromosome replication. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1578–1601. [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Mitta M, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 33.Mizushima T, Nishida S, Kurokawa K, Katayama T, Miki T, Sekimizu K. Negative control of DNA replication by hydrolysis of ATP bound to DnaA protein, the initiator of chromosomal DNA replication in Escherichia coli. EMBO J. 1997;16:3724–3730. doi: 10.1093/emboj/16.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Roger I, Macian F, Armengod M E. Transcription termination in the Escherichia coli dnaA gene is not mediated by the internal DnaA box. J Bacteriol. 1995;177:1896–1899. doi: 10.1128/jb.177.7.1896-1899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaus N A, O’Day K, Peters W, Wright A. Isolation and characterization of amber mutations in gene dnaA of Escherichia coli K-12. J Bacteriol. 1981;145:904–913. doi: 10.1128/jb.145.2.904-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekimizu K, Bramhill D, Kornberg A. Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J Biol Chem. 1988;263:7124–7130. [PubMed] [Google Scholar]
- 37.Skarstad K, Boye E, Steen H B. Timing of initiation of chromosome replication in individual E. coli cells. EMBO J. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skarstad K, Steen H B, Boye E. Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J Bacteriol. 1985;163:661–668. doi: 10.1128/jb.163.2.661-668.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skarstad K, von Meyenburg K, Hansen F G, Boye E. Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J Bacteriol. 1988;170:852–858. doi: 10.1128/jb.170.2.852-858.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sprengart M L, Fuchs E, Porter A G. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 41.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Freiesleben U, Rasmussen K V, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14:763–772. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- 43.Yung B Y M, Kornberg A. Membrane attachment activates dnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:7202–7205. doi: 10.1073/pnas.85.19.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]