Significance
Cerebellar development relies on a precise coordination of metabolic signaling, epigenetic signaling, and transcriptional regulation. Here, we reveal that O-GlcNAc transferase (OGT) regulates cerebellar neurogenesis and medulloblastoma growth via a Sonic hedgehog (Shh)-Smo-Gli2 pathway. We identified Gli2 as a substrate of OGT, and unveiled cross-talk between O-GlcNAc and epigenetic signaling as a means to regulate Gli2 transcriptional activity. Moreover, genetic ablation or chemical inhibition of OGT significantly suppresses tumor progression and increases survival in a mouse model of Shh subgroup medulloblastoma. Taken together, the data in our study provide a line of inquiry to decipher the signaling mechanisms underlying cerebellar development, and highlights a potential target to investigate related pathologies, such as medulloblastoma.
Keywords: O-GlcNAcylation, Gli2, GNPs, neurogenesis, cerebellum
Abstract
Sonic hedgehog (Shh) signaling plays a critical role in regulating cerebellum development by maintaining the physiological proliferation of granule neuron precursors (GNPs), and its dysregulation leads to the oncogenesis of medulloblastoma. O-GlcNAcylation (O-GlcNAc) of proteins is an emerging regulator of brain function that maintains normal development and neuronal circuitry. Here, we demonstrate that O-GlcNAc transferase (OGT) in GNPs mediate the cerebellum development, and the progression of the Shh subgroup of medulloblastoma. Specifically, OGT regulates the neurogenesis of GNPs by activating the Shh signaling pathway via O-GlcNAcylation at S355 of GLI family zinc finger 2 (Gli2), which in turn promotes its deacetylation and transcriptional activity via dissociation from p300, a histone acetyltransferases. Inhibition of OGT via genetic ablation or chemical inhibition improves survival in a medulloblastoma mouse model. These data uncover a critical role for O-GlcNAc signaling in cerebellar development, and pinpoint a potential therapeutic target for Shh-associated medulloblastoma.
The cerebellum controls balance, coordination, movement, and motor skills, with impact on memory and cognitive ability (1–3). Its normal development relies on an intricate spatial and temporal distribution of signaling molecules, the activity of which are often controlled via posttranslational modifications. Disruption of these molecular processes during development causes abnormalities in morphogenesis and circuit function (4, 5). Sonic hedgehog (Shh) signaling is crucial for regulating cerebellum development by promoting the proliferation and differentiation of granule neuron precursors (GNPs) (6, 7). Recent studies suggest that Shh regulates key processes in the proliferation and self-renewal of neural stem cells (NSCs) and neural patterning in the dentate gyrus of the hippocampus (8–10). However, the mechanisms underlying Shh signaling to regulate cerebellum morphology and its potential impact on medulloblastoma, a malignant brain tumor, remains unclear.
O-GlcNAcylation (O-GlcNAc) has recently emerged as a critical regulator of neurodevelopment, synaptic maturation, circuit excitability, and cognition (11–13). As a rapid and reversible posttranslational modification, O-GlcNAc is controlled by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) (14). OGT adds O-GlcNAc to the Ser and Thr residues of proteins, while OGA removes the O-GlcNAc modification. O-GlcNAc impacts adult neurogenesis, evidenced by that decreased O-GlcNAc modification in the dentate gyrus correlates with reduced adult neural stem cells (NSCs) and increased glial differentiation (15). O-GlcNAc also controls the proliferation and differentiation of NSCs in the cortex through endoplasmic reticulum stress and the Notch signaling pathway (16, 17). However, despite its high expression level in the embryonic and adult cerebellum (18–20), little is known about its role with respect to cerebellar development and dysregulation.
Here, we report that O-GlcNAc regulates the neurogenesis of GNPs during cerebellum development, and aberrant O-GlcNAc signaling in GNPs may contribute to medulloblastoma oncogenesis. Specifically, we identify OGT as a key signaling molecule in regulating the proliferation of GNPs through the Shh signaling pathway. O-GlcNAc of the GLI family zinc finger 2 protein (Gli2), a transcription factor in the Shh-signaling pathway, regulates GNP neurogenesis and cerebellum development via inhibition of its interaction with histone acetyltransferase (HAT). Depletion or pharmacological inhibition of OGT in a medulloblastoma mouse model significantly inhibited tumor proliferation and increased survival via inhibiting Gli2 transcriptional activity. This was further supported by abnormally elevated O-GlcNAc levels in human samples of Shh-subtype medulloblastoma. Our results illustrate a fundamental role of O-GlcNAc in cerebellum development, and established OGT as a potential target against Shh-subtype medulloblastoma.
Results
Ablation of OGT in GNPs Disrupts Cerebellar Development.
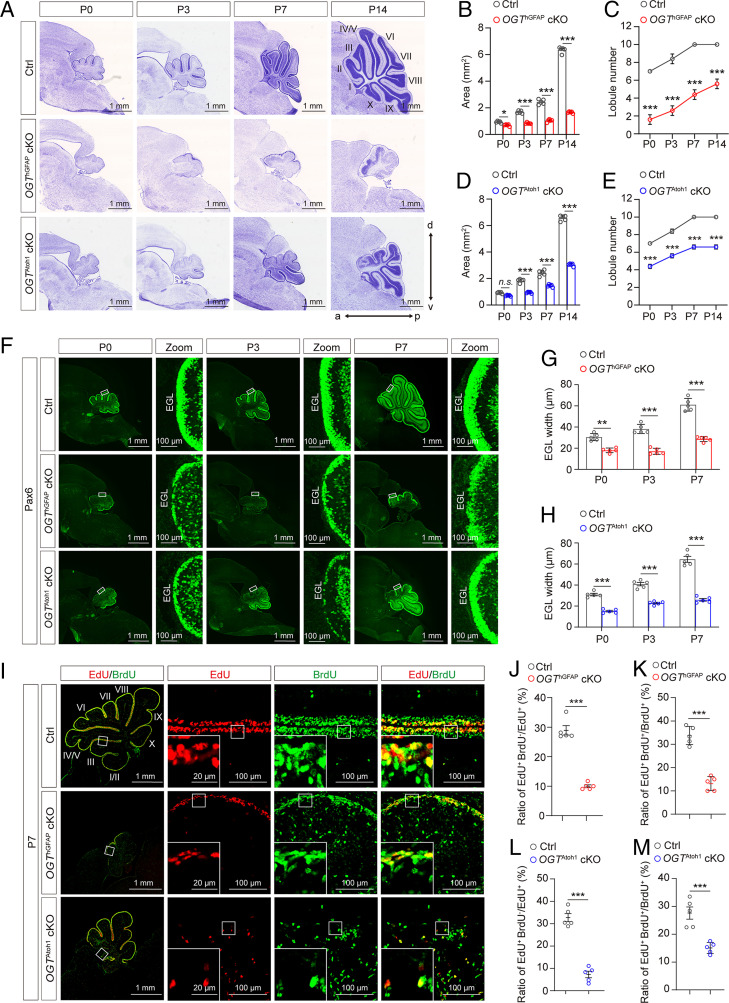
The expression of OGT and O-GlcNAc–modified proteins was previously demonstrated throughout the cerebellar cortex in the adult rat (19). The cerebellar cortex is a multilayered structure, containing the outer molecular layer (ML), the middle Purkinje cell layer (PCL), and the internal granular layer (IGL). Here, we observed that both OGT and overall O-GlcNAc levels increased from postnatal day 0 (P0) to P14 throughout the cerebellar cortex, which coincides with the developmental peak of the cerebellum (SI Appendix, Fig. S1A). At P7, Pax6+ GNPs displayed high levels of O-GlcNAc, particularly in the outer layer of the external granule layer (EGL) (SI Appendix, Fig. S1B, Zoom), a transient proliferation zone during cerebellar development (21, 22). In addition, at P14, NeuN+ mature granule cells in the IGL, brain lipid-binding protein (BLBP)+ Bergmann glial cells (BGs), and Calbindin+ Purkinje cells (PCs) in the PCL also show ubiquitous expression of O-GlcNAc (SI Appendix, Fig. S1C). To pinpoint the contributions of O-GlcNAc and OGT in cerebellar development, we established two types of OGT conditional knockout mouse (cKO) models, in which the Ogt gene was deleted in multipotent NSCs (OGThGFAP cKO) and GNPs (OGTAtoh1 cKO) using well-characterized human GFAP-Cre (hGFAP-Cre) and Atoh1-Cre (also termed as Math1-Cre) mouse lines (7, 23, 24), respectively.
Successful OGT deletion in OGThGFAP cKO mice was evidenced by largely decreased levels of OGT and O-GlcNAc in the cerebellum (SI Appendix, Fig. S2). From P3, the developing cerebellum in both OGThGFAP and OGTAtoh1 cKO mutant mice displayed significantly decreased sagittal area and lobule numbers of the medial cerebellar vermis compared with control mice (Fig. 1 A–E), indicating that OGT is required for postnatal cerebellar compartmentation and morphogenesis. To test whether OGT regulates cell production and divisions of the NSCs and GNPs during cerebellar development, we analyzed immunofluorescent staining of Pax6+ GNPs in the EGL in postnatal control and indicated mutant mice from P0 to P7 (Fig. 1F). We found that the width of Pax6+ EGL in both OGThGFAP and OGTAtoh1 cKO mutants were significantly reduced compared with controls at P0, P3, and P7, respectively (Fig. 1 G and H), indicating impaired proliferation or survival of GNPs in OGT KO mutants.
Fig. 1.
Loss of OGT in multipotent NSCs or GNPs impairs cerebellar morphogenesis. (A) Nissl staining of sagittal histological sections of the cerebellar vermis, showing considerable foliation defects in OGThGFAP cKO and OGTAtoh1cKO mutant mice at the indicated postnatal developmental stages. (Scale bars, 1 mm.) (B and C) Quantification of the area (B) and lobule number (C) of sagittal sections of the cerebellar vermis in Ctrl and OGThGFAP cKO mutant mice at indicated developmental stages (n = 5). Two-way ANOVA and Sidak’s multiple comparisons test. (D and E) Quantification of the area (D) and lobule number (E) of sagittal sections of the cerebellar vermis in Ctrl and OGTAtoh1 cKO mutant mice at indicated developmental stages (n = 5). Two-way ANOVA and Sidak’s multiple comparisons test. (F) Anti-Pax6 labeling of GNPs in sagittal sections of the vermis at the indicated postnatal developmental stages in Ctrl, OGThGFAP cKO, and OGTAtoh1 cKO mutant mice, respectively. (Scale bars, 1 mm and 100 μm.) (G and H) Quantification of EGL width in Ctrl, OGThGFAP cKO (G), and OGTAtoh1 cKO (H) mice at indicated postnatal stages (n = 5). Two-way ANOVA and Sidak’s multiple comparisons test. (I) Immunofluorescent staining of EdU (red) and BrdU (green) to assess GNP proliferation in the cerebellum of Ctrl, OGThGFAP cKO, and OGTAtoh1 cKO mice at P7. Mice were injected with BrdU at P6 and with EdU after 23 h. (Scale bars 100 μm.) Boxed areas are shown at higher magnification (Bottom Left corner) to illustrate EdU+BrdU+ cells remaining in the cell cycle. (Scale bar, 20 μm.) (J and K) Quantification of the ratio of EdU+BrdU− cells to the total number of EdU+ cells (J), or EdU+BrdU+ cells to the total number of BrdU+ cells (K) within the EGL, in the cerebellum of Ctrl and OGThGFAP cKO mice at P7, respectively (n = 5). Unpaired t test. (L and M) Quantification of the ratio of EdU+BrdU− cells to the total number of EdU+ cells (L), or EdU+BrdU+ cells to the total number of BrdU+ cells (M) within the EGL in the rostral cerebellum from Ctrl and OGTAtoh1 cKO mice at P7, respectively (n = 5). Unpaired t test. All data represent mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, n.s., no significance. a, anterior; d, dorsal; p, posterior; v, ventral. See also SI Appendix, Figs. S1–S4.
Furthermore, immunofluorescent staining of cerebellar vermis sections of control mice at P14 exhibited normal cortical lamination and foliation, with NeuN+ granule cells restricted within the IGL and present in the separated fissures, Calbindin+ PCs restricted within the PCL, and BLBP+ BGs localized in the PCL near the pial surface (SI Appendix, Fig. S3A). In contrast, OGThGFAP and OGTAtoh1 cKO mutants exhibited notable disrupted histoarchitecture of the cerebellar cortex (SI Appendix, Fig. S3A). Although NeuN+ granule cells, Calbindin+ PCs, and BLBP+ BGs were detectable in OGThGFAP and OGTAtoh1 cKO mutants, the mutants displayed significantly increased numbers of aberrant ectopic granule cells, PCs, and BGs in the disordered ML (SI Appendix, Fig. S3 B–G), which further illustrates disorganization in the developing cerebellar cortex (Fig. 1A). Together, these data suggest that OGT regulates cerebellar neurogenesis, cortical lamination, and foliation.
The decreased numbers of GNPs observed following OGT KO may be due to impaired production of progenitor cells or decreased cell survival. To further delineate the mechanism, we colabeled Pax6 with Ki67, a cellular proliferation marker, or cleaved-caspase-3 for apoptosis. Results show that GNP proliferation was dramatically impaired in both OGThGFAP and OGTAtoh1 cKO mutants (SI Appendix, Fig. S4 A–C), whereas the level of GNP apoptosis was indistinguishable between control and mutants (SI Appendix, Fig. S4 D–G). These data suggest decreased GNP production as the main reason underlying cerebellar cortical lamination and foliation impairment in mutants. To further assess the effects of OGT loss on postnatal GNP proliferation, we performed sequential pulse-chase experiments by injecting two thymidine analogs, 5-bromo-2′-deoxyuridine (BrdU) and 5-ethynyl-2′-deoxyuridine (EdU) spaced 23 h apart (Fig. 1I). If a progenitor cell enters S-phase more than once during this period, it would incorporate both BrdU and EdU. Compared with control littermates, both mutant mice strains exhibited a significantly lower EdU+BrdU−/EdU+ ratio, which marks newly proliferating cells within 1 h. Similarly, the EdU+BrdU+/BrdU+ ratio was also decreased in the whole vermis of OGThGFAP mutants and in the rostral vermis of OGTAtoh1 mutants, suggesting consistent impaired neurogenesis of GNPs (Fig. 1 J–M). Together, these data suggest that OGT is required for regulating GNP proliferation during cerebellar development.
OGT Is Required for Shh Signaling Activation in the GNPs.
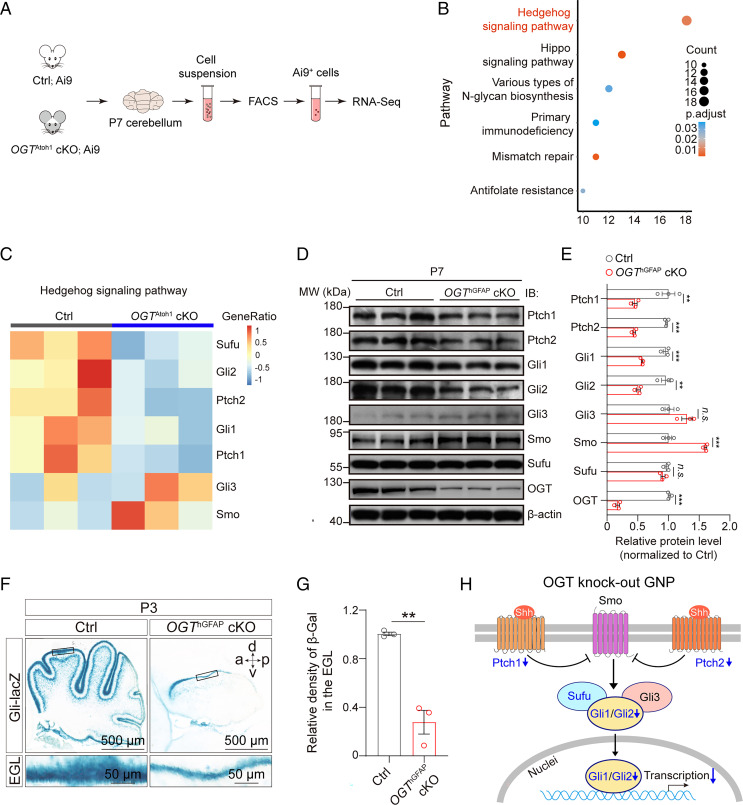
Having established that OGT promotes GNP proliferation, we next sought to understand its molecular mechanism using RNA-sequencing (RNA-seq) analysis. We dissected cerebella from Ai9 reporter control mice and OGTAtoh1 cKO;Ai9 reporter mutant mice at P7, and purified Ai9+ GNPs with flow sorting (Fig. 2A). RNA-seq of Ai9+ GNPs revealed 1,693 significant differentially expressed genes in OGTAtoh1 cKO mice (up-regulated genes: 1,072; down-regulated genes: 621 absolute log2 fold-change > 0.5 and false-discovery rate [FDR] < 0.05), compared with Ai9 control mice. Enrichment analysis of those differentially expressed genes identified notable changes in the Shh signaling pathway in OGTAtoh1 cKO mice (Fig. 2B). Within the Shh signaling pathway, the expression of vital genes—such as Gli1, Gli2, Ptch1, Ptch2, and Suppressor of Fused (Sufu)—was significantly down-regulated in OGTAtoh1 cKO mice (Fig. 2C). The Shh pathway is a master regulator for maintaining cerebellar GNPs in an undifferentiated state. Inhibition of this pathway allows GNPs to exit the cell cycle, differentiate and mature (25). Hence, we hypothesized that OGT might mediate GNP proliferation by regulating Shh pathway activity.
Fig. 2.
OGT-deficient GNPs exhibit disrupted Shh signaling pathway activation in vivo. (A) Schematic diagram of the experimental procedure for RNA-seq. The cerebellum from Ai9Atoh1 reporter Ctrl (Ctrl; Ai9) mice and OGTAtoh1; Ai9Atoh1 reporter cKO (OGTAtoh1 cKO; Ai9) mice at P7 were digested for FACS sorting of Ai9+ GNPs for RNA-seq analysis. (B) Gene ontology terms of differentially expressed genes in cKO compared with Ctrl mice, showing enriched signaling pathways in cKO mice. Color indicates the adjusted P value. The number of genes in each pathway are indicated by circle size. (C) Heatmap of the expression for Ptch1, Ptch2, Gli1, Gli2, Gli3, Smo, and Sufu genes in Shh signaling pathway. The mean expression levels in the heatmap were standardized by taking the z-score within each row (n = 3). The top bar indicates the genotypes of the samples. (D and E) Representative immunoblot showing the levels of proteins in the Shh signaling pathway (Ptch1, Ptch2, Gli1, Gli2, Gli3, Smo, Sufu) and OGT levels from Ctrl and OGThGFAP cKO mice, normalized to β-actin (D), and quantification (E) (n = 3). Unpaired t test. (F) X-gal staining of Gli signaling reporter in the sagittal section through the cerebellar vermis of Gli-LacZ Ctrl and OGThGFAP cKO mice at P3. (Scale bars, 500 μm and 50 μm.) a, anterior; d, dorsal; p, posterior; v, ventral. (G) Quantification of the relative X-gal signaling density in the EGL in Ctrl and OGThGFAP cKO mice at P3 (n = 3). Unpaired t test. (H) Schematic diagram showing altered Shh signaling pathway in OGT knockout GNPs. Down-regulated signaling molecules (with log2 fold-change < −0.5 and FDR < 0.05) are indicated in blue in the diagram. All data represent mean ± SEM **P < 0.01, ***P < 0.001; n.s., no significance. See also SI Appendix, Figs. S5 and S6.
To test this hypothesis, we first performed Western blotting to analyze the expression of proteins in the Shh pathway from control and OGThGFAP cKO cerebellum at P7. Consistent with the transcriptome analysis results, the protein levels of Gli1, Gli2, but not Gli3 were dramatically decreased in the cKO mice (Fig. 2 D and E). Moreover, Ptch1 and Ptch2 were also significantly down-regulated in the cKO mice. Notably, compared to control mice, the expression of Smo was relatively higher in the cKO mice, although it was not significantly changed at the transcriptional level (Fig. 2 C–E), which may represent a feedback regulation to elevate Shh signaling activity in cKO mice. In addition, the protein level of Sufu, a negative regulator of the Shh pathway, was not changed in cKO compared to control mice (Fig. 2 D and E). Moreover, in control and OGThGFAP cKO mice crossed with Gli-lacZ reporter mice at P3 (26), we found significantly lower LacZ expression in the EGL of mutants compared with controls (Fig. 2 F and G). RNA in situ hybridization using RNAscope further confirmed that both Gli1 and Gli2 transcripts were significantly decreased in the EGL of OGThGFAP cKO mice at P3 and P7, compared to control mice (SI Appendix, Fig. S5).
To further verify that depletion of OGT deregulates Shh signaling, we dissected and cultured primary GNP neurospheres from homozygous OGT floxed mice at P5 (27), and then treated with control GFP or Cre-GFP lentivirus for 6 d (SI Appendix, Fig. S6A). We found that both Gli1 and Gli2 were significantly reduced in Cre-GFP lentivirus-treated GNPs (SI Appendix, Fig. S6 B–E). OGT is a multidomain protein that exerts it biological effects through enzymatic activity and protein–protein interactions. Next, to determine whether the decreased expression of Gli1 and Gli2 in OGT-depleted GNPs is dependent on its enzymatic activity, GNPs were treated with the OGT enzyme-inhibitor OSMI-1 or the OGA enzyme-inhibitor Thiamet-G (TM-G), respectively (28, 29). Consistent with the transcriptome analysis results (Fig. 2 C–E), suppression of O-GlcNAc by OSMI-1 in GNPs significantly decreased the expression of Gli1 and Gli2 without altering the level of OGT (SI Appendix, Fig. S6 F–J). In contrast, augmenting O-GlcNAc by TM-G enhanced the expression of Gli1 and Gli2 (SI Appendix, Fig. S6 G and H). To further determine whether the enzymatic activity of OGT is required for Shh signal pathway activation, we treated NIH 3T3 cells with either OSMI-1 or TM-G and examined endogenous Gli transcriptional activity (SI Appendix, Fig. S6K). Results show administration of OSMI-1 suppressed luciferase activity in NIH 3T3 cells pretreated with Smoothened agonist (SAG) in a dose-dependent manner, suggesting a down-regulation of Shh pathway activity, whereas TM-G enhanced luciferase activity (SI Appendix, Fig. S6L). Taken together, these data demonstrate that the enzymatic activity of OGT is essential for Shh signaling activation.
O-GlcNAc of Gli2 at S355 Regulates Its Transcriptional Activity.
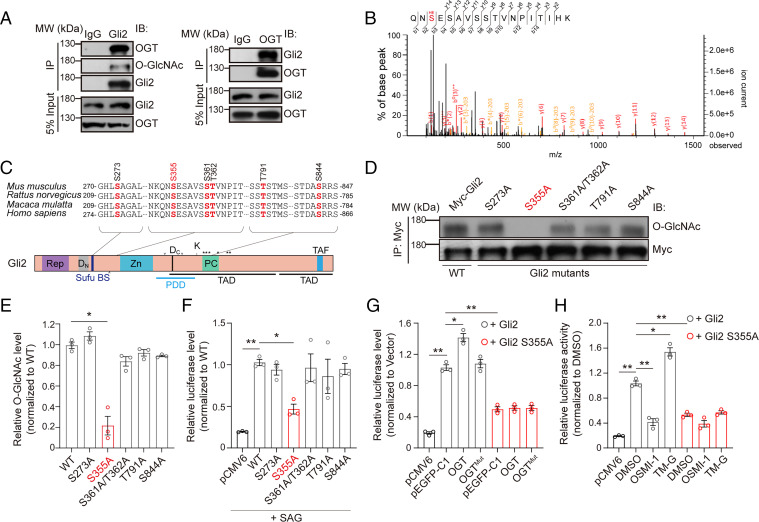
We next asked if Shh pathway factors can be directly O-GlcNAcylated. In the mouse cerebellum, OGT formed endogenous complexes with Gli1/2 and Ptch1/2, but not other Shh signaling components (Fig. 3A and SI Appendix, Fig. S7). However, O-GlcNAcylated Ptch1 were barely detected in the cerebellar lysates (SI Appendix, Fig. S7A). Interestingly, O-GlcNAcylated Ptch2 was detected, albeit at lower levels compared to Gli1 and Gli2, which showed notable O-GlcNAc modification (SI Appendix, Fig. S7C). In contrast, both Gli1 and Gli2 showed much higher O-GlcNAc modification (Fig. 3A and SI Appendix, Fig. S7K). Previous studies have demonstrated that Gli1 is not required for the ectopic activation of the Shh signaling pathway and loss of Gli1 does not affect cerebellar development (26, 30, 31), with Gli1-null mice displaying normal cerebellar histoarchitecture. In contrast, Gli2 plays an indispensable role in cerebellar development (30, 32). Therefore, we emphasized on Gli2 regulation for further study.
Fig. 3.
O-GlcNAc of mouse Gli2 occurs at S355. (A) Coimmunoprecipitation of OGT and Gli2 from the cerebellum of WT mice at P7. (B) O-GlcNAc of Gli2 on serine 355 in vivo. Gli2 was purified from mice cerebellum and analyzed by MS. (C) Cross-species sequence alignment of Gli2 revealed conserved O-GlcNAc sites at S273, S355, S361, T362, T791, and S844, respectively. (D and E) Representative immunoblot (D) and quantification (E) of the O-GlcNAcylation levels of immunoprecipitated WT and mutant Gli2 constructs. HEK293T cells were transfected for 48 h with Myc-tagged WT or the indicated mutant Gli2 construct (n = 3). One-way ANOVA and Dunnett’s multiple comparisons test. (F) Luciferase readout in NIH 3T3 cells treated with 0.1 μM SAG for 36 h and cotransfected for 48 h with pCMV6-vector (pCMV6), WT Gli2, or Gli2 mutants S273A, S355A, S361/T362A, T791A, S844A with Bcl2 promoter luciferase reporter plasmid and Renilla luciferase reporter plasmid pRL-TK as internal reference (n = 3). One-way ANOVA and Dunnett’s multiple comparisons test. (G) Relative luciferase activity in HEK293T cells cotransfected for 48 h with pCMV6-vector (pCMV6), WT Gli2 or Gli2 S355A mutant, the Bcl2 promoter luciferase reporter plasmid and Renilla luciferase reporter plasmid pRL-TK, with pEGFP-C1 vector (pEGFP-C1), pEGFP-C1-OGT (OGT), or pEGFP-C1-OGTMut (H498A/H558A double mutant, termed as OGTMut), respectively (n = 3). One-way ANOVA and Dunnett’s multiple comparisons test. (H) Relative luciferase activity in HEK293T cells cotransfected for 48 h with pCMV6-vector (pCMV6), WT Gli2 or Gli2 S355A mutant, the Bcl2 promoter luciferase reporter plasmid and Renilla luciferase reporter plasmid pRL-TK. Transfected cells were treated with OGT inhibitor OSMI-1 (5 μM), OGA inhibitor TM-G (80 μM), and DMSO as control for 36 h (n = 3). One-way ANOVA and Dunnett’s multiple comparisons test. All data represent mean ± SEM *P < 0.05, **P < 0.01. See also SI Appendix, Figs. S7–S10.
Gli2 is a zinc-finger transcription factor. High-resolution mass spectrometry (MS) of purified Gli2 from the mouse cerebellum revealed six endogenous O-GlcNAcylation sites (Fig. 3B and SI Appendix, Fig. S8 A–D), which were conserved in mammals (Fig. 3C). We constructed a series of Gli2 mutants and detected the overall O-GlcNAc level of the WT Gli2 and indicated mutants, and found that Gli2 S355A dramatically decreased the overall O-GlcNAc abundance of Gli2 (Fig. 3 D and E). This indicates that S355 is the primary O-GlcNAc site of Gli2. To ensure whether the S355 is a functional O-GlcNAc site, we used a luciferase reporter to assess Gli2 downstream effectors (8GBS, Bcl2 promoter, Bcl2 enhancer, and Foxm1) (33–35) (SI Appendix, Fig. S9A). The Gli2 mutant S355A, but not others, demonstrated a sharp decrease in Gli2 transcriptional activity in transfected HEK293T cells compared to WT, suggesting S355 as a critical site for regulating Gli2 activity (Fig. 3F and SI Appendix, Fig. S9 B–E). Moreover, given that serine residue can also be phosphorylated, to exclude the potential role of phosphorylation, we mutated serine 355 to aspartic acid (S355D) to mimic constitutional phosphorylation. We found that, like Gli2 S355A, the Gli2 S355D mutant also could not keep Gli2 activity as WT Gli2 (SI Appendix, Fig. S9 F–I). Hence, these results indicate that S355 of Gli2 is a functional O-GlcNAcylation site, and O-GlcNAcylation is required for mediating the transcriptional activity of Gli2.
To further explore how the O-GlcNAc enzymatic activity of OGT is required for the transcriptional activity of Gli2, we detected the transcriptional activity of the Gli2 and Gli2 S355A mutant, when coinfected with OGT or OGT H498A/H558A double-point mutant (OGTMut), which inactivates the enzyme (36). Consistent with the result in Fig. 3F, the transcriptional activity of the S355A mutant reduced sharply compared to WT Gli2, when coinfected with pEGFP-C1 control vector (Fig. 3G). Meanwhile, OGT, but not OGTMut, enhanced the transcriptional activity of WT Gli2 (Fig. 3G). This shows that the enzymatic activity of OGT is necessary for the transcriptional activity of Gli2. Furthermore, neither cotransfection of OGT nor OGTMut affected transcriptional activity of Gli2 S355A compared with control (Fig. 3G). This indicates that enzymatic activity of OGT has no effect on the transcriptional activity of Gli2 S355A, because it could not be O-GlcNAcylated. In addition, pharmacological treatment of OSMI-1 or TM-G in HEK293T cells transfected with WT or S355A mutant were carried out to analyze the transcriptional activity of Gli2. OSMI-1 sharply reduced the transcriptional activity of WT but not S355A (Fig. 3H). In contrast, TM-G increased WT, but not S355A, transcriptional activity (Fig. 3H). These results indicate that the enzymatic activity of OGT/OGA regulates Gli2 transcriptional activity by changing the O-GlcNAc level of Gli2. Together, these data suggest that OGT promotes the transcriptional activity of Gli2 through its enzyme activity.
O-GlcNAc of Gli2 S355 Promotes the Proliferation of GNPs.
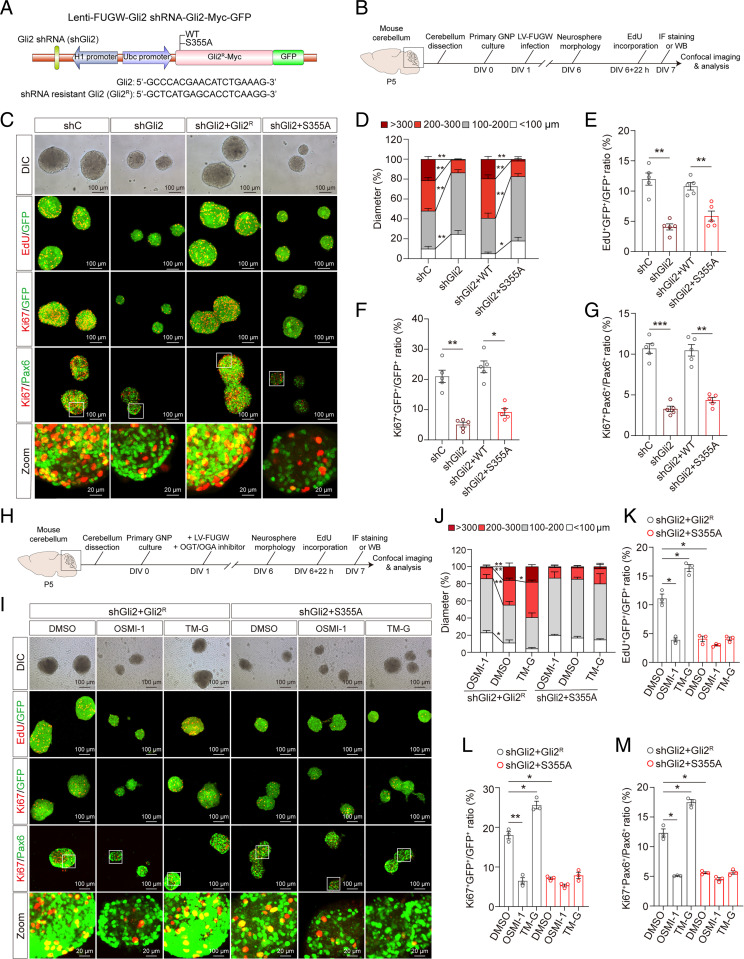
Gli2 has been implicated in cellular proliferation and neurogenesis (37, 38). We next asked if O-GlcNAc of Gli2 S355 regulates GNP proliferation. Using the FUGW vector (39), we generated constructs for H1 promotor-driven short-hairpin RNA (shRNA)-mediated endogenous mouse Gli2 knockdown, and Ubc promotor-driven heterologous WT (Gli2R) or S355A Gli2 (S355A) overexpression constructs resistant to Gli2-targeting shRNA (Fig. 4A). We separately packaged four groups of plasmids into lentivirus, including an shRNA control (shC), shRNA Gli2 knockdown (shGli2), and shRNA Gli2 knockdown with either WT resistant Gli2 (shGli2 + Gli2R) or resistant Gli2 S355A (shGli2 + S355A), and infected primary GNP neurospheres (Fig. 4B). Decreasing endogenous Gli2 levels reduced the diameter of GNP neurospheres compared to shC, which confirmed the role of Gli2 in promoting GNP proliferation (Fig. 4 C and D). Overexpression of Gli2R, but not S355A, rescued the diameter of GNP neurospheres (Fig. 4 C and D). In addition, Gli2 knockdown dramatically decreased EdU+ and Ki67+ cells in GFP+ lentiviral-infected GNPs, which was rescued by Gli2R but not S355A Gli2R overexpression (Fig. 4 C, E, and F). Similar results were observed for Ki67+/Pax6+ cells (Fig. 4 C and G). Together, these results suggest that Gli2 S355 is crucial for GNP proliferation in vitro.
Fig. 4.
O-GlcNAc of Gli2 S355 promotes the proliferation of GNPs in vitro. (A) Schematic diagram of the Lenti-FUGW-Gli2 shRNA-Gli2-Myc-GFP plasmid with endogenous mouse Gli2 knockdown and overexpression of exogenous shRNA-resistant mouse Gli2 or Gli2 S355A mutant. Four constructs were utilized: shRNA control (shC), Gli2 shRNA knockdown (shGli2), Gli2 shRNA knockdown, and overexpression of shRNA-resistant mouse Gli2 (shGli2+Gli2R), or shRNA-resistant mouse Gli2 S355A mutant (shGli2+S355A). (B) Schematic diagram of experimental timeline and procedure for C–G. (C) Immunofluorescent staining with anti-EdU, anti-Ki67, and anti-Pax6 antibodies, overlayed on GFP+ or Pax6+ GNP neurospheres at day in vitro (DIV) 6. (Scale bars, 100 μm.) (D) The distribution of the diameter of GNP neurospheres at DIV6 (n = 4). Two-way ANOVA and Tukey’s multiple comparisons test. (E–G) Quantification of EdU+GFP+/GFP+ (E), Ki67+GFP+/GFP+ (F), and Ki67+Pax6+/Pax6+ (G) in GNP neurospheres at DIV6 (n = 5). One-way ANOVA and Dunnett’s multiple comparisons test. (H) Schematic diagram of experimental procedures for I–M. (I) Immunofluorescent staining with anti-EdU, anti-Ki67, and anti-Pax6 antibodies to label proliferating GNP neurospheres at DIV6. (Scale bars, 100 μm.) (J) The distribution of the diameter of GNP neurospheres infected with lentivirus and treated with OSMI-1 (10 μM) or TM-G (80 μM) at DIV6 (n = 3). Two-way ANOVA and Tukey’s multiple comparisons test. (K–M) Quantification of EdU+GFP+/GFP+ (K), Ki67+GFP+/GFP+ (L), and Ki67+Pax6+/Pax6+ (M) in GNP neurospheres at DIV6 (n = 3). One-way ANOVA and Dunnett’s multiple comparisons test. All data represent mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001. See also SI Appendix, Fig. S11.
To further examine if the effect of Gli2 S355 on GNP proliferation is mediated by O-GlcNAc, we treated shGli2 + Gli2R and shGli2 + S355A lentiviral-infected GNP neurospheres with DMSO, OSMI-1, or TM-G, respectively (Fig. 4 H and I). Consistent with the result in Fig. 4D, the diameter of S355A Gli2R GNP neurospheres reduced sharply compared to Gli2R neurospheres. OSMI-1 decreased the diameter of Gli2R GNP neurospheres, whereas TM-G increased the diameter of Gli2R neurospheres. In contrast, neither OSMI-1 nor TM-G significantly changed the diameter of S355A Gli2R neurospheres (Fig. 4 I and J). Furthermore, staining for Ki67+ and EdU+ in GFP+ lentiviral-infected cells showed decreased ratio of proliferating cells in S355A Gli2R neurospheres compared to WT Gli2R controls (Fig. 4 I, K, and L). In Gli2R neurospheres, OSMI-1 significantly reduced Ki67+ and EdU+ cells, whereas TM-G had the opposite effect. In contrast, the inhibitors did not significantly affect proliferating cells in S355A Gli2R neurospheres (Fig. 4 I, K, and L). Similar results were observed with Ki67+/Pax6+ GNPs (Fig. 4 I and M). Together, these results show that Gli2 requires O-GlcNAc at S355 to promote GNP proliferation.
O-GlcNAc of GLI2 S358 Is a Conserved Functional Site in Human Cells.
Gli2 S355 is well conserved among mammalian species (Fig. 3C), suggesting that O-GlcNAc of Gli2 S355 may be an evolutionarily conserved mechanism promoting Gli2 activity. To verify this hypothesis, we screened for O-GlcNAc sites on human GLI2 purified from Daoy cells and found several potential sites shared between mouse and human GLI2 (SI Appendix, Figs. S8 E–H and S10A). By analyzing O-GlcNAc levels in human GLI2 mutants, we pinpointed human S358, an analog of the mouse S355 as a potential O-GlcNAc target (SI Appendix, Fig. S10 B and C). To further verify S358 as a functional O-GlcNAc site, we used a luciferase reporter to assess GLI2 downstream effectors (8GBS, Bcl2 promoter, Bcl2 enhancer, and Foxm1). Human GLI2 S358A, but not other GLI2 mutants, resulted in dramatically decreased GLI2 activity in transfected HEK293T cells compared to WT (SI Appendix, Fig. S10 D–G). These results suggest S358 as a critical site for regulating GLI2 transcriptional activity in human cells. To exclude the role of phosphorylation, we mutated S358 to aspartic acid (S358D) to mimic constitutional phosphorylation. Consistent with results from the mouse Gli2 S355D mutant, the human S358D mutant could not sustain GLI2 activity compared to WT (SI Appendix, Fig. S10 H–K). The result above indicates that human GLI2 S358 is a functional O-GlcNAcylation site and necessary for mediating GLI2 transcriptional activity.
To examine functional links between OGT and GLI2, similar to Fig. 4A, we generated constructs for shRNA-mediated endogenous mouse Gli2 knockdown and overexpression of heterologous WT or S358A human GLI2 resistant to Gli2-targeting shRNA, which were packaged into lentivirus and used to infect primary GNP neurospheres (SI Appendix, Fig. S11 A and B). shGli2 sharply decreased endogenous Gli2 protein level, which was rescued by heterologous overexpression of either WT or S358A GLI2 to similar levels (SI Appendix, Fig. S11 C–E. In agreement with Fig. 4C, decreasing endogenous Gli2 levels reduced the diameter of GNP neurospheres compared to shC, which confirmed the role of Gli2 in promoting GNP proliferation (SI Appendix, Fig. S11 F and G). In contrast, overexpression of GLI2R, but not the S358A mutant, rescued the diameter of GNP neurospheres (SI Appendix, Fig. S11G). In addition, the decreased ratios of EdU+ and Ki67+ cells in GFP+ lentiviral-infected cells or Pax6+ cells in shGli2 GNP spheres were rescued by GLI2R but not S358A GLI2R overexpression (SI Appendix, Fig. S11 F and H–J). Taken together, our results suggest an evolutionary conserved mechanism of O-GlcNAc in regulating Gli2 activity and GNP proliferation in mice and humans.
O-GlcNAc of Gli2 S355 Deacetylates Gli2 via Dissociation from HAT.
We next sought to understand the molecular mechanisms by which O-GlcNAc controls Gli2 transcriptional activity and its role in promoting GNP proliferation. First, O-GlcNAc of Gli2 at S355 may degrade Gli2 protein levels to alter its transcriptional activity. Cotransfection of HEK293T cells with OGT did not change ubiquitinated Gli2 protein levels compared to Gli2 transfection alone (SI Appendix, Fig. S12A). WT or S355A Gli2 resulted in similar total and ubiquitinated Gli2 levels (SI Appendix, Fig. S12B), suggesting O-GlcNAc of Gli2 does not alter its degradation.
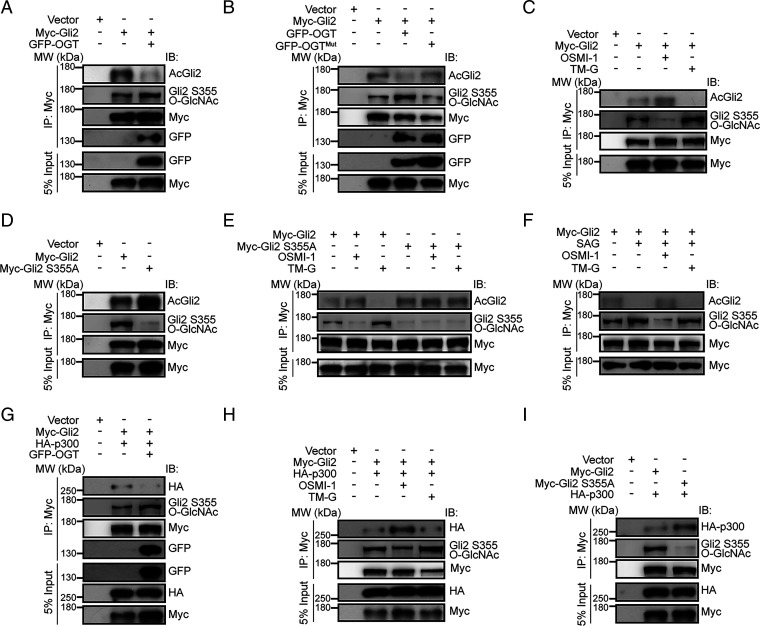
Deacetylation of Gli2 induced by histone deacetylases (HDACs) promotes its transcriptional activity (40). We observed that overexpression of OGT-induced O-GlcNAc of Gli2 S355 significantly decreased Gli2 acetylation (AcGli2) (Fig. 5A). Conversely, overexpression of OGTMut, a catalytically inactive variant of OGT, resulted in higher levels of AcGli2 compared to WT OGT (Fig. 5B). Moreover, OSMI-1 increased, whereas TM-G decreased AcGli2 levels in cells overexpressing WT Gli2 (Fig. 5C), suggesting that OGT regulates Gli2 acetylation through its enzymatic function. Consistently, we detected higher levels of Gli2 acetylation in the S355A mutant compared to WT (Fig. 5D). In contrast, neither OSMI-1 nor TM-G significantly altered AcGli2 levels for the S355A Gli2 mutant (Fig. 5E). Furthermore, OSMI-1 increased AcGli2 levels in mouse NIH 3T3 cells treated with the Smo agonist SAG, whereas TM-G reduced AcGli2 levels (Fig. 5F). Together, these findings indicate that O-GlcNAc of Gli2 S355 regulates Gli2 acetylation and transcriptional activity.
Fig. 5.
O-GlcNAc of Gli2 S355 suppresses Gli2 acetylation by inhibiting binding to HAT. (A and B) Representative immunoblot of AcGli2 and O-GlcNAc of Gli2 S355 (Gli2 S355 O-GlcNAc) wtih (A) WT OGT or (B) H498A/H558A double mutant OGT (OGTMut), respectively. (C) Representative immunoblot of AcGli2 and Gli2 S355 O-GlcNAc with Myc-tagged Gli2. Transfected cells were treated with OSMI-1 (10 μM) or TM-G (80 μM) for 36 h. (D) Representative immunoblot of AcGli2 and Gli2 S355 O-GlcNAc with Myc-tagged Gli2 or Myc-tagged Gli2 S355A (Myc-Gli2 S355A), respectively. (E) Representative immunoblot of AcGli2 and Gli2 S355 O-GlcNAc with Myc-tagged Gli2 or Myc-Gli2 S355A, treated with OSMI-1 (10 μM) or TM-G (80 μM) for 36 h, respectively. (F) Representative immunoblot AcGli2 and Gli2 S355 O-GlcNAc immunoprecipitated from NIH 3T3 cells transfected with Myc-tagged Gli2 in the presence of SAG (0.1 μM) for 36 h, followed by OSMI-1 (10 μM) or TM-G (80 μM) treatment for 24 h. (G) Representative immunoblot of HA-p300 with Myc-tagged Gli2, with or without GFP-tagged OGT. (H) Representative immunoblot of HA-p300 with Myc-tagged Gli2. Transfected cells were treated with OSMI-1 (10 μM) or TM-G (80 μM) for 36 h. (I) Representative immunoblot of HA-p300 with of Myc-tagged Gli2, or Myc-Gli2 S355A mutant, respectively. See also SI Appendix, Fig. S12.
AcGli2 levels are fine-tuned by HATs and HDACs (40). To investigate how O-GlcNAc of Gli2 S355 involves its acetylation, we coimmunoprecipitated Gli2 with HDAC1 or p300 (a member of the HAT family) from HEK293T cells transfected with or without OGT. We found overexpression of OGT inhibited Gli2 binding with p300 (Fig. 5G), but not HDAC1 (SI Appendix, Fig. S12C). In addition, OSMI-1 promoted Gli2 binding with p300, whereas TM-G decreased the association (Fig. 5H). In contrast, neither inhibitor altered Gli2-HDAC1 binding (SI Appendix, Fig. S12D). Moreover, compared with WT Gli2, the S355A mutant showed much higher binding to p300 (Fig. 5I) but not HDAC1 (SI Appendix, Fig. S12E). Together, these data suggest that O-GlcNAc of Gli2 S355 inhibits Gli2 binding to p300, but not HDAC1, to reduce AcGli2 levels.
Gli2 also interacts with Sufu, a scaffolding protein that directly inhibits its function (41). To exclude the possibility that O-GlcNAc of Gli2 at S355 affects its binding to Sufu, we further coimmunoprecipitated WT or S355A Gli2 with Sufu and found similar binding levels (SI Appendix, Fig. S12F), indicating that O-GlcNAc of Gli2 S355 does not interfere with Gli2-Sufu binding. Altogether, these data suggest O-GlcNAc of Gli2 at S355 inhibits its binding to p300, resulting in reduced AcGli2 and increased transcriptional activity.
O-GlcNAcylation Mediates Shh Subgroup Medulloblastoma Progression.
Medulloblastoma is a malignant pediatric tumor of the cerebellum (42). Transcriptional profiling studies reveal that medulloblastomas exist as four main molecular subgroups (43), with approximately one-third associated with misactivation of the Shh signaling pathway (44). Aberrant or sustained Shh signaling in GNPs usually leads to medulloblastoma oncogenesis (25).
Given that O-GlcNAc of Gli2 S355 promotes GNP proliferation, we next asked if OGT and O-GlcNAc are involved in the occurrence of Shh-subtype medulloblastoma. We first treated the human Shh-subtype medulloblastoma cell line Daoy cells (45) with DMSO, OSMI-1, or TM-G. Cellular proliferation was subsequently assessed using multiple approaches, including the Cell Counting Kit-8 (CCK8) assay, the formation of single cell colonies, and EdU and Ki67 immunofluorescence staining. OSMI-1 reduced OD450 in the CCK8 assay, colony formation, and EdU+ and Ki67+ cells in a dose-dependent manner (SI Appendix, Fig. S13 A–E). In contrast, TM-G enhanced colony formation and EdU+ and Ki67+ cells (SI Appendix, Fig. S13 B–E). These results suggest O-GlcNAc promotes the malignant proliferation of Daoy cells in vitro. To further determine whether Shh signaling is correspondingly altered, the expression of GLI1-3 was detected in Daoy cells treated with OSMI-1 and TM-G, respectively. We found that OSMI-1 inhibited the expression of GLI1/2 without altering the level of GLI3. In contrast, TM-G significantly increased the expression of GLI1/2, but not GLI3 (SI Appendix, Fig. S13 F–I), which was also in agreement with the pharmacological effects on primary GNPs (SI Appendix, Fig. S6 F–H).
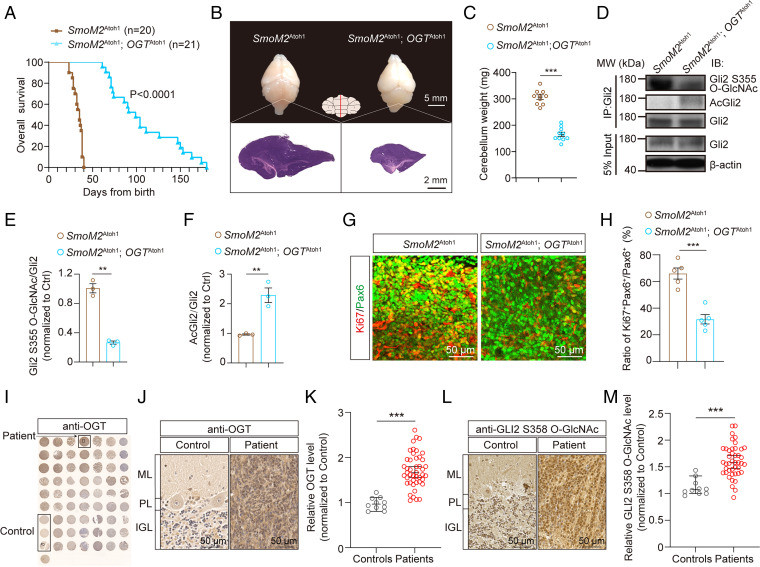
To investigate the involvement of OGT in medulloblastoma progression in vivo, we genetically ablated OGT specifically in cerebellar GNPs in the SmoM2Atoh1 medulloblastoma mouse model (SmoM2Atoh1; OGTAtoh1), with SmoM2Atoh mice as control. KO of OGT in GNPs prolonged median survival compared to SmoM2Atoh1 mice (110 d vs. 37 d) (Fig. 6A) and dramatically reduced the weight and size of tumors (Fig. 6 B and C). We next asked if pharmaceutical inhibition of OGT recapitulates these effects. OSMI-1 was previously shown to decrease O-GlcNAc levels in the brain following systemic delivery (46), and we found that OSMI-1 readily passes the blood–brain barrier (SI Appendix, Fig. S14). OSMI-1 treatment reduced O-GlcNAc levels and prolonged survival and reduced tumor size and weight of SmoM2Atoh1 mice (SI Appendix, Fig. S15 A–D), suggesting a role for OGT for medulloblastoma growth.
Fig. 6.
Disruption of O-GlcNAc inhibits the oncogenesis of Shh-subtype medulloblastoma. (A) Kaplan–Meier curves of SmoM2Atoh1 and SmoM2Atoh1; OGTAtoh1 rescue mice (n = 20 or 21). Log-rank test. (B) Representative images of the whole brain and H&E stained micrographs of midsagittal sections through the cerebellar vermis at P30. (Scale bars, 5 mm and 2 mm.) (C) Cerebellar weight from Ctrl, SmoM2Atoh1 and SmoM2Atoh1; OGTAtoh1 rescue mice at P30 (n = 10). Unpaired t test. (D–F) Representative immunoblots (D) and quantification of (E) Gli2 S355 O-GlcNAc and (F) AcGli2 immunoprecipitated from SmoM2Atoh1 or SmoM2Atoh1; OGTAtoh1 rescue cerebellum at P30 (n = 3). Unpaired t test. (G and H) Immunofluorescent staining (G) and quantification (H) of Ki67+ GNPs in the cerebellar vermis from SmoM2Atoh1 and SmoM2Atoh1;OGTAtoh1 rescue mice at P30 (n = 5). (Scale bars, 50 μm.) Unpaired t test. (I–K) Tissue microarray analysis (TMA) of OGT. Immunohistochemistry images of TMA stained with anti-OGT antibody (I), enlarged IHC images of OGT staining from the TMA (J), and data analyzed using an Unpaired t test (K) (n = 10 in healthy controls and 45 in medulloblastoma patients, respectively). (Scale bars, 50 μm.) (L and M) Enlarged immunohistochemistry images of O-GlcNAcylated GLI2 S358 from the TMA (L) and quantification using an Unpaired t test (M) (n = 10 in healthy controls and 43 in medulloblastoma patients, respectively). (Scale bars, 50 μm.) All data represent mean ± SEM **P < 0.01, ***P < 0.001. See also SI Appendix, Figs. S13–S18.
To further examine the signaling pathways involved, we quantified the levels of O-GlcNAcylated Gli2 S355 and AcGli2 from the cerebellum of SmoM2Atoh1 and SmoM2Atoh1;OGTAtoh1 mice, respectively. Compared to SmoM2Atoh1 control mice, O-GlcNAcylated Gli2 S355 protein level decreased and AcGli2 level increased in SmoM2Atoh1;OGTAtoh1 rescue mice (Fig. 6 D–F). Similar results were observed in SmoM2Atoh1 mice treated with OSMI-1 compared with DMSO-treated controls (SI Appendix, Fig. S15 E–G). These results suggest that O-GlcNAc of Gli2 S355 promotes medulloblastoma growth by decreasing its acetylation and therefore increasing Gli2 transcriptional activity. Moreover, Ki67+ cells in GNPs were also significantly reduced in both SmoM2Atoh1;OGTAtoh1 rescue mice (Fig. 6 G and H) and OSMI-1–treated SmoM2Atoh1 mice (SI Appendix, Fig. S15 H and I) compared with their respective controls, suggesting inhibition of OGT significantly reduces the progression and proliferation of Shh-subtype of medulloblastoma in vivo.
We next asked whether O-GlcNAc levels are altered in clinical medulloblastoma samples from patients. The specificity of both anti-OGT and anti-GLI2 S358 O-GlcNAc for immunostaining was first validated (SI Appendix, Fig. S16). We then tested 45 medulloblastoma samples and 10 healthy samples via the combination of immunohistochemistry and tissue microarray technology. GAB1 was used to identify Shh-subtype medulloblastomas (45). The results showed significantly increased OGT levels in medulloblastoma samples compared with control samples from healthy people (Fig. 6 I–K). Increased O-GlcNAcylated GLI2 S358 levels was also found in 43 medulloblastoma samples (Fig. 6 L and M) compared to 10 healthy samples. We further found a positive correlation of the expression of OGT and GLI2 S358 O-GlcNAc with the GAB1+ Shh-subtype of medulloblastoma (47) (SI Appendix, Fig. S17). Taken together, these data suggest that O-GlcNAc of GLI2 S358 is associated with medulloblastoma growth in humans.
Discussion
Shh regulates the development of the vertebrate central nervous system, with Gli1-3 as downstream factors that drive the transcription of target genes (48). The transcriptional activity of both Gli1 and Gli2 is repressed by acetylation (40, 49). A previous study showed that O-GlcNAcylation of GLI1 augments its activity in vitro (50), but little is known about O-GlcNAc and acylation interactions for modulating neurogenesis. In this study, we demonstrated that OGT-mediated O-GlcNAcylation of Gli2 impairs its association with p300 and promotes Gli-mediated Shh signaling activation, which in turn affects GNP neurogenesis, cerebellar development, and the growth of Shh-subtype medulloblastoma (SI Appendix, Fig. S18).
Although our results here identified Gli2 as a direct O-GlcNAc target in the cerebellum, it cannot be ruled out that O-GlcNAc may regulate other proteins in the Shh signaling pathway. Gli2 controls downstream activity of target genes involved in cellular proliferation (e.g., CDK, Cyclins, Myc, and so forth), and Shh pathway autoregulated genes (Ptch1 and Gli itself) (33, 35, 51–54). Our data revealed decreased expression of Gli1, Gli2, and Ptch1 in OGT KO GNPs (Fig. 2 C–E). Notably, we found low O-GlcNAcylation levels of other Shh signaling pathway members, such as Ptch1 (SI Appendix, Fig. S7A). Interestingly, we detected O-GlcNAcylated Ptch2 in the mouse cerebellum (SI Appendix, Fig. S7C). However, Ptch2-deficient mice are viable and fertile with normal cerebellar architecture in contrast to embryonic-lethal Ptch1 mutants, suggesting that Ptch1 is the major receptor for Shh signaling and Ptch2 plays a compensatory role for Ptch1 (55, 56). Despite the exact role of O-GlcNAcylated Ptch2 in the context of cerebellar development remains elusive, our results together with previous studies (26, 31, 55) suggest that Gli2 is a central hub in regulating GNP growth and cerebellar development for the Shh signaling pathway. It should be noted that over 5,000 proteins and 7,000 sites have been identified in the human O-GlcNAcome (57). Given that the Shh-Ptch-Smo-Gli axis extensively interconnects with several critical developmental pathways—including Wnt, Notch, EGF, and TGF-β during organogenesis (7, 37, 38, 53, 58–60)—and that Wnt and Notch signaling pathways can also be O-GlcNAcylated during neural development (16, 61), O-GlcNAc may regulate cerebellar neurogenesis through further means that remains to be investigated.
GNP proliferation in the EGL determines the production, migration, and location of granule neurons and affects cerebellar morphology and function (4, 6, 62). Hence, the cross-talk of molecules involved in regulating GNP proliferation—such as Atoh1, Zic1, Ccnd2, Wnt, and members of the Shh pathway—are essential for a fully developed and functional cerebellum (4, 63, 64). Our previous results implicated the receptor for activated C kinase 1 (Rack1) in cerebellum development by opposing Wnt/β-catenin and Shh signaling pathways (7). Here, in both mouse and human tissue, we identified O-GlcNAc as a posttranslational modification mechanism to control Shh pathway activation and GNP proliferation during cerebellar development. O-GlcNAc has been proposed as a nutrient sensor. Of note, nutrient availability is essential for the regulation of cell cycle progression and tumorigenesis (65–67). Therefore, the influence of Gli2 O-GlcNAcylation-mediated metabolic signaling in the regulation of GNP neurogenesis and medulloblastoma oncogenesis deserves further study.
Medulloblastoma is a malignant nervous system tumor arising from disruptions in cerebellar development (68). Dysfunctional Shh signaling in GNPs are associated with about 30% of medulloblastoma in humans (69, 70), with particularly poor prognosis in children (71). Therefore, defining the extracellular and intracellular signals that coordinate the Shh pathway to control cerebellar neurogenesis, especially the proliferation of GNPs, has been critical in unraveling the mechanisms underlying medulloblastoma formation and growth (72). Here, we found that aberrantly increased expression of OGT and GLI2 O-GlcNAcylation were positively correlated with the Shh-subtype medulloblastoma. We also showed that inhibiting OGT, either through genetic or chemical means, greatly extended survival time in an Shh-subtype medulloblastoma mouse model. OGT serves as a key molecular target connecting glucose metabolism with proteins involved in DNA transcription and cell proliferation (73). Tumor cells derive energy from reprogrammed glucose metabolism to sustain their uncontrolled replication (74, 75), suggesting that reducing O-GlcNAc of GLI2 might be a new means to suppress its transcriptional activity and the malignant proliferation of cancer cells by targeting nutrient availability. Although OGT expression is high in medulloblastoma (Fig. 6 and SI Appendix, Fig. S17), little is known about the regulatory mechanisms involved. Previous studies have shown that E3 ubiquitin ligase XIAP is modified by O-GlcNAc and in turn promotes OGT degradation (76). In breast cancer, the mTOR/c-MYC axis positively regulates OGT expression and O-GlcNAc level (77). Interestingly, c-MYC serves as one of the target genes of Gli signaling in vivo (78, 79), indicating the potentially mutual regulation and positive feedback between Gli transcriptional activity and OGT expression.
In sum, our results illustrate a connection between O-GlcNAcylation and acetylation of Gli2 and suggest O-GlcNAc is a crucial mechanism to control neurogenesis and patterning in cerebellum development, and may function as a target for therapies against Shh-subtype medulloblastoma.
Materials and Methods
Mice, Immunofluorescence Staining, RNA-Seq, and Liquid Chromatography-MS/MS Assay.
All animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Beijing Institute of Basic Medical Sciences (#SYXK2019-0004). Detailed information for genotyping, immunofluorescence staining, RNA-seq, and liquid chromatography-MS/MS assay for O-GlcNAc site identification are provided in SI Appendix, Materials and Methods.
Primary GNPs Isolation, Plasmid Construction, Lentivirus Infection, and Luciferase Assay.
The primary GNPs were isolated from the cerebellum of P5 Ctrl and OGThGFAP cKO mice according to our previously published work (7), and infected with the indicated lentivirus for further studies. Detailed information for plasmid construction, lentiviral packaging and infection, OGT/OGA inhibitor treatment for pharmacological, and luciferase assay are provided in SI Appendix, Materials and Methods.
Immunohistochemistry of Human Medulloblastoma Tissue Microarray Slides.
The tissue microarray slides of human medulloblastoma were purchased from Alenabio (BC17012c), Avilabio (DC-Bra01022), and Bioaitech (N035Cb01). Detailed methods for immunohistochemistry staining and imaging assay are provided in SI Appendix, Materials and Methods.
Quantification and Statistical Analysis.
All experiments were repeated at least three separate times. All data between independent repeats are presented as mean ± SEM. For associations between gene-expression values, significance was evaluated by Pearson product moment correlation coefficient. For other statistical analysis, unpaired t test, one-way or two-way ANOVA, and multiple comparisons test were used and analyzed using GraphPad Prism 9 and SPSS. All images were analyzed by ImageJ (NIH) and NDP.View2 software (Hamamatsu). P < 0.05 were considered significant; P < 0.01 were considered extremely significant.
A more detailed description of materials and methods is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Shiwen Luo at Nanchang University, Dr. Zilong Qiu at the Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences (CAS), Dr. Jun Yao at Tsinghua University, and Dr. Huiyan Li at National Center of Biomedical Analysis for providing the valuable reagents for this work; Dr. Hong Zhang at Institute of Biophysics, CAS, for providing the OGTF/F mice; Dr. Yizheng Wang at Beijing Institute of Basic Medical Sciences for providing the valuable Gli lacZ reporter (Gli1lz/+) mice; Dr. Xiaomei Zhuang at Beijing Institute of Pharmacology and Toxicology for helping with the LC-MS/MS analysis; Dr. Wantao Ying and Dr. Jie Ma at Beijing Institute of Lifeomics for helping with the proteomics raw data uploading; Dr. Xiang Yu and Dr. Jian Zhu at Peking University for valuable input; and all members of the H.W. laboratory for discussion. This work was supported by the National Key Research and Development Program of China (Grants 2021ZD0202500 and 2021YFA1101801), the National Natural Science Foundation of China (32171148, 31770929, and 31522029), and the Beijing Municipal Science and Technology Commission (Z181100001518001 and Z161100000216154) of China.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. F.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202821119/-/DCSupplemental.
Data, Materials, and Software Availability
The original RNA-seq data in this study have been deposited in the National Center for Biotechnology Information Sequence Read Achieve (accession no. PRJNA798583). MS data reported in this paper have been deposited to the ProteomeXchange Consortium via the iProX partner repository (dataset identifier PXD034270/IPX0004530000). The entire datasets are publicly available at https://www.iprox.cn//page/project.html?id=IPX0004530000. All other study data are included in the main text and SI Appendix.
References
- 1.De Zeeuw C. I., Lisberger S. G., Raymond J. L., Diversity and dynamism in the cerebellum. Nat. Neurosci. 24, 160–167 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Stoodley C. J., Schmahmann J. D., Functional topography of the human cerebellum. Handb. Clin. Neurol. 154, 59–70 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Therrien A. S., Bastian A. J., The cerebellum as a movement sensor. Neurosci. Lett. 688, 37–40 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Hatten M. E., Roussel M. F., Development and cancer of the cerebellum. Trends Neurosci. 34, 134–142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sillitoe R. V., Joyner A. L., Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu. Rev. Cell Dev. Biol. 23, 549–577 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Roussel M. F., Hatten M. E., Cerebellum development and medulloblastoma. Curr. Top. Dev. Biol. 94, 235–282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H., et al. , Opposite regulation of Wnt/β-catenin and Shh signaling pathways by Rack1 controls mammalian cerebellar development. Proc. Natl. Acad. Sci. U.S.A. 116, 4661–4670 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favaro R., et al. , Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 12, 1248–1256 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Dias J. M., et al. , A Shh/Gli-driven three-node timer motif controls temporal identity and fate of neural stem cells. Sci. Adv. 6, eaba8196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn S., Joyner A. L., In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 437, 894–897 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Hwang H., Rhim H., Functional significance of O-GlcNAc modification in regulating neuronal properties. Pharmacol. Res. 129, 295–307 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Lagerlöf O., O-GlcNAc cycling in the developing, adult and geriatric brain. J. Bioenerg. Biomembr. 50, 241–261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma X., Li H., He Y., Hao J., The emerging link between O-GlcNAcylation and neurological disorders. Cell. Mol. Life Sci. 74, 3667–3686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart G. W., Nutrient regulation of signaling and transcription. J. Biol. Chem. 294, 2211–2231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White C. W. III, et al. , Age-related loss of neural stem cell O-GlcNAc promotes a glial fate switch through STAT3 activation. Proc. Natl. Acad. Sci. U.S.A. 117, 22214–22224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., et al. , Ogt controls neural stem/progenitor cell pool and adult neurogenesis through modulating Notch signaling. Cell Rep. 34, 108905 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Cheng J., et al. , Loss of O-GlcNAc transferase in neural stem cells impairs corticogenesis. Biochem. Biophys. Res. Commun. 532, 541–547 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Rex-Mathes M., et al. , O-GlcNAc expression in developing and ageing mouse brain. Biochimie 83, 583–590 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Akimoto Y., et al. , Localization of the O-GlcNAc transferase and O-GlcNAc-modified proteins in rat cerebellar cortex. Brain Res. 966, 194–205 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Rexach J. E., Clark P. M., Hsieh-Wilson L. C., Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat. Chem. Biol. 4, 97–106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alder J., Cho N. K., Hatten M. E., Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron 17, 389–399 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Altman J., Postnatal development of the cerebellar cortex in the rat. I. The external germinal layer and the transitional molecular layer. J. Comp. Neurol. 145, 353–397 (1972). [DOI] [PubMed] [Google Scholar]
- 23.Zhuo L., et al. , hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31, 85–94 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Matei V., et al. , Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 234, 633–650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee E. Y., et al. , Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 9736–9741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai C. B., Auerbach W., Lee J. S., Stephen D., Joyner A. L., Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Shafi R., et al. , The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Meoz R. F., et al. , A small molecule that inhibits OGT activity in cells. ACS Chem. Biol. 10, 1392–1397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuzwa S. A., et al. , A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Corrales J. D., Rocco G. L., Blaess S., Guo Q., Joyner A. L., Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development 131, 5581–5590 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Park H. L., et al. , Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Corrales J. D., Blaess S., Mahoney E. M., Joyner A. L., The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development 133, 1811–1821 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Katoh Y., Katoh M., Hedgehog target genes: Mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 9, 873–886 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Regl G., et al. , Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 64, 7724–7731 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Katoh M., Genomic testing, tumor microenvironment and targeted therapy of Hedgehog-related human cancers. Clin. Sci. (Lond.) 133, 953–970 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Lazarus M. B., Nam Y., Jiang J., Sliz P., Walker S., Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palma V., Ruiz i Altaba A., Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 131, 337–345 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Palma V., et al. , Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development 132, 335–344 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng J., et al. , Loss of O-GlcNAcylation on MeCP2 at threonine 203 leads to neurodevelopmental disorders. Neurosci. Bull. 38, 113–134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canettieri G., et al. , Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 12, 132–142 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Han Y., Shi Q., Jiang J., Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc. Natl. Acad. Sci. U.S.A. 112, 6383–6388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khatua S., Song A., Citla Sridhar D., Mack S. C., Childhood medulloblastoma: Current therapies, emerging molecular landscape and newer therapeutic insights. Curr. Neuropharmacol. 16, 1045–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Archer T. C., Mahoney E. L., Pomeroy S. L., Medulloblastoma: Molecular classification-based personal therapeutics. Neurotherapeutics 14, 265–273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellison D. W., et al. , Medulloblastoma: Clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 121, 381–396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higdon R., et al. , Integrated proteomic and transcriptomic-based approaches to identifying signature biomarkers and pathways for elucidation of Daoy and UW228 subtypes. Proteomes 5, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S. M., et al. , REM sleep deprivation impairs learning and memory by decreasing brain O-GlcNAc cycling in mouse. Neurotherapeutics 18, 2504–2517 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaur K., et al. , Integrating molecular subclassification of medulloblastomas into routine clinical practice: A simplified approach. Brain Pathol. 26, 334–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., et al. , The role of Shh signalling pathway in central nervous system development and related diseases. Cell Biochem. Funct. 39, 180–189 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Zhou F., et al. , Nek2A/SuFu feedback loop regulates Gli-mediated Hedgehog signaling pathway. Int. J. Oncol. 50, 373–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das S., et al. , O-GlcNAcylation of GLI transcription factors in hyperglycemic conditions augments Hedgehog activity. Lab. Invest. 99, 260–270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutter M., Wang J., Huang Z., Kuliszewski M., Post M., Gli2 influences proliferation in the developing lung through regulation of cyclin expression. Am. J. Respir. Cell Mol. Biol. 42, 615–625 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Regl G., et al. , Human GLI2 and GLI1 are part of a positive feedback mechanism in basal cell carcinoma. Oncogene 21, 5529–5539 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Pandolfi S., Stecca B., Cooperative integration between HEDGEHOG-GLI signalling and other oncogenic pathways: Implications for cancer therapy. Expert Rev. Mol. Med. 17, e5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikram M. S., et al. , GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J. Invest. Dermatol. 122, 1503–1509 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Nieuwenhuis E., et al. , Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia. Mol. Cell. Biol. 26, 6609–6622 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee Y., et al. , Patched2 modulates tumorigenesis in patched1 heterozygous mice. Cancer Res. 66, 6964–6971 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Wulff-Fuentes E., et al. , The human O-GlcNAcome database and meta-analysis. Sci. Data 8, 25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarkar L., et al. , Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proc. Natl. Acad. Sci. U.S.A. 97, 4520–4524 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie G., et al. , Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice. Hepatology 58, 1801–1813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo X., Wang X. F., Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 19, 71–88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen H., et al. , O-GlcNAc transferase Ogt regulates embryonic neuronal development through modulating Wnt/β-catenin signaling. Hum. Mol. Genet. 31, 57–68 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Delgado-García J. M., [Structure and function of the cerebellum] [in Spanish]. Rev. Neurol. 33, 635–642 (2001). [PubMed] [Google Scholar]
- 63.Aruga J., Inoue T., Hoshino J., Mikoshiba K., Zic2 controls cerebellar development in cooperation with Zic1. J. Neurosci. 22, 218–225 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flora A., Klisch T. J., Schuster G., Zoghbi H. Y., Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science 326, 1424–1427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foster D. A., Yellen P., Xu L., Saqcena M., Regulation of G1 cell cycle progression: Distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s). Genes Cancer 1, 1124–1131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bohnsack B. L., Hirschi K. K., Nutrient regulation of cell cycle progression. Annu. Rev. Nutr. 24, 433–453 (2004). [DOI] [PubMed] [Google Scholar]
- 67.González A., Hall M. N., Lin S. C., Hardie D. G., AMPK and TOR: The yin and yang of cellular nutrient sensing and growth control. Cell Metab. 31, 472–492 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Marino S., Medulloblastoma: Developmental mechanisms out of control. Trends Mol. Med. 11, 17–22 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Gilbertson R. J., Ellison D. W., The origins of medulloblastoma subtypes. Annu. Rev. Pathol. 3, 341–365 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Barakat M. T., Humke E. W., Scott M. P., Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol. Med. 16, 337–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sidaway P., Medulloblastoma: Prognostic subtypes revealed. Nat. Rev. Clin. Oncol. 18, 131 (2021). [DOI] [PubMed] [Google Scholar]
- 72.De Luca A., et al. , Sonic hedgehog patterning during cerebellar development. Cell. Mol. Life Sci. 73, 291–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu D., Jin J., Qiu Z., Liu D., Luo H., Functional analysis of O-GlcNAcylation in cancer metastasis. Front. Oncol. 10, 585288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bose S., Le A., Glucose metabolism in cancer. Adv. Exp. Med. Biol. 1063, 3–12 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Hay N., Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 16, 635–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seo H. G., et al. , Mutual regulation between OGT and XIAP to control colon cancer cell growth and invasion. Cell Death Dis. 11, 815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sodi V. L., et al. , mTOR/MYC axis regulates O-GlcNAc transferase expression and O-GlcNAcylation in breast cancer. Mol. Cancer Res. 13, 923–933 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim Y., et al. , Selective down-regulation of glioma-associated oncogene 2 inhibits the proliferation of hepatocellular carcinoma cells. Cancer Res. 67, 3583–3593 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Milla L. A., et al. , Yeast-based assay identifies novel Shh/Gli target genes in vertebrate development. BMC Genomics 13, 2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original RNA-seq data in this study have been deposited in the National Center for Biotechnology Information Sequence Read Achieve (accession no. PRJNA798583). MS data reported in this paper have been deposited to the ProteomeXchange Consortium via the iProX partner repository (dataset identifier PXD034270/IPX0004530000). The entire datasets are publicly available at https://www.iprox.cn//page/project.html?id=IPX0004530000. All other study data are included in the main text and SI Appendix.