Significance
Tight junctions form selectively permeable seals that limit paracellular flux. Increased tight junction permeability has been associated with intestinal disease. Previous studies indicate that casein kinase 2 (CK2) phosphorylates S408 within an unstructured region of the cytoplasmic tail of the tight junction protein occludin. Moreover, CK2 inhibition and consequent S408 dephosphorylation reduce paracellular permeability in vitro and in vivo and attenuate experimental immune-mediated intestinal disease in vivo. Here, we show that S408 phosphorylation enhances intramolecular interactions between the unstructured region and the distal α-helical bundle to limit binding to the tight junction protein zonula occludens-1 (ZO-1). Conversely, S408 dephosphorylation limits intramolecular interactions, promotes occludin binding to ZO-1 in vitro, and enhances occludin anchoring at the tight junction in vivo.
Keywords: tight junction, barrier, permeability, intestine
Abstract
Occludin is a tetramembrane-spanning tight junction protein. The long C-terminal cytoplasmic domain, which represents nearly half of occludin sequence, includes a distal bundle of three α-helices that mediates interactions with other tight junction components. A short unstructured region just proximal to the α-helical bundle is a phosphorylation hotspot within which S408 phosphorylation acts as molecular switch that modifies tight junction protein interactions and barrier function. Here, we used NMR to define the effects of S408 phosphorylation on intramolecular interactions between the unstructured region and the α-helical bundle. S408 pseudophosphorylation affected conformation at hinge sites between the three α-helices. Further studies using paramagnetic relaxation enhancement and microscale thermophoresis indicated that the unstructured region interacts with the α-helical bundle. These interactions between the unstructured domain are enhanced by S408 phosphorylation and allow the unstructured region to obstruct the binding site, thereby reducing affinity of the occludin tail for zonula occludens-1 (ZO-1). Conversely, S408 dephosphorylation attenuates intramolecular interactions, exposes the binding site, and increases the affinity of occludin binding to ZO-1. Consistent with an increase in binding to ZO-1, intravital imaging and fluorescence recovery after photobleaching (FRAP) analyses of transgenic mice demonstrated increased tight junction anchoring of enhanced green fluorescent protein (EGFP)-tagged nonphosphorylatable occludin relative to wild-type EGFP-occludin. Overall, these data define the mechanisms by which S408 phosphorylation modifies occludin tail conformation to regulate tight junction protein interactions and paracellular permeability.
The discovery of zonula occludens-1 (ZO-1) over 35 y ago marked the beginning of a new era of tight junction biology that extended beyond morphology to focus on tight junction components (1). Cingulin, ZO-2, and other peripheral membrane, or cytoplasmic plaque, proteins were subsequently identified, but none of these could be the critical integral membrane proteins that define the intercellular junctions. This situation changed with the discovery of occludin (2), a tetraspanning transmembrane protein and component of tight junction strands. Occludin expression enhances intercellular adhesion, increases tight junction strand complexity (3), and augments the paracellular barrier to macromolecular flux (4–7). Occludin knockout in mice leads to abnormalities that include features of TORCH syndrome (8), male infertility (9), and hearing loss (10) as well as resistance to epithelial apoptosis and colitis (11). Conversely, transgenic occludin overexpression in intestinal epithelial cells attenuates acute cytokine-induced barrier loss (12). Despite these observations, the absence of spontaneous renal or intestinal disease in occludin knockout mice and subsequent discovery of claudins (13) have led many to conclude that occludin loss is inconsequential.
Alternatively, many studies indicate that occludin is a regulatory protein. These functions often require the C-terminal cytoplasmic tail (14–20), which comprises almost half of occludin. The membrane-distal portion of occludin includes three α-helices, which form a coiled coil that mediates interactions with ZO-1 and other proteins (21–23) and is required for tumor necrosis factor–induced barrier loss (24). Multiple studies have identified phosphorylation sites within the cytoplasmic C-terminal occludin tail (15–20, 25–31) and linked them to regulation of interprotein interactions, epithelial barrier function, vasculogenesis, monolayer maturation, and colitis in vitro and in vivo (14–20, 25, 28–30, 32).
A short cluster of 11 amino acids, Y398 to S408, within an unstructured region just proximal to the α-helical bundle that includes conserved phosphorylation sites for c-Src, PKC isoforms, and casein kinase 2 (CK2) has received significant attention (26, 28, 33, 34). In particular, CK2-mediated phosphorylation of occludin S408 acts as a molecular switch that limits occludin binding to ZO-1 (16, 19, 27). The bulk of tight junction–associated occludin is phosphorylated at S408, but CK2 inhibition and subsequent S408 dephosphorylation trigger assembly of a tripartite complex in which ZO-1 links occludin to claudins (19). The interactions mediated by this complex inactivate claudin-2 channels in vitro and in vivo (19, 32), and either claudin-2 knockout or CK2 inhibition is sufficient to attenuate immune-mediated colitis (32). Disruption of occludin S408 phosphorylation may therefore be a therapeutic approach to enhancing epithelial barrier function and limiting disease.
Here, we report our studies of the structural and dynamic consequences of charge changes at the S408 position. This residue is within the unstructured region proximal to the α-helical bundle, which is not seen in the X-ray crystallographic structure (21). We used NMR techniques including paramagnetic relaxation enhancement (PRE) to identify and characterize dynamic interactions between this unstructured region and the α-helical bundle. Effects of S408 phosphorylation were most prominent at sites where the three helices are linked by flexible hinges. The data further indicate that, when phosphorylated at S408, the unstructured region interferes with ZO-1 binding to the α-helical bundle. These data explain how S408 phosphorylation functions as a molecular switch to modulate occludin interactions within the tight junction to regulate paracellular permeability.
Results
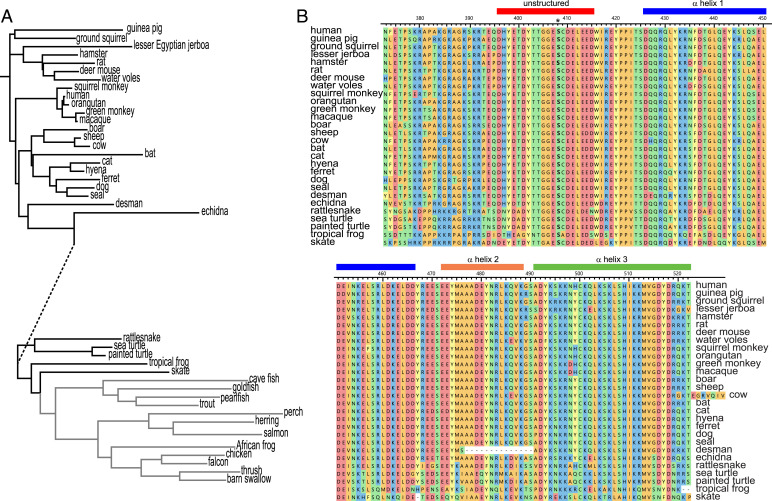
The primary sequence of occludin has been preserved throughout evolution (Fig. 1A). This preservation is best exemplified by the 257 residue C-terminal cytoplasmic domain, which can be broken into membrane-proximal (residues 266–372) and membrane-distal (residues 373–522) regions. The membrane-distal portion includes the α-helices (426–466, 472–488, and 491–522), which form the ZO-1 binding site. The unstructured sequence proximal to the first α-helix includes a short anionic region (396–415, DHYETDYTTGGESCDELEED) with eight acidic residues and six additional residues that can be phosphorylated to further increase the charge density of this region (33). This sequence is 100% identical throughout mammalian evolution (35) and more than 90% conserved in birds, amphibians, and fish (Fig. 1). Within this region, S408 phosphorylation has been implicated as a regulatory mechanism and potential a therapeutic target (16, 19, 27, 32).
Fig. 1.
The primary sequence of occludin is conserved throughout evolution. (A) Dendrogram showing evolution of the complete occludin sequence. Line lengths indicate degree of conservation, with the exception of the dashed segment, whose length has been reduced by ∼50% for presentation. (B) Sequence alignment of the membrane-distal portion of the occludin C-terminal cytoplasmic domain for species shown in black in panel A. Sequences corresponding to the unstructured region (396–415) and each of the three α-helices are indicated.
Global Occludin 383–522 Structure Is Unaffected by S408 Pseudophosphorylation.
Despite the biological importance of S408 phosphorylation and dephosphorylation, it is not known how modification of this site affects occludin structure. This question cannot be addressed with crystallography because residues 383–415 are unstructured and are not seen in crystal structures (21). To help define the mechanisms by which S408 phosphorylation status affect occludin structure and dynamics, we studied recombinant occludin 383–522 in which S408 was substituted by aspartic acid or alanine to form pseudophosphorylated and nonphosphorylatable tails, respectively (SI Appendix, Table S1). Circular dichroic spectra (SI Appendix, Fig. S1A) showed that these proteins were essentially identical with largely α-helical structure (56.6%). Moreover, both proteins demonstrated sigmoidal melting curves with reversible thermal melting behavior and similar Tm values of ∼50 °C (SI Appendix, Fig. S1B). Further analysis by size exclusion chromatography showed that both proteins eluted earlier than expected, with an apparent molecular mass of ∼70 kDa (SI Appendix, Fig. S1C). If it were spherical, occludin 383–512 (∼16.5 kDa) would have a volume of ∼2.8 × 104 Å3, radius of ∼17.5 Å, and density of 1.34 g/cm3 and would elute at the expected position. However, the occludin α-helical bundle is a prolate ellipsoid with an axial ratio of a:b = 5:1 (21). This value corresponds to a major semiaxis (a) of ∼51.1 Å, minor semiaxis (b) of ∼10.2 Å, and hydrodynamic radius of ∼27.5 Å. A spherical protein of this radius and the same density would have a molecule weight of ∼70 kDa, thereby explaining the elution of both occludin structures at that apparent molecular weight. Some small, earlier eluting peaks, consistent with oligomers, were detected for both 383–522S408A and 383–522S408D (SI Appendix, Fig. S1C). The overall chromatographic profiles of 383–522S408A and 383–522S408D were indistinguishable, however. Thus, despite previous reports suggesting occludin tail homotypic interactions (19, 22, 36, 37), the chromatographic profiles indicate that both pseudophosphorylated and nonphosphorylatable occludin 383–522 are primarily monomeric under these mildly reducing conditions and, more importantly, that S408 pseudophosphorylation does not induce oligomerization. We conclude that in terms of α-helical content and overall conformation, S408 pseudophosphorylation does not significantly affect occludin 383–522 structure.
NMR Spectra of 383–522S408A and 383–522S408D Demonstrate Only Small Differences.
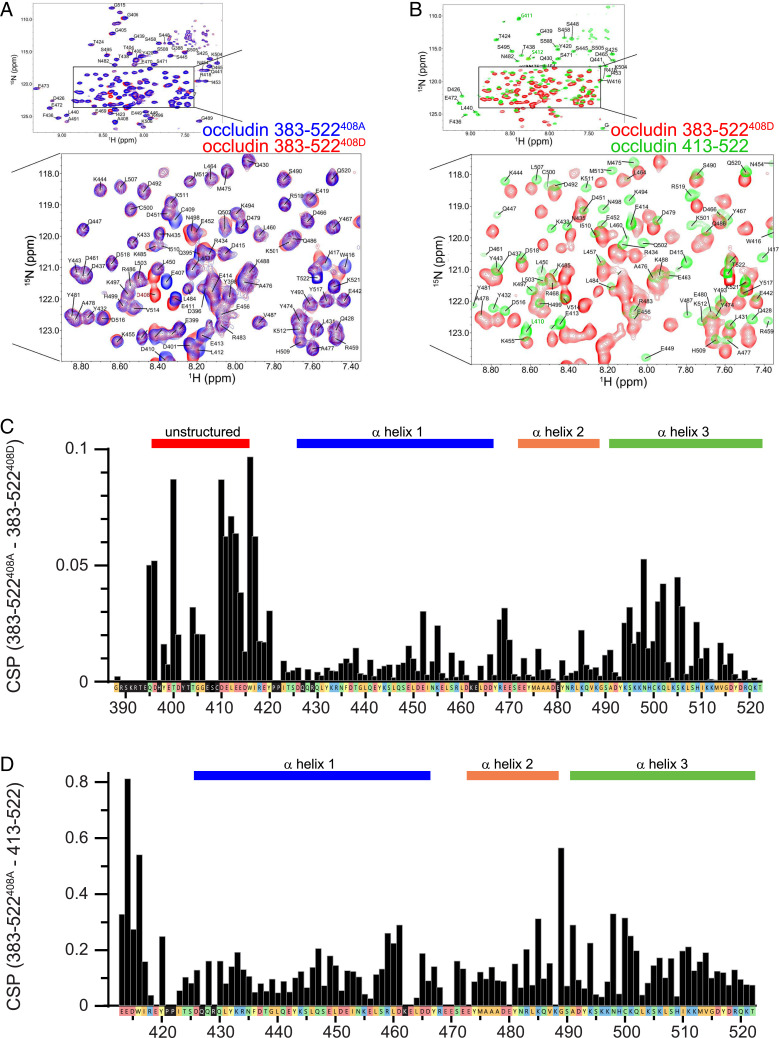
Standard triple resonance techniques (including N-TROSY, HNCO, HNcaCO, HNcoCACB, HNCACB, HNCA, and HNcoCA) were used to assign 15N,13C-labeled occludin 383–522S408A and 383–522S408D (Fig. 2A and SI Appendix, Figs. S2 and S3). With the exception of local changes around the S408 substitution by alanine or aspartic acid, only small chemical shift perturbations (CSPs) were observed (Fig. 2C and SI Appendix, Tables S2 and S3). These CSPs were detected primarily within the distal end of the first α-helix and the center of the third α-helix (Fig. 2B), which are adjacent to one another within the α-helical bundle (21). Heteronuclear 1H–15N nuclear Overhauser effect spectra (SI Appendix, Fig. S4) and CLEANEX-PM (SI Appendix, Fig. S5) experiments showed that the unstructured region residues are highly dynamic and solvent exposed. In contrast, the α-helical bundles are not dynamic and are largely solvent protected; residues at bends within or turns between the α-helices were somewhat more dynamic and solvent exposed.
Fig. 2.
Effects of the unstructured region (383–412) on the α-helical bundle are largely independent of S408 phosphorylation. (A) 15N-TROSY spectra comparing occludin 383–522S408A (blue) and 383–522S408D (red). The Inset shows a higher-magnification view of the boxed region. (B) 15N-TROSY spectra comparing occludin 383–522S408D (red) and 413–522 (green). The Inset shows a higher-magnification view of the boxed region. Occludin 413–522 was synthesized with a short N-terminal extension of GPLGS (SI Appendix, Table S1). Residues L410, G411, and S412 are part of this nonnative sequence and a labeled in green font for clarity. (C) Chemical shift perturbations between occludin 383–522S408A and 383–522S408D. Sequences corresponding to the unstructured region (396–415) and the three α-helices are indicated. The color of each box indicates small or hydrophobic (orange), nucleophilic or amide (green), aromatic (yellow), acidic (red), and basic (blue) residues. Black boxes indicate residues for which no signal was detected. (D) Chemical shift perturbation differences between occludin 383–522S408A and 413–522. Note the eightfold greater range of the ordinate axis relative to panel C.
In crystallographic studies of variants of occludin, the α-helices were isomorphous, but the angle between the second and third α-helices in occludin413-522 was 9° narrower than in occludin383-522 (21). We compared NMR spectra of occludin413-522 (i.e., lacking residues 383–412, which forms the unstructured region) and both occludin383-522 variants. In contrast to comparisons between occludin 383–522S408A and 383–522S408D, there were large chemical shift differences between occludin413-522 and either form of occludin383-522 (Fig. 2 C and D and SI Appendix, Table S4). Sites showing large CSPs relative to occludin383-522 were present throughout occludin413-522 but were most prominent surrounding the bend between the first two α-helices and within the proximal portion of third α-helix. These data indicate that the proximal unstructured region modifies relationships within the distal α-helical bundle.
The Unstructured Region Interacts Dynamically with the α-Helical Bundle.
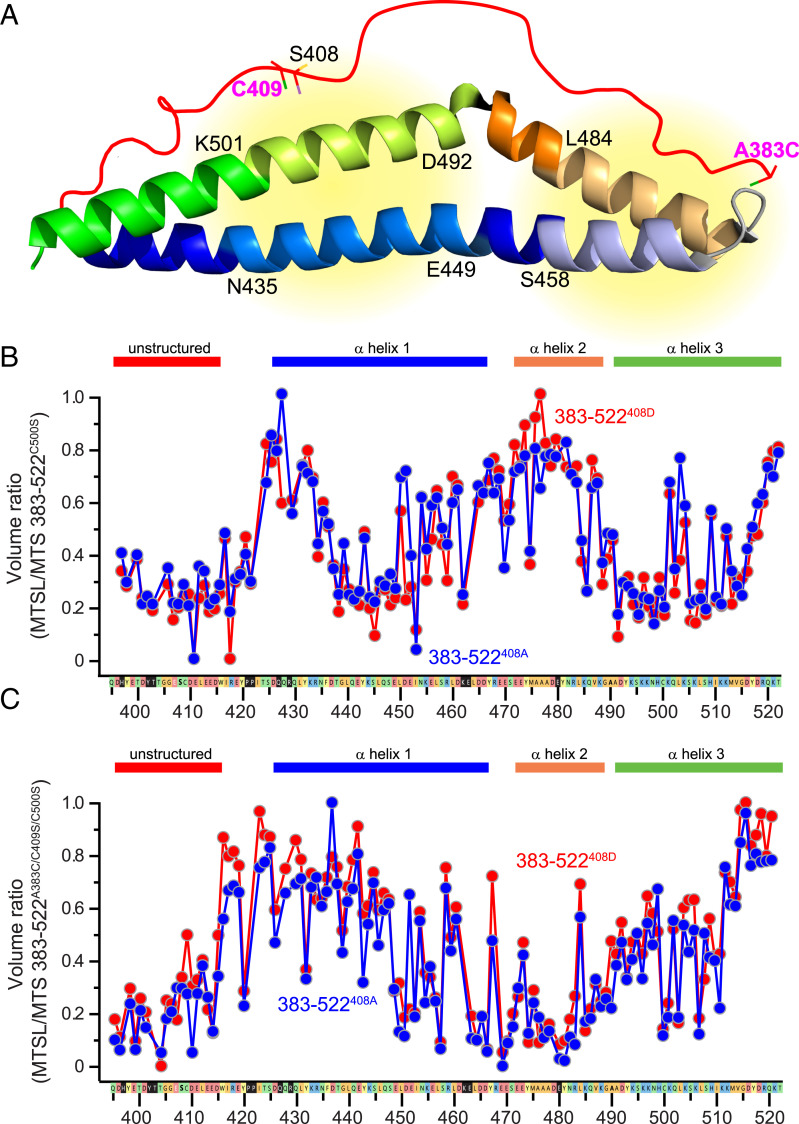
The differences between occludin413-522 and occludin383-522 spectra suggest that the unstructured domain may interact directly with the α-helical bundle. To test this hypothesis, we turned to PRE methods. We initially studied occludin 383–522S408A and 383–522S408D with C500S mutations, so that C409 was the only cysteine residue present. Either S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL) or its diamagnetic variant 1-acetyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl-methanethiosulfonate (MTS) was then used to label C409, and PRE was determined as the ratio of MTSL-occludin to MTS-occludin peak volumes. Consistent with the circular dichroic spectroscopy and NMR differences induced by pseudophosphorylation, the overall PRE patterns of labeled occludin 383–522S408A and 383–522S408D were similar. In general, two regions of relaxation enhancement, as indicated by reduced peak values, were detected from N435 to E449 and from D492 to K501 (Fig. 3 A and B). These represent regions within the first and third α-helices, respectively (Fig. 3A).
Fig. 3.
The unstructured region demonstrates dynamic intramolecular interactions with the α-helical bundle. (A) PRE analyses took advantage of C409 (Left) or an A383C mutation (Right). The areas in which PRE was detected are indicated (yellow). (B) PRE analysis of occludin 383–522S408A (blue) and 383–522S408D (red) after labeling of C409. Sequences corresponding to the unstructured region (396–415) and each of the three α-helices are indicated. (C) PRE analysis of occludin 383–522S408A (blue) and 383–522S408D (red) after labeling A383C.
In order to analyze the N terminus of the unstructured region, we generated A383C, C409S mutants of occludin 383–522. With the MTSL or MTS label in position 383, the strongest PRE effects were seen in the region of residues S458 to L484 (Fig. 3C), which corresponds to the distal part of the first α-helix and proximal half of the second α-helix (Fig. 3A). These data indicate that residues along the full length of the unstructured region interact with the α-helical bundle.
Occludin S408 Phosphorylation Modifies Interactions between the Unstructured Region and the α-Helical Bundle.
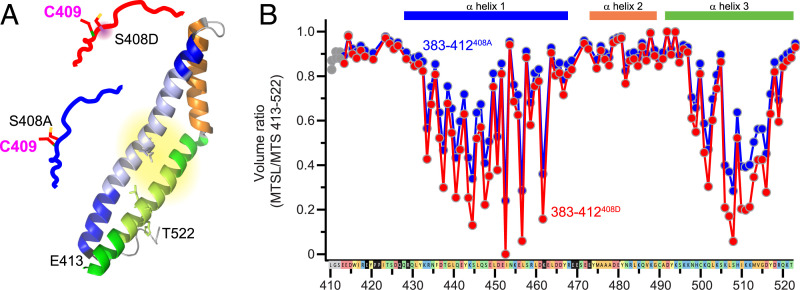
To define effects of occludin S408 phosphorylation more rigorously, occludin 383–412S408A or 383–412S408D unstructured region peptides were labeled at C409 with MTSL and added to the unlabeled occludin413-522 α-helical bundle in trans. The pattern of relaxation enhancement generated was generally similar to that observed when C409 was labeled within occludin 383–522 constructs, but areas the signal generated by the pseudophosphorylated 383–412S408D peptide was greater than that of the 383–412S408A peptide in two areas (Fig. 4A). The first area, D437–D461, is within the first α-helix. The second area, H499–M513, is within the distal half of the third α-helix (Fig. 4B). Specific residues in which 383–412S408D induced greater PRE signals than 383–412S408A included L446, Q447, D461, L507, H509, and I510, all of which are within the occludin α-helical bundle.
Fig. 4.
S408 pseudophosphorylation modifies intermolecular interactions between the free unstructured region and the α-helical bundle. (A) PRE analyses took advantage of C409 (Left) within occludin 383–412S408A (blue) and 383–412S408D (red). (B) Intermolecular interactions between each spin-labeled peptide and the occludin413-522 α-helical bundle are shown. Residues L40, G411, and S412 derive from the occludin 413–522 N-terminal extension adding during synthesis and are labeled in gray. These were not affected by S408 pseudophosphorylation.
Occludin 383–412S408D Binds the α-Helical Bundle and Reduces Affinity for ZO-1.
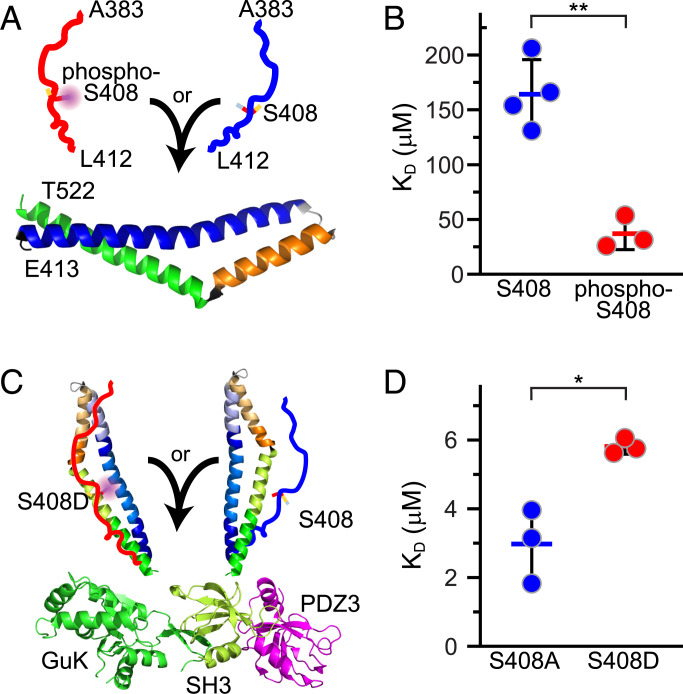
The PRE data indicate that interactions of the α-helical bundle with pseudophosphorylated 383–412S408D are somewhat greater than those of 383–412S408A. We hypothesized that this difference might correlate with the affinities of 383–412S408A and 383–412S408D for the α-helical bundle (Fig. 5A). To test this idea, we assessed binding between fluorescent-tagged occludin413-522 and synthetic occludin383-412 in which S408 was either phosphorylated or nonphosphorylated via microscale thermophoresis. S408-phosphorylated peptide bound to occludin413-522 with a Kd of 37 ± 15 μM, while the Kd of nonphosphorylated peptide binding was 175 ± 27 μM (Fig. 5B, P = 0.0015).
Fig. 5.
S408 phosphorylation enhances intermolecular binding between the free unstructured region and the α-helical bundle but reduces binding to ZO-1 SH3-GuK. (A) Synthetic occludin 383–412 peptides with (red) or without (blue) phosphorylation at S408 were added to recombinant the occludin 413–522 α-helical bundle. Binding was determined by MST. (B) S408 phosphorylation markedly enhanced binding of occludin 383–412 peptides to the α-helical bundle. (C) Occludin 383–522S408A (blue) or 383–522S408D (red) were added to recombinant ZO-1 PDZ3-SH3-GuK fusion construct, and binding was determined by microscale thermophoresis (MST). (D) S408 pseudophosphorylation reduced occludin 383–522 binding to ZO-1 PDZ3-SH3-GuK. **P < 0.01. *P < 0.05.
Our previous glutathione S-transferase (GST) pulldown data show that occludin 383–522S408A interacts more strongly with the ZO-1 occludin binding region than occludin 383–522S408D. To quantify this interaction, we assessed interactions between the U5-GuK occludin binding region of ZO-1 and fluorescent-tagged occludin 383–522S408A and 383–522S408D via microscale thermophoresis (Fig. 5C). ZO-1 PDZ3-SH3-GuK bound to occludin 383–412S408A with a Kd of 3.0 ± 1.1 μM, while the Kd of PDZ3-SH3-GuK binding to occludin 383–522S408D was 5.8 ± 0.2 μM (Fig. 5D, P = 0.011). S408 pseudophosphorylation is therefore sufficient to modestly reduce the affinity of the occludin α-helical bundle for the ZO-1 U5-GuK domain.
S408 Dephosphorylation Stabilizes Occludin at the Tight Junction In Vivo.
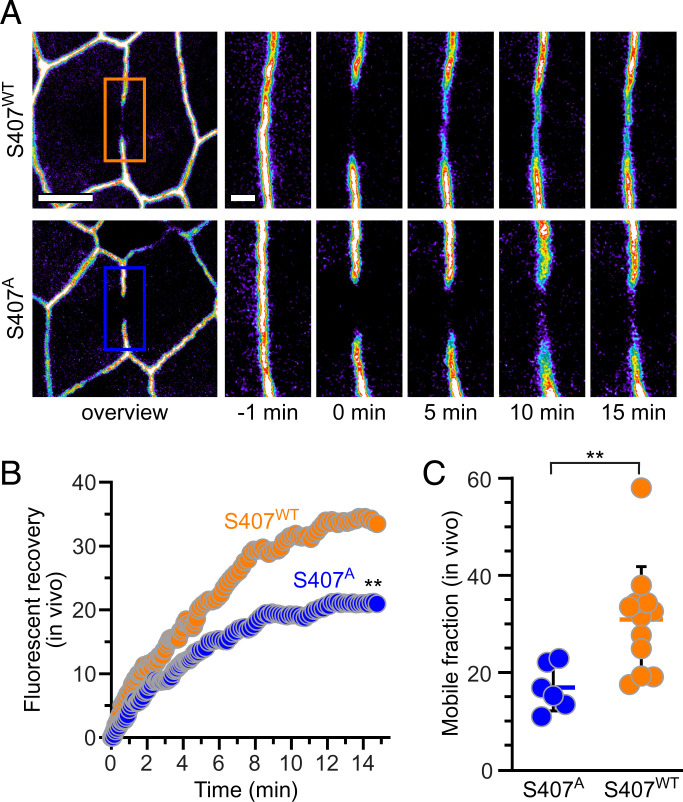
Our previous in vitro and in vivo data suggest that the bulk of occludinS408 is phosphorylated in living cells (19, 32). In order to prevent this phosphorylation, we mutated S407 within mouse occludin, which is equivalent to S408 within the human occludin sequence, and generated transgenic mice expressing enhanced green fluorescent protein (EGFP)-occludinS407A from the intestinal epithelial specific villin promoter. These and transgenic mice expressing EGFP-occludinWT from the villin promoter were crossed to intestinal epithelial-specific occludin knockout mice and analyzed by fluorescence recovery after photobleaching (FRAP) via intravital imaging (Fig. 6 A and B) (11, 12). As we have previously reported (19), EGFP-occludinWT was highly mobile, with a mobile fraction of 31 ± 3% (Fig. 6C). In contrast, the mobile fraction of EGFP-occludinS407A was only 17 ± 2% (P < 0.01). Thus, genetic inhibition of S408 (human) phosphorylation enhances occludin anchoring at the tight junction. Our previous in vitro analyses have shown that increased tight junction anchoring of S408 dephosphorylated or S408A occludin, relative to phosphorylated or S408D occludin, requires the presence of ZO-1 (19). Thus, the in vivo FRAP data reported here support the hypothesis that S408 dephosphorylation stabilizes interactions between the α-helical bundle and ZO-1 to increase anchoring and restrict occludin diffusion within tight junction membranes.
Fig. 6.
Inhibition of S408 phosphorylation stabilizes tight junction–associated occludin in vivo. (A) Low- and high-magnification views from intravital imaging of intestinal epithelia within villin–Cre x Oclnf/f mice expressing transgenic EGFP-occludinWT and EGFP-occludinS407A from the villin promoter. Individual high-magnification frames represent images taken at indicated times after photobleaching. Bars = 5 μm and 1 μm. (B) Representative recovery curves of EGFP-occludinWT (orange) and EGFP-occludinS407A (blue). (C) Mobile fraction of EGFP-occludinWT (orange, n = 12) and EGFP-occludinS407A (blue, n = 6). **P < 0.01.
Discussion
The function of occludin as a tight junction component has remained enigmatic, but abundant data indicate it serves regulatory, rather than essential structural, functions (3, 4, 6, 8, 9, 11, 19, 24, 30, 38–41). Consistent with this function, the α-helical bundle, or coiled-coil OCEL domain, within the distal C-terminal tail mediates occludin binding to multiple proteins (21–24, 36, 37, 42). These interactions may be regulated by phosphorylation within a short unstructured region just proximal to the α-helical bundle (14–20, 43). Here, we focused on S408, which is located within the unstructured region and acts as a molecular switch that regulates binding to ZO-1 (16, 19, 21). S408 dephosphorylation enhances occludin binding to ZO-1 and assembly of an occludin–ZO-1–claudin-2 complex that inhibits claudin-2 channel function (16, 19, 32). This mechanism allows CK2 inhibition to inactivate claudin-2 channels and reverse cytokine-induced, claudin-2-dependent intestinal permeability increases in vitro and in vivo (19, 32). The observation that CK2 inhibition limits progression of immune-mediated colitis in wild-type but not claudin-2 knockout mice indicates that the same mechanism of claudin-2 channel inhibition may be an effective therapeutic intervention. Molecular understanding of the means by which occludin S408 phosphorylation regulates intraprotein and interprotein interactions is an essential prerequisite to development of specific therapeutic agents that exploit this mechanism.
Although the crystal structure of the C-terminal α-helical bundle, which forms the ZO-1 binding domain, has been solved, the precise sites at which occludin interacts with ZO-1 remain controversial (21, 23, 24). Nevertheless, analyses of occludin mutants suggest that phosphorylation of residues within the unstructured region, particularly Y398, Y402, or S408, negatively regulates ZO-1 binding (17, 19). In cells, tight junction localization of occludinY398D/Y402D is impaired (17), and either CK2 inhibition or alanine substitution at S408 enhance occludin anchoring at tight junctions (19). Here, we have demonstrated that inhibition of S408 (S407 in mice) phosphorylation by alanine substitution enhances occludin anchoring at the tight junction in vivo (19). When combined with the in vitro data showing that S408 phosphorylation reduces the affinity of occludin 383–522 for ZO-1 PDZ3-SH3-GuK and previous work showing that modulation of occludin tight junction anchoring by S408 phosphorylation requires ZO-1, these results indicate that S408 dephosphorylation enhances in vivo interactions between occludin and ZO-1. Although S408 phosphorylation or dephosphorylation is sufficient, but there are five potential phosphorylation sites within the unstructured region. Previous work shows that phosphorylation of Y398 and Y402 also reduces occludin binding to ZO-1 (17) and many studies have linked other occludin phosphorylation sites to function (15, 18, 44). Thus, multiple phosphorylation events may contribute to occludin regulation.
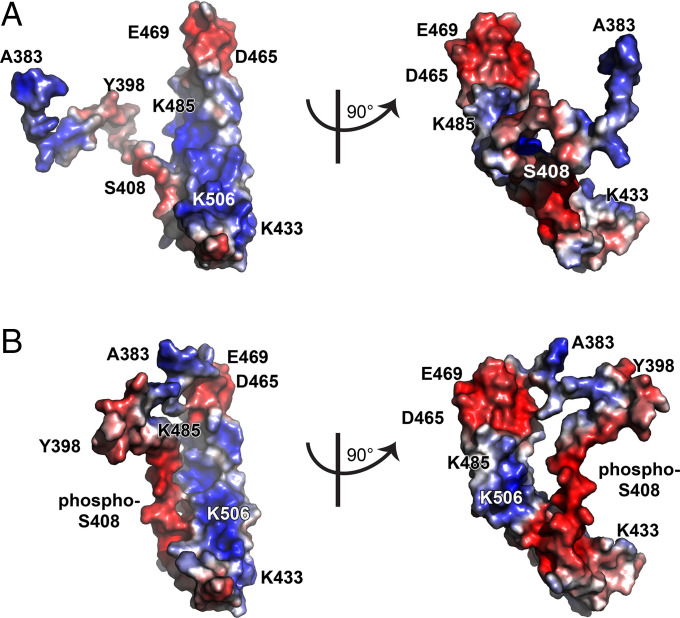
The NMR PRE experiments showed that the unstructured domain associates dynamically with the α-helical bundle and that these interactions can be modified by S408 phosphorylation or pseudophosphorylation. Specifically, the data suggest that increasing anionic charge density within the unstructured region enhances interactions with the putative ZO-1 binding site composed of basic residues within the α-helical bundle, including K433, K444, K485, K488, K501, K504, K506, and K511 (Fig. 7A), which has been proposed to be the ZO-1 binding site (21). In particular, S408 is in close proximity to K501 within the α-helical bundle. Moreover, the adjacent region ES408CDELEED (410–415) could form a charge-neutralizing interface with KQLKSK (501–506). Finally, PRE comparing occludinA383C/S408A/C500S and occludinA383C/S408D/C500S showed that S408 pseudophosphorylation enhanced interactions between A383 and the acidic tip of the bundle where the distal portion of the first α-helix is adjacent to the proximal portion of the second α-helix. Remarkably, this region has also been reported to represent a ZO-1 binding site (23). S408 phosphorylation therefore enhances interactions between the unstructured region and the α-helical bundle, particularly in areas postulated to represent ZO-1 binding sites.
Fig. 7.
S408 phosphorylation may cause the unstructured region to mask the ZO-1 binding site within the occludin C-terminal α-helical bundle. (A) Structure of occludin 383–522S408A. The structure of the α-helical bundle is based on reported X-ray diffraction data (21). The position of the unstructured region (383–413) was generated in MUSTER software (45). (B) Structure of occludin 383–522S408D. Two views oriented at right angles to one another are shown for each structure. Colors (blue, red) indicate the electrostatic potential as determined via Poisson–Boltzmann equation.
The PRE data indicate that S408 phosphorylation causes the unstructured region to interact with the exposed ZO-1-binding face within α-helical bundle. This finding is consistent with the observations that A383-L412S408P binds the α-helical bundle with greater affinity than A383-L412S408A while, conversely, occludin 383–522408D bound ZO-1 PDZ3-SH3-GuK with reduced affinity relative to occludin 383–522408A. Together, the structural and binding data support the hypothesis that S408 phosphorylation–enhanced binding of the unstructured region obstructs the ZO-1 binding domain within α-helical bundle. To test this idea in silico, we modeled occludin 383–522 structure by using MUlti-Sources ThreadER (MUSTER) software (45). This model predicted that increased anionic charge density at the distal end of the unstructured region can cause it to extend over and mask, or at least limit access to, the basic face of the α-helical bundle (Fig. 7B). While unproven, this would provide a mechanism by which S408 phosphorylation regulates interactions between the α-helical bundle, the unstructured region, and ZO-1. It does not, however, explain how occludin S408 phosphorylation leads to increased interactions between ZO-1 and claudin-2 (19). Nevertheless, our in vivo analysis showing enhanced anchoring of occludinS407A at the tight junction, relative to occludinWT, is consistent with the proposed model and provides evidence that this process is active in vivo. Moreover, the approximately twofold mobile fraction reduction triggered by S407A mutation is similar to that induced by S408A mutation of human occludin in vitro (19). Both in vitro and in vivo CK2 inhibition had comparable effects on wild-type occludin mobile fraction (19). Finally, in vitro (19) and in vivo (32) studies have shown that this increased anchoring is sufficient to reverse interleukin-13-induced barrier loss by inhibiting claudin-2 channels. Thus, although the changes in ZO-1 binding to occludin and occludin mobile fraction triggered by S408 dephosphorylation are small, they are sufficient to impact function dramatically and are consistent with the molecular mechanism proposed here.
As a whole, our data provide a compelling model for understanding how phosphorylation events within the unstructured region just proximal to the α-helical bundle regulate binding to ZO-1 and specifically implicate S408 phosphorylation as a key signaling event within that process. Although other kinases are not excluded, the results presented here, together with those of previous studies (16, 19, 26, 27, 32), are consistent with CK2 as the critical kinase that phosphorylates S408. Nevertheless, the phosphatase that opposes CK2-mediated occludin phosphorylation remains unidentified. Moreover, physiologically and pathophysiologically relevant stimuli that regulate S408 phosphorylation and dephosphorylation have not been elucidated. Further work is therefore needed to fully understand the regulation and impact of S408 phosphorylation. These data help to build a foundation that will support pursuit of answers to these questions and, potentially, development of novel therapeutic strategies to regulate epithelial and endothelial tight junction barriers.
Materials and Methods
Protein and Peptide Expression.
Clones for GST-occludin 383–522 wild-type, S408D, and S408A constructs were generated by site-directed mutagenesis as previously described (19). Occludin 383–412 peptides were expressed as GST fusion proteins with an intervening TEV site. All were purified by affinity and gel chromatography.
NMR Assignments.
2H, 13C, and 15N-labeled proteins were analyzed using TROSY sequences (e.g., 15N-TROSY) for NMR assignments.
PRE Experiments.
For PRE experiments (46), used occludin constructs that had undergone mutagenesis so that only a single cysteine was present. These were labeled with MTSL or its diamagnetic variant MTS (47).
Microscale Thermophoresis.
For microscale thermophoresis, His-tagged occludin constructs were labeled with a MonolithNT His-Tag Labeling Kit (NanoTemper). Thermophoresis was performed with a Monolith NT.115 (NanoTemper). Data were analyzed in MO Affinity Analysis software.
Mice, Intravital Imaging, and FRAP.
Mice 7–12 wk old maintained on a C57BL/6 genetic background were used for all experiments. EGFP-occludinS407A transgenic mice were created and imaged as previously described for villin–EGFP-occludin (wild-type) mice (12, 19).
Complete details for all materials and methods are provided in the SI Appendix.
Supplementary Material
Acknowledgments
Supported by R01 DK061931 (J.R.T.), R01 DK068271 (J.R.T.), and R21 AG067036 (S.C.M.).
Footnotes
Competing interest statement: J.R.T. is a founder and shareholder of Thelium Therapeutics and has served as a consultant for Entrinsic, Immunic, and Kallyope.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204618119/-/DCSupplemental.
Data, Materials, and Software Availability
Data that support the findings of this study are included in the main text or SI Appendix. Requests for additional information or materials may be addressed to the corresponding author.
References
- 1.Stevenson B. R., Siliciano J. D., Mooseker M. S., Goodenough D. A., Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 103, 755–766 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse M., et al. , Occludin: A novel integral membrane protein localizing at tight junctions. J. Cell Biol. 123, 1777–1788 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito A. C., et al. , Occludin and tricellulin facilitate formation of anastomosing tight-junction strand network to improve barrier function. Mol. Biol. Cell 32, 722–738 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furuse M., et al. , Overexpression of occludin, a tight junction-associated integral membrane protein, induces the formation of intracellular multilamellar bodies bearing tight junction-like structures. J. Cell Sci. 109, 429–435 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Van Itallie C. M., Anderson J. M., Occludin confers adhesiveness when expressed in fibroblasts. J. Cell Sci. 110, 1113–1121 (1997). [DOI] [PubMed] [Google Scholar]
- 6.McCarthy K. M., et al. , Occludin is a functional component of the tight junction. J. Cell Sci. 109, 2287–2298 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Yu A. S., et al. , Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. Am. J. Physiol. Cell Physiol. 288, C1231–C1241 (2005). [DOI] [PubMed] [Google Scholar]
- 8.O’Driscoll M. C., et al. , Recessive mutations in the gene encoding the tight junction protein occludin cause band-like calcification with simplified gyration and polymicrogyria. Am. J. Hum. Genet. 87, 354–364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saitou M., et al. , Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell 11, 4131–4142 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitajiri S., et al. , Deafness in occludin-deficient mice with dislocation of tricellulin and progressive apoptosis of the hair cells. Biol. Open 3, 759–766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo W. T., et al. , Inflammation-induced occludin downregulation limits epithelial apoptosis by suppressing caspase-3 expression. Gastroenterology 157, 1323–1337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchiando A. M., et al. , Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J. Cell Biol. 189, 111–126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S., Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141, 1539–1550 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreeva A. Y., Krause E., Müller E. C., Blasig I. E., Utepbergenov D. I., Protein kinase C regulates the phosphorylation and cellular localization of occludin. J. Biol. Chem. 276, 38480–38486 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Bolinger M. T., et al. , Occludin S471 phosphorylation contributes to epithelial monolayer maturation. Mol. Cell. Biol. 36, 2051–2066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dörfel M. J., et al. , CK2-dependent phosphorylation of occludin regulates the interaction with ZO-proteins and tight junction integrity. Cell Commun. Signal. 11, 40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elias B. C., et al. , Phosphorylation of Tyr-398 and Tyr-402 in occludin prevents its interaction with ZO-1 and destabilizes its assembly at the tight junctions. J. Biol. Chem. 284, 1559–1569 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., et al. , Occludin S490 phosphorylation regulates vascular endothelial growth factor-induced retinal neovascularization. Am. J. Pathol. 186, 2486–2499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raleigh D. R., et al. , Occludin S408 phosphorylation regulates tight junction protein interactions and barrier function. J. Cell Biol. 193, 565–582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakakibara A., Furuse M., Saitou M., Ando-Akatsuka Y., Tsukita S., Possible involvement of phosphorylation of occludin in tight junction formation. J. Cell Biol. 137, 1393–1401 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Fanning A. S., Anderson J. M., Lavie A., Structure of the conserved cytoplasmic C-terminal domain of occludin: Identification of the ZO-1 binding surface. J. Mol. Biol. 352, 151–164 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Nusrat A., et al. , The coiled-coil domain of occludin can act to organize structural and functional elements of the epithelial tight junction. J. Biol. Chem. 275, 29816–29822 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Tash B. R., et al. , The occludin and ZO-1 complex, defined by small angle X-ray scattering and NMR, has implications for modulating tight junction permeability. Proc. Natl. Acad. Sci. U.S.A. 109, 10855–10860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buschmann M. M., et al. , Occludin OCEL-domain interactions are required for maintenance and regulation of the tight junction barrier to macromolecular flux. Mol. Biol. Cell 24, 3056–3068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong V., Phosphorylation of occludin correlates with occludin localization and function at the tight junction. Am. J. Physiol. 273, C1859–C1867 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Cordenonsi M., et al. , Xenopus laevis occludin. Identification of in vitro phosphorylation sites by protein kinase CK2 and association with cingulin. Eur. J. Biochem. 264, 374–384 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Smales C., et al. , Occludin phosphorylation: Identification of an occludin kinase in brain and cell extracts as CK2. FEBS Lett. 545, 161–166 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Dörfel M. J., Westphal J. K., Huber O., Differential phosphorylation of occludin and tricellulin by CK2 and CK1. Ann. N. Y. Acad. Sci. 1165, 69–73 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T., et al. , PKC eta regulates occludin phosphorylation and epithelial tight junction integrity. Proc. Natl. Acad. Sci. U.S.A. 106, 61–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seth A., Sheth P., Elias B. C., Rao R., Protein phosphatases 2A and 1 interact with occludin and negatively regulate the assembly of tight junctions in the CACO-2 cell monolayer. J. Biol. Chem. 282, 11487–11498 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Siddiqui M. R., Mayanil C. S., Kim K. S., Tomita T., Angiopoietin-1 regulates brain endothelial permeability through PTPN-2 mediated tyrosine dephosphorylation of occludin. PLoS One 10, e0130857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raju P., et al. , Inactivation of paracellular cation-selective claudin-2 channels attenuates immune-mediated experimental colitis in mice. J. Clin. Invest. 130, 5197–5208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dörfel M. J., Huber O., A phosphorylation hotspot within the occludin C-terminal domain. Ann. N. Y. Acad. Sci. 1257, 38–44 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Cordenonsi M., et al. , Occludin dephosphorylation in early development of Xenopus laevis. J. Cell Sci. 110, 3131–3139 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Sundstrom J. M., et al. , Identification and analysis of occludin phosphosites: A combined mass spectrometry and bioinformatics approach. J. Proteome Res. 8, 808–817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter J. K., et al. , Redox-sensitivity of the dimerization of occludin. Cell. Mol. Life Sci. 66, 3655–3662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter J. K., et al. , The oligomerization of the coiled coil-domain of occludin is redox sensitive. Ann. N. Y. Acad. Sci. 1165, 19–27 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Mir H., et al. , Occludin deficiency promotes ethanol-induced disruption of colonic epithelial junctions, gut barrier dysfunction and liver damage in mice. Biochim. Biophys. Acta 1860, 765–774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edelblum K. L., et al. , Dynamic migration of γδ intraepithelial lymphocytes requires occludin. Proc. Natl. Acad. Sci. U.S.A. 109, 7097–7102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber D., Balda M. S., Matter K., Occludin modulates transepithelial migration of neutrophils. J. Biol. Chem. 275, 5773–5778 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Chen Y. H., Lu Q., Goodenough D. A., Jeansonne B., Nonreceptor tyrosine kinase c-Yes interacts with occludin during tight junction formation in canine kidney epithelial cells. Mol. Biol. Cell 13, 1227–1237 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt A., et al. , Occludin binds to the SH3-hinge-GuK unit of zonula occludens protein 1: Potential mechanism of tight junction regulation. Cell. Mol. Life Sci. 61, 1354–1365 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manda B., et al. , Phosphorylation hotspot in the C-terminal domain of occludin regulates the dynamics of epithelial junctional complexes. J. Cell Sci. 131, jcs206789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami T., Frey T., Lin C., Antonetti D. A., Protein kinase cβ phosphorylates occludin regulating tight junction trafficking in vascular endothelial growth factor-induced permeability in vivo. Diabetes 61, 1573–1583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S., Zhang Y., MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins 72, 547–556 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fawzi N. L., Ying J., Torchia D. A., Clore G. M., Probing exchange kinetics and atomic resolution dynamics in high-molecular-weight complexes using dark-state exchange saturation transfer NMR spectroscopy. Nat. Protoc. 7, 1523–1533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sjodt M., Clubb R. T., Nitroxide labeling of proteins and the determination of paramagnetic relaxation derived distance restraints for NMR Studies. Bio Protoc. 7, e2207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are included in the main text or SI Appendix. Requests for additional information or materials may be addressed to the corresponding author.