Significance
Glioblastoma multiforme (GBM) has been impervious to immune interventional therapies. Here, we analyzed the transcriptome of highly pure CD4+ and CD8+ T cells from the tumor bed, normal-appearing brain tissue, and peripheral blood of treatment-naive GBM patients. While the transcriptome of tumor-infiltrating CD8+ T cells was consistent with a potentially robust antitumor response, tumor-infiltrating CD4+ T cells showed a strong commitment to the TH17 lineage. Since intratumoral TH17 cells might exert a dominant-negative function as to a productive antitumor response, our data suggest that a site-directed anti-TH17 intervention may be a prerequisite for efficient antitumor immunity in GBM.
Keywords: glioblastoma multiforme, TH17 cells, tissue residency, RNA sequencing, gene set enrichment analysis
Abstract
Although glioblastoma multiforme (GBM) is not an invariably cold tumor, checkpoint inhibition has largely failed in GBM. In order to investigate T cell–intrinsic properties that contribute to the resistance of GBM to endogenous or therapeutically enhanced adaptive immune responses, we sorted CD4+ and CD8+ T cells from the peripheral blood, normal-appearing brain tissue, and tumor bed of nine treatment-naive patients with GBM. Bulk RNA sequencing of highly pure T cell populations from these different compartments was used to obtain deep transcriptomes of tumor-infiltrating T cells (TILs). While the transcriptome of CD8+ TILs suggested that they were partly locked in a dysfunctional state, CD4+ TILs showed a robust commitment to the type 17 T helper cell (TH17) lineage, which was corroborated by flow cytometry in four additional GBM cases. Therefore, our study illustrates that the brain tumor environment in GBM might instruct TH17 commitment of infiltrating T helper cells. Whether these properties of CD4+ TILs facilitate a tumor-promoting milieu and thus could be a target for adjuvant anti-TH17 cell interventions needs to be further investigated.
While immune therapies have been changing the field of oncology for many cancer entities, glioblastoma multiforme (GBM) has appeared impervious to many immune therapeutic approaches and the standard of care has not essentially changed for GBM in the last 15 y, with survival times in the range of 15 mo (1). However, GBM may not be an invariably “cold” tumor. In fact, current approaches to use the adaptive immune system to generate an efficient antitumor response, for example, with anti-PD1 checkpoint inhibitors, improved survival in a fraction of GBM patients (2). However, while extensive data are available on the phenotype and function of T cells infiltrating solid tumors elsewhere in the body, in particular in melanoma and lung cancer (3, 4), we are only beginning to investigate the biology of T cells infiltrating GBM tissue in the context of the specific circumstances of adaptive immune responses in the central nervous system (CNS).
In fact, in untreated GBM tissue, myeloid cells are the dominating immune cell type. Only recently, using single-cell RNA-sequencing (RNA-seq) approaches, tumor-associated macrophages (TAMs) have been characterized in terms of their heterogeneity in brain metastases (BrMs) and gliomas with mutations in IDH1 and IDH2 (IDHmut) as well as with wild-type IDH (IDHWT), namely GBM (5–7). TAMs comprise monocyte-derived macrophages (Mo-TAMs) as well as brain-resident microglial cells that are instructed by the tumor microenvironment (TME) to become tumor-associated microglia (MG-TAM). While IDHmut gliomas comprise higher fractions of MG-TAMs than Mo-TAMs, Mo-TAMs are the prevailing myeloid cell type in GBM and constitute one-third of the absolute tumor mass (8, 9). Lymphocytes, and in particular T cells, are present in gliomas but at lower fractions than in BrM (6). It has been speculated that the higher abundance of T cells in BrM might be responsible for the more favorable response of BrM (in particular those from melanoma and lung cancer) than GBM to anti-PD1 therapy (10). However, other reasons intrinsic to GBM, including the profound heterogeneity and plasticity of GBM and thus the resulting lack of “quality” neoantigens (11, 12) but also the exquisite capacity of GBM (and less so BrM) to shape the phenotype of infiltrating immune cells (7), might be relevant for the profound resistance of GBM to therapy.
Although rarer than in BrM, tumor-infiltrating T cells (TILs) in GBM still constitute a fraction of about 10 to 15% of all tumor-associated leukocytes, and it is possible that a dysfunctional T cell phenotype contributes to the failure of T cell–directed therapies in GBM (9). While an exhausted phenotype has been described in GBM TILs (often with simultaneous up-regulation of PD1, TIM3, and LAG3) (7, 13), less is known about aberrant T helper cell responses in GBM. For instance, in melanoma, a preexisting subset of type 1 T cell (TH1) TILs is a predictor of a favorable response to anti–CTLA-4 treatment (3). Conversely, TH17 TILs have been associated with dysfunctional adaptive antitumor responses due to the induction of tissue-destructive inflammation (14) and enhanced angiogenesis (15) that may support the tumor niche.
Here, we focused on the analysis of the T cell phenotype outside and within the tumor niche in the CNS of a highly selected cohort of treatment-naive GBM patients. By flow cytometry and bulk RNA-seq, we provide an in-depth characterization of the CD8+ and CD4+ TIL compartment. Our observation of a pronounced TH17 commitment of CD4+ TILs in human GBM may inform testable hypotheses for a novel set of immune interventional therapies in patients with GBM.
Results
CD4+ T Cells and CD8+ T Cells Are Enriched in the GBM Tumor Bed as Compared with Normal-Appearing Brain.
Adaptive immune responses against GBM are being harnessed in a variety of therapeutic vaccination approaches (16). Since a population of T cells has been found in the tumor bed of untreated GBM patients, stalled T cell responses in the TME have been hypothesized to be responsible for inefficient tumor control. Therefore, checkpoint inhibitors have been tried in GBM. However, checkpoint inhibition has largely failed, and a variety of reasons have been proposed (17).
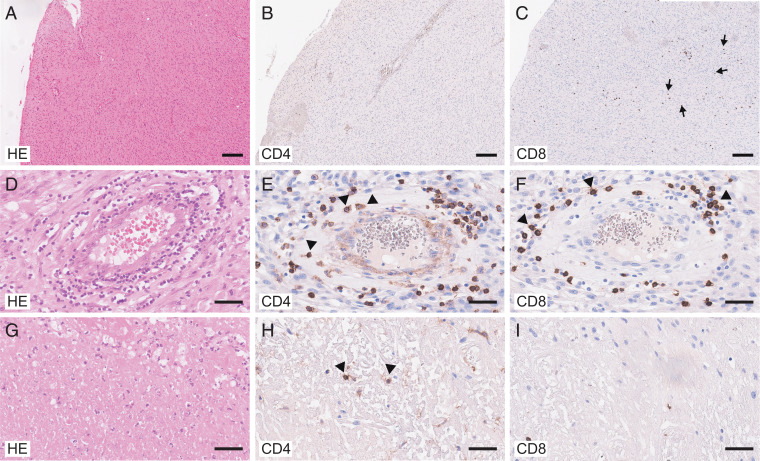
Here, we performed an in-depth immunologic analysis of the T cell compartment in the tumor bed in normal-appearing brain tissue and in the peripheral blood of untreated GBM patients. We identified nine patients who were referred to our center with suspected GBM (based on MRI; SI Appendix, Table S1). All patients were treatment-naive and underwent tumor resection. Tissue specimens from normal-appearing brain and the tumor bed were collected and peripheral blood was drawn. In all patients, the diagnosis of GBM was confirmed by histology and further molecular analysis (Fig. 1 and SI Appendix, Table S1). While CD4+ T cells were found in association with the wall of brain vessels and in necrotic areas (Fig. 1 A, B, D, E, G, and H), CD8+ T cells were largely located either perivascularly or scattered in the tumor bed (Fig. 1 A, C, D, F, G, and I).
Fig. 1.
Localization of CD4+ T cells and CD8+ T cells in the GBM site. Histologic analysis of GBM tissue (overview; A–C) comparing perivascular (D–F) and necrotic areas (G–I). Hematoxylin and eosin (HE) staining (A, D, and G) and immunohistologic staining for CD4 (B, E, and H) and CD8 (C, F, and I). (Scale bars, 200 μm [A–C] and 50 μm [D–I].) Note that CD4+ T cells were present in the vessel walls (arrowheads in E) and in necrotic tissue (arrowheads in H) while CD8+ T cells were largely localized in perivascular areas (arrowheads in F) or scattered in GBM tissue (arrows in C).
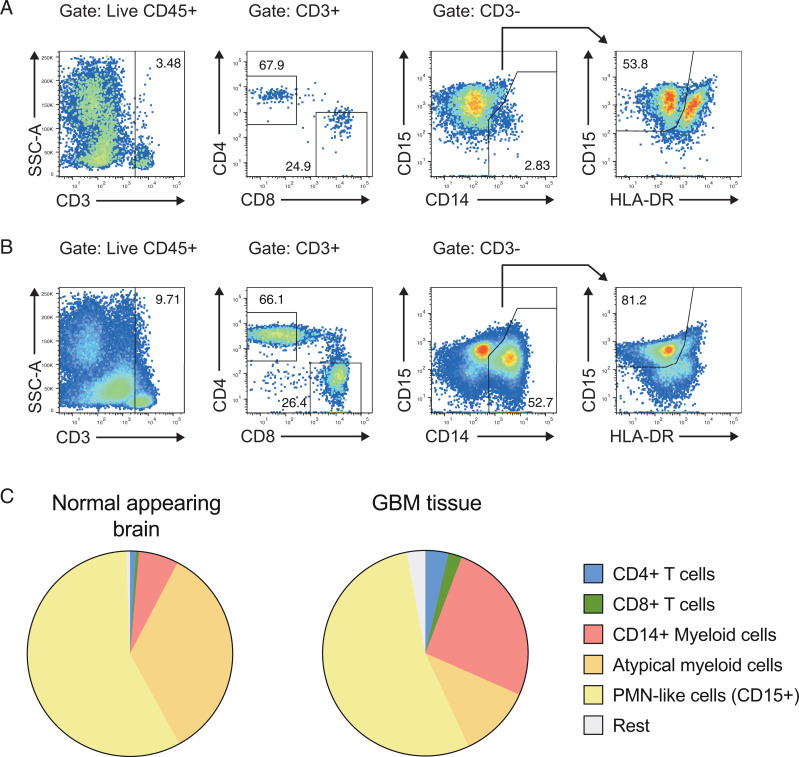
In flow cytometric analysis, the fractions of CD4+ and CD8+ T cells in the tumor bed were larger than in normal-appearing brain tissue (Fig. 2). The CD4+/CD8+ ratios in the tumor and in normal-appearing brain tissue were 1.3 as compared with 1.9 in the blood of untreated GBM patients. The fraction of typical monocytes (macrophages) expressing CD14 was increased in the tumor bed as compared with normal-appearing brain (Fig. 2) while equal fractions of CD15int myeloid cells were present in the GBM situs and normal-appearing brain tissue (Fig. 2). Overall, treatment-naive GBM sites showed intense infiltration with adaptive immune cells, in particular CD4+ and CD8+ TILs.
Fig. 2.
CD4+ T cells and conventional myeloid cells are enriched in the GBM bed as compared with normal-appearing brain. (A and B) Single-cell suspensions from normal-appearing brain tissue (A) and the GBM situs (B) were prepared, and the indicated immune cell populations were analyzed by flow cytometry. (C) The frequencies of CD4+ T cells, CD8+ T cells, CD14+ myeloid cells (CD45+CD3–CD14+), “atypical myeloid cells” (CD45+CD3–CD14–CD15–), and “PMN-like cells” (CD45+CD3–CD14–CD15+HLA-DR–) were analyzed by flow cytometry.
The Transcriptomes of CD4+ and CD8+ TILs and Lymphocytes Residing in Normal-Appearing CNS Are Overlapping.
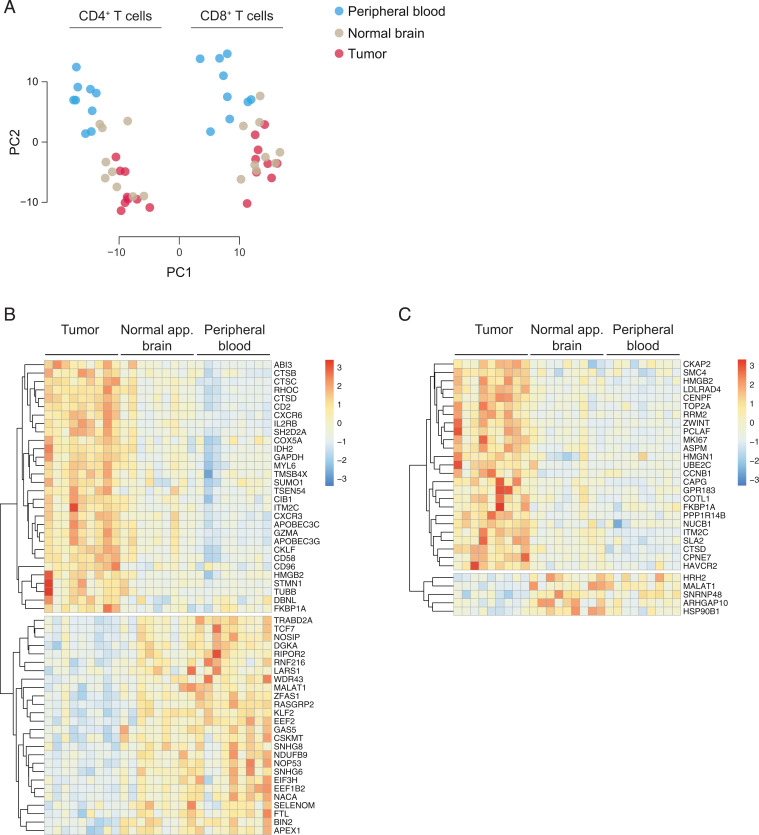
To assess the functional phenotypes of TILs at the GBM site on the population level, we sorted CD4+ T cells and CD8+ T cells by flow cytometry from different anatomical compartments, namely peripheral blood, normal-appearing CNS tissue, as well as GBM tissue, and performed RNA-seq. In the principal-component analysis (PCA), PC1 loading was determined by the cell type (CD4+ vs. CD8+ T cells), while the anatomical niche of tissue sampling was largely reflected in PC2. On the whole-transcriptome level, PC2 of peripheral blood T cells was distinct from CNS-derived T cells. However, within the CNS, TILs and T cells from normal-appearing brain tissue did not segregate from each other (Fig. 3A). These data suggested that the genome-wide transcriptome of T cells in the CNS was profoundly shaped by the specific milieu in the CNS rather than by their location in the tumor bed vs. normal brain tissue. Whether the phenotype of CNS T cells was built by differential recruitment or local imprinting in the CNS cannot be decided based on bulk RNA-seq data.
Fig. 3.
GBM TILs exhibit a specific signature that is independent of their localization in the brain. Bulk RNA-seq was performed on highly pure CD4+ and CD8+ T cells isolated from blood, normal-appearing brain, and GBM. (A) Principal-component analysis. (B and C) Heatmaps of DEGs in the CD4+ T cell compartment (B) and the CD8+ T cell compartment (C).
CD4+ TILs and CD8+ TILs Exhibit a Distinct Tumor Bed Signature.
Although the CNS milieu was the dominating determinant for the global transcriptome of CNS T cells irrespective of their localization in normal or GBM-infiltrated CNS tissue, we sought to identify a group of genes that were differentially expressed in TILs vs. T cells in normal-appearing brain tissue to define a GBM-specific T cell signature. Therefore, we compared the transcriptomes of TILs with the transcriptome of their counterparts derived from normal-appearing CNS tissue. Due to the clear segregation of CD4+ and CD8+ T cells in the PCA, the differentially expressed genes (DEGs) were analyzed separately in the CD4+ and CD8+ T cell compartments (Fig. 3 B and C). As compared with CD4+ T cells in normal-appearing brain, CD4+ TILs were characterized by up-regulated costimulatory or coinhibitory molecules (CD2, CD58, CD96), chemokine receptors (CXCR3, CXCR6), and enzymes of the intermediary metabolism (IDH2, GAPDH). A series of transcriptional regulators (including TCF7, KLF2) were down-regulated in CD4+ TILs as compared with CD4+ T cells sorted from normal-appearing brain tissue (Fig. 3B). Therefore, in addition to the CNS-specific imprinting of the transcriptome, the tumor bed was associated with the regulation of a distinct set of genes in CD4+ TILs. In CD8+ TILs, EBI2 (GPR183) and the coinhibitory molecule TIM3 (HAVCR2), but not LAG3 or PD1, were up-regulated as compared with CD8+ T cells isolated from normal-appearing brain tissue (Fig. 3C). Overall, the CD8+ TIL transcriptome was reminiscent of exhausted T cells (18).
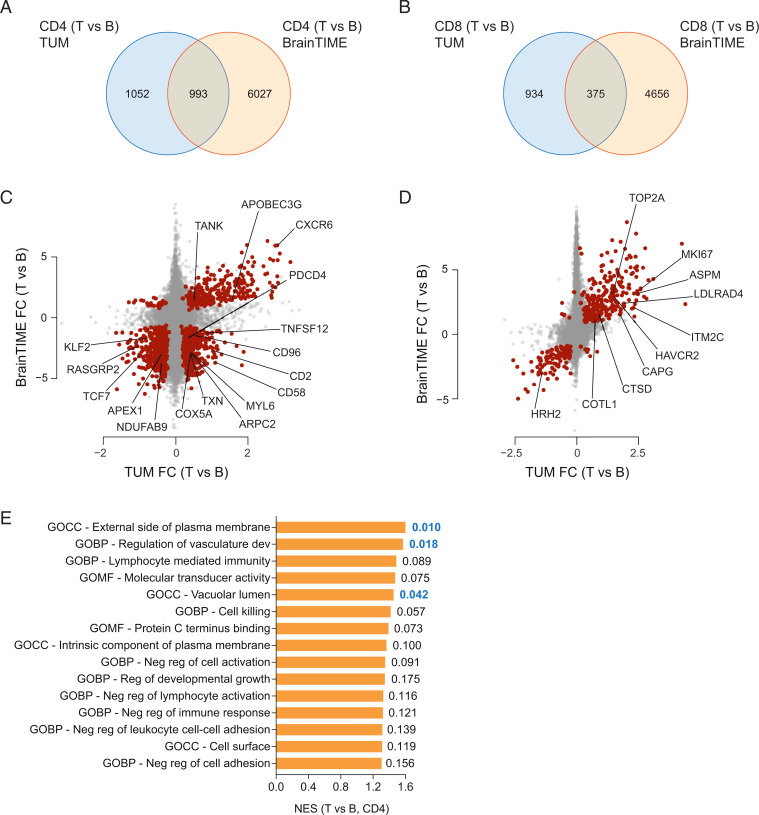
We compared the GBM TIL signature with published expression data that reported the DEGs of CD4+ or CD8+ T cells of GBM patients in brain vs. peripheral blood (6). Here, we found an overlap of 993 and 375 genes between our dataset and the BrainTIME dataset (6) for the CD4+ and CD8+ T cell compartments, respectively (Fig. 4 A and B and Dataset S1). The CD4 overlap contained most of the genes that were also differentially expressed in the CD4+ T cells isolated from the GBM bed as compared with normal-appearing brain tissue, including CD2, CD58, CD96, CXCR6, KLF2, PDCD4, RASGRP2, TCF7, and TWEAK (Fig. 4C and Dataset S2). The CD8 overlap was less instructive due to the limited number of genes differentially expressed in CD8+ GBM TILs and CD8+ T cells isolated from normal-appearing brain tissue (Fig. 4D and Dataset S2). To globally assess the biologic significance of the core signatures of CD4+ and CD8+ TILs extracted from our and the BrainTIME datasets, we performed gene set enrichment analyses (GSEAs) for the Gene Ontology (GO) term collections available at https://www.gsea-msigdb.org. The 15 most enriched GO terms and their normalized enrichment scores illustrated that CD4+ TILs might contribute to creating an environment that is favorable for GBM growth and dysfunctional for productive immune responses (Fig. 4E).
Fig. 4.
Comparison with an independent dataset reveals a core signature of CD4+ and CD8+ TILs. The CD4+ (A) and CD8+ (B) TIL signatures (as related to blood) were compared with publicly available TIL-specific transcriptomes (6). Overlay of our TIL transcriptomes with the DEGs (tumor [T] vs. blood [B]) in CD4+ and CD8+ T cells from the BrainTIME dataset (6). Fold change (FC) (tumor vs. blood) differential gene expression in our cohort vs. the BrainTIME cohort in the CD4+ compartment (C) and the CD8+ compartment (D). The overlapping DEGs (see also A and B) are depicted as red dots. Some relevant genes in the intersection are highlighted both for CD4+ TILs and CD8+ TILs. GO term analyses were performed on the core CD4+ signature (E) by running GSEAs using validated GO term gene sets (https://www.gsea-msigdb.org). Normalized enrichment scores (NESs) and corresponding nominal P values (Right; significant values are in bold) are depicted for the 15 most enriched GO term signatures in CD4+ TILs (E).
CD4+ TILs Develop into TH1 Cells but Also Comprise an Exaggerated TH17 Trajectory.
In order to assess the potential significance of major cell biologic pathways in CD4+ TILs, we performed GSEAs of known hallmark pathways in CD4+ T cells isolated from the tumor bed, normal-appearing brain, or blood (Fig. 5A). IL-2–STAT5 signaling but also other hallmark pathways including “apoptosis” were strongly overrepresented in the transcriptome of CD4+ TILs vs. blood-derived CD4+ T cells. When comparing CD4+ TILs with CD4+ T cells from neighboring normal-appearing brain tissue, a striking pathway that—besides IL-2–STAT5 and PI3K–AKT–mTOR signaling—remained enriched in CD4+ TILs was the IL-6–JAK–STAT3 signaling pathway (Fig. 5A), which is intricately associated with the development and maintenance of TH17 cells but not TH1 cells (19).
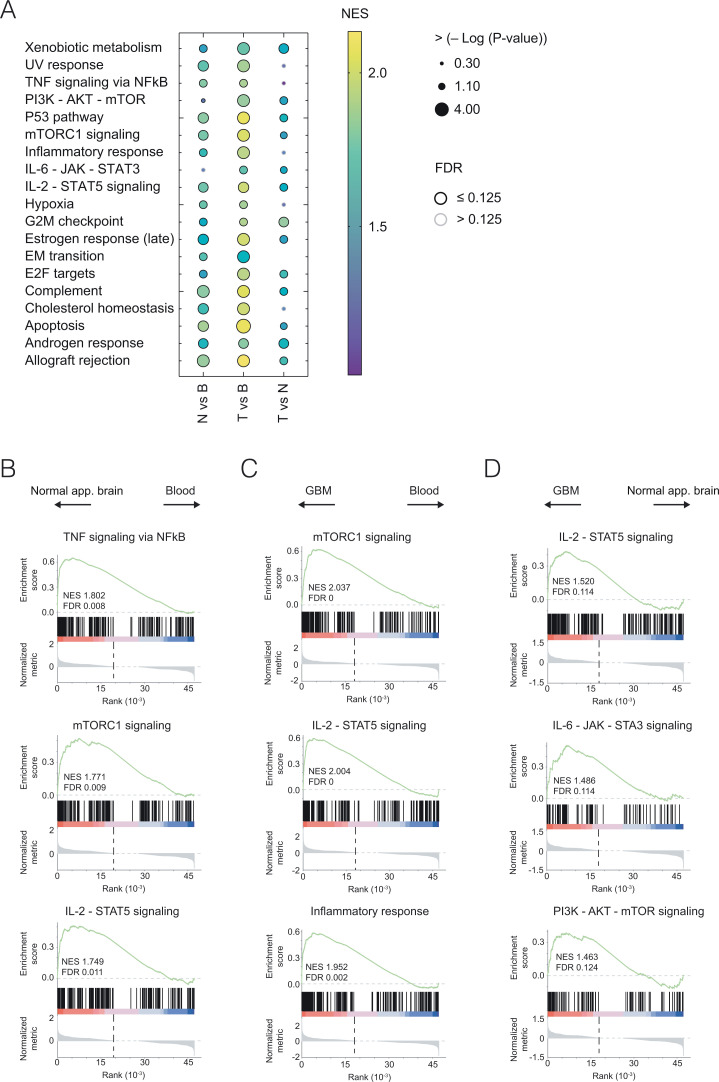
Fig. 5.
Transcriptome of CD4+ TILs is enriched for genes associated with TH17 hallmark molecular pathways. (A) Gene expression ratios of normal-appearing brain vs. blood CD4+ T cells (N vs. B), of GBM-residing CD4+ T cells vs. blood CD4+ T cells (T vs. B), and of GBM-residing CD4+ T cells vs. normal-appearing brain CD4+ T cells (T vs. N) were ranked and probed for the enrichment of known molecular hallmark pathway signatures (https://www.gsea-msigdb.org). NESs are color-coded. GSEAs with a false discovery rate (FDR) of ≤0.125 are indicated by black circles. Examples of GSEAs for the comparison of N vs. B (B), T vs. B (C), and T vs. N (D) are depicted. Note that several pathways, including but not limited to IL-6–JAK–STAT3 and PI3K–AKT–mTOR, were enriched in T vs. N, suggesting that the TME had an impact on the molecular imprinting of GBM-residing CD4+ T cells beyond the CNS-driven changes in the CD4+ T cell transcriptome.
Failed commitment of CD4+ T cells to the TH1 lineage or lack of cytotoxic T cells (CTLs) in the tumor bed or a fixed exhausted phenotype of CTLs (TIM3, PD1, LAG3 triple-positive) have been associated with an insufficient antitumor response (3, 18). In order to position the transcriptomes of CD4+ and CD8+ TILs of our GBM patients in the landscape of known T cell subsets, we mined multiple known gene sets of helper cell lineages including regulatory T (TREG) cells within the CD4+ T cell compartment and of effector T (TEFF) cells and various types of memory T cells within the CD8+ T cell compartment. We categorized these public domain gene sets into gene sets associated with TH1 cells, TH2 cells, TH17 cells, T follicular helper (TFH) cells, and TREG cells in the CD4+ T cell universe, and into gene sets associated with TEFF cells, central memory T (TCM) cells, effector memory T (TEM) cells, and tissue-resident memory T (TRM) cells in the CD8+ T cell universe. Then, we applied these curated gene sets to the ranked DEGs derived from pairwise comparisons (normal-appearing brain vs. peripheral blood, tumor bed vs. peripheral blood, and tumor bed vs. normal-appearing brain) of our CD4+ and CD8+ T cell transcriptomes. The normalized enrichment scores of these GSEAs were color-coded (SI Appendix, Figs. S1 and S2). In the CD4+ T cell compartment, TH1 signatures were enriched in TILs vs. normal-appearing brain T cells. Yet, strikingly and consistent with the enrichment of the IL-6–JAK–STAT3 pathway, TH17 signatures were also overrepresented in CD4+ TILs as compared with T cells from normal-appearing brain (Fig. 6A), indicating that an aberrant commitment of CD4+ T cells to the TH17 lineage in the tumor bed was a robust feature of GBM. In the CD8+ T cell compartment, CTL signatures (TEFF) and TEM signatures but also TCM signatures were overrepresented in TILs as compared with either blood or normal-appearing brain CD8+ T cells (Fig. 6B).
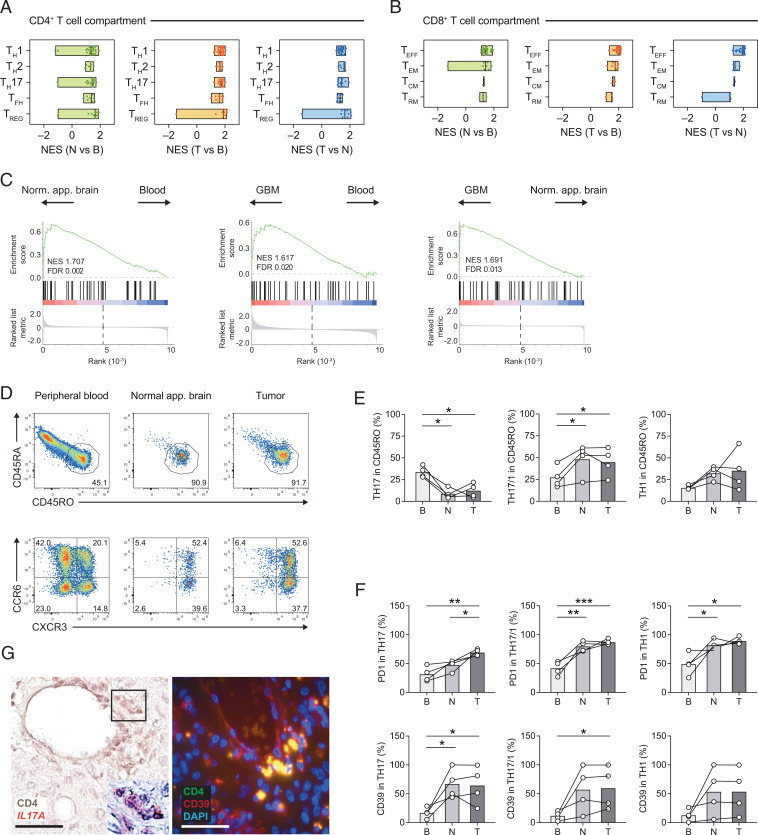
Fig. 6.
CD4+ TILs develop a robust TH17 cell signature. (A and B) Pairwise compared gene expression between two anatomical compartments was tested for enrichment of curated gene signatures for CD4+ TILs (TH1, TH2, TH17, TFH, and TREG cell signatures) (A) and CD8+ TILs (TEFF, TEM, TCM, and TRM signatures) (B); see Materials and Methods for details. The minimum and maximum NESs as well as the median for each canonical signature are depicted. (C) GSEA of a published AHR signature of human CD4+ T cells (23) based on the ranked fold changes for the indicated pairwise comparisons. NESs and FDRs are indicated in each enrichment plot. (D–F) Flow cytometric analysis of CD4+ T cells isolated from blood, normal-appearing brain, and GBM tissue. (D) Activated T cells were gated based on CD45RO expression and subclassified into T helper cell subsets according to the expression of distinct chemokine receptors: TH17 cells (CCR6+CXCR3–), TH17/1 cells (CCR6+CXCR3+), and TH1 cells (CCR6–CXCR3+). (E) Fraction of TH17, TH17/1, and TH1 cells among CD4+CD45RO+ activated/memory T cells in tissue compartments as indicated (B, blood; N, normal-appearing brain; T, GBM [tumor] site). (F) Flow cytometric assessment of PD1 and CD39 in TH17, TH17/1, and TH1 cells in the tissue compartments as indicated. One-way ANOVA (repeated-measures) with Tukey post test, *P < 0.05, **P < 0.003, ***P < 0.0003. (G) IL17A messenger RNA detection (by RNAscope) in CD4+ T cells (frame, magnified region with CD4+ T cells showing punctate IL17A signals; Left) and immunofluorescence staining for CD4 (green) and CD39 (red) (merge: yellow) at the GBM site (Right). (Scale bars, 50 μm.)
Signaling through the aryl hydrocarbon receptor (AHR) is a prominent feature in TH17 cells but not TH1 cells (20, 21). Moreover, the GBM site has been shown to be rich in kynurenine that acts as a natural ligand of AHR (22). Therefore, we tested our CD4+ T cell transcriptomes for the enrichment of an AHR signature (23). The AHR signature was significantly enriched in T cells isolated from the brain as compared with peripheral blood T cells (Fig. 6C). Importantly, the AHR signature was also enriched in GBM-derived vs. normal-appearing brain-derived CD4+ T cells (Fig. 6C), consistent with the overrepresentation of xenobiotic metabolism pathways in tumor-derived T cells (Fig. 5A) and supporting the idea that AHR ligands at the tumor site might be sensed by T helper cells in this micromilieu.
In order to validate the prevailing commitment of CD4+ T cells to the TH17 lineage at the GBM site, we performed flow cytometric analysis in four treatment-naive GBM patients additionally recruited to the study (SI Appendix, Table S2). We designed a flow panel to approximate the fractions of human TH17 cells, TH17/1 cells, and TH1 cells in CD45RO+ activated/memory cells based on the expression of the chemokine receptors CCR6 and CXCR3 (Fig. 6D), as has been reported and validated before for ex vivo isolated human T cell subsets (24). While the fraction of CCR6+ single-positive TH17 cells, which have been associated with decreased inflammatory potential as compared with CCR6+CXCR3+ double-positive TH17/1 cells (25–27), decreased in the brain vs. the peripheral blood, we found a significant increase in TH17/1 cells in the CNS (Fig. 6E). Importantly, the expression of PD1 was higher in TH17 lineage cells (TH17 and TH17/1) at the tumor site as compared with peripheral blood (Fig. 6F). CD39 is an ectonucleotidase expressed in TH17 cells at tumor sites (28) and, indeed, CD39 was significantly more highly expressed in TH17 cells and TH17/1 cells derived from the GBM site than in their peripheral blood counterparts (Fig. 6F). Interestingly, IL-17–producing CD4+ T cells and CD4+CD39+-coexpressing T cells were frequently detected in the proximity of vessels in GBM tissue (Fig. 6G).
In summary, T cell commitment to TH1 cells and CTLs with potentially efficient antitumor capacity did not appear to be impaired at the GBM site. However, at the same time, TH17 cell–associated T cell signatures were markedly enriched in GBM TILs, giving rise to the hypothesis that exaggerated TH17 responses in the GBM bed might block productive adaptive antitumor immunity and prevent the effect of checkpoint-inhibiting approaches.
TH17 Cell Commitment Is a Feature of a Major Subset of GBM Cases.
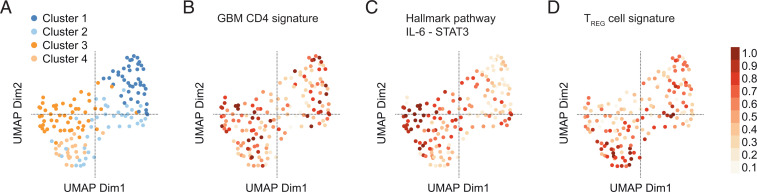
Finally, we sought to test our hypothesis on the public The Cancer Genome Atlas (TCGA) dataset of GBM samples. The available transcriptome data refer to the bulk transcriptome of all cells in the tumor site including CNS-intrinsic cells and hematopoietic cells. Clustering of the GBM transcriptomes yielded four clusters (Fig. 7A). Interestingly, our CD4+ GBM signature (as derived from the tumor vs. normal-appearing brain CD4+ T cell DEGs) essentially projected on clusters 1 and 3 of the TCGA samples when gene set variation analysis (GSVA) was performed (Fig. 7B). Notably, while the IL-6–STAT3 hallmark pathway was strongly associated with cluster 3 (Fig. 7C), cluster 1 partly coincided with a signature of resting TREG cells (Fig. 7D), suggesting that the expression profile of distinct GBM subsets might be determined by a TH17 cell or TREG cell response. Therefore, we wondered whether our CD4+ GBM signature that reflects important features of a TH17 cell response would coincide with particular cellular states in GBM that were recently described based on single-cell RNA-seq analyses (29). Yet, when assigning a score derived from our CD4+ GBM signature to each measured cell in the dataset and projecting these on the metamodule score matrix as suggested by ref. 29, no significant overrepresentation of our T cell signature in either oligodendrocytic precursor-like, neural progenitor cell-like, astrocyte-like, or mesenchymal-like GBM states was observed (SI Appendix, Fig. S3), indicating that the GBM-associated CD4+ T cell signature may not segregate with a distinct GBM cellular state but represent universal properties of CD4+ TILs in GBM tissue.
Fig. 7.
GBM transcriptomes of the TCGA dataset are clustered according to TH17 and TREG cell signatures. (A) UMAP representation of the transcriptomes of the TCGA GBM cohort (n = 154). (B–D) GSVA was run on the variance-stabilized expression data of the TCGA cohort for the indicated gene sets. Resulting GSVA scores were normalized to the interval 0 to 1 and projected onto the UMAP representation. (B) Projection of our GBM CD4+ T cell signature (i.e., DEGs in GBM vs. normal-appearing brain CD4+ T cells). (C) Projection of the IL-6–JAK–STAT3 hallmark pathway signature. (D) Projection of a published TREG cell signature (GSE15659). Note that the GBM CD4+ TIL signature coincides with clusters 1 and 3. The IL-6–JAK–STAT3 signature projected on cluster 3, and the TREG cell signature projected on cluster 1 (and 4).
Discussion
The immune cell infiltrate of GBM is dominated by myeloid cells. However, in order to induce robust antitumor immunity, the T cell compartment must be targeted. Therefore, we performed an in-depth analysis of the CD4+ and CD8+ T cell compartment in the GBM tumor bed and normal-appearing brain tissue and in the systemic immune compartment of treatment-naive patients with GBM. CD4+ T cells and CD8+ T cells comprised about 10 and 7% of the immune cell infiltrate in GBM, respectively. Comparing the T cell compartments in the GBM bed and the normal-appearing CNS tissue, we focused on GBM-instructed and not CNS-instructed properties of T cells. GBM-associated CD8+ T cells showed a partially exhausted phenotype with TIM3 (but not PD1) differentially expressed in GBM vs. normal-appearing brain tissue. Notably, the most striking feature of GBM-associated CD4+ T cells was a vigorous TH17 cell commitment, which is known to create a dominant-negative environment for productive TH1 and CTL responses.
Stable neoantigens as potential targets of T cell responses are not well-established in GBM; for example, the widely studied EGFR variant III is only expressed in about 10% of newly diagnosed GBM (30). In addition, the lymphatic drainage from the CNS parenchyma is still a matter of debate and may result in tolerogenic rather than immunogenic priming of T cells (31). Therefore, controlling the GBM-specific immunomodulatory TME has been envisioned as a potential strategy. Yet checkpoint inhibition only has a limited effect in GBM, suggesting that a better understanding of the “stalled” T cell response in GBM is required for more efficient T cell–directed immune interventions, in particular since it has been observed that a high fraction of CD4+ T cells in GBM is associated with a worse clinical outcome (32).
In our analysis of purely sorted CD4+ T cells, we observed that the GBM niche did not prevent the accumulation of T helper cells with type 1 features that are believed to be efficient responders to tumor antigens. The fraction of TH1 cells in the tumor bed is a predictor of a favorable response to anti–CTLA-4 treatment in melanoma (3). However, the CD4+ T cell compartment in GBM was also characterized by a substantial differentiation into TH17 cells—a phenomenon also observed in a series of solid cancers including hepatocellular carcinoma, renal cancer, and melanoma (33). In fact, we found a strong overrepresentation of the TH17-associated IL-6–STAT3 axis in intratumoral CD4+ T cells as compared with their counterparts in normal-appearing brain tissue. Since TH17 responses generate a highly inflammatory micromilieu, it is possible that it is not the lack of TH1 response capacity of GBM-residing CD4+ T cells in the first place but a hostile TH17 cell–instructed tumor micromilieu, which counterregulates efficient antitumor TH1 and CTL responses. Evidence from preclinical models, in which the elimination of IL-17 responses in addition to checkpoint blockade yielded superior antitumor responses as compared with checkpoint inhibition alone (34, 35), supports this concept. TH17 cell responses are diverse, and specific subsets of TH17 cells might be particularly impervious to generate a milieu that is permissive for antitumor TH1 and CTL responses. For instance, the expression of CD96 may be a marker for such a TH17 cell subset (36). Notably, CD96 is among the top up-regulated genes in GBM CD4+ T cells. CD96 is a type I transmembrane protein with an intracellular immunoreceptor tyrosine-based inhibitory motif that upon engagement of its ligand (CD155) might convey a negative signal into the cell. Interestingly, CD96 is also discussed as a novel target for checkpoint inhibition in CD4+ T cells (37, 38). In addition, CD39 expression at the GBM site has been associated with an unfavorable immunosuppressive milieu. While CD39 is expressed in myeloid cells recruited to the GBM site (39), our data suggest that also a substantial fraction of TH17 and TH17/1 cells express CD39 in the CNS and in particular at the GBM site. Whether CD39 is a direct target of AHR activation in GBM-residing TH17 cells—as has been described for macrophages (39)—remains to be determined. Notably, a substantial fraction of CD4+ T cells in the GBM appears to be located in proximity to vessel structures. Further studies need to investigate the concept of whether and how tumor-residing TH17 cells are involved in angiogenesis at the GBM site. Finally, it is currently an open question whether TH17 cells that are located in nonlymphoid tissues need to continuously sense IL-6 in order to maintain their functional phenotype (40). The identification of relevant sources of IL-6 at the GBM site might reveal important commonalities to solid tumors in other organs. Interestingly, within GBMs, endothelial cells have been reported to produce IL-6 (41) and might thus create a vascular niche that reinforces not only alternative macrophage polarization but also the maintenance of TH17 cells and TH17/1 cells.
Similar to the situation that efficient antitumor TH1 cells are, in principle, present at the GBM site, in the GBM-associated CD8+ T cell compartment, CTL development is not impaired per se and, more importantly, the exhaustion phenotype of GBM-residing CD8+ T cells would not predict an irreversibly paralyzed cytotoxic response. For instance, we did not observe a combined up-regulation of PD1, TIM3, and LAG3 in GBM CD8+ T cells. Also, bona fide TRM signatures that have, in principle, been associated with a favorable response to checkpoint inhibition in lung cancer (4) are found in GBM CD8+ T cells, again suggesting that immune therapeutic strategies in GBM may not fundamentally be bound to fail. Rather, a TH17 cell–guided microenvironment at the GBM site may set the scene for adapted immune therapeutic concepts in GBM.
In summary, our study established that the CD4+ T cell phenotype in GBM is unfavorably biased into the TH17 lineage, which might not only prevent a more efficient endogenous adaptive antitumor response but also render interventional strategies frustrating that target known checkpoints. Novel checkpoints (including CD96) might have to be explored in GBM and therapeutic interventions combined with anti-TH17 strategies. A key interpretation of our data would suggest that the presence of a robust TH17 response in the GBM situs is a dominant-negative determinant for a productive adaptive antitumor response, for example, by directly (e.g., via CD39-mediated adenosine production) or indirectly (through recruitment of myeloid cells) inducing a state of terminal exhaustion in CTLs (34, 35, 42). Here, our study might help to raise informed and testable hypotheses for improved immunotherapy of GBM.
Materials and Methods
Patients.
Patients with suspected GBM were recruited into the study in the Department of Neurosurgery at the Technical University of Munich School of Medicine between 2017 and 2022. Patients were included when they were over 18 y of age and had a histology-confirmed diagnosis of GBM. Exclusion criteria were the previous administration of any antitumor therapy including radiation therapy. During resection of the tumors, tumor tissue and tissue from normal-appearing brain within the operative channel were collected. Blood was drawn during the surgical procedure. All patients gave written informed consent. Only patients with a complete set of specimens (CD4+ TILs, CD8+ TILs, CD4+ T cells from normal-appearing brain, CD8+ T cells from normal-appearing brain, blood-derived CD4+ and CD8+ T cells) containing a minimum of 1,000 cells in each sorted sample were further analyzed. The study was approved by the local ethics committee (Technical University of Munich [TUM] School of Medicine) and conducted following the Declaration of Helsinki.
Preparation of Blood, Tumor Tissue, and Normal-Appearing Brain.
All samples were immediately processed. Briefly, tissue samples were minced with a scalpel and samples were then digested for 30 min with collagenase D and DNase I (Sigma). After preparation of single-cell suspensions using a 70-μm cell strainer, cells were isolated using a 37 or 37/70% Percoll gradient centrifugation. If needed, erythrocytes were lysed.
Antibodies and Sorting.
Staining was performed with human FC block, LIVE/DEAD Fixable Near-IR Stain Kit (Invitrogen), and fluorochrome-conjugated anti-human surface markers (CD3, CD4, CD8, CD14, CD15, CD19, CD33, CD39, CD45, CD45RA, CD45RO, CD96, CD161/KLRB1, CD183/CXCR3, CD185/CXCR5, CD194/CCR4, CD196/CCR6, CD197/CCR7, CD279/PD1, HLA-DR, Lox-1, and TCR γδ), all from BioLegend except for CD4, CD161 (BD Biosciences), and CD45RO (Beckman Coulter). Fluorescence minus one (FMO) staining was performed with all blood samples. Advanced cell-surface characterization of T cell subsets was performed on an Aurora spectral analyzer (Cytek). For RNA analysis, cells were sorted into TCL buffer (Qiagen) with a FACSAria III machine (BD Biosciences) and stored at −80 °C until further use.
Immunohistochemistry and RNAscope.
Immunohistochemistry and immunofluorescence were performed using ethylenediaminetetraacetate buffer and steam cooking for antigen retrieval followed by standard protocols. The antibodies used were anti-CD39 (EPR26473-58) (rabbit monoclonal, Abcam, 1:1,000), anti-CD4 (4B12) (mouse, monoclonal, Invitrogen, 1:10), and anti-CD4 (SP35) (rabbit, monoclonal, Invitrogen, 1:50).
For RNAscope with an IL-17A probe (ACDBio) and CD4 double staining, 4-μm serial sections were cut from formaldehyde-fixed paraffin-embedded (FFPE) tumor tissues. Subsequently, deparaffinization, pretreatment, and hybridization were performed according to the manufacturer’s protocol using the provided pretreatment solutions and wash buffer. Incubation steps were performed in a humidity control tray and a HybEZ oven (Advanced Cell Diagnostics). After hybridization, the signal was detected using Fast Red as chromogen provided by the manufacturer (RedB:RedA, 1:60). Immunohistochemistry was then performed according to standard protocols. Counterstaining was with 50% Gill’s hematoxylin 1 (American MasterTech), bluing with tap water, and 0.02% ammonium hydroxide in water. Slides were mounted with xylene and EcoMount (Biocare Medical).
Flow Cytometric Analysis.
Flow cytometry data were plotted with FlowJo (BD Biosciences). Statistical analysis of surface marker expression was performed by Prism v9.2.0 (GraphPad Software). Unless otherwise indicated, an unpaired t test was used if values of two groups were normally distributed and a nonparametric Mann–Whitney U test was applied otherwise. One-way ANOVA with correction for multiple comparisons was applied for the analysis of more than two groups. Significance was established at P < 0.05.
RNA-Seq.
Total RNA was isolated from sorted cell populations using the RNAeasy Plus Micro Kit (Qiagen, 74034). Quality and integrity of total RNA were controlled on a Bioanalyzer 2100 (Agilent Technologies). Library preparation for bulk sequencing of poly(A)-RNA was done as described previously (43). Briefly, barcoded complementary DNA (cDNA) of each sample was generated with a Maxima RT polymerase (Thermo Fisher Scientific, EP0742) using oligo-dT primer–containing barcodes, unique molecular identifiers (UMIs), and an adaptor. Ends of the cDNAs were extended by a template switch oligo (TSO) and full-length cDNA was amplified with primers binding to the TSO site and the adaptor. The NEB Ultra II FS Kit was used to fragment cDNA. After end repair and A tailing, a TruSeq adaptor was ligated and 3′-end fragments were finally amplified using primers with Illumina P5 and P7 overhangs. In comparison with previous descriptions (43), the P5 and P7 sites were exchanged to allow sequencing of the cDNA in read 1 and barcodes and UMIs in read 2 to achieve a better cluster recognition. The library was sequenced on a NextSeq 500 (Illumina) with 59 cycles for the cDNA in read 1 and 16 cycles for the barcodes and UMIs in read 2. Data were processed using the published Drop-seq pipeline (v1.0) to generate sample- and gene-wise UMI tables (44). A reference genome (GRCh38) was used for alignment. Transcript and gene definitions were used according to Gencode annotation version 35.
Data from RNA-seq were processed in R version 4.1.0 (2021-05-18) using DESeq2 (v1.32.0), openxlsx (v4.2.4), ggplot2 (v3.3.5), and dplyr (v1.0.7). Required packages were BiocParallel (v1.26.0), SummarizedExperiment (v1.22.0), Biobase (v2.52.0), MatrixGenerics (v1.4.0), matrixStats (v0.59.0), GenomicRanges (v1.44.0), GenomeInfoDb (v1.28.0), IRanges (v2.26.0), S4Vectors (v0.30.0), and BiocGenerics (v0.38.0). Sequencing data were processed using the DESeq2 default pipeline: estimation of size factors, estimation of dispersion, negative binomial generalized linear model fitting, and Wald statistics. lfcShrink with apeglm (45) was used to obtain the shrunken log fold change for each of the comparisons. PCAs were created using the variance-stabilizing transformation (function DESeq2::vst) with the option blind = TRUE. The 500 most variable genes after VST were used to calculate the PCA in BiocGenerics::plotPCA on the vst data. We also used stats::prcomp as a comparison to explore PC3.
GSEA was performed with ranked ratios of gene expression in CD4+ T cells or CD8+ T cells from normal-appearing brain vs. blood, GBM vs. blood, and GBM vs. normal-appearing brain. The gene sets were selected from Molecular Signatures Database version 7.5.1 (https://www.gsea-msigdb.org/gsea/msigdb/) according to the following criteria: All available gene sets were searched for “TH1” or “TH2” or “TH17” or “TFH” or “TREG” in order to analyze the RNA-seq data of CD4+ T cells and for “effector” or “memory” in order to analyze the RNA-seq data of CD8+ T cells. The gene sets were manually curated to fit gene signatures associated with TH1, TH2, TH17, TFH, or TREG cells in the CD4+ T cell compartment or TEFF cells, TEM cells, TCM cells, or TRM cells in the CD8+ T cell compartment. All selected gene sets are listed in SI Appendix, Figs. S1 and S2. GSEA was performed using GSEA version 4.2.2 (46, 47).
RNA data from the TCGA glioblastoma dataset were downloaded from the Genomic Data Commons data portal (https://portal.gdc.cancer.gov/). Primary tumor samples were selected for analysis and lowly expressed genes were removed from the dataset. Data were variance-stabilized without prior information about experimental groups. PCA was conducted on the mean-centered variance-stabilized data after selecting the 10% most variable genes based on SD. The function RunUMAP from the Seurat package (https://doi.org/10.1038/nbt.3192) was used to calculate the uniform manifold approximation and projection (UMAP) representation of the data based on the first 20 PCs. To identify subclusters, the 20 PCs were hierarchically clustered and the computed dendrogram was partitioned into four subtrees. GSVA (https://doi.org/10.1186/1471-2105-14-7) was run on the variance-stabilized expression data for the gene sets “HALLMARK_IL6_JAK_STAT3_SIGNALING” and “GSE15659_CD45RA_NEG_CD4_TCELL_VS_RESTING_TREG_DN” from the MSig database collection as well as with the marker genes determined by the pairwise comparisons of CD4+ T cells. Resulting GSVA scores were normalized to the interval 0 to 1 and projected onto the UMAP representation.
Analysis of metamodule score GBM data (29) was as follows. Processed data from Neftel et al. (29) were derived from the Broad Institute Single-Cell Portal. Normalized expression data were used to calculate cell-specific scores for the CD4+ GBM signature with UCell (48). Cells in the precomputed t-SNE representation of the data and the cell state hierarchy plot were then colored according to the score.
Supplementary Material
Acknowledgments
We thank Sandra Baur for excellent technical assistance. This study was supported by the Deutsche Forschungsgemeinschaft (SFB1054-B06 [ID 210592381], TRR128-A07 [ID 213904703], TRR128-A12 [ID 213904703], TRR128-Z02 [ID 213904703], TRR274-A01 [ID 408885537], and EXC 2145 [SyNergy, ID 390857198]), European Research Council (CoG 647215), and Hertie Network of Clinical Neuroscience.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206208119/-/DCSupplemental.
Data, Materials, and Software Availability
The RNA-seq data reported in this article have been deposited in the European Nucleotide Archive (accession no. PRJEB51618) (49).
All study data are included in the article and/or supporting information.
References
- 1.Stupp R., et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group, Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Zhao J., et al. , Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 25, 462–469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei S. C., et al. , Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170, 1120–1133.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganesan A. P., et al. , Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat. Immunol. 18, 940–950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon C. C., et al. , Differential microglia and macrophage profiles in human IDH-mutant and -wild type glioblastoma. Oncotarget 10, 3129–3143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klemm F., et al. , Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643–1660.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebel E., et al. , Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 181, 1626–1642.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Sørensen M. D., Dahlrot R. H., Boldt H. B., Hansen S., Kristensen B. W., Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol. Appl. Neurobiol. 44, 185–206 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Andersen B. M., et al. , Glial and myeloid heterogeneity in the brain tumour microenvironment. Nat. Rev. Cancer 21, 786–802 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg S. B., et al. , Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: Early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol. 17, 976–983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng L., et al. , Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell 153, 139–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schäfer N., et al. , Longitudinal heterogeneity in glioblastoma: Moving targets in recurrent versus primary tumors. J. Transl. Med. 17, 96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woroniecka K., et al. , T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin. Cancer Res. 24, 4175–4186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., et al. , IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J. Exp. Med. 206, 1457–1464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Numasaki M., et al. , Interleukin-17 promotes angiogenesis and tumor growth. Blood 101, 2620–2627 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Platten M., et al. , A vaccine targeting mutant IDH1 in newly diagnosed glioma. Nature 592, 463–468 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson C. M., Choi J., Lim M., Mechanisms of immunotherapy resistance: Lessons from glioblastoma. Nat. Immunol. 20, 1100–1109 (2019). [DOI] [PubMed] [Google Scholar]
- 18.McLane L. M., Abdel-Hakeem M. S., Wherry E. J., CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Korn T., Bettelli E., Oukka M., Kuchroo V. K., IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Veldhoen M., et al. , The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Quintana F. J., et al. , Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Opitz C. A., et al. , An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011). [DOI] [PubMed] [Google Scholar]
- 23.McAleer J. P., Fan J., Roar B., Primerano D. A., Denvir J., Cytokine regulation in human CD4 T cells by the aryl hydrocarbon receptor and Gq-coupled receptors. Sci. Rep. 8, 10954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acosta-Rodriguez E. V., et al. , Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8, 639–646 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Becattini S., et al. , T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science 347, 400–406 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Aschenbrenner D., et al. , An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells. Nat. Immunol. 19, 1126–1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghoreschi K., et al. , Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thibaudin M., et al. , Human ectonucleotidase-expressing CD25high Th17 cells accumulate in breast cancer tumors and exert immunosuppressive functions. OncoImmunology 5, e1055444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neftel C., et al. , An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan C. W., et al. ; TCGA Research Network, The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Na S. Y., et al. , Oligodendrocytes enforce immune tolerance of the uninfected brain by purging the peripheral repertoire of autoreactive CD8+ T cells. Immunity 37, 134–146 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Han S., et al. , Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br. J. Cancer 110, 2560–2568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou W., Restifo N. P., T(H)17 cells in tumour immunity and immunotherapy. Nat. Rev. Immunol. 10, 248–256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim B. S., et al. , Type 17 immunity promotes the exhaustion of CD8+ T cells in cancer. J. Immunother. Cancer 9, e002603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagaoka K., et al. , Deep immunophenotyping at the single-cell level identifies a combination of anti-IL-17 and checkpoint blockade as an effective treatment in a preclinical model of data-guided personalized immunotherapy. J. Immunother. Cancer 8, e001358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heink S., et al. , Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells. Nat. Immunol. 18, 74–85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgiev H., Ravens I., Papadogianni G., Bernhardt G., Coming of age: CD96 emerges as modulator of immune responses. Front. Immunol. 9, 1072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q., et al. , Immune and clinical features of CD96 expression in glioma by in silico analysis. Front. Bioeng. Biotechnol. 8, 592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takenaka M. C., et al. , Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci. 22, 729–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korn T., Hiltensperger M., Role of IL-6 in the commitment of T cell subsets. Cytokine 146, 155654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q., et al. , Vascular niche IL-6 induces alternative macrophage activation in glioblastoma through HIF-2α. Nat. Commun. 9, 559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C., et al. , Blocking IL-17A enhances tumor response to anti-PD-1 immunotherapy in microsatellite stable colorectal cancer. J. Immunother. Cancer 9, e001895 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parekh S., Ziegenhain C., Vieth B., Enard W., Hellmann I., The impact of amplification on differential expression analyses by RNA-seq. Sci. Rep. 6, 25533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macosko E. Z., et al. , Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu A., Ibrahim J. G., Love M. I., Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreatta M., Carmona S. J., UCell: Robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Mitsdoerffer et al., The glioblastoma multiforme tumor site promotes the commitment of tumor-infiltrating lymphocytes to the TH17 lineage in humans. European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB51618. Accessed 25 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this article have been deposited in the European Nucleotide Archive (accession no. PRJEB51618) (49).
All study data are included in the article and/or supporting information.