Abstract
Carbon monoxide dehydrogenases (CO-DH) are the enzymes responsible for the oxidation of CO to carbon dioxide in carboxydobacteria and consist of three nonidentical subunits containing molybdopterin flavin adenine dinucleotide (FAD), and two different iron-sulfur clusters (O. Meyer, K. Frunzke, D. Gadkari, S. Jacobitz, I. Hugendieck, and M. Kraut, FEMS Microbiol. Rev. 87:253–260, 1990). The three structural genes of CO-DH in Hydrogenophaga pseudoflava were cloned and characterized. The genes were clustered on the chromosome in the transcriptional order cutM-cutS-cutL. The cloned cutM, cutS, and cutL genes had open reading frames of 864, 492, and 2,412 nucleotides, coding for proteins with calculated molecular weights of 30,694, 17,752, and 87,224, respectively. The overall identities in the nucleotide sequence of the genes and the amino acid sequence of the subunits with those of other carboxydobacteria were 64.5 to 74.3% and 62.8 to 72.3%, respectively. Primer extension analysis revealed that the transcriptional start site of the genes was the nucleotide G located 47 bp upstream of the cutM start codon. The deduced amino acid sequences of the three subunits of CO-DH implied the presence of molybdenum cofactor, FAD, and iron-sulfur centers in CutL, CutM, and CutS, respectively. Fluorometric analysis coupled with denaturing polyacrylamide gel electrophoresis of fractions from hydroxyapatite column chromatography in the presence of 8 M urea of active CO-DH and from gel filtration of spontaneously inactivated enzyme revealed that the large and medium subunits of CO-DH in H. pseudoflava bind molybdopterin and FAD cofactors, respectively. Iron-sulfur centers of the enzyme were identified to be present in the small subunit on the basis of the iron content in each subunit eluted from the denaturing polyacrylamide gels.
Carbon monoxide dehydrogenase (CO-DH) is the key enzyme for CO oxidation in carboxydobacteria, which grow on CO as the sole source of carbon and energy (21, 29). The enzymes purified from several bacteria consist of three nonidentical subunits (L, M, and S) and contain molybdopterin cytosine dinucleotide, flavin adenine dinucleotide (FAD), and two different [2Fe-2S] clusters as cofactors (21, 29). The enzyme is induced by CO and repressed by organic compounds, except for those of Hydrogenophaga pseudoflava (formerly Pseudomonas carboxydoflava [43]) (18) and Acinetobacter sp. strain JC1 (36), which are expressed constitutively also under heterotrophic growth conditions.
The structural genes for CO-inducible CO-DHs have been cloned from Pseudomonas thermocarboxydovorans (named cutA, cutB, and cutC for large, medium, and small subunits, respectively) (34) and Oligotropha carboxidovorans (named coxL, coxM, and coxS for large, medium, and small subunits, respectively) (29, 40). The three genes were clustered in the transcriptional order cox(cut)BCA(MSL). The two reports, however, did not describe the transcription of CO-DH genes, except that a sequence identical to a part of the consensus sequence for the −10 region of ς54-type promoters is present in the upstream region of the cutB gene in P. thermocarboxydovorans (34).
The deduced amino acid sequences of CO-DH subunits revealed significant homologies with the three corresponding domains of xanthine dehydrogenases from several eukaryotic organisms (12, 17, 25, 35). The sequence data proposed that iron-sulfur centers of the two CO-DHs are within the small subunits. The molybdopterin and FAD were assumed to be present in the large and medium subunits, respectively, in P. thermocarboxydovorans (34) like other three-subunit or three-domain molybdenum-containing hydroxylases (MCHs) (3, 8, 9, 17). The two cofactors, however, were proposed to bind to the large subunit in O. carboxidovorans (40), implying that the location of FAD in CO-DHs, possibly in all MCHs, is controversial. X-ray crystallography analysis of the three-dimensional structure of aldehyde oxidoreductase of Desulfovibrio gigas, a two-domain MCH, clearly identified the large and small domains as molybdopterin- and iron-sulfur center-binding sites, respectively (37), which supports through analogy the current assumptions on the corresponding CO-DH cofactor-binding sites. The structure, however, has suggested no solutions on the FAD-binding site, since the enzyme contains neither FAD nor a domain corresponding to the medium subunit or domain in other MCHs. In xanthine dehydrogenase, the intermediate domain has generally been accepted to bind FAD, but direct evidence is lacking (1, 5, 11, 17, 33).
In this study, we cloned and characterized the genes for the constitutive CO-DH of H. pseudoflava to explain the diversity and basic transcriptional characteristics of CO-DH genes. We also tried to identify the location of the three CO-DH cofactors and found direct evidence indicating that the molybdopterin, FAD, and iron-sulfur centers are present in the large, medium, and small subunits of CO-DH in H. pseudoflava, respectively.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and cultivation.
The bacterial strains, phages, and plasmids used in this work are described in Table 1. H. pseudoflava DSM 1084 was grown at 30°C in Luria-Bertani medium (LB) or in standard mineral base medium with a gas mixture of 30% CO–70% air as described previously (20). Growth was measured by turbidity determined at 650 nm with a Hitachi U-2000 spectrophotometer. Escherichia coli strains were cultivated at 37°C in LB. For phage infection, LB containing 0.2% (wt/vol) maltose and 10 mM MgSO4 was used.
TABLE 1.
Bacterial strains, phages, and plasmids used in this study
| Bacterial strain, phage, or plasmid | Description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| H. pseudoflava | Wild type (DSM 1084) | 43 |
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene |
| XL1-Blue MRA (P2) | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 gyrA96 relA1 lac (P2 lysogen) | Stratagene |
| Phages | ||
| Lambda EMBL3 | BamHI-digested arms | Stratagene |
| Lambda clone 311 | Lambda EMBL3 clone containing cut genes of H. pseudoflava | This study |
| Plasmids | ||
| pUC18 | 2.69-kb cloning or sequencing vector; Ampr | 44 |
| pBKS2 | pUC18 containing 1.5-kb SalI fragment from clone 311 | This study |
| pBKX1 | pUC18 containing 3.5-kb SmaI fragment from clone 311 | This study |
DNA manipulations.
Plasmid DNAs were isolated from E. coli by the alkaline lysis method described by Sambrook et al. (38) and from H. pseudoflava by the method of Kado and Liu (16). Chromosomal DNA from H. pseudoflava was isolated from cells harvested at early stationary growth phase following the method of Marmur (26). Electrotransformation of E. coli was carried out with a Gene Pulser apparatus (Bio-Rad). Competent cells were prepared as described by Dower et al. (6). Phage lysate preparation and phage DNA isolation were performed as described elsewhere (34a).
PCR.
Two synthetic primers, SL and LS, were used to amplify the region covering the CO-DH small-subunit gene under the assumption that the transcriptional order of H. pseudoflava CO-DH (HpsCut) genes is the same as those of O. carboxidovorans (40) and P. thermocarboxydovorans (34) enzymes. The primer SL was a 1,024-fold degenerate 23-mer (5′-GCNCARGARAARGCNGTNGARCC-3′) derived from the N-terminal protein sequence AQEKAVEP of the HpsCut small subunit (29), and the primer LS was a 128-fold degenerate 17-mer (5′-TGNACNGGNGCRTTCAT-3′) based on the N-terminal amino acid sequence MNAPVQ of the HpsCut large subunit (29). The PCR mixture contained a 200 μM concentration of each deoxynucleoside triphosphate, 50 pmol of each primer, template DNA, and 0.4 U of Taq polymerase in 10 μl of reaction buffer (50 mM Tris-HCl [pH 8.3], bovine serum albumin [250 μg/ml], 1% Ficoll, 2.5 mM MgCl2). Amplification was carried out in a Fast Air Thermal Capillary Cycler (Daehan Medical System Co., Seoul, Korea) as follows: after primary denaturation for 10 s at 95°C, 45 cycles of annealing for 1 s at 62°C, elongation for 15 s at 72°C, and denaturation for 1 s at 95°C were performed, and then postelongation was performed for 30 s at 72°C. The PCR products were eluted from agarose gel after electrophoresis and used as templates to synthesize the random primed probes labeled with digoxigenin-11-dUTP for Southern blot and plaque hybridizations.
Southern hybridization.
DNAs digested with restriction enzymes were subjected to electrophoresis and then transferred to Hybond-N+ membranes (Amersham) by capillary blotting. Hybridization and washing were carried out at 68°C following the instructions supplied by the manufacturer. Colorimetric detection of positive bands was performed according to reference 3a.
Cloning and DNA sequencing.
The H. pseudoflava DNA partially digested with Sau3AI was ligated to lambda EMBL3 DNA, packaged by using Gigapack III gold packaging extracts (Stratagene), and transduced to E. coli XL1-Blue MRA (P2) cells following the manufacturer’s instruction. Plaque replicas were screened with the labeled random primed probes.
DNA fragments in subclones and nested deletion clones were sequenced by the dideoxy chain termination method (39) using a Sequenase DNA sequencing kit (version 2.0; United States Biochemical Corp.). DNA and deduced amino acid sequences were analyzed by using DNASIS and PROSIS programs (version 7.00; Hitachi Software Engineering Co., Yokohama, Japan). The BLAST program was used to search the protein sequence database at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, Md.).
RNA isolation and primer extension.
Total RNA was isolated from cells growing at the early exponential growth phase on CO with the Ultraspec RNA isolation system (Biotech Laboratories Inc., Houston, Tex.) according to the manufacturer’s instruction (1a). A primer extension experiment was performed with the avian myoblastosis virus reverse transcriptase primer extension system (Promega) and a 30-mer oligonucleotide primer, 5′-TCGCCGACCGACTTGGGCGCGTGGTACTCG-3′, which is complementary to nucleotide positions 15 to 44 bp downstream of the cutM start codon, according to the method of Sambrook et al. (38).
Protein determination.
Protein was determined by the method described by Bradford (4). Amounts of proteins in crude cell extracts were estimated by the same method after the extracts were boiled in 20% NaOH for 10 min (31).
Enzyme assay.
CO-DH activity was assayed photometrically at 30°C by measuring CO-dependent reduction of 2-(4-indophenyl)-3-(4-nitrophenyl)-2H-tetrazolium chloride (ɛ496 = 17.981 mM−1 cm−1) in 50 mM potassium phosphate buffer (pH 7.2) at 496 nm by the method of Kraut et al. (22).
Electrophoresis.
Nondenaturing polyacrylamide gel electrophoresis (PAGE) of the native enzyme was performed by the method of Laemmli (24) but without sodium dodecyl sulfate (SDS), as described previously (20). SDS-PAGE was carried out following the method of Laemmli (24) with several modifications as described by Kim et al. (19). Proteins were stained with Coomassie brilliant blue R-250 by the method (20) modified from that of Weber and Osborn (42).
Enzyme purification.
Cells of H. pseudoflava grown on CO were harvested in the midexponential growth phase and washed once in 50 mM Tris hydrochloride buffer (pH 7.5; standard buffer). All purification steps were carried out at 4°C except when noted. Samples were concentrated, if needed, with an ultrafiltration membrane (Amicon YM 10) under an atmosphere of nitrogen gas.
A 30-g portion of the washed cells was suspended in 116 ml of standard buffer and disrupted by sonic treatment (20 s/ml) in portions of 20 ml. The suspension was centrifuged at 15,000 × g for 30 min. The resulting supernatant was then treated with protamine sulfate to a final concentration of 0.054%, left in ice for 1 h, and then centrifuged at 112,000 × g for 90 min. The supernatant fluid was next made 30% saturated with respect to ammonium sulfate. After 1 h, this fraction was centrifuged at 15,000 × g for 30 min. The resulting supernatant was further treated with ammonium sulfate to achieve a final concentration of 40% saturation. After 1 h, the solution was centrifuged again at 15,000 × g for 30 min, and the precipitate was suspended in a small volume of standard buffer. The suspension was then dialyzed against three 3-liter changes of standard buffer for 18 h. The dialysate was then applied to a DEAE Sepharose CL-6B column (2 by 15 cm) preequilibrated with standard buffer. Elution was carried out with 150 ml of standard buffer followed by a linear NaCl gradient (500 ml; 0 to 0.5 M) in standard buffer. Fractions were collected at a flow rate of 9.2 ml/cm2 per h, and fractions with high specific activity were pooled. The pooled fractions were concentrated with an ultrafiltration membrane and then applied to a Sephacryl S-300 column (1.6 by 100 cm) preequilibrated with standard buffer. Elution was performed with standard buffer at a flow rate of 3.8 ml/cm2 per h. Fractions with the highest specific activity were pooled and stored at −20°C.
The purified enzyme revealed only a single band on nondenaturing polyacrylamide gel and was judged to be >95% homogeneous after densitometric analysis of the gel stained with Coomassie brilliant blue R-250 (data not shown).
Subunit separations.
To separate subunits from active CO-DH, 9 mg of active enzyme was applied to a hydroxyapatite column (1 by 3 cm; type III; Sigma Chemical Co.) preequilibrated with standard buffer. Elution was performed successively with 7 ml of standard buffer and then with 9 ml of standard buffer containing 8 M urea followed by elution with 8 M urea in 7 and 5 ml of 50 and 500 mM potassium phosphate buffer (pH 7.5), respectively, under 15 cm of hydrostatic pressure.
Separation of CO-DH subunits was also performed with 36 mg of spontaneously inactivated enzyme and a Sephacryl S-300 column (1.6 by 100 cm) preequilibrated with standard buffer. Elution was carried out with the same buffer at a rate of 3.8 ml/cm2 per h.
Cofactor analysis.
Molybdopterin was measured by fluorescence of emission at 460 nm after excitation at 360 nm with a fluorometer (model F2000; Hitachi) after being converted to form A fluorescent oxidation products. The form A derivative of molybdopterin was prepared by boiling of the purified CO-DH for 20 min at pH 2.5 in the presence of I2 plus KI following the method of Johnson and Rajagopalan (15). FAD was detected by fluorescence of emission at 525 nm after excitation at 370 nm (7). The iron content in CO-DH subunits prepared by electroelution from slices of polyacrylamide gel after SDS-PAGE of purified enzyme was calculated from the molar extinction coefficient of ferrous iron-o-phenanthroline (ɛ510 = 1.11 × 104 M−1 cm−1 [27]) separated from trichloroacetic acid- and o-phenanthroline-treated subunit solutions as described previously (32).
Nucleotide sequence accession number.
The cut sequences of H. pseudoflava have been assigned accession number U80806 in the GenBank database.
RESULTS AND DISCUSSION
CO-DH genes are located on the chromosome.
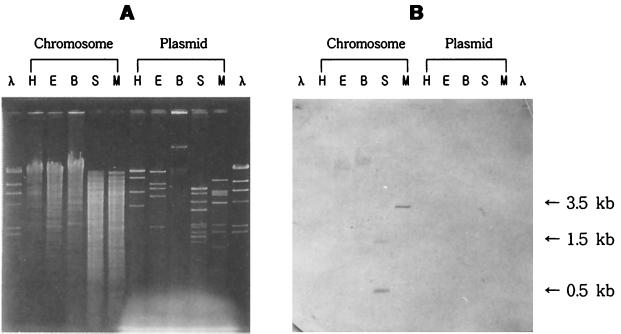
By using the synthetic primers LS and SL, a 0.5-kb DNA fragment, which is appropriate in size for encoding the small subunit of CO-DH, was amplified by PCR with H. pseudoflava genomic DNA as a template (data not shown). Part of the DNA was sequenced and found to be highly homologous to the sequences of genes for small subunits of CO-DHs from other carboxydobacteria. Southern hybridization of chromosomal and plasmid DNAs with random primed probes synthesized based on the PCR product revealed that the probes hybridized to two SalI (0.5- and 1.5-kb) fragments and a SmaI (3.5-kb) fragment of chromosomal DNA but not to any plasmid DNA fragments (Fig. 1). This result, together with the observation that the 0.5-kb DNA fragment was not obtained when H. pseudoflava plasmid DNA was used as a template for PCR, indicated that the genes for HpsCut are present on the chromosomal DNA as are those of P. thermocarboxydovorans (2). The genes of HpsCut, however, were previously judged to be plasmid borne after dot blot and Southern hybridizations employing oligonucleotide probes synthesized based on the N-terminal amino acid sequences of CO-DH subunits from O. carboxidovorans and Pseudomonas carboxydohydrogena (13, 22).
FIG. 1.
Southern hybridization of H. pseudoflava DNA. Southern hybridization was performed with random primed probes synthesized by using a 0.5-kb PCR product covering the CO-DH small subunit as a template. Shown are gels subjected to agarose gel electrophoresis (A) and Southern hybridization with random primed probes (B). H. pseudoflava chromosomal and plasmid DNAs were digested with HindIII (H), EcoRI (E), BamHI (B), SalI (S), and SmaI (M). λ DNA digested with HindIII (λ) was used as a size marker.
Cloning and characteristics of the cutMSL gene cluster.
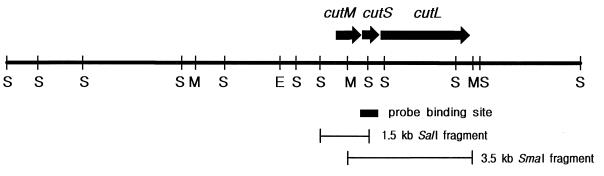
Twenty-five positive clones containing the 3.5-kb SmaI fragment were obtained by plaque hybridization of the lambda library with the random primed probes. Among the positive clones, lambda clone 311, which contains a 17-kb insert DNA, was selected for restriction mapping. Based on the restriction map of the 17-kb insert (Fig. 2), two subclones, pBKX1 and pBKS2, were constructed in pUC18 for sequence analysis using the 3.5-kb SmaI fragment and 1.5-kb SalI fragment, respectively, which hybridized with the random primed probes.
FIG. 2.
Restriction map of a 17-kb insert DNA in lambda clone 311 containing H. pseudoflava CO-DH genes. EcoRI (E), SmaI (M), and SalI (S) were used for restriction mapping. The probe binding site, a 1.5-kb SalI fragment in pBKS2, and a 3.5-kb SmaI fragment in pBKX1 are indicated under the map. ORFs for large (cutL), medium (cutM), and small (cutS) subunits of CO-DH are shown by arrows.
Sequencing of the two DNA fragments revealed the presence of three complete open reading frames (ORFs) of 864, 492, and 2,412 bp, coding for proteins with calculated molecular weights of 30,694, 17,752, and 87,224, respectively. Isoelectric points (pIs) of large, medium, and small subunits were calculated as 5.88, 8.43, and 7.77, respectively. The pIs suggest that most of the surface area of CO-DH is occupied by a negatively charged large subunit under neutral conditions, since the CO-DH band was detected on polyacrylamide gel after nondenaturing PAGE of the enzyme at pH 6.8 and the enzyme bound to DEAE-Sepharose CL-6B at pH 7.5 during purification (data not shown).
The deduced amino acid sequences derived from the long, medium, and short reading frames matched completely with portions of the N-terminal sequences of the large, medium, and small subunits of HpsCut (29), respectively, except the 13th residue of the medium subunit, S, which was previously reported to be H after N-terminal sequencing (29). This, together with the calculated molecular weights of the three genes, suggests that the cloned genes are HpsCut genes which are clustered in the order cutM-cutS-cutL like those of other carboxydobacteria (34, 40). Western blot analysis using antisera to large and medium subunits of HpsCut revealed that CutL and CutM were produced in E. coli harboring pBKX1 and a derivative of pBKS2, respectively, further supporting this suggestion (data not shown). The molecular weights of CutM and CutS, but not cutL, are comparable to those obtained by denaturing PAGE of purified CO-DH (33,000 and 17,000 for medium and small subunits, respectively [30]): the molecular weight of CutL was estimated to be 70,000 previously (30), which is much smaller than that calculated in the present study. The numbers of amino acids present in the CutM and CutS of H. pseudoflava are the same as those in the corresponding subunits of P. thermocarboxydovorans but 1 and 3 residues less than those in the CoxM and CoxS of O. carboxidovorans, respectively. H. pseudoflava CutL also contains fewer amino acid residues than those of other carboxydobacteria: the CutL consists of 30 and 6 fewer residues than the large subunits of P. thermocarboxydovorans and O. carboxidovorans, respectively.
All the three HpsCut genes begin with the standard translational start codon ATG, as do those in P. thermocarboxydovorans (34). The coxM gene of O. carboxydovorans, however, begins with alternative translational start codon GTG (40). Potential Shine-Dalgarno sequences, GGAGA, were found 8 bp upstream of the translation initiation codons of the three genes. Two directly repeated sequences (underlined), TTCGTGCAGCCAGGCGTGCAGTTCGTGC and CGTGCAGCCAGGCGTGCAG, were found 156 to 129 and 154 to 136 bp upstream of the cutM translational start site, respectively. In addition, three inverted repeats (underlined), AAAACGGTTGTTTT, CCGACGATGGCGTCGG, and ATTGACAAAAGTCAAT, were identified 125 to 112, 90 to 75, and 45 to 30 bp upstream of the cutM start codon, respectively. The average G+C ratio of the cloned cut genes was 66.6%, which is higher than those of P. thermocarboxydovorans (61.4% [34]) and O. carboxidovorans (57.2% [40]) cut genes but is in good agreement with the total G+C content of H. pseudoflava (66 to 68% [28]). Codon usage analysis showed a strong codon bias for Gs and Cs in the third position.
Transcription of cut genes.
The cutM and cutS genes and the cutS and cutL genes are separated by 17- and 31-bp intergenic regions, respectively. The regions are too narrow to work as promoters and contain no special sequences except ribosome-binding sites. These, together with the fact that CO-DHs consist of three subunits in 2:2:2 ratio, suggested that the three genes constitute an operon. Northern blot analysis using a 1.5-kb SalI insert in pBKS2 and a 2.0-kb SalI fragment from pBKX1 labeled with [α-32P]dCTP to probe cutM and cutL transcripts, however, has not been succeeded yet. It has been shown that the three genes for PthCut are separated by a 22-bp intergenic region (34). In the case of O. carboxidovorans, coxS started 18 bp downstream from the translation stop codon of coxM, and an overlap of 4 bp was observed between coxS and coxL (40).
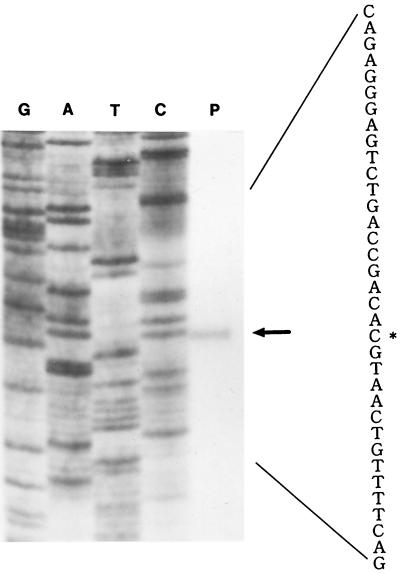
Primer extension revealed that the transcriptional start site of HpsCut genes is the G nucleotide located 47 bp upstream of the cutM start codon (Fig. 3). Transcriptional start sites were not determined for OcaCox and PthCut genes (34, 40).
FIG. 3.
Identification of transcriptional start site of cut genes. The transcriptional start of cut genes was identified by primer extension mapping using a 32P-labeled 30-mer oligonucleotide primer which is complementary to codons 7 to 16 of the HpsCut medium subunit. Extension products (lane P) were analyzed in parallel with the sequencing ladder (lanes G, A, T, and C) primed with the same primer. The asterisk shows the transcriptional start site.
Though Pearson et al. (34) found a part (TTGCA) of the consensus sequence for the −10 region of a ς54-type promoter upstream of cutB in P. thermocarboxydoflava, the consensus sequences of ς54, GC and GG in the −12 and −24 regions, respectively, was not observed upstream of the transcriptional start site of HpsCut genes. This is quite understandable, since the expression of CO-DH genes in the two organisms is controlled by different regulatory mechanisms; i.e., genes for PthCut are CO inducible and repressed by organic compounds, while those of HpsCut are expressed constitutively even under heterotrophic conditions. The consensus sequence of known promoters in other bacteria was also not found in −10 and −35 regions, suggesting that a new kind of promoter sequence may work for cut genes in H. pseudoflava. The presence of several direct repeats and inverted repeats found upstream of cutM suggests that these repeated sequences are involved in the binding of regulatory molecules. Analysis of the 200-bp region downstream from the cutL termination codon revealed no sequences capable of forming a transcriptional termination structure but identified a putative reading frame. The frame is homologous to the N-terminal region of P. thermocarboxydovorans ORF4 (34) and starts with ATG located 48 bp downstream of the cutL stop codon TAA. O. carboxidovorans was also found to have ORF4 downstream of the cox genes (40).
Sequences of CutMSL are highly conserved in CO-DHs and other molybdenum-containing hydroxylases.
The nucleotide sequences of large-, medium-, and small-subunit genes of HpsCut were 79.5, 69.0, and 74.5% identical to those of P. thermocarboxydovorans CO-DH (PthCut [34]), respectively, and 67.6, 57.1, and 68.8% identical to those of O. carboxidovorans CO-DH (OcaCox [40]), respectively. The amino acid sequences of large, medium, and small subunits of HpsCut were 84.4, 58.4, and 74.2% identical to those of PthCut (34), respectively, and 67.7, 52.6, and 68.2% identical to those of OcaCox (40), respectively (Fig. 4 to 6), indicating that HpsCut is more closely related to PthCut than to OcaCox. The identities in the nucleotide sequences of CO-DH genes (66.3, 58.1, and 66.2% for large-, medium-, and small-subunit genes, respectively) and the amino acid sequences of CO-DH subunits (69.0, 47.7, and 65.1% for large-, medium-, and small-subunit genes, respectively) between P. thermocarboxydovorans and O. carboxydovorans were less than those between H. pseudoflava and the two bacteria (Fig. 4 to 6).
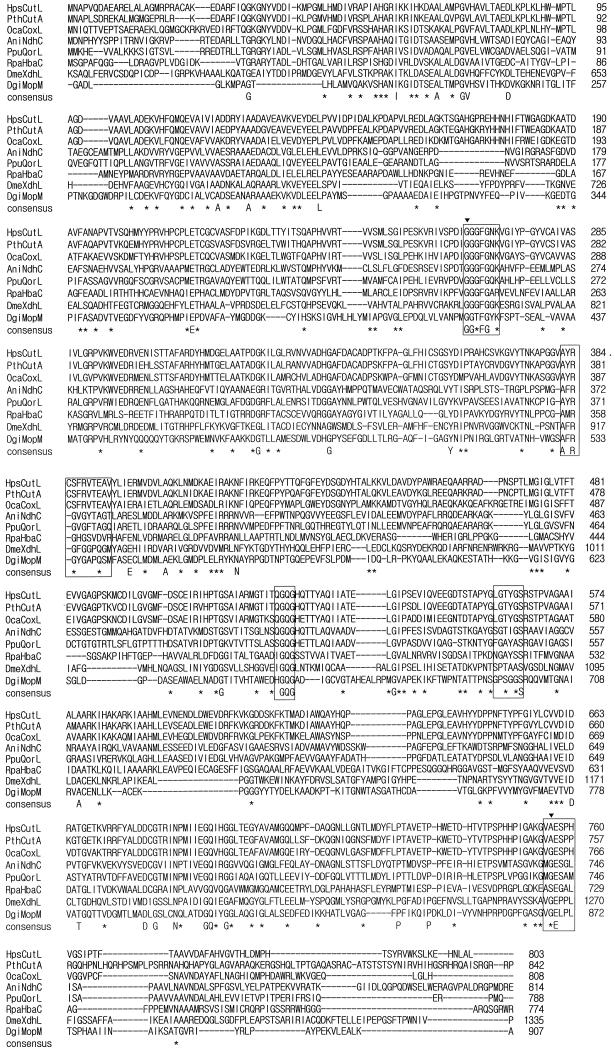
FIG. 4.
Comparison of amino acid sequence of the HpsCut large subunit with those of other molybdopterin-containing subunits and domains. The sequences compared are those of large subunits of HpsCut (HpsCutL), PthCut (PthCutA) (34), OcaCox (OcaCoxL) (40), AniNdh (AniNdhC) (9), PpuQor (PpuQorL) (3), and RpaHba (RpaHbaC) (8) and molybdopterin-containing domains of DmeXdh (DmeXdhL) (17) and DgiMop (DgiMopM) (41). Conserved residues are indicated by a capital letter (identical) or an asterisk (similar). Marked amino acids (▾) are those changed in the DmeXdh large domain from rosy mutants of D. melanogaster (14). Five molybdopterin contacting regions in the DgiMop molybdopterin-containing domain are boxed. Sequences were aligned by using the PCGENE program.
FIG. 6.
Comparison of amino acid sequence of the HpsCut small subunit with other [Fe-S]-containing subunits and domains. The sequences compared are those of small subunits of HpsCut (HpsCutS), PthCut (PthCutC) (34), OcaCox (OcaCoxS) (40), AniNdh (AniNdhB) (9), PpuQor (PpuQorS) (3), and RpaHba (RpaHbaB) (8) and iron-sulfur center-containing domains of DmeXdh (DmeXdhS) (17) and DgiMop (DgiMopF) (41). Conserved residues are indicated by a capital letter (identical) or an asterisk (similar). Cysteines bound to Fe-S clusters are shown as white letters on a black background. Marked amino acids (▾) are those changed in the DmeXdh small domain from rosy mutants of D. melanogaster (14). Sequences were aligned by using the PCGENE program.
The amino acid sequences were highly conserved over the whole ranges of the three CO-DH subunits except the C-terminal region of large subunits (Fig. 4 to 6). Residues 45 to 72 in the medium subunit of PthCut exhibited no homologies, except the G at position 62, with the corresponding residues in other CO-DHs (Fig. 5).
FIG. 5.
Comparison of amino acid sequence of the HpsCut medium subunit with those of other medium subunits of FAD-containing proteins. The sequences compared are those of medium subunits of HpsCut (HpsCutM), PthCut (PthCutB) (34), OcaCox (OcaCoxM) (40), AniNdh (AniNdhA) (9), and PpuQor (PpuQorM) (3). Conserved residues are indicated by a capital letter (identical) or an asterisk (similar). Marked amino acids (▾) are those changed in DmeXdh medium domain from rosy mutants of D. melanogaster (DmeXdh) (14). Sequences were aligned by using the PCGENE program.
A BLAST search revealed that amino acid sequences of CO-DHs are highly conserved in several other MCHs. The proteins were nicotine dehydrogenase from Arthrobacter nicotinovorans (AniNdh [9]), quinoline 2-oxidoreductase from Pseudomonas putida (PpuQor [3]), 4-hydroxybenzoyl coenzyme A reductase from Rhodopseudomonas palustris (RpaHba [8]), xanthine dehydrogenase from Drosophila melanogaster (DmeXdh [17]), and aldehyde oxidoreductase from D. gigas (DgiMop [41]) (Fig. 4 to 6). Amino acid sequences of the large, medium, and small subunits of AniNdh (9) exhibited 31.7, 33.7, and 50.3% identity with the corresponding subunits of HpsCut. Among the five hydroxylases, AniNdh (9), PpuQor (3), and RpaHba (8) consist of three subunits in the order M-S-L, M-S-L, and S-L-M, respectively, and contain molybdopterin, FAD, and Fe-S centers. DmeXdh, on the other hand, has three distinct domains in the order S-M-L and also contains molybdopterin, FAD, and Fe-S centers (17). DgiMop, however, consists of two domains, a large molybdopterin-containing domain and a small Fe-S center-binding domain in the order S-L (41). Dendrograms of the pairwise scores plotted by the PCGENE program revealed that the orders of similarity to HpsCut large, medium, and small subunits of the corresponding subunits or domains of other MCHs are PthCut-OcaCox-AniNdh-PpuQor-RpaHba-DmeXdh-DgiMop, PthCut-OcaCox-AniNdh-PpuQor-RpaHba-DmeXdh, and PthCut-OcaCox-RpaHba-AniNdh-PpuQor-DgiMop-DmeXdh, respectively, indicating that HpsCut is most closely related to CO-DHs, next most closely related to other subunit enzymes, and least closely related to nonsubunit proteins. The above results imply that MCHs may be evolutionarily and structurally related to each other and have common motifs for cofactor binding.
CutL contains a molybdopterin-binding site.
The five regions known to be the molybdopterin-contacting sites in DgiMop after X-ray crystallography (37) were conserved in all MCHs examined (Fig. 4), suggesting that the molybdopterin cytosine dinucleotide in HpsCut (29) binds to CutL through these regions. Localization of two amino acid residues of CutL, G65 and A756, in a hypothetical molybdopterin-contacting region at positions identical to those of two amino acids in the DmeXdh large domain, G800 and G1266, also supports this assumption, since replacing G800 and G1266 with E and D, respectively, is known to be responsible for rosy mutations in D. melanogaster (14).
Schübel et al. (40) reported that the OcaCox CoxL is the putative FAD-binding site, since the GLGTYG sequence in CoxL (Fig. 4 [residues 564 to 569]) is identical to dinucleotide-binding motif GXGXX(G/A). This region, however, may not be the FAD-binding site, since the domain containing the proposed sequence is similar to the molybdopterin-binding domain of DgiMop (37) and the G569 in CutL corresponds to the G696 of DgiMop involved in the formation of a hydrogen bond with the Mo(IV)-carried oxygen ligand in molybdopterin molecule.
CutM contains a FAD-binding site.
The DmeXdh medium domain is known to contain three internal regions that bind FAD and NAD+: K3, which binds FAD, and K5 and N, which bind NAD+ (14). Among the three regions, the K3 region is most highly conserved in CO-DH, which does not use NAD+ as a coenzyme, and also in other MCHs containing both FAD and the medium subunit or domain (Fig. 5). The K3 sequence is absent in DgiMop containing no medium domain and FAD (41). These together with the finding that two amino acid residues of CutM, G114 and G119, in the K3 region align at positions identical to those of two amino acids in the DmeXdh medium domain, G348 and G353, strongly suggest that HpsCut CutM binds FAD, since replacing the amino acids G348 and G353 with E and D, respectively, is known to result in rosy mutants of D. melanogaster (14).
CutS has two [2Fe-2S] center-binding sites.
The amino acid sequence of CutS was very similar to the small subunits and domains of other MCHs containing Fe-S clusters (Fig. 6). The regions which contain eight cysteines responsible for binding two [2Fe-2S] clusters in the DgiMop small domain (37) were conserved at identical positions in all MCHs examined. The first region, containing four cysteines in positions 42, 47, 50, and 62 of CutS, is a typical [2Fe-2S] binding region, C-X4-C-X2-C-Xn-C, found in bacterial and plant ferredoxins (10). The second four-cysteine-containing region, C-X2-C-Xn-C-X-C, which binds another type of [2Fe-2S] cluster, is more conserved than the first one in MCHs. Both Fe-S motifs are absent in CutL and CutM. These considerations together with the observation that the G41 and E78 of CutS located at positions corresponding to those of the two changed residues of xanthine dehydrogenase in D. melanogaster rosy mutants (G42→E and E89→K) (14) imply CutS as the iron-sulfur protein of HpsCut.
Molybdopterin, FAD, and iron-sulfur centers bind to CutL, CutM, and CutS, respectively.
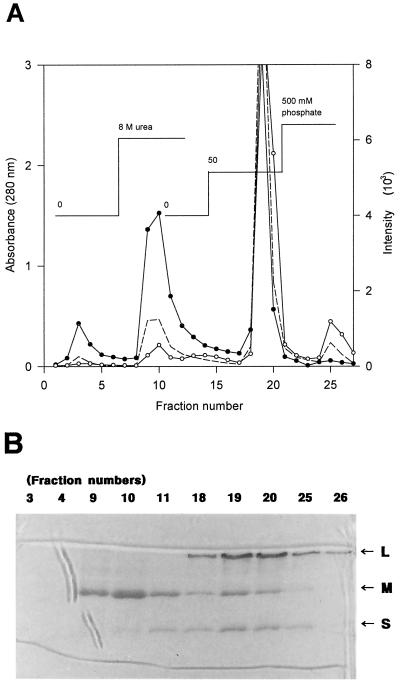
In a preliminary test, most of the active purified CO-DH appeared as a single brownish-yellow band on nondenaturing polyacrylamide gels even after incubation for 1 h at 20°C in standard buffer containing 8 M urea (data not shown), indicating that CO-DH is stable and maintains its cofactors under the conditions tested. The enzyme, however, began to lose the typical band after treatment with 2 M guanidine-HCl for 1 h. The band disappeared completely when the enzyme was incubated for 1 h in 4 M guanidine-HCl solution. These results supported previous observations that the molybdopterin and FAD were released from CO-DH when the enzyme was treated with guanidine-HCl, SDS, heat, or acid but not when it was incubated in the presence of urea (23). We, however, assumed that treatment of active CO-DH with 8 M urea affects the binding strength within the three CO-DH subunits to a certain degree even though it does not force the CO-DH to release its cofactors. We, therefore, tried to prepare subunits containing bound cofactors from the active CO-DH, employing hydroxyapatite column chromatography in the presence of 8 M urea to localize molybdopterin and FAD in the enzyme. It was found that a large amount of medium subunits, but almost no large subunits, was eluted with standard buffer containing 8 M urea (Fig. 7), as expected from the calculated pIs of the large (5.88) and medium subunits (8.43). The fractions containing medium subunits exhibited a strong signal for FAD but a weak one for molybdopterin (Fig. 7A). The weak fluorescence for molybdopterin in fractions 9 and 10 may be due to the background from the FAD signal. When the enzyme was eluted with 50 mM phosphate buffer (pH 7.5) in the presence of 8 M urea, all three CO-DH subunits appeared in the collected fractions (Fig. 7B), which showed strong signals for both molybdopterin and FAD (Fig. 7A). Among the fractions obtained with 8 M urea-containing 500 mM phosphate buffer (pH 7.5), fraction 26 contained only the large subunit and exhibited a signal only for molybdopterin (Fig. 7). These results support the assumption that 8 M urea hardly affects the binding between subunits and cofactors but may reduce the binding strength between each subunit. It also indicates that molybdopterin and FAD bind to the large and medium subunits of H. pseudoflava CO-DH, respectively, which was suggested from the deduced amino acid sequences of CutL and CutM. The coincidence of the signal patterns of the two cofactors with the elution pattern of the proteins further supports this indication (Fig. 7). It is also plausible to assume from the protein profile of each fraction (Fig. 7B) that the medium subunit in the native CO-DH interacts with both the large and the small subunits of the enzyme.
FIG. 7.
(A) Hydroxyapatite column chromatography of active CO-DH. Active purified CO-DH was applied to a hydroxyapatite column (1 by 3 cm) and eluted successively with standard buffer, standard buffer containing 8 M urea, 8 M urea-containing 50 mM phosphate buffer, and 8 M urea-containing 500 mM phosphate buffer under 15 cm of hydrostatic pressure as described in Materials and Methods. Protein was monitored by absorbance at 280 nm (–––). Cofactors were analyzed by fluorescence of emission at 460 nm after excitation at 360 nm and emission at 525 nm after excitation at 370 nm for molybdopterin (○) and FAD (●), respectively. (B) CO-DH subunits in fractions from hydroxyapatite chromatography. Denaturing PAGE of CO-DH subunits present in fractions obtained from hydroxyapatite chromatography was carried out on a 12.5% polyacrylamide gel in the presence of 0.1% SDS following the method of Laemmli (24). Twenty microliters each of several fractions eluted with standard buffer (fractions 3 and 4), standard buffer containing 8 M urea (fractions 9, 10, and 11), 50 mM phosphate buffer containing 8 M urea (fractions 18, 19, and 20), and 500 mM phosphate buffer containing 8 M urea (fractions 25 and 26) were analyzed together with active purified enzyme. Arrows indicate large (L), medium (M), and small (S) subunits of CO-DH.
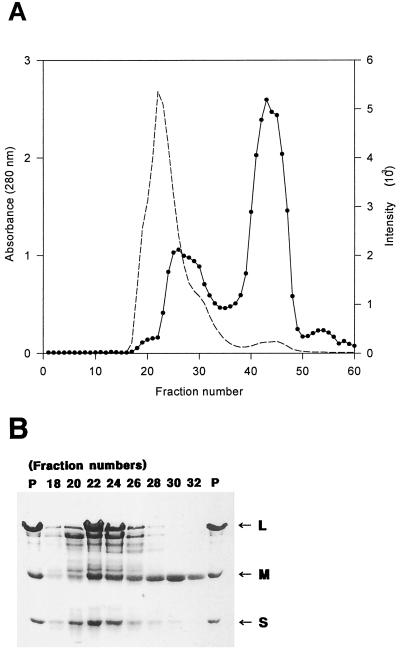
It was found that the purified CO-DH of H. pseudoflava was inactivated after incubation for a month at 4°C, suggesting spontaneous denaturation of the enzyme. The location of FAD was further identified with the inactive enzyme. When the inactive enzyme was subjected to gel filtration, it did not elute as a sharp peak (Fig. 8A). The signal for FAD appeared as two peaks and did not coincide with the elution profile of the enzyme. The intensity of the two signals and the elution pattern indicated that the first and second signals are due to protein-bound and protein-free FADs, respectively. These results indicate that the enzyme may have been inactivated partly due to loss of FAD cofactor during spontaneous denaturation of the enzyme. Analysis of the protein pattern in each fraction revealed that fraction 32, which exhibits the FAD signal, contains the medium subunit almost exclusively (Fig. 8), strongly supporting the above indication deduced from analysis of fractions from hydroxyapatite column chromatography that the medium subunit in H. pseudoflava CO-DH binds FAD. The sharp decrease in the density of large and small subunits (Fig. 8B) and the sharp increase in that of the FAD signal (Fig. 8A) in fractions 22 to 26 suggest that the FAD-binding medium subunit may occupy the innermost part of native CO-DH, resulting in quenching of the FAD signal by the large and, possibly, the small subunits surrounding it. This suggestion was strongly supported by the observation that the intensity of the FAD signal of fractions 22 and 24 increased after treatment of the fractions with trichloroacetic acid or high temperature (data not shown). The acidic nature of native CO-DH at neutral pH together with the estimated pI of the medium subunit (8.43) also supports this suggestion.
FIG. 8.
(A) Elution pattern of inactive CO-DH on a Sephacryl S-300 column. Spontaneously inactivated purified CO-DH was subjected to gel filtration on a Sephacryl S-300 column (1.6 by 100 cm) with standard buffer as described in Materials and Methods. Protein and FAD were monitored by absorbance at 280 nm (–––) and fluorescence of emission at 525 nm after excitaion at 370 nm (●), respectively. (B) Denaturing PAGE of fractions from Sephacryl S-300 column chromatography. CO-DH subunits in 20 μl each of fractions 18, 20, 22, 24, 26, 28, 30, and 32 from Sephacryl S-300 chromatography of the inactivated CO-DH were subjected to denaturing PAGE together with inactive purified enzyme (7 μg) (P) on a 12.5% polyacrylamide gel in the presence of 0.1% SDS by the method of Laemmli (24). The large (L), medium (M), and small (S) subunits of CO-DH are indicated by arrows.
Analysis of iron content in CO-DH subunits prepared from active purified CO-DH revealed the presence of 0.58, 0.67, and 1.96 atoms of iron per mol of large, medium-, and small-subunit proteins, respectively, indicating that the two iron-sulfur centers in H. pseudoflava CO-DH are associated with the small subunit as expected from the deduced amino acid sequence of CutS. Part of the iron in the small subunit seems to be dissociated during boiling of CO-DH in the presence of SDS for denaturing PAGE, resulting in a value lower than the expected one (4 atoms per mol of subunit). The iron detected in the large and medium subunits may be due to contamination during PAGE and electroelution.
The present results clearly indicate through analogy with sequence homology data (3, 9, 17, 34, 40) that the molybdopterin, FAD, and iron-sulfur centers bind to the large, medium, and small subunits, respectively, of all three-subunit or three-domain molybdenum-containing hydroxylases studied, including the CO-DH from O. carboxidovorans.
ACKNOWLEDGMENT
This study was partly supported by a nondirected research fund from the Korea Research Foundation (1996) to Y.M.K.
REFERENCES
- 1.Amaya Y, Yamazaki K, Sato M, Noda K, Nishino T, Nishino T. Proteolytic conversion of xanthine dehydrogenase from the NAD-dependent type to the O2-dependent type. J Biol Chem. 1990;265:14170–14175. [PubMed] [Google Scholar]
- 1a.Biotech Laboratories Inc. Biotech Laboratories bulletin no. 27. Houston, Tex: Biotech Laboratories Inc.; 1992. [Google Scholar]
- 2.Black G W, Lyons C M, Williams E, Colby J, Kehoe M, O’Reilly C. Cloning and expression of the carbon monoxide dehydrogenase genes from Pseudomonas thermocarboxydovorans strain C2. FEMS Microbiol Lett. 1990;70:249–254. doi: 10.1016/s0378-1097(05)80003-5. [DOI] [PubMed] [Google Scholar]
- 3.Blase M, Bruntner C, Tshisuaka B, Fetzner S, Lingens F. Cloning, expression, and sequence analysis of the three genes encoding quinoline 2-oxidoreductase from Pseudomonas putida 86. J Biol Chem. 1996;271:23068–23079. doi: 10.1074/jbc.271.38.23068. [DOI] [PubMed] [Google Scholar]
- 3a.Boehringer-Mannheim. The Dig system user’s guide. Indianapolis, Ind: Boehringer-Mannheim; 1993. [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Coughlan M P, Betcher-Lange S L, Rajagopalan K V. Isolation of the domain containing the molybdenum, iron-sulfur I, and iron-sulfur II centers of chicken liver xanthine dehydrogenase. J Biol Chem. 1979;254:10694–10699. [PubMed] [Google Scholar]
- 6.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghisla S. Fluorescence and optical characteristics of reduced flavins and flavoproteins. Methods Enzymol. 1980;66:360–373. doi: 10.1016/0076-6879(80)66481-7. [DOI] [PubMed] [Google Scholar]
- 8.Gibson J, Dispensa M, Harwood C. 4-Hydroxybenzoyl coenzyme A reductase (dehydroxylating) is required for anaerobic degradation of 4-hydroxybenzoate by Rhodopseudomonas palustris and shares features with molybdenum-containing hydroxylases. J Bacteriol. 1997;179:634–642. doi: 10.1128/jb.179.3.634-642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grether-Beck S, Igloi G L, Pust S, Schilz E, Decker K, Brandsch R. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol Microbiol. 1994;13:929–936. doi: 10.1111/j.1365-2958.1994.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 10.Harayama S, Polissi A, Rekik M. Divergent evolution of chloroplast-type ferredoxins. FEBS Lett. 1991;285:85–88. doi: 10.1016/0014-5793(91)80730-q. [DOI] [PubMed] [Google Scholar]
- 11.Hille R, Nishino T. Xanthine oxidase and xanthine dehydrogenase. FASEB J. 1995;9:995–1003. [PubMed] [Google Scholar]
- 12.Houde M, Tiveron M-C, Brégégère F. Divergence of the nucleotide sequences encoding xanthine dehydrogenase in Calliphora vicina and Drosophila melanogaster. Gene. 1989;85:391–402. doi: 10.1016/0378-1119(89)90432-0. [DOI] [PubMed] [Google Scholar]
- 13.Hugendieck I, Meyer O. The structural genes encoding CO dehydrogenase subunits (cox L, M and S) in Pseudomonas carboxydovorans OM5 reside on plasmid pHCG3 and are, with the exception of Streptomyces thermoautotrophicus, conserved in carboxydotrophic bacteria. Arch Microbiol. 1992;157:301–304. doi: 10.1007/BF00245166. [DOI] [PubMed] [Google Scholar]
- 14.Hughes R K, Dotle W A, Chovnick A, Whittle J R S, Burke J F, Bray R C. Use of rosy mutant strains of Drosophila melanogaster to probe the structure and function of xanthine dehydrogenase. Biochem J. 1992;185:507–513. doi: 10.1042/bj2850507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson J L, Rajagopalan K V. Structural and metabolic relationship between the molybdenum cofactor and urothine. Proc Natl Acad Sci USA. 1982;79:6856–6860. doi: 10.1073/pnas.79.22.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keith T P, Riley M A, Lewontin R C, Curtis D, Chambers G. Sequence of the structural gene for xanthine dehydrogenase (rosy locus) in Drosophila melanogaster. Genetics. 1987;116:67–73. doi: 10.1093/genetics/116.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiessling M, Meyer O. Profitable oxidation of carbon monoxide and hydrogen during heterotrophic growth of Pseudomonas carboxydoflava. FEMS Microbiol Lett. 1982;13:333–338. [Google Scholar]
- 19.Kim K S, Ro Y T, Kim Y M. Purification and some properties of carbon monoxide dehydrogenase from Acinetobacter sp. strain JC1 DSM 3803. J Bacteriol. 1989;171:958–964. doi: 10.1128/jb.171.2.958-964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y M, Hegeman G D. Purification and some properties of carbon monoxide dehydrogenase from Pseudomonas carboxydohydrogena. J Bacteriol. 1981;148:904–911. doi: 10.1128/jb.148.3.904-911.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y M, Hegeman G D. Oxidation of carbon monoxide by bacteria. Int Rev Cytol. 1983;81:1–32. doi: 10.1016/s0074-7696(08)62333-5. [DOI] [PubMed] [Google Scholar]
- 22.Kraut M, Hugendieck I, Herwig S, Meyer O. Homology and distribution of CO dehydrogenase structural genes in carboxydotrophic bacteria. Arch Microbiol. 1989;152:335–341. doi: 10.1007/BF00425170. [DOI] [PubMed] [Google Scholar]
- 23.Krüger B, Meyer O. Structural elements of bactopterin from Pseudomonas carboxydoflava carbon monoxide dehydrogenase. Biochim Biophys Acta. 1987;912:357–364. doi: 10.1016/0167-4838(87)90040-9. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee C S, Curtis D, McCarron M, Love C, Gray M, Bender W, Chovnick A. Mutations affecting expression of the rosy locus in Drosophila melanogaster. Genetics. 1987;116:55–66. doi: 10.1093/genetics/116.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 27.Massey V. Studies on succinic dehydrogenase. VII. Valency state of the iron in beef heart succinic dehydrogenase. J Biol Chem. 1957;229:763–770. [PubMed] [Google Scholar]
- 28.Meyer O, Frunzke K, Gadkari D, Jacobitz S, Hugendieck I, Kraut M. Utilization of carbon monoxide by aerobes—recent advances. FEMS Microbiol Rev. 1990;87:253–260. [Google Scholar]
- 29.Meyer O, Frunzke K, Mörsdorf G. Biochemistry of the aerobic utilization of carbon monoxide. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Andover, Mass: Intercept Limited; 1993. pp. 433–459. [Google Scholar]
- 30.Meyer O, Jacobitz S, Krüger B. Biochemistry and physiology of aerobic carbon monoxide-utilizing bacteria. FEMS Microbiol Rev. 1986;39:161–179. [Google Scholar]
- 31.Meyer O, Schlegel H G. Reisolation of the carbon monoxide-utilizing hydrogen bacterium Pseudomonas carboxydovorans (Kistner) comb. nov. Arch Microbiol. 1978;118:35–43. doi: 10.1007/BF00406071. [DOI] [PubMed] [Google Scholar]
- 32.Miller R W, Massey V. Dihydroorotic dehydrogenase. I. Some properties of the enzyme. J Biol Chem. 1965;240:1453–1465. [PubMed] [Google Scholar]
- 33.Nishino T, Nishino T. The nicotinamide adenine dinucleotide-binding site of chicken liver xanthine dehydrogenase. J Biol Chem. 1989;264:5468–5473. [PubMed] [Google Scholar]
- 34.Pearson D M, O’Reilly C, Colby J, Black G W. DNA sequence of the cut A, B and C genes, encoding the molybdenum containing hydroxylase carbon monoxide dehydrogenase, from Pseudomonas thermocarboxydovorans strain C2. Biochim Biophys Acta. 1994;1188:432–438. doi: 10.1016/0005-2728(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 34a.Promega. Promega protocols and application guide. 2nd ed. Madison, Wis: Promega; 1995. [Google Scholar]
- 35.Riley M A. Nucleotide sequence of the xdh region in Drosophila pseudoobscura and an analysis of the evolution of synonymous codons. Mol Biol Evol. 1989;6:33–52. doi: 10.1093/oxfordjournals.molbev.a040529. [DOI] [PubMed] [Google Scholar]
- 36.Ro Y T, Chung I K, Lee J, Kim D, Suh J W, Kim S W, Kim Y M. Activity and expression of carbon monoxide dehydrogenase in Acinetobacter sp. strain JC1 growing with different growth substrates. Microorganisms Ind. 1995;21:252–260. [Google Scholar]
- 37.Romão A, Moura I, Moura J J G, LeGall J, Engh R, Schneider M, Hof P, Huber R. Crystal structure of the xanthine oxidase-related aldehyde oxido-reductase from D. gigas. Science. 1995;270:1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schübel U, Kraut M, Mörsdorf G, Meyer O. Molecular characterization of the gene cluster coxMSL encoding the molybdenum-containing carbon monoxide dehydrogenase of Oligotropha carboxidovorans. J Bacteriol. 1995;177:2197–2203. doi: 10.1128/jb.177.8.2197-2203.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoenes U, Flores O L, Neves A, Devreese B, Van Beeumen J J, Huber R, Romao M J, LeGall J, Moura J J, Rodrigues-Pousada C. Molecular cloning and sequence analysis of the gene of the molybdenum-containing aldehyde oxido-reductase of Desulfovibrio gigas. The deduced amino acid sequence shows similarity to xanthine dehydrogenase. Eur J Biochem. 1994;220:901–910. doi: 10.1111/j.1432-1033.1994.tb18693.x. [DOI] [PubMed] [Google Scholar]
- 42.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 43.Willems A, Busse J, Goor M, Pot B, Falsen E, Jantzen E, Hoste B, Gillis M, Kersters K, Auling G, De Ley J. Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga palleronii (formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (formerly Pseudomonas pseudoflava and Pseudomonas carboxydoflava), and Hydrogenophaga taeniospiralis (formerly Pseudomonas taeniospiralis) Int J Syst Bacteriol. 1989;39:319–333. [Google Scholar]
- 44.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]