Significance
Molecular mechanisms that give rise to the remarkable dynamics of the actin and tubulin subunits have been a focus of extensive research. The tightly regulated import/export of tubulin is particularly important for ciliogenesis, given that tubulins form the axoneme “backbone” of all cilia. The interaction with the IFT-74/81 subunits helps load tubulin onto the intraflagellar transport (IFT) machinery for ciliary import. We found that the conserved DYF-5/MAK kinase localizing at the ciliary tip phosphorylates IFT-74, reducing the binding affinity between tubulin and IFT-74/81 sixfold. In vivo manipulation phosphorylation status of IFT-74 changed ciliary length. We propose that DYF-5/MAK–dependent phosphorylation promotes tubulin unloading at the ciliary tip and contributes to site-specific regulation of tubulin dynamics to maintain axonemal integrity.
Abstract
Cilia are microtubule-based organelles that power cell motility and regulate sensation and signaling, and abnormal ciliary structure and function cause various ciliopathies. Cilium formation and maintenance requires intraflagellar transport (IFT), during which the kinesin-2 family motor proteins ferry IFT particles carrying axonemal precursors such as tubulins into cilia. Tubulin dimers are loaded to IFT machinery through an interaction between tubulin and the IFT-74/81 module; however, little is known of how tubulins are unloaded when arriving at the ciliary tip. Here, we show that the ciliary kinase DYF-5/MAK phosphorylates multiple sites within the tubulin-binding module of IFT-74, reducing the tubulin-binding affinity of IFT-74/81 approximately sixfold. Ablation or constitutive activation of IFT-74 phosphorylation abnormally elongates or shortens sensory cilia in Caenorhabditis elegans neurons. We propose that DYF-5/MAK–dependent phosphorylation plays a fundamental role in ciliogenesis by regulating tubulin unloading.
Cilia are microtubule (MT)-based organelles responsible for cell motility and sensory signaling (1), and ciliary dysfunction leads to a wide range of human disorders, collectively termed ciliopathies (2). Intraflagellar transport (IFT) is essential for cilium construction and maintenance, dependent on the IFT particle complex consisting of ∼22 subunits and kinesin-2 and dynein-2 motors (3–6). IFT particles carrying diverse ciliary cargoes reach the ciliary tip via kinesin-2–mediated anterograde transport, and upon arrival they are proposed to unload cargo molecules and activate dynein-2–mediated retrograde transport for recycling the anterograde IFT machinery and ciliary turnover products (5, 6). More than 600 ciliary proteins are thought to be delivered by IFT machinery for cilium formation and maintenance (7); however, how the spatiotemporal intraciliary cargo turnover is implemented and regulated is largely unknown (8–10).
Previous studies have indicated axonemal MTs are assembled at ciliary tips via IFT-mediated tubulin transport (11–14) and suggested a “balance-point model” (i.e., a balance of assembly and disassembly) for cilium length control (12–15). A recent study showed that IFT-74, an IFT-B complex subunit, functions with IFT-81 to bind and transport ciliary tubulins (16). It remains mysterious how to weaken the interaction between tubulins and IFT-74/81, allowing tubulin release from the IFT particle at the ciliary tip. Ciliary protein kinases, including male germ cell-associated kinase (MAK), have been identified as critical ciliary regulators (2). The Chlamydononas reinhardtii MAK homolog IF4 and Caenorhabditis elegans MAK/DYF-5 are involved in cilium assembly and length regulation (17). The deficiency of the mammalian MAKs disrupted IFT and abnormally elongated cilia (8–10), contributing to the pathogenesis of human retinitis pigmentosa (18, 19). Here, we combined the in vitro biochemistry with C. elegans genetics and imaging to show that DYF-5/MAK–mediated IFT-74 phosphorylation significantly reduces its tubulin-binding affinity, thereby providing an unloading mechanism to release tubulin from IFT machinery in regulating ciliogenesis in C. elegans sensory cilia.
Results
DYF-5 Kinase Phosphorylates Multiple Sites of IFT-74 N Terminus.
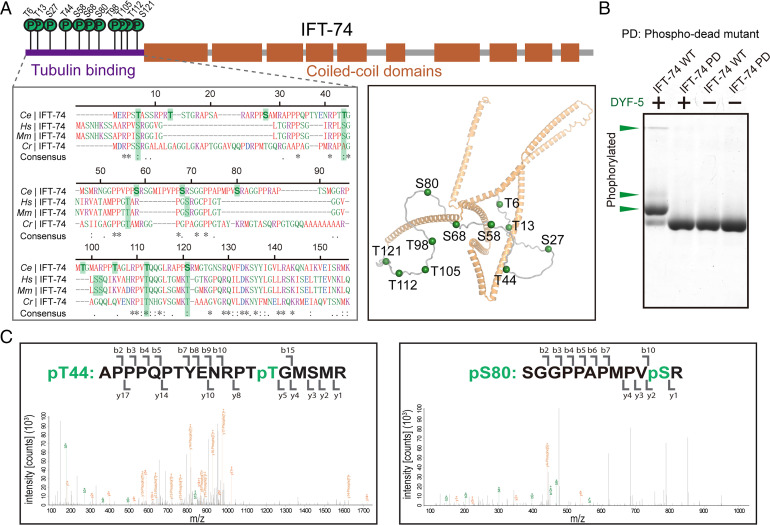
We first sought to dissect the DYF-5 phosphorylation sites on IFT-74 by liquid chromatography–tandem mass spectrometry (LC-MS/MS). We treated the recombinant IFT-74 protein (GST-tagged IFT-74, 1–372) with purified DYF-5 kinase. LC-MS/MS analysis of digested IFT-74 phosphopeptides identified 11 phosphorylation sites (Thr6, Thr13, Ser27, Thr44, Ser58, Ser68, Ser80, Thr98, Thr105, Thr112, and Ser121) in the N-terminal flexible tubulin-associating loop (IFT-74N) (Fig. 1A). Remarkably, the result indicated PX[S/T]*, particularly RPX[S/T]* (7 of 11 phosphorylation sites), as the substrate motif of DYF-5 kinase. To further confirm the LC-MS/MS result, Phos-tag kinase assay (20) was performed using wild-type (WT) and phosphorylation-dead mutant (PD; Ser-Ala/Thr-Ala mutant of all phosphorylation sites) IFT-74 proteins (SI Appendix, Fig. S1). Compared to the negative controls, phosphorylations of WT IFT-74 by DYF-5 were observed as stoichiometric upward bandshifts in Phos-tag sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), whereas IFT-74 PD mutant exhibited no phosphorylation by DYF-5 treatment (Fig. 1A), which verified the LC-MS/MS determination of IFT-74 phosphorylation sites.
Fig. 1.
DYF-5 phosphorylates the tubulin-binding region of IFT-74. (A) Schematic diagrams (Top), homologous sequence alignment (Bottom Left), and cartoon structural representation (Bottom Right) showing the identified phosphorylation sites in C. elegans IFT-74. The phosphorylation residues and their possible corresponding homologous sites were labeled with green shadows in the alignment. The structural coordinate of IFT-74 from AlphaFold Protein Structure Database (ID: A0A2C9C2L6) was used to generate the structural figure. (B) Phos-tag kinase assays validating the determined phosphorylation sites. Purified WT and phospho-dead (PD; Ser-Ala/Thr-Ala) mutant IFT-74 (1-372) protein was treated with DYF-5 kinase and evaluated by Phos-tag SDS-PAGE. The phosphorylation-induced bandshifts are indicated by green arrowheads. (C) Representative LC-MS/MS results of the determination of IFT-74 phosphorylation sites.
IFT-74 Phosphorylation by DYF-5 Reduces Its Tubulin-Binding Affinity.
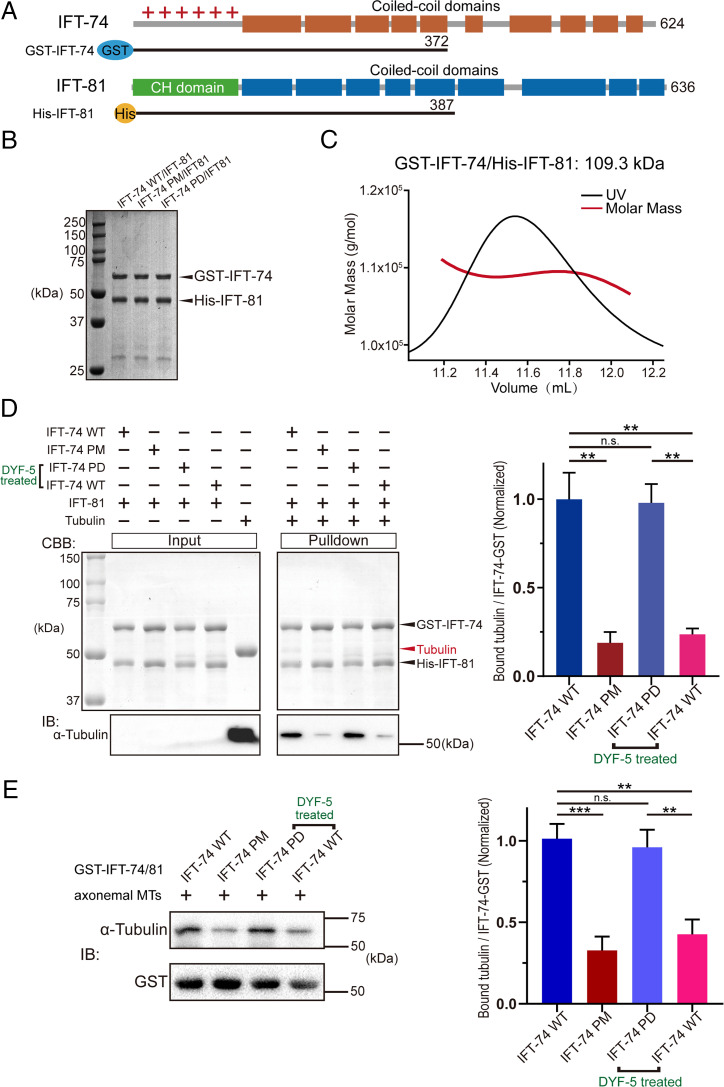
Previous studies described that IFT-74N and IFT-81 form a tubulin-binding module responsible for intraciliary tubulin transport during ciliogenesis (16, 21). Given all the determined phosphorylation sites of IFT-74 concentratively reside in IFT-74N, we next investigated the effects of DYF-5–mediated IFT-74 phosphorylation on tubulin binding. We constructed, coexpressed, and purified an IFT-74/81 (GST-IFT-74/His-IFT-81) heterodimer complex for biochemical assessments (Fig. 2 A and B). The purified complex exhibited a single peak with the molecular weight of 109.3 kDa in size-exclusion chromatography–multiangle light scattering (SEC-MALS) analysis (Fig. 2C), which is close to the theoretical calculation with a 1:1 stoichiometric ratio. Then, pull-down assays were performed to evaluate the effects of phosphorylation on IFT-74–tubulin interaction using purified porcine brain tubulin. We found that phosphorylation-mimic mutant (PM; Ser-Asp/Thr-Glu of all phosphorylation sites) protein showed significantly reduced binding to tubulin compared to WT IFT-74/81 protein in the presence of IFT-81 (Fig. 2D). Meanwhile, WT IFT-74/81 protein significantly decreased tubulin binding upon DYF-5 treatment compared to IFT-74 PD/IFT-81 protein (Fig. 2D). In the pull-down assay, we obtained consistent results using axonemal tubulin samples enriched from C. reinhardtii (Fig. 2E).
Fig. 2.
IFT-74 phosphorylation by DYF-5 disrupts the binding of tubulin with IFT-74/81. (A) Schematic diagrams showing the IFT-74/81 constructs used for biochemical analysis, which are tagged by GST (GST-IFT-74 1–372) or 6×His (His-IFT-81 1–387). (B) SDS-PAGE result of the purified IFT-74/81 heterodimer proteins. (C) SEC-MALS analysis of the IFT-74/81 recombinant protein. (D) Pull-down assay evaluating the interaction of tubulin with WT, phospho-mutant, and DYF-5-treated IFT-74/81 proteins. (E) Pull-down assay analyzing tubulin-IFT-74/81 binding using C. reinhardtii axonemal MTs. The assays were performed with three replicates and the quantifications are shown at the right. PM: phospho-mimic mutant; PD: phospho-dead mutant. Statistical significance compared with indicated genotype is based on Student's t-test, **P < 0.01, ***P < 0.001, n.s., no significance.
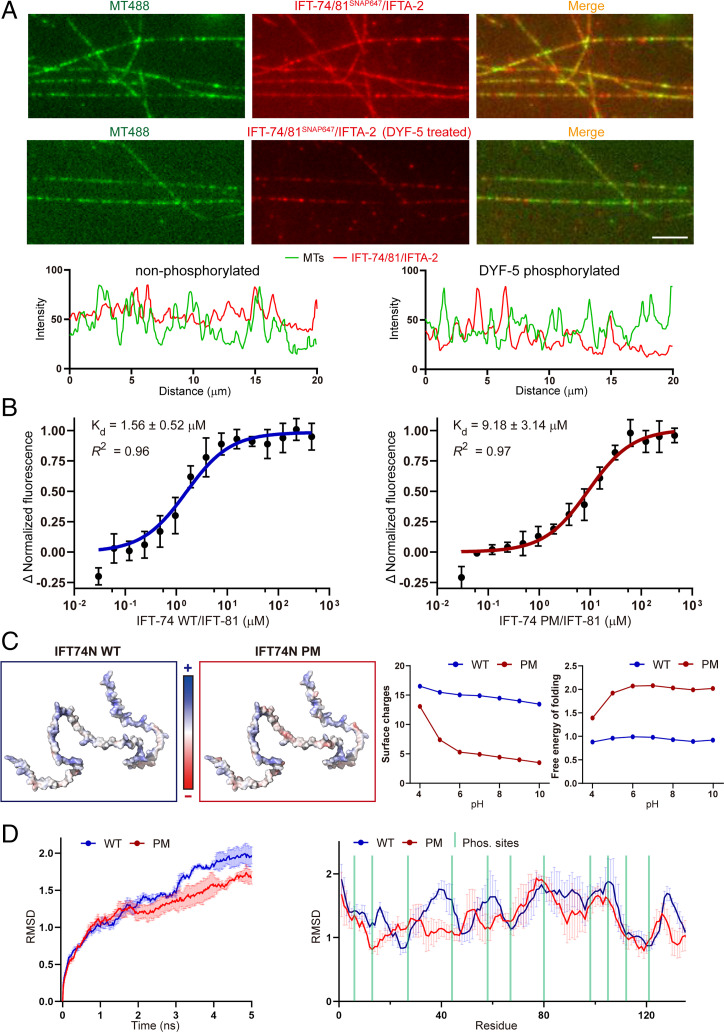
To further confirm the biochemical results, we performed total internal reflection fluorescence (TIRF) microscopic assays to assess the colocalization of MTs with WT and DYF-5–phosphorylated IFT-74/81/IFTA-2 complex proteins. We found that the colocalization between IFT-74/81/IFTA-2 and MTs was reduced by DYF-5 phosphorylation (Fig. 3A). We determined the affinity constant of IFT-74/81–tubulin interaction using microscale thermophoresis assays. The PM mutant showed an approximately sixfold reduction of tubulin-binding affinity compared to the WT protein (Fig. 3B). Together, these results indicated phosphorylation-induced release of tubulin from the IFT-74/81 module.
Fig. 3.
IFT-74 phosphorylation diminishes its tubulin-binding affinity via rearrangement of surface potentials. (A) TIRF MT-colocalization assay evaluating the interaction between MTs and IFT-74/81/IFTA-2 protein with or without DYF-5 treatment. Representative fluorescence intensity shown (Bottom) was measured by a 20-μm line randomly drawn on MTs. Scale bar, 5 µm. (B) Microscale thermophoresis assay dissecting the binding affinity of tubulin with WT and phospho-mimic IFT-74/81 proteins. (C) Analysis of the phosphorylation-induced changes of IFT-74N charges and folding free energy. (D) Molecular dynamic analysis of the phosphorylation-induced changes of IFT-74N structural stability/flexibility. AlphaFold2-based structural coordinate of IFT-74 (ID: A0A2C9C2L6) was used for the analysis. IFT-74N: IFT-74 N terminus.
Phosphorylation Disrupts Surface Charges of IFT-74N Responsible for Tubulin Association.
Next, we conducted structural and molecular dynamic analyses to assess the effects of phosphorylation on IFT-74N. The AlphaFold2-predicted IFT-74N and the generated phospho-mimic mutant structural models were used. The phosphorylation triggered a considerable increase of negative side-chain charges, which neutralized the positive surface charges crucial for its association with negatively charged tubulin C-terminal tails (Fig. 3C). In addition, the free energy of folding of the PM mutant model was approximately twofold higher than the WT model, suggesting the lower structural stability of phosphorylated IFT-74N (Fig. 3C, Right). Moreover, 5-ns time-lapse molecular dynamic simulations were performed with the physiological solvent condition. The results indicated a lower structural variation of PM mutant than WT IFT-74N, which suggests that the phosphorylation may perturb the structural flexibility of IFT-74N, which is vital for tubulin interaction (Fig. 3D).
Disruption of IFT-74N Phosphorylation Leads to Abnormal Cilium Length in Sensory Neurons.
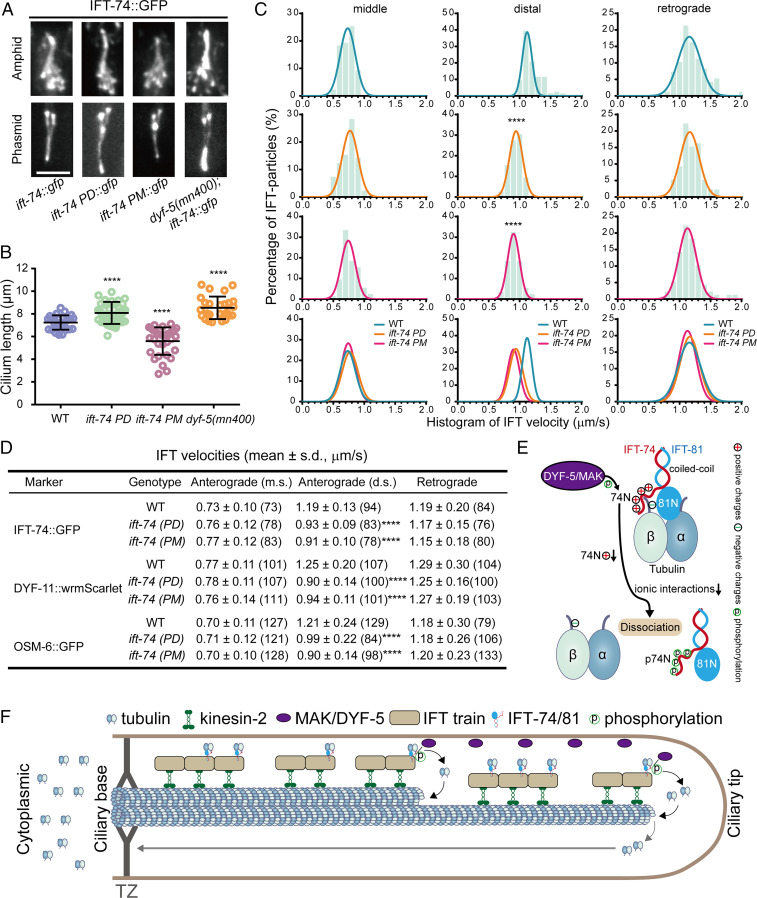
Next, we wondered whether DYF-5–mediated IFT-74N phosphorylation regulates ciliogenesis in vivo. Using CRISPR-Cas9–based genome editing methods, we constructed ift-74 phospho-mimic (ift-74 PM) and phospho-dead (ift-74 PD) C. elegans strains starting from the WT (N2) genetic background or ift-74::gfp(cas499) knock-in animals, respectively (SI Appendix, Fig. S1 and Table S1). We compared their cilia morphology with WT or dyf-5(mn400) null allele. Consistent with previous studies (8, 22), dyf-5(mn400) exhibited elongated cilia with a severe accumulation of IFT particles at the ciliary tip (Fig. 4 A and B). Compared to WT animals, ift-74 PD or ift-74 PM mutant animals developed longer or shorter cilia (Fig. 4 A and B). Tracking the motility of IFT-74::GFP in the respective mutant strains revealed an impact on IFT velocities: Although the speeds at the middle ciliary segments or the retrograde direction remain unchanged, the anterograde IFT velocities along the distal segments were markedly reduced in the ift-74 PD and PM mutants (Fig. 4 C and D). By introducing another two IFT-B subunits, OSM-6 and DYF-11, into ift-74 PD or ift-74 PM mutant animals, we validated defects of ciliary length and IFT (Fig. 4D and SI Appendix, Figs. S2 and S3). These data suggest that the phosphorylation of IFT-74 by DYF-5 impairs cilium formation and IFT in C. elegans neurons.
Fig. 4.
Phosphorylation of IFT-74 controls cilium length via regulating intraciliary tubulin transport. (A) Amphid (Top) and phasmid (Bottom) cilia morphology in ift-74 PM and PD mutants in comparison with WT (N2) and dyf-5 null mutant (mn400) animals. Cilia were imaged with GFP-tagged WT or mutated IFT-74; IFT-74 PD::GFP and IFT-74 PM::GFP were tracked, and the IFT-74::GFP was tracked in the WT animals or dyf-5(mn400) null mutant worms. (Scale bar: 5 μm.) (B) Cilium length in WT, dyf-5(mn400), and ift-74 PM/PD animals (mean ± SD; n = 30; ****P < 0.0001). IFT-74::GFP was used as the cilium marker. (C) Histogram of IFT velocities in WT and ift-74 mutant animals. (Left) Anterograde IFT along the middle segments. (Middle) Anterograde IFT along the distal segments. (Right) Retrograde IFT. IFT-74::GFP was used as the IFT marker for the measurement. Each plot was fit by a Gaussian distribution. Comparisons were performed between the WT and mutants. ****P < 0.0001. (D) Statistics of IFT velocities of WT and mutant animals measured independently using IFT-74::GFP, DYF-11::wrmScarlet, and OSM-6::GFP as ciliary IFT markers. m.s.: middle segment; d.s.: distal segment; ****P < 0.0001. The n values are given in the parentheses. PM: phospho-mimic mutant; PD: phospho-dead mutant. (E) Model representation of DYF-5/MAK–induced IFT-74/81–tubulin dissociation via IFT-74N phosphorylation. (F) Model illustrating the regulation of intraciliary tubulin transport by DYF-5/MAK–mediated IFT-74 phosphorylation. IFT-74/81 binds and transports tubulin toward ciliary middle and tip, where DYF-5/MAK phosphorylates the IFT-74 tubulin-binding region to release bound tubulin for ciliary construction and maintenance. TZ: transition zone.
Discussion
Our in vitro and in vivo findings suggest a model in which the DYF-5/MAK–dependent phosphorylation of IFT-74 reduces the interaction between tubulin and IFT particle subunits, resulting in tubulin unloading, a prerequisite step for free tubulins incorporating at the ciliary tip. In addition to tubulin concentration and IFT frequency, ciliary kinase DYF-5/MAK–mediated phosphorylation of the tubulin-binding module on the IFT complex serves as a previously undocumented but critical regulatory pathway for cilium length control (Fig. 4 E and F).
The specific ciliary tip localization and the optimal activity of DYF-5/MAK are essential for cilium formation (8–10, 22), but the underlying mechanisms are unclear, partially due to the limited knowledge of its functional substrates. Previous studies have suggested that tubulin subunits assemble onto the axonemes at the tips of the middle and distal segments within worm sensory cilia. Our early work showed that the endogenous DYF-5 protein distributes from the tip of the middle ciliary segment, the distal segment to the distal tip, and that the ciliary localization requires the C terminus of the DYF-5 protein (22). This pattern of kinase localization would explain the spatial localization of tubulin unloading observed in these cilia. The significant disruption of IFT-74N–tubulin binding by DYF-5 via phosphorylating 11 sites on IFT-74N (Figs. 1–3) confers the efficient dissociation and turnover of tubulin at the ciliary tip (Fig. 4 E and F). In agreement with the dyf-5-null mutant, ift-74 PD mutants abnormally elongated their cilia (Fig. 4 A–D and SI Appendix, Figs. S2 and S3), likely resulting from the reduced tubulin unloading and continuous delivery of tubulin to the ciliary tip. By contrast, the shortened cilia in ift-74 PM mutants may reflect a reduced capacity of IFT machinery to ferry tubulin since the phosphorylation-dependent decrease in tubulin affinity for the IFT particles leads to a decrease in the delivery of tubulin subunits to the MS and DS segment tips, thereby shortening cilia, which is in line with the short cilia phenotype observed from overexpressed DYF-5/MAK localizing at the ciliary middle or base (8, 9). The dyf-5-null mutant exhibited more severe ciliary aggregations than ift-74 PD mutant, suggesting that IFT turnaround or other cargo unloading events are perturbed. In the absence of IFT-74 N-terminal region phosphorylation, tubulin affinity for the IFT rafts will remain high so that tubulin subunits will remain sequestered rather than being released for polymerization onto the axoneme tips, which will generate the shorter cilia due to the lack of tubulin for assembly. However, this is opposite to what we observed. We cannot exclude the possibility that the longer cilia might have resulted from more assembly or less disassembly of axonemal tubulin. Measurement of the ciliary axonemal assembly and turnover rate will be crucial for examining the model in the future. DYF-5–dependent IFT74N phosphorylation may be involved in IFT reorganization and kinesin handover at ciliary tips, and thus IFT-74 PD probably causes defects in kinesin-II removing, IFT handover, and bidirectional transport switching that was suggested to result in elongated cilia in dyf-5 mutants (8, 10). Hence, the longer cilia phenotype may result from the integrated effects of dyf-5 loss of function or IFT-74 PD. In the dyf-5 null allele, kinesin-II ectopically enters the distal ciliary segment, abnormally elongating cilia, suggesting that other DYF-5 substrates or mechanisms might contribute to the long cilia phenotype. The early work showed that, in dyf-5 null allele, kinesin-II abnormally entered and moved along the distal segments and that OSM-3 was dissociated from IFT particles and moved slowly in the distal ciliary domain, which indicates that DYF-5 is required for the undocking of kinesin-II at the tip of the middle segment but the docking of OSM-3 with IFT particles in the distal ciliary segments. Considering that the DYF-5 kinase must have multiple substrates, we argue that IFT-74 is one of its functional substrates and that identifying additional substrates can be essential for understanding the undocking of kinesin-II and docking of OSM-3 onto IFT particles.
Our results are consistent with the balance-point model regarding dynamic tubulin delivery via IFT and axonemal MT assembly (12–15). Considering tubulin unloading at ciliary tips can be a critical intermediate step between tubulin transport and tubulin incorporation into the axoneme, disrupting tubulin unloading affects the equilibrium of ciliary tubulin turnover, likely resulting in aberrant ciliary length. Thus, our identified DYF-5/IFT-74 pathway is responsible for cilium length control via its direct function on tubulin unloading (Fig. 4 E and F) and perhaps its indirect impact on IFT-motor handover.
The role of multiple tubulin isotypes has been a major focus during the past decades but remains unresolved (23). The C. elegans tubulin family consists of nine α-, six β-, and one γ-tubulin, among which three α-tubulins, TBA-5, TBA-6, TBA-9, and one β-tubulin, TBB-4, localize in sensory cilia (24, 25). Despite the potential functional redundancy among these isotypes, the specific tubulin group optimizes the functional properties of C. elegans sensory cilia (13, 24, 26). In particular, TBB-4 assembles into MTs at the tips of the middle and distal segments and distributes along the entire ciliary length, whereas TBA-5 concentrates in distal singlets, suggesting that distinct tubulin isotypes might be delivered differently. Moreover, postmodifications of tubulin, such as glutamylation, are involved in ciliogenesis (27–29). Thus, understanding how tubulin isotypes and modifications contribute to distinct ciliary segment formation can be fascinating questions for future studies. In addition, our verification of the DYF-5/MAK kinase substrate motif, (R)PX[S/T]*, indicates the selectivity of DYF-5/MAK phosphorylation and will aid in discovering additional DYF-5/MAK downstream substrates regulating ciliogenesis.
Materials and Methods
Constructs, Protein Expression, and Purification.
Encoding genes of C. elegans WT and mutant IFT-74 (WormBase ID: CE52310) residues 1 to 372 and WT IFT-81 (WormBase ID: CE41565) residues 1 to 387 were commercially synthesized (BGI) and cloned into pGEX-6p-1 (with an N-terminal GST tag) and pET-21a (with an N-terminal 6×His tag) vectors, respectively. Cells from a single transformed clone were incubated in lysogeny broth medium at 37 °C until the optical density at 600 nm reached 0.6 to 0.8 and then were induced using 0.5 mM isopropyl β-d-1-thiogalactopyranoside at 16 °C for 20 h. The cells were collected by centrifugation and stocked.
For purification of WT and mutant IFT-74 proteins, the cells were lysed by sonication and centrifuged at 15,000 × g for 30 min. The supernatants were loaded onto the balanced Glutathione Sepharose 4B beads (GE Healthcare) with buffer A (20 mM Tris⋅HCl, pH 8.0, 300 mM NaCl, and 5% [vol/vol] glycerol) and eluted with the supplementation of 10 mM reduced glutathione (Sigma). The eluate was subsequently purified by gel filtration chromatography using a 16/60 Superdex 200 Increased column (GE Healthcare) with buffer B (20 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 1 mM dithiothreitol). For reconstruction of the WT and mutant IFT-74/81 complex proteins (GST-IFT-74 1–372/His-IFT-81 1–387), the cells expressing each of the complex components were premixed, lysed, and centrifuged, and the supernatants were loaded on a Ni-NTA affinity column (Qiagen) with buffer A and eluted with 400 mM imidazole. The eluate was further purified by Glutathione Sepharose beads and a Superdex 200 Increased column. IFT-74/81/IFTA-2 complex was prepared as previously described (30). The purified proteins were concentrated using ultrafiltration to 5 to 20 mg/mL and then quick-frozen in liquid nitrogen and stored at −80 °C for biochemical use.
In Vitro Kinase Assay and Phos-Tag SDS-PAGE.
DYF-5 kinase was prepared as previously described (22). Each of 20 μM WT and mutant IFT-74 proteins was mixed with 0.5 μM DYF-5 kinase in Kinase Buffer (CST) supplemented with 10 μM ATP and incubated for 1 h at 30 °C. Then the samples were analyzed by Phos-tag SDS-PAGE (20). Electrophoresis was performed with 10 mM Mn2+ Phos-tag (Wako) on acrylamide gels with a constant current of 30 mA, followed by Coomassie brilliant blue (CBB) staining.
LC-MS/MS.
IFT-74 proteins treated by DYF-5 were separated by SDS-PAGE with CBB staining, and the target bands were excised from the gel and reduced, alkylated, and digested with trypsin overnight. The resulting tryptic peptides were analyzed using an UltiMate 3000 RSLCnano System (Thermo Fisher Scientific), which was directly interfaced with an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). The MS/MS data were searched against the selected database using an in-house proteome discovery searching algorithm. The phosphorylation sites were identified by the PhosphoRS algorithm and validated manually.
SEC-MALS Analysis.
MALS was performed in line with SEC by using an Alliance 2695 high-performance liquid chromatography system equipped with Dawn Heleos II 18-angle MALS detectors (Wyatt) and a 2414 Refractive Index (RI) detector (Waters). Fifty-micromolar purified IFT-74/81 samples were applied onto a Superdex 200 Increase 10/300 column and examined on the RI-MALS system under the flow rate of 0.5 mL/min. The obtained data were analyzed by the ASTRA6.1 software (Wyatt). The data were plotted using GraphPad Prism7 (GraphPad Software).
Pull-Down Assays.
Tubulin was purified from a porcine brain, and taxol-stabilized polymerized MTs were prepared using standard methods. C. reinhardtii axoneme samples were prepared as previously described (31). The WT and mutant IFT-74/81 proteins, with or without DYF-5 kinase pretreatment, were incubated with the balanced Glutathione Sepharose 4B beads for 1 h at 4 °C in a buffer containing 50 mM Pipes, pH 6.8, 150 mM KCl, 1 mM MgCl2, 1 mM EGTA, and phosphatase inhibitors. Then, the beads were washed three times in the same buffer, followed by the incubation with taxol-stabilized MTs or C. reinhardtii axoneme samples for 1 h at room temperature. The beads were washed three times to remove unbound proteins, and the bound proteins were eluted with an elution buffer consisting of 50 mM Pipes, pH 6.8, 150 mM KCl, and 10 mM reduced glutathione. The eluates were analyzed using SDS-PAGE followed by CBB staining and Western blotting. Three replicates were conducted for each group and the quantification analysis was performed using ImageJ (NIH) and GraphPad Prism7.
Immunoblotting.
The eluates from the pull-down assays were separated on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (Thermo Fisher Scientific). The membranes were blocked for 30 min at room temperature with the blocking buffer (5% bovine serum albumin [mass/vol] and 0.05% [vol/vol] Tween in Tris-buffered saline). For each specific analysis, the blocked membranes were incubated with the corresponding primary antibody (mouse monoclonal anti-GST antibody, 1/2000, Research Resource Identifier [RRID]: AB_86554; mouse monoclonal anti-α-tubulin, 1/5000, RRID: AB_521686) overnight at 4 °C followed by incubation with the anti-mouse immunoglobulin G, horseradish peroxidase–conjugated secondary antibody (1/5000, RRID: AB_330924) for 30 min at room temperature. Then, the membranes were washed and processed with SuperSignal Western Blot Enhancer reagent (Sigma) for 1 min, and the signals were detected by chemiluminescence using ChemiDoc MP Imaging System (Bio-Rad).
MT Decoration Analysis and TIRF Microscopy.
The SNAP-tagged IFT-74/81/IFTA-2 were fluorescently labeled by SNAP-Surface Alexa Fluor 647 and Alexa Fluor 488–labeled MTs were prepared, and the MT decoration assays were performed as described previously (30, 32). MT–protein mixtures were mounted onto a glass slide and images were taken using TIRF microscopy. The acquired images were further processed using ImageJ software.
Microscale Thermophoresis.
Porcine tubulin was labeled on lysine side chains using the Cy3 dye (GE Healthcare) according to the manufacturer’s instructions; 200 nM of Cy3-labeled tubulin was titrated with 0.02 to 400 μM of IFT-74/81 proteins after 5-min preincubation in a total volume of 20 µL and 20 thermophoresis measurements were recorded. Thermophoresis measurements were carried out using the NanoTemper Monolith NT.115 instrument (NanoTemper) using 50% light-emitting diode and 65% laser power with the laser on for 40 s followed by an off period of 10 s. The resulting raw data were analyzed using the NanoTemper software to obtain binding curves and dissociation constants were calculated using GraphPad Prism7.
Structural and Molecular Dynamic Analysis.
The structural coordinate of C. elegans IFT-74 was obtained from AlphaFold Protein Structure Database (ID: A0A2C9C2L6), and its N-terminal tubulin-binding region (IFT-74N, residues 1 to 132) was used for analysis. Surface atomic potentials and the free energy of folding were calculated using the PDB2PQR (33) programs, and the electrostatic distribution surface was calculated and generated using the APBS tool (RRID: SCR_008387). The PM mutant structure and structural figures were generated using the program PyMOL (RRID: SCR_000305). Molecular dynamics analysis was performed as described previously (34) using GROMACS (RRID: SCR_014565) version 4.6.7 using the OPLS-AA force field parameter set. Each system was solvated in 150 mM NaCl solvent with SPC/E water models in a cubic box. Then, neutralizing counterions were introduced, and the steepest descent energy minimization was conducted, followed by a two-step equilibration consisting of 100 ps of isochoric–isothermal equilibration and 100 ps of isothermal–isobaric equilibration. All position restraints were removed, and simulations were performed for 5 ns. Root mean square fluctuation and deviation analysis calculate the SD of the atomic positions of the specified amino acids compared to their starting position within the energy minimized and equilibrated structure. Each simulation was performed in triplicate.
Strain Maintenance.
C. elegans strains were grown and maintained as previously described (35, 36). The N2 strain was obtained from the CGC (Caenorhabditis Genetics Center). All strains were cultured on the nematode growth medium (NGM) plates seeded with Escherichia coli OP50 at 20 °C. Adult hermaphrodite worms were used in the live cell imaging experiments. C. elegans strains used in this study are listed in SI Appendix, Table S1.
Strain Construction.
To generate the ift-74 CRISPR knock-in strains, single guide (sg) sequences were designed by the CRISPR design tool (https://zlab.bio/guide-design-resources). IFT-74 phosphorylation-dead mutant strain (ift-74 PD) and IFT-74 PM mutant strain (ift-74 PM), were generated by Suny Biotech (https://www.sunybiotech.com) using CRISPR-Cas9. Mutant or knock-in strains were back-crossed three times to remove background mutations.
Live-Cell Imaging.
Young adult C. elegans hermaphrodites were anesthetized with 0.1 mmol/L levamisole in M9 buffer, mounted on 3% agar pads, and maintained at 20 °C. Our regular imaging system includes an Axio Observer Z1 microscope (Carl Zeiss) equipped with a 100×, 1.46 numerical aperture (NA) objective lens, an electron-multiplying charge-coupled device camera (iXon+ DU-897D-C00-#BV-500; Andor Technology), and the 405-nm, 488-nm, and 561-nm lines of a Sapphire CW CDRH USB Laser System (Coherent) with a spinning disk confocal scan head (CSU-X1 Spinning Disk Unit; Yokogawa Electric Corporation). Our high-resolution live imaging system includes an Olympus IX83 microscope equipped with a 150×, 1.45 NA objective lens, a Neo 5.5 sCMOS Camera (DC-152Q-C00-FI; Andor Technology), and the same spinning disk confocal modules as mentioned above. Time-lapse images were acquired by μManager (https://www.micro-manager.org) at an exposure time of 200 ms. Image stacks were z-projected using maximum projection. All the images were taken using identical settings.
Image Processing and Analysis.
Images and movies are processed as previously described (37). We used ImageJ software (https://rsbweb.nih.gov/ij/) to process images, generate kymographs, and collect the original data for quantifications of cilium length and velocity. We used a ciliary marker IFT-74::GFP to measure the length of the cilia in WT, ift-74 PD, and ift-74 PM mutant animals. Considering that ciliary markers DYF-11::wrmScarlet and OSM-6::GFP showed the identical distribution pattern with IFT-74::GFP in ift-74 PD and ift-74 PM mutant animals, all the markers can faithfully reflect the length and morphology of the cilia. To ensure the quality of images used for velocity calculation, movies only in stable focal planes that cover the whole cilium structures were used to generate kymographs. For the detailed analysis of IFT speeds, we used the same processing method as before (38).
Sequence Alignment.
Sequence alignment was performed using Clustal X2.1 (www.clustal.org/). Protein sequences were obtained from Wormbase (https://www.wormbase.org/) or UniProt (https://www.uniprot.org/). Conserved domains were identified by ESPript (RRID: SCR_006587) online tools. Sequence IDs used include CE52310 (CeIFT-74), NP_079379.2 (HsIFT-74), NP_080595.2 (MmIFT-74), and XM_001689511.2 (CrIFT-74). The full-length sequences were used to perform alignment.
Supplementary Material
Acknowledgments
We thank the staffs of the Mass Spectrometry Center and Protein Research Center at Tsinghua University for their kind assistance in biochemical and LC-MS experiments. We also thank J. Pan and C. Xie for their discussions. This work was supported by the following funding programs: the National Natural Science Foundation of China (grants 31991190, 32070706, 32021002, 31730052, 31525015, 31861143042, 31561130153, 31671444, 31671451, 31970180, 31871352, and 31900535), the National Key R & D Program of China (2017YFA0503501, 2019YFA0508401, and 2017YFA0102900), and the European Research Council grant 335623 to Z.Ö. In addition, X.J. was supported by the International Postdoctoral Exchange Fellowship Program of China Postdoc Council (Talent-Introduction Program; YJ20210107).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. W.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207134119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Mitchell D. R., Evolution of cilia. Cold Spring Harb. Perspect. Biol. 9, a028290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiter J. F., Leroux M. R., Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L., A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U.S.A. 90, 5519–5523 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbaum J. L., Witman G. B., Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Taschner M., Lorentzen E., The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol. 8, a028092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prevo B., Scholey J. M., Peterman E. J. G., Intraflagellar transport: Mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 284, 2905–2931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pazour G. J., Agrin N., Leszyk J., Witman G. B., Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burghoorn J., et al. , Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 104, 7157–7162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omori Y., et al. , Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc. Natl. Acad. Sci. U.S.A. 107, 22671–22676 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi P., Xie C., Ou G., The kinases male germ cell-associated kinase and cell cycle-related kinase regulate kinesin-2 motility in Caenorhabditis elegans neuronal cilia. Traffic 19, 522–535 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Johnson K. A., Rosenbaum J. L., Polarity of flagellar assembly in Chlamydomonas. J. Cell Biol. 119, 1605–1611 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall W. F., Rosenbaum J. L., Intraflagellar transport balances continuous turnover of outer doublet microtubules: Implications for flagellar length control. J. Cell Biol. 155, 405–414 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao L., et al. , Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat. Cell Biol. 13, 790–798 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craft J. M., Harris J. A., Hyman S., Kner P., Lechtreck K. F., Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J. Cell Biol. 208, 223–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel B. D., Ludington W. B., Marshall W. F., Intraflagellar transport particle size scales inversely with flagellar length: Revisiting the balance-point length control model. J. Cell Biol. 187, 81–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhogaraju S., et al. , Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341, 1009–1012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman S. A., Wilson N. F., Haas N. A., Lefebvre P. A., A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 13, 1145–1149 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Ozgül R. K., et al. ; European Retinal Disease Consortium, Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am. J. Hum. Genet. 89, 253–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tucker B. A., et al. , Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 108, E569–E576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T., Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5, 749–757 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Kubo T., et al. , Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J. Cell Sci. 129, 2106–2119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D., et al. , RNA editing restricts hyperactive ciliary kinases. Science 373, 984–991 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Nsamba E. T., Gupta M. L., Tubulin isotypes – functional insights from model organisms. J. Cell Sci. 135, jcs259539 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Hurd D. D., Miller R. M., Núñez L., Portman D. S., Specific alpha- and beta-tubulin isotypes optimize the functions of sensory Cilia in Caenorhabditis elegans. Genetics 185, 883–896 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurd D. D., Tubulins in C. elegans. WormBook 2018, 1–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva M., et al. , Cell-specific α-tubulin isotype regulates ciliary microtubule ultrastructure, intraflagellar transport, and extracellular vesicle biology. Curr. Biol. 27, 968–980 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Hagan R., et al. , The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr. Biol. 21, 1685–1694 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Hagan R., Barr M. M., Regulation of tubulin glutamylation plays cell-specific roles in the function and stability of sensory cilia. Worm 1, 155–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Hagan R., et al. , Glutamylation regulates transport, specializes function, and sculpts the structure of cilia. Curr. Biol. 27, 3430–3441.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohamed M. A. A., Stepp W. L., Ökten Z., Reconstitution reveals motor activation for intraflagellar transport. Nature 557, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma M., et al. , Structure of the decorated ciliary doublet microtubule. Cell 179, 909–922.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberhofer A., et al. , Myosin Va’s adaptor protein melanophilin enforces track selection on the microtubule and actin networks in vitro. Proc. Natl. Acad. Sci. U.S.A. 114, E4714–E4723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolinsky T. J., et al. , PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao R., et al. , Inhibition of histone H3-H4 chaperone pathways rescues C. elegans sterility by H2B loss. PLoS Genet. 18, e1010223 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byerly L., Scherer S., Russell R. L., The life cycle of the nematode Caenorhabditis elegans. Dev. Biol. 51, 23–33 (1976). [DOI] [PubMed] [Google Scholar]
- 37.Ou G., Blacque O. E., Snow J. J., Leroux M. R., Scholey J. M., Functional coordination of intraflagellar transport motors. Nature 436, 583–587 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Yi P., Li W. J., Dong M. Q., Ou G., Dynein-driven retrograde intraflagellar transport is triphasic in C. elegans sensory cilia. Curr. Biol. 27, 1448–1461.e7 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.