Significance
Current messenger RNA (mRNA) vaccines in the clinic were reported to induce side effects in the liver, such as reversible hepatic damages and T cell–dominant immune-mediated hepatitis, which might be caused by the undesired expression of antigens in the liver. Therefore, exploring a lymphoid-organ–specific mRNA vaccine could be a promising strategy for developing next-generation mRNA vaccines. Herein, we reported a lymph-node–targeting mRNA vaccine based on lipid nanoparticles named 113-O12B for cancer immunotherapy. The targeted delivery of the mRNA vaccine elicits robust CD8+ T cell responses, exhibiting excellent protective and therapeutic effects on B16F10 melanoma. Notably, 113-O12B can efficiently deliver both a full-length protein and a short-peptide–based, antigens-encoded mRNA, thus providing a universal platform for mRNA vaccines.
Keywords: lipid nanoparticles, lymph node-targeting mRNA delivery, mRNA vaccine, cancer immunotherapy, melanoma
Abstract
The targeted delivery of messenger RNA (mRNA) to desired organs remains a great challenge for in vivo applications of mRNA technology. For mRNA vaccines, the targeted delivery to the lymph node (LN) is predicted to reduce side effects and increase the immune response. In this study, we explored an endogenously LN-targeting lipid nanoparticle (LNP) without the modification of any active targeting ligands for developing an mRNA cancer vaccine. The LNP named 113-O12B showed increased and specific expression in the LN compared with LNP formulated with ALC-0315, a synthetic lipid used in the COVID-19 vaccine Comirnaty. The targeted delivery of mRNA to the LN increased the CD8+ T cell response to the encoded full-length ovalbumin (OVA) model antigen. As a result, the protective and therapeutic effect of the OVA-encoding mRNA vaccine on the OVA-antigen–bearing B16F10 melanoma model was also improved. Moreover, 113-O12B encapsulated with TRP-2 peptide (TRP2180–188)–encoding mRNA also exhibited excellent tumor inhibition, with the complete response of 40% in the regular B16F10 tumor model when combined with anti–programmed death-1 (PD-1) therapy, revealing broad application of 113-O12B from protein to peptide antigens. All the treated mice showed long-term immune memory, hindering the occurrence of tumor metastatic nodules in the lung in the rechallenging experiments that followed. The enhanced antitumor efficacy of the LN-targeting LNP system shows great potential as a universal platform for the next generation of mRNA vaccines.
Messenger RNA (mRNA) vaccines have achieved great success amid the pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-COV-2), attracting increasing attention to this field (1, 2). Compared with other types of vaccines, mRNA vaccines show advantages in several aspects, including rapid production, safety, and high immune response (3). mRNA vaccines only result in the transient expression of tumor antigens, therefore avoiding possible mutations caused by DNA vaccines (4). Moreover, mRNA cancer vaccines can encode various antigens, including full proteins and peptides, in a similar process, showing the flexibility in integrating all required tumor antigens together (5). Furthermore, compared with traditional inactivated pathogen or protein-based vaccines, mRNA cancer vaccines can induce stronger humoral and cellular response, leading to an improved therapeutic outcome (6). Inspired by the superiority of mRNA vaccines, industries have been expanding the applications of mRNA technology to cancer treatment. To date, more than 20 mRNA cancer vaccines have enrolled in clinical trials (7).
mRNA and delivery system are the two key factors of the mRNA vaccine. Enormous efforts have been made on optimizing both mRNA production and various delivery systems. The major limitation of mRNA is the high immunogenicity, which has been addressed and mitigated by modification of nucleic acids (8). The addition of a cap structure and polyA tail further stabilizes the mRNA and facilitates transfection. Additionally, the development of novel mRNA delivery systems, especially lipid nanoparticles (LNPs), has significantly improved the stability and transfection efficiency of mRNA in humans. The most commonly used LNPs for RNA delivery can be classified into three generations based on their properties (9, 10). The first generation is nondegradable, e.g., 1,2-dioleoyl-3-dimethyaminopropane and 1,2-dilinoleyloxy-N,N-dimethyl-3-aminopropane, showing modest transfection effect but notable in vivo toxicity (11–13). The second generation, such as 4-(dimethylamino)-butanoic acid, (10Z,13Z)-1-(9Z,12Z)-9,12-octadecadien-1-yl-10,13-nonadecadien-1-yl ester (DLin-MC3-DMA) with biodegradable ester linkers, effectively delivers small RNAs, such as small interfering RNA (siRNA) to liver, leading to high and durable knockdown of targeted serum proteins (14). The third generation, including ALC-0315 and SM-102, exhibits high transfection effects of long-chain mRNA in vivo and was used in the production of COVID-19 mRNA vaccines (15).
Although the rapid development of LNPs brings major advancements to mRNA delivery, a majority of the reported LNPs administered intravenously (IV) or intramuscularly (IM) show very strong mRNA expression in the liver. As reported in pharmacokinetics data provided by Pfizer to the European Medicines Agency (EMA), the COVID-19 mRNA vaccine (BNT162b2) administrated by IM injection distributed mainly in the liver and injected site, leading to reversible hepatic damages in animals (16). Moreover, the BNT162b2 mRNA could be reverse-transcribed intracellularly into DNA in human liver cell line (Huh7) as fast as 6 h by an endogenous reverse transcriptase, resulting in a great threat to human health (16). More importantly, the vaccination by BNT162b2 was also reported to elicit CD8+ T-cell-dominant hepatitis (17). Therefore, the targeting expression of mRNA in vivo can minimize side effects and improve efficacy, which is considered as the key point of the next-generation LNPs (18). For mRNA cancer vaccines, the targeted delivery and expression of mRNA-encoding tumor antigens in lymphoid organs are considered to be a promising strategy to improve the efficacy and reduce the side effects of mRNA vaccines (19). Though many nanosystems can deliver cargos to specific organs by introducing active-targeting ligands, there are still some limitations of their clinic applications. First, the targeting ligands increase the complexity of delivery system, hindering the rapid production of the mRNA vaccine. Second, as it is challenging to transfect immune cells in vivo, the successful mRNA delivery and transfection to lymphoid organs are still rarely reported (20, 21).
Our group has developed a series of LNPs with targeting specificity to the liver, spleen, and lung without the use of targeting ligands (22, 23). In this work, LNP 113-O12B, with lymph node (LN)-targeting specificity, was explored and applied for a therapeutic mRNA cancer vaccine. Compared with LNPs formulated with ALC-0315, a key component in the US Food and Drug Administration (FDA)-approved Comirnaty, 113-O12B showed significantly reduced mRNA expression in the liver and higher expression in LNs after subcutaneous (SC) injection. The targeted delivery of full-length ovalbumin (OVA)-encoding mRNA vaccine showed remarkably enhanced CD8+ T cell response and thereby excellent protective and therapeutic effect against OVA-transduced B16F10 tumor model. Moreover, the mRNA vaccine encoding a tumor-associated peptide antigen TRP2180–188 achieved great therapeutic effect on established B16F10 tumor models, revealing that the 113-O12B platform can be generalized to multiple antigen types. Notably, the combination with anti–programmed death-1 (PD-1) antibody further improved the complete response (CR) to these established tumor models. All the surviving mice from the therapeutic experiments resisted the rechallenging of lung metastatic model, revealing long-term antitumor immunity generated by our mRNA cancer vaccine.
Results
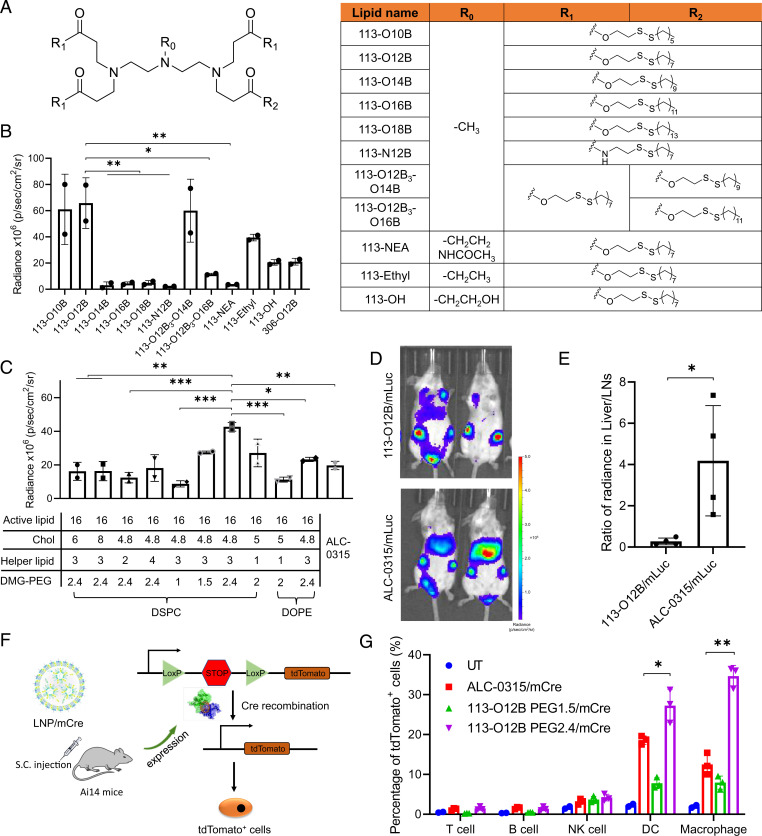
Screening and Optimization of LNPs for LN-Targeting Delivery.
The lipids used for in vivo screening were synthesized via Michael addition between amine-bearing heads and acryloyl groups containing aliphatic chains as reported in previous work (SI Appendix, Fig. S1) (24). Our group developed a library of reduction-responsive lipids, showing liver-specific mRNA delivery in previous work (22). Based on previous results, we further expanded the library based on structures of the head, tail, and linker, such as side group in head amines, linker types, tail lengths, and tail combinations (Fig. 1A). The LNPs were formulated with cholesterol (Chol), dioleoylphosphatidylcholine (DOPC), and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG) at the weight ratio of 16:4:2:1. Luciferase mRNA (mLuc) was used as a model mRNA for tracking the in vivo distribution of the expressed protein. The total intensity within LNs after SC injection of LNP/mLuc mRNA for 6 h was calculated in Fig. 1B, showing the influence of the chemical structure on mRNA expression in LNs. First, the importance of the tail length was proven by analyzing delivery efficiency of lipids containing different length tails. The shorter length tails, O10B and O12B, exhibited a higher expression in the LNs compared with longer tails that showed almost no efficacy at all. Additionally, combining tails of different lengths also proved the importance of tail length; that is, the combination of longer tails in the lipid decreased the delivery efficacy. Second, the ester linker was proven to play an important role in mRNA delivery. The replacement of the ester bond to an amide bond significantly decreased the transfection in LNs. Third, replacing the methyl groups of the amine head to hydroxyl, ethyl, or N-(1,2-ethanediyl)acetamide groups also reduced the signal. Resultantly, 113-O12B was selected as the top lipid for mRNA delivery to LNs in this library. Additionally, the pKa and size of the LNPs were further analyzed to explore the in-depth relationship between the properties of LNPs and mRNA expression (SI Appendix, Fig. S2). However, no obvious correlation was observed.
Fig. 1.
Screening and optimization of LNPs with targeting ability to LNs. (A) The chemical structure of lipids used in this study. (B) The bioluminescence within inguinal LNs after treatment with LNP/mLuc subcutaneously at the tail base for 6 h. (C) The bioluminescence within inguinal LNs after treatment by LNP/mLuc with different formulations for 6 h. (D) Representative images of bioluminescence distribution in mice treated with 113-O12B/mLuc and ALC-0315/mLuc for 6 h. (E) Ratio of radiance in liver and inguinal LNs after SC injection of mLuc for 6 h. (F) Mechanism of subcellular analysis of mRNA expression in Ai14 reporter mice. (G) Percentage of tdTomato-positive cells in different types of immunocytes after treatment with LNP/mCre subcutaneously at tail base for 48 h. The error bar around each data point is the SD. Tukey's multiple comparisons test was used to calculate the statistical significance. *P < 0.05 was considered statistically significant. **P < 0.01 and ***P < 0.001 were considered highly significant.
The 113-O12B formulation was further optimized for targeted delivery to the LNs. We selected ALC-0315 in Pfizer/BioNTech’s COVID-19 mRNA vaccine Comirnaty as a comparison. As shown in Fig. 1C, the active lipid, Chol, helper lipid, and DMG-PEG all impacted the mRNA transfection in LNs. The optimized weight ratio of the above four components was determined to be 16:4.8:3:2.4, showing much stronger signal in LNs compared with ALC-0315/mLuc. The results of the bioluminescence distribution in mice after SC injection of LNP/mLuc are shown in Fig. 1D. The obvious signal in draining LNs could be observed in both 113-O12B and ALC-0315, but ALC-0315 showed significantly higher expression in liver. In Fig. 1E, the radiance of luminescence in the liver after treatment with ALC-0315/mLuc was about four times higher than that in LNs. In contrast, the intensity in the liver decreased to only 32% of that in LNs in the 113-O12B group, confirming the superior LN-targeting ability of 113-O12B. The organ-targeting delivery of mRNA by LNPs might be related to the protein corona adsorbed on the LNPs during systemic circulation (25). However, the detailed mechanism of the protein corona and biodistribution of LNPs still remains unknown.
To explore which cell types could be transfected by LNP within LNs, the gene-engineered Ai14 mice were employed due to the activatable and stable expression of red florescence protein, tdTomato, after Cre mRNA (mCre) expression (Fig. 1F). Briefly, the gene encoding tdTomato was blocked by a stop gene between two LoxP segments. When the mRNA encoding Cre mRNA is delivered and expressed in cells, the LoxP gene is cut. Subsequently, the tdTomato gene is activated and the protein is expressed with red fluorescence, which can be detected by flow cytometry. As shown in Fig. 1G, both 113-O12B and ALC-0315 successfully delivered Cre mRNA to antigen-presenting cells (APCs), including macrophages and dendritic cells (DCs). 113-O12B/mCre showed positive mRNA expression in ∼27% DCs and ∼34% macrophages, both higher than those levels resulting from ALC-0315/mCre. The enhanced expression of mRNA in APCs is important to the following activation of adaptive immunity. In addition, we discovered that the mRNA expression in all cell types was diminished as the amount of DMG-PEG in LNP decreased, suggesting the necessity of including PEG in formulations for in vivo delivery.
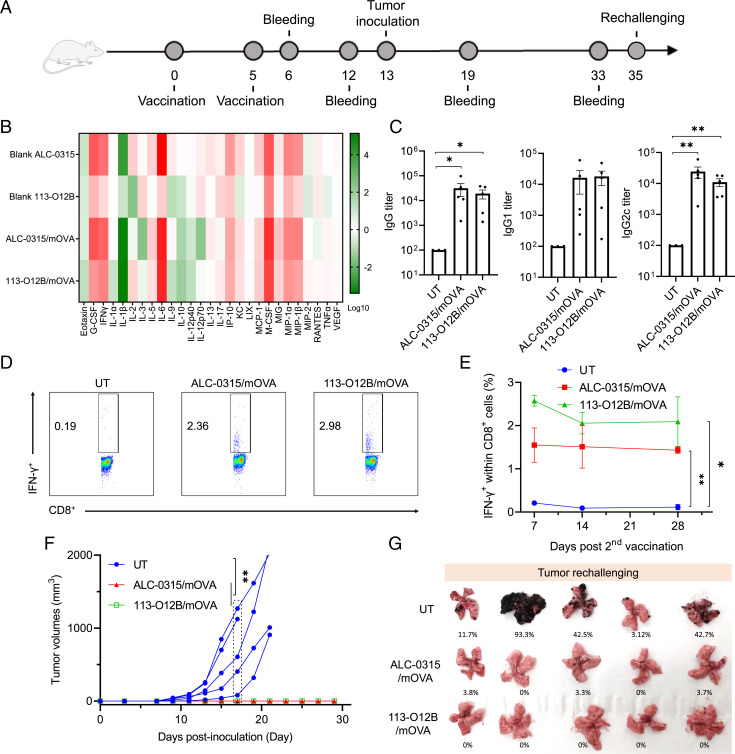
113-O12B/mOVA Elicited a Robust CD8+ T Cell Response and Protective Effect on the B16F10-OVA Tumor Model.
OVA was chosen as a model antigen, and OVA-transduced B16F10 (B16F10-OVA) cells were used as the model tumor cells. The vaccination followed the timeline in Fig. 2A, with the prime dose on day 0 and the boost dose on day 5. The levels of cytokines and chemokines after the treatment of blank LNPs and LNP/mOVA for 24 h were shown in Fig. 2B. Blank ALC-0315 showed stronger activation of innate immune system with significantly increased levels of proinflammatory cytokines (G-CSF, M-CSF, IFN-γ, and IL-6) and proinflammatory chemokines (MCP-1, MIP-1α, and MIP-1β). The formulation with OVA mRNA (mOVA) further increased the acute inflammatory response upon the expression of the foreign protein OVA. Though the activation of the innate immunity induced by blank 113-O12B was much weaker than blank ALC-0315, 113-O12B/mOVA still expressed comparable proinflammatory cytokines and chemokines compared with those of ALC-0315/mOVA, which might be due to the rapid expression of OVA in LNs. Notably, the level of IL-6, which plays an important role in the proliferation and differentiation of T cells for adaptive immunity, was significantly elevated in 113-O12B/mOVA-treated groups compared with the blank 113-O12B, which might induced by the strong expression of OVA antigen in LNs (26).
Fig. 2.
T cell response and protective effect after vaccination. (A) Timeline for vaccination and blood withdrawal. (B) Changes of cytokines and chemokines in the mice treated by blank or OVA mRNA-formulated LNPs for 24 h. (C) OVA-specific antibody titers in the mice treated by 113-O12B/mOVA and ALC-0315/mOVA on day 12. (D) Representative flow cytometry diagrams of IFN-γ-positive cells within CD3+ CD8+ T cells 7 d after second vaccination. (E) Time-dependent changes of IFN-γ-positive cells 7, 14, and 28 d after second vaccination. (F) Tumor volumes of B16F10-OVA tumor model. (G) Lungs collected 18 d after the intravenous injection of B16F10-OVA cells. UT: Untreteated. The error bar around each data point is the SD. Tukey's multiple comparisons test was used to calculate the statistical significance. *P < 0.05 was considered statistically significant. **P < 0.01 and ***P < 0.001 were considered highly significant.
The antibody levels induced by both mRNA vaccines were evaluated 14 d after the second dose (Fig. 2C). Three types of immunoglobulin G (IgG), including total IgG, IgG1, and IgG2c, were measured by enzyme-linked immunosorbent assay (ELISA). 113-O12B/mOVA showed comparable antibody response of all three antibodies compared with those induced by ALC-0315/mOVA. However, the highest antibody level was observed in the ALC-0315 group, which might result from the capture and presentation of secreted OVA protein expressed in the liver by APCs. The percentage of OVA-specific CD8+ T cells was evaluated by intracellular cytokine staining. As shown in Fig. 2 D and E, 14 d after the second vaccination, the percentage of IFN-γ+ cells within CD8+ T cells stimulated by OVA peptide (SIINFEKL) in 113-O12B group reached about 2.57%, which was significantly higher than that of ALC-0315 group (∼1.55%). Moreover, the percentage of IFN-γ+ cells remained above 2% in 113-O12B group after 4 weeks after the second vaccination (Fig. 2E), indicating the long-lasting T cell response induced by the mRNA vaccine.
The protection effect of the mRNA vaccine was evaluated in B16F10-OVA tumor model. One million tumor cells were injected SC at the right flank of the untreated or vaccinated mice on day 13. As shown in Fig. 2F, the tumor grew rapidly in the mice without vaccination, until it reached the humane endpoint within 20 d. However, no obvious tumor growth was observed in both the 113-O12B/mOVA- and ALC-0315/mOVA-vaccinated mice, indicating the superior protection effect of the mRNA vaccines. To further confirm these findings, we established a metastatic model by IV injecting 1 million cells into the control and vaccinated mice. After 18 d of the injection, the mice were killed, and the lungs were isolated for comparison. As shown in Fig. 2G, four out of five mice from the nontreated group showed obvious metastatic nodules, while zero mice in 113-O12B/mOVA-vaccinated group had apparent metastatic nodules. All these results demonstrated that the LNP/mOVA has excellent protection to the B16F10-OVA tumor model.
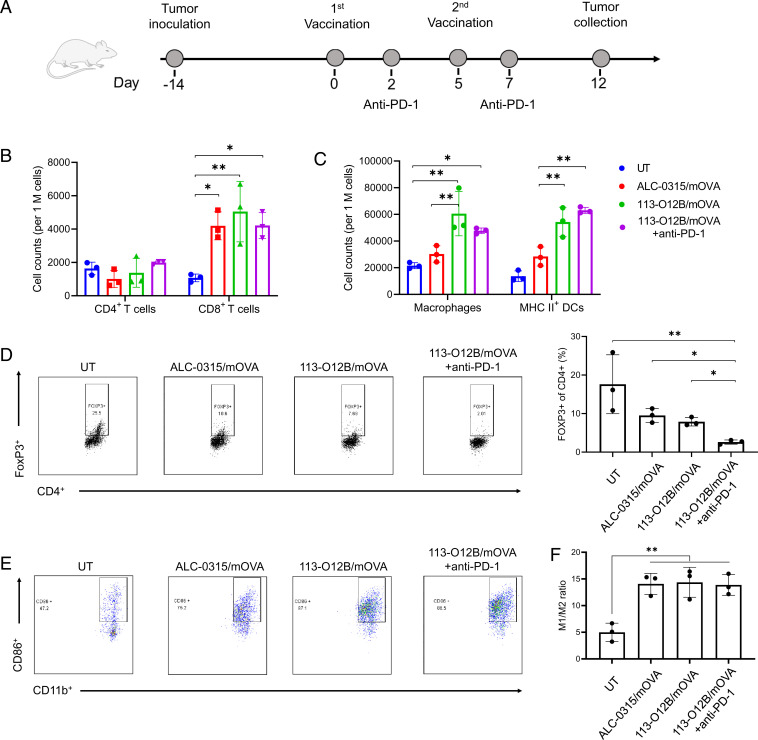
113-O12B/mOVA Shifted the Immune Cell Composition in the Established B16F10-OVA Tumor Model.
The superior protection effect of the mRNA cancer vaccine encouraged us to further evaluate the therapeutic effect on the established tumors. First, the impact of the mRNA vaccine on immune cell composition of established tumor was studied in B16F10-OVA tumor model. One million B16F10-OVA cells were inoculated SC on day −14. Two weeks later, the mice received the prime and boost vaccinations on day 0 and day 5, respectively (Fig. 3A). To inhibit immunosuppression, the check point inhibitor anti–PD-1 antibody was injected intraperitoneally on days 2 and 7. Tumors were collected on day 12, that is 1 week after the second dose, and analyzed by flow cytometry in Fig. 3 B and C. All the vaccinated mice exhibited a significantly increased no. of CD8+ T cells within tumors compared with the untreated group, while there was no significant difference in CD4+ T cells (Fig. 3B). Interestingly, 113-O12B/mOVA group showed a greater increase in infiltration of macrophages and activated DCs within the tumor compared with the ALC-0315 group, suggesting an enhanced therapeutic effect (Fig. 3C). The promoted migration of both CD8+ T cells and APCs is important to the therapeutic outcome of the mRNA vaccine. Moreover, the combination of anti–PD-1 did not result in a significant difference in the no. of T cells and APCs compared with those in the 113-O12B group.
Fig. 3.
Changes of the immune cell composition in established B16F10-OVA tumor after vaccination. (A) Timeline for tumor inoculation and vaccination. (B and C) Changes of T cells and APCs within the tumor 7 d after the second vaccination. (D) Representative flow cytometry diagrams and percentages of FoxP3+ cells within CD4+ T cells 7 d after the second vaccination. (E) Typical flow cytometry results of CD86+ cells within CD11b+ F4/80+ cells. (F) Ratio of M1 to M2 macrophages after vaccination. The error bar around each data point is the SD. Tukey's multiple comparisons test was used to calculate the statistical significance. *P < 0.05 was considered statistically significant. **P < 0.01 and ***P < 0.001 were considered highly significant.
To further understand the subcellular types of the infiltrated T cells, the CD4+ T cells were stained with FoxP3, which distinguishes regulatory T (Treg) cells and conventional T helper cells (Fig. 3D). Roughly 18% of CD4+ cells were determined to be Treg cells in the tumors of untreated mice, indicating a strong immunosuppressive environment of B16F10-OVA tumor (Fig. 3D). After the two vaccinations, both Treg cells in the 113-O12B/mOVA and ALC-0315/mOVA groups decreased to roughly 10%. Impressively, the anti–PD-1 treatment significantly reduced the percentage of Treg cells compared with other groups. A similar phenomenon was observed in many other works, indicating that the combination of anti–PD-1 and mRNA cancer vaccine is critical for overcoming tumor immunosuppression (27).
The polarization of macrophages is also important to antitumor immunity. M1-like macrophage benefits antitumor response, while M2-like macrophage suppresses adaptive immunity (28). We further evaluated the polarization of macrophages within tumors by flow cytometry. M1-like macrophages were marked as F4/80+, CD11b+, and CD86+. M2-like macrophages were marked as F4/80+, CD11b+, and CD163+. As shown in Fig. 3E, less than 50% of macrophages were M1-like macrophages in untreated groups. After two doses of the vaccination, the percentage of M1-like macrophages increased to more than 80%. The ratio of M1/M2-like macrophages increased significantly in all vaccinated mice compared with the untreated group. The driven polarization to M1-like macrophages by vaccination further indicates the generation of a strong antitumor immunity.
Therapeutic Effect of 113-O12B/mOVA on the Established B16F10-OVA Tumor Model.
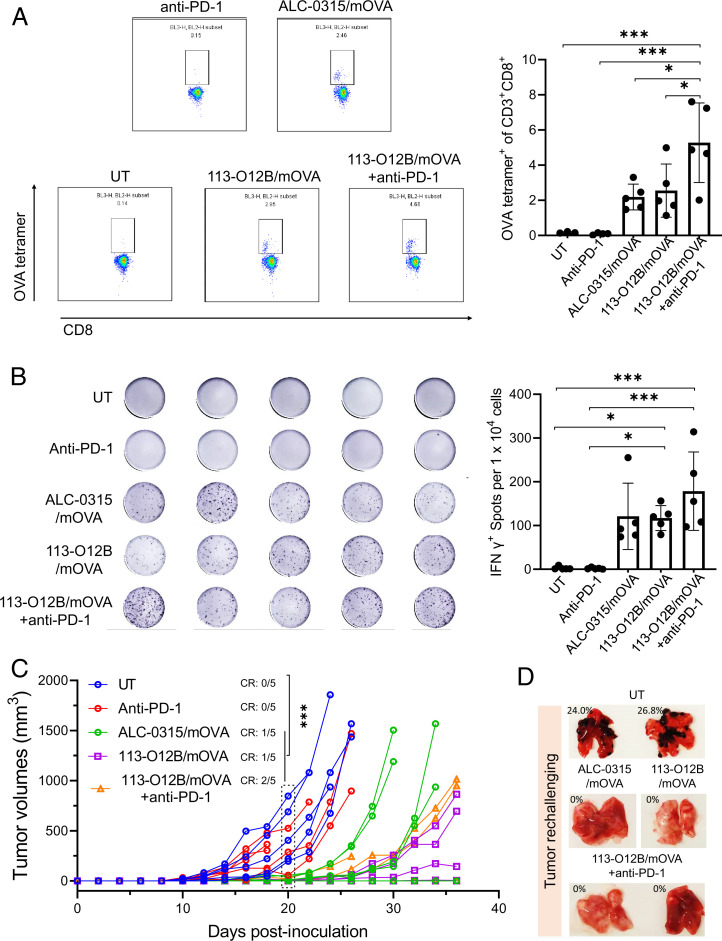
The therapeutic effect of 113-O12B-based mRNA vaccine was firstly evaluated in the B16F10-OVA tumor model. One million B16F10-OVA cells were injected SC at the right flank of C57/BL6 mice. After the tumor inoculation, three groups of mice received the prime and boost dose of the mRNA vaccine on days 5 and 12. Anti–PD-1 antibody was administrated by intraperitoneal injection with or without 113-O12B/mOVA on days 7, 11, and 15. The percentages of CD3+ CD8+ T cells bearing T cell receptors binding to H-2Kb OVA tetramer-SIINFEKL within peripheral blood mononuclear cells (PBMCs) were measured on day 19. In Fig. 4A, almost no SIINFEKL-specific CD8+ T cells could be detected, while the vaccination with ALC-0315/mOVA and 113-O12B/mOVA increased the percentage to 2.2% and 2.5%, respectively, revealing the generation of tumor-killing CD8+ T cells after vaccination. Moreover, the combination with anti–PD-1 further significantly increased the percentage of SIINFEKL-specific CD8+ T cells to 5.3%, suggesting the important role of checkpoint inhibition therapy. The levels of IFN-γ-secreting T cells within PBMCs were evaluated by an ELISpot assay in Fig. 4B. Similar to the percentage of SIINFEKL-specific CD8+ T cells, the mice without vaccination showed no response to the stimulation of SIINFEKL. However, all the vaccinated groups generated IFN-γ-secreting T cells to some extent, indicating the robust T cells response generated by the mRNA vaccine.
Fig. 4.
Therapeutic effect on B16F10-OVA tumor after vaccination by mRNA cancer vaccine. (A) Representative flow cytometry diagrams and percentages of CD3+ CD8+ T cells bearing T cell receptors binding to H-2Kb OVA tetramer-SIINFEKL within PBMCs 7 d after the second vaccination. (B) ELISpot images and spot nos. of IFN-γ-secreting T cells within PBMCs of vaccinated mice. (C) Tumor volumes of B16F10-OVA model posttreatment. (D) Lungs collected 18 d after the intravenous injection of B16F10-OVA cells. The error bar around each data point is the SD. Tukey's multiple comparisons test was used to calculate the statistical significance. *P < 0.05 was considered statistically significant. **P < 0.01 and ***P < 0.001 were considered highly significant.
The tumor volumes monitored posttreatment are demonstrated in Fig. 4C. The untreated mice exhibited rapid growth of the tumor, reaching an endpoint within 25 d (CR = 0/5). Single administration of anti–PD-1 antibody failed to inhibit tumor growth (CR = 0/5). However, in the group treated with ALC-0315/mOVA, the eradication of tumor was observed in one mouse, whereas rapid growth of tumor was still observed in two out of five mice. 113-O12B/mOVA exhibited a more-effective tumor inhibition compared with that of ALC-0315. All the mice treated by 113-O12B/mOVA survived longer than 35 d, and one of the mice showed no tumor growth during the whole treatment. The combination of anti–PD-1 did not significantly improve the overall tumor inhibition but increased the CR to 2/5, which might be caused by the individual variation in response to the anti–PD-1 therapy. To further evaluate the long-term immune memory of the mRNA vaccine, the mice that did not develop tumors after 30 d were injected with 1 million B16F10-OVA cells IV. Eighteen days after this injection, the lungs of the mice were collected and imaged; the photograph of the lungs is shown in Fig. 4D. Obvious metastatic tumor nodules were found in the lungs of the mice without the vaccination. However, all the mice that survived the therapeutic experiment showed no metastatic tumors, suggesting the generation of long-term immune response.
Therapeutic Effect of 113-O12B/TRP2180–188 on the Established B16F10 Tumor Model.
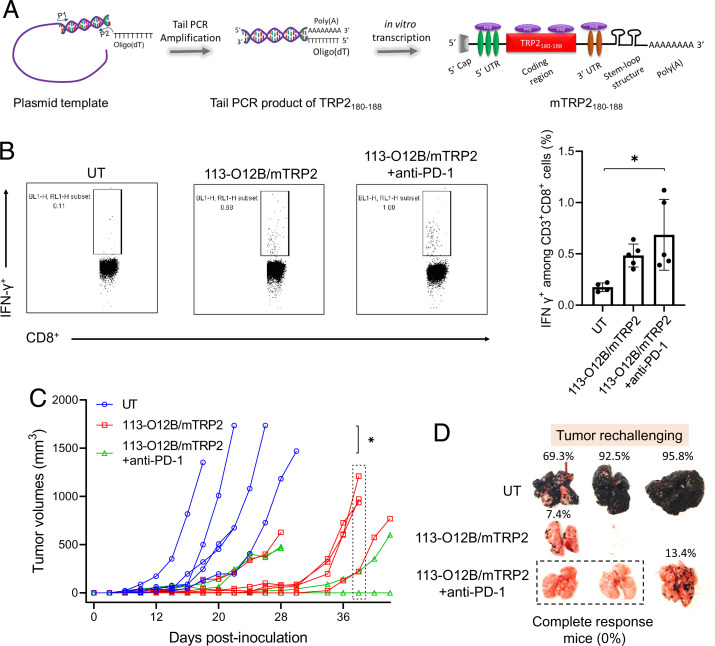
The engineered tumor B16F10-OVA expressed OVA antigens, making it easier to be recognized by the adaptive immune response generated by the mRNA vaccine encoding the OVA antigen. However, the therapeutic effect on regular B16F10 is more meaningful for future clinical applications. Tyrosinase-related protein-2 (TRP2) is a tumor-associated antigen overexpressed in murine and human melanomas with a weak immunogenicity. Therefore, induction of a strong antibody response and T cell immunity to TRP2 is necessary to generate a strong anticancer immune response in the TRP2-based cancer vaccine (29). Herein, we chose the TRP2180–188 peptide (SVYDFFVWL), a major histocompatibility complex (MHC) class I H-2Kb restricted epitope in mice, as the model peptide antigen for the design of the mRNA cancer vaccine against regular melanoma (30). N1-methylpseudouridine (N1mψ)-modified TRP2180–188 mRNA (mTRP2) was synthesized by in vitro transcription (IVT) with an ARCA cap and 120 nt poly(A) tail (Fig. 5A).
Fig. 5.
Therapeutic effect on normal B16F10 tumor model after vaccination by the mRNA cancer vaccine. (A) IVT of TRP2180–188 mRNA. (B) Representative flow cytometry diagrams and percentages of IFN-γ-positive cells within CD3+ CD8+ T cells in PBMCs 7 d after the second vaccination. (C) Tumor volumes of B16F10 model during the experiments. (D) Lungs collected 18 d after the intravenous injection of B16F10 cells. The error bar around each data point is the SD. Tukey's multiple comparisons test was used to calculate the statistical significance. *P < 0.05 was considered statistically significant. **P < 0.01 and ***P < 0.001 were considered highly significant.
The mice were inoculated with one million B16F10 cells at the right flank on day 0. Afterward, two groups of mice received two doses of the mRNA vaccine on days 5 and 12. Anti–PD-1 therapy was also dosed in one group on days 7, 11, and 15. First, the percentage of IFN-γ+ cells within CD8+ T cells in PBMCs stimulated by TRP2180–188 peptide was evaluated and is shown in Fig. 5B. The vaccination with the mRNA vaccine significantly increased the percentage of IFN-γ-secreting cells 7 d after the second vaccination. Although the administration with anti–PD-1 antibody did not result in a significant difference in the IFN-γ+ cells 7 d after the second vaccination, two mice achieved relatively higher responses than the mice treated with 113-O12B/mTRP2, suggesting the individual variation in the response to anti–PD-1 therapy (Fig. 5B). The tumor inhibition is correlated to the T cell response (Fig. 5C). The mice without treatment reached the endpoint within 28 d, while the mRNA vaccine extended the endpoint to more than 36 d. Notably, the two mice showing strongest T cell response exhibited CR in the group combined with anti–PD-1 therapy, indicating the excellent therapeutic effect of the mRNA vaccine in combination with the check point inhibitor.
The long-term antitumor immunity was also evaluated using the B16F10 metastatic model. Similarly, one million B16F10 cells were IV injected into the untreated and surviving mice on day 30. After 18 d, the lungs were collected and photographed. As shown in Fig. 5D, metastatic nodules appeared in almost all the lungs of mice without treatment, even reaching up to more than 95% of the lung area. In vaccinated mice, the growth of tumor nodules was not observed as extensively. More importantly, no obvious metastatic nodules were observed in the mice with a CR.
Discussion
Although the development of the COVID-19 mRNA vaccines has shown great success in the protection against SARS-COV-2 and the advancement of mRNA vaccines, there currently are no cancer vaccines approved for clinical use. This may be due to the weak immunogenicity of tumor antigens that requires stronger and more specific activation of the immune system or the undesired expression of tumor antigens in other nonlymphoid organs, such as liver, increases the risk of mRNA cancer vaccine. Therefore, the targeted delivery of mRNA in lymphoid organs might not only improve the antitumor immunity but also reduce undesired side effects, providing a promising strategy for the development of next-generation mRNA cancer vaccines. In this work, we developed a LN-targeting lipid, 113-O12B, which was used as a delivery vehicle in the therapeutic mRNA cancer vaccine against a melanoma mouse model. We used ALC-0315 used in Comirnaty as our chosen standard for comparison.
First, we evaluated the influence of the lipid structure on mRNA expression in the LNs. The tail length, linker bond, and amine head all impacted the delivery efficiency to the LNs. The lipids with shorter tails (≤ 12 carbons) exhibit greater efficacy compared with the lipids with longer tails (> 12 carbons). Additionally, the lipids with an ester bond linker proved to have greater efficiency than those with an amide bond linker. Furthermore, replacing the methyl group in the head amine also significantly decreased the mRNA expression in the LNs. Second, the optimized formulation of 113-O12B/mLuc was obtained by changing the weight ratio of different components and replacing the helper lipids, to resultantly exhibit a better mRNA expression in the LNs compared with ALC-0315. Notably, ALC-0315/mLuc exhibited a strong signal in the liver, while 113-O12B/mLuc delivered mRNA more specifically to the LNs. The expression of mRNA in liver was also observed in other vaccines by IM injection (23). As the undesired transfection of mRNA in the liver might induce side effects, 113-O12B showed great advantages regarding the safety for in vivo applications. 113-O12B also promoted mRNA expression in APCs within the LNs compared with ALC-0315, owing to the superior LN-targeting ability. Compared with other targeting systems with active targeting ligands, 113-O12B with endogenous LN-targeting ability is more practical for clinical applications.
Administration of blank ALC-0315 showed significantly up-regulated cytokines and chemokines related to proinflammation, indicating the highly inflammatory effect of blank ALC-0315. It was reported that LNPs were capable of inducing the inflammatory cell death (31). Then the inflammatory response induced by blank LNPs is caused by the damage-associated molecular patterns released from these cells. The exact role of the inherent immunogenicity of mRNA or LNPs in the generation of adaptive immunity is still unknown (32, 33). On one hand, the immunogenicity of the mRNA or LNPs activates the innate immune response, benefiting the subsequent activation of the adaptive immunity. On the other hand, the high immunogenicity of the mRNA or LNPs hinders the expression of mRNA-encoding antigens, thereby weakening the generation of the adaptive immunity. The LNPs encapsulating OVA mRNA (LNP/mOVA) showed increased secretion of proinflammatory factors, indicating the successful activation of the innate and adaptive immunity after the expression of the antigen. The vaccination with both 113-O12B/mOVA and ALC-0315/mOVA showed a strong antibody response. Moreover, 113-O12B/mOVA elicits stronger CD8+ T cell response compared with ALC-0315/mOVA and is still maintained at a high level 4 weeks after the second vaccination. The vaccinations with 113-O12B/mOVA and ALC-0315/mOVA both exhibit a full protection effect from the B16F10-OVA tumor over 40 d, confirming the generation of a strong antitumor immunity.
One advantage of LN-targeting 113-O12B/mOVA is the shift of the immune cell composition, which is confirmed by the up-regulated infiltration of APCs compared with that of ALC-0315/mOVA. The mRNA vaccine reduces the population of Treg cells by activating the adaptive immunity. More importantly, the combination of anti–PD-1 therapy significantly decreases the percentage of Treg cells to 2.6%, suggesting the importance of the check point inhibitor. The macrophages within the tumor of the vaccinated mice also exhibited M1 polarization. All these results show that the vaccination significantly changed the immune cell composition to inflammatory types.
The therapeutic efficacy of 113-O12B was evaluated in two tumor models, including OVA-engineered B16F10-OVA and regular B16F10 tumor model. Although the vaccination by 113-O12B/mOVA and ALC-0315/mOVA both achieve similar T cell responses, the 113-O12B/mOVA elicits a prolonged survival time compared with ALC-0315/mOVA. Notably, the integration of the mRNA vaccine and anti–PD-1 antibody eradicate the tumor in two of five mice. The improved therapeutic outcome may be attributed to the activation of cytotoxicity T cells and the inhibition of Treg cells.
There are two major challenges of regular B16F10 tumor model compared with the model antigen OVA-engineered cell line, including low immunogenicity of tumor-associated antigens and down-regulation of the antigens on the tumor surface (34). TRP2 is proven to be an effective tumor-associated antigen and TRP2180–188 peptide, which is why it was chosen in this study. Different from the full protein antigen, the LN-targeting delivery of peptide antigens might be superior to that of the untargeted ones. The LN-targeted delivery of TRP2180–188 mRNA to APCs in the LNs might lead to the higher presentation of TRP2180–188 peptide on MHC class I molecules, subsequently generating more TRP2180–188-specific tumor-killing T cells. When combined with anti–PD-1 therapy, 113-O12B/mTRP2180–188 vaccine shows significant tumor inhibition with a 40% rate of CR. The long-term memory of mRNA-based vaccines is also evaluated in the metastatic tumor model. In all the protection and therapeutic experiments, metastatic nodule was not observed in all mice with CR, implying the long-term efficacy of the mRNA vaccine.
In summary, 113-O12B LNP, an LN-targeting LNP delivery system, is developed for a mRNA cancer vaccine. The 113-O12B/mRNA shows enhanced expression in APCs compared with ALC-0315/mRNA, indicating the LN-specific targeting ability. The vaccination with 113-O12B/mOVA elicits a comparable antibody response and CD8+ T cell response compared with ALC-0315/mOVA. Moreover, 113-O12B/mOVA induces greater infiltration of APCs to the tumor site, leading to improved therapeutic efficacy on the established tumor model compared with ALC-0315/mOVA. In addition to the full OVA antigen, the mRNA encoding a peptide epitope TRP2180–188 is also successfully delivered by 113-O12B, suggesting that the LNP/mRNA system may provide a universal platform for processing multiple types of tumor antigens. The vaccination with 113-O12B/mTRP2180–188 in combination with anti–PD-1 therapy significantly suppresses and even eradicates the established B16F10 tumor. Finally, all mice surviving from the therapeutic experiment show no growth of metastatic nodules, indicating that the mRNA cancer vaccine shows great promise in providing long-term antitumor efficacy.
Materials and Methods
Synthesis and Formulation of LNPs.
Lipids were synthesized by Michael addition between amine-bearing head and acryloyl group containing aliphatic chain. The head and tail were mixed in the molar ratio of 1:4.8 and reacted at 70 °C for 3 d. Then the mixture was purified by flash chromatography (CombiFlash, USA). The lipids were further characterized by electrospray ionization-mass spectrometry.
The LNPs were prepared by dropwise adding the ethanol solution containing the mixture of active lipid, Chol, helper lipid, and DMG-PEG at the defined weight ratio to 25 mM sodium acetate solution. Then the mixture was dialyzed with Slide-A-Lyzer MINI Dialysis Device (3.5K molecular weight cutoff, Thermo Scientific, USA). The LNP/mRNA was prepared by simply mixing blank LNP with mRNA at the weight ratio of 10:1 in aqueous solution.
Synthesis of TRP2180–188 mRNA.
The coding sequence for TRP2180–188 was amplified by PCR and introduced into a pMRNAxp vector (System Biosciences, USA) using primers A109/A110 (SI Appendix, Table S1). The pMRNA-TRP2180–188 plasmid was used as templates for gene polyadenylation using the Tail PCR Primer Mix (System Biosciences, USA), of which reverse primer contains 120 oligodT. The Tail PCR (50 µL) was performed in a Phusion High-Fidelity DNA Polymerase Kit following the manufacturer’s protocol (New England Biolab Inc., USA). The reaction was then applied to a PCR program: 98 °C 3 min, 98 °C 30 s, 64 °C 30 s, 72 °C 10 s, 72 °C 10 min, and 4 °C hold for 30 cycles. The PCR mixture was further treated with Proteinase K and purified with a GeneJET PCR Purification Kit (Thermo Scientific, USA). N1mψ-modified TRP2180–188 mRNA was synthesized through IVT reaction. The reaction mixture was treated with DNase I and Antarctic Phosphatase (New England Biolabs, USA) and purified using MegaClear Kit (Life Technologies, USA). N1mψ was incorporated to completely substitute the natural counterparts in TRP2180–188 mRNA synthesis.
In Vivo Expression of Luc mRNA.
BALB/c mice (4–6 wk old) were injected with LNPs containing 5 μg mRNA and 50 μg active lipid SC at the tail base. Six hours after the injection, 100 μL of luciferin, at a concentration of 15 mg/mL, was intraperitoneally injected into the mice. After 10 min, the mice were imaged using the In Vivo Imaging System (PerkinElmer).
Delivery of Cre mRNA to LNs in Ai14 Reporter Mice.
Ai14 mice were injected with LNPs/mCre containing 10 μg mRNA and 100 μg active lipid SC at tail base. Forty-eight hours after the injection, the mice were killed and inguinal LNs were collected. The cell suspensions were prepared by grinding and filtrating through a 70-μm strainer. Then 2 × 106 cells were incubated in 100 μL flow cytometry staining buffer (eBioscience) containing fluorophore-conjugated antibody of interest listed in SI Appendix, Table S2 at the recommended concentration at 4 °C for 1 h. Then the cells were kept at 4 °C for analysis after washing twice with staining buffer. Data were collected by LSR-II flow cytometer (BD Biosciences) and analyzed by FlowJo-v10. Gating information is shown in SI Appendix, Fig. S3.
ELISA for Antibody Titer.
The antibody titer was measured by indirect ELISA. The high binding ELISA plates (Greiner Bio-One, USA) were covered with 50 μL of OVA at 20 μg mL−1 in sodium carbonate solution (pH 8.0) at 4 °C overnight. The plates were then washed with PBS containing 0.5% Tween-20 and blocked by 5% bovine serum albumin solution (Sigma-Aldrich). The serum collected from immunized mice was initially diluted in 1:100. After performing a serial dilution in triplicate, the diluted serum was added into the plates for 2 h at room temperature. Then the plates were washed and incubated with horseradish-1:10,000-diluted-peroxidase-conjugated anti-IgG, IgG1, and IgG2c antibodies for 1 h. The plates were washed and incubated with 100 μL of 3,3′,5,5′-tetramethylbenzidine substrate (Sigma-Aldrich). The reaction was stopped by 0.16 M sulfuric acid solution. The optical density at 450 nm was measured by BioTex microplate reader. The endpoint titer is defined as the reciprocal of the highest dilution of a serum that gives a reading above the cutoff (four times the PBS group).
Intracellular Cytokine Staining.
A total of 2 × 106 spleen cells or PBMCs were isolated and suspended in 200 μL RPMI-1640 medium containing 10% fetal bovine serum. GolgiPlug protein transport inhibitor (BD Biosciences) was added to inhibit the intracellular trafficking of cytokines. The cells were then stimulated with respective peptides at 2 μg/mL for 6 h. Then the cells were washed by flow cytometry staining buffer (eBioscience) and then incubated with fluorescent antibodies against surface markers at 4 °C for 1 h. The cells were washed and fixed by Fixation Buffer (Biolegend) in the dark for 20 min at room temperature. The fixed cells were washed with Intracellular Staining Perm Wash Buffer (Biolegend) twice and labeled with an optimum concentration of fluorophore-conjugated antibodies of interest (e.g., APC anti–IFN-γ and APC anti-FoxP3) for 20 min in the dark at room temperature. After washing two more times, the cells were measured by Attune NxT Flow Cytometer. Data were analyzed by FlowJo-10. Gating information is shown in SI Appendix, Fig. S4.
Tumor Immune Cell Composition Experiment.
C57/BL6 mice (4–6 wk old) were inoculated with 1 × 106 B16F10-OVA cells at the right flank on day −14. On days 0 and 5, LNPs formulated containing 50 μg active lipid and 5 μg OVA mRNA were injected SC at tail base as the prime and boost vaccination. Moreover, one of the 113-O12B/mOVA-vaccinated groups was treated with anti–PD-1 antibody on days 2 and 7. Tumors were collected on day 12 and suspended into cells using 70 μm cell strainer (Corning, USA). Then 2 × 106 cells were stained with fluorophore-conjugated antibodies listed in SI Appendix, Table S2 at 4 °C for 1 h. Then the cells were washed and analyzed by Attune NxT Flow Cytometer. Gating information is shown in SI Appendix, Fig. S5.
ELISpot Assay.
One week after the second vaccination, PBMCs were isolated and suspended in 200 μL RPMI-1640 medium containing 10% fetal bovine serum. The ELISpot assay was conducted using the Mouse Interferon gamma ELISPOT Kit (ab64029, Abcam, USA). Then 2 × 104 PBMCs cells were incubated in complete RPMI-1640 medium with or without 2 μg/mL of SIINFEKL peptide at 37 °C for 12 h. Then the plates were washed and incubated with biotinylated anti–IFN-γ antibody and then streptavidin-alkaline phosphatase conjugate according to the manufacturer’s protocol. The pictures were taken, and spot nos. of each mouse were calculated automatically.
Immunization and Tumor Therapy.
To establish B16F10-OVA tumor model, 1 × 106 B16F10-OVA cells were injected SC at the right flank of C57/BL6 mice (4–6 wk old, n = 5) on day 0. The mice received two doses of vaccination at the dose equivalent to 50 μg active lipid and 5 μg OVA mRNA on days 5 and 12. Mice without any treatment are used as control group. For one group vaccinated by 113-O12B/mOVA, the mice were also treated with anti–PD-1 on days 7, 11, and 15. The length (L) and width (W) of the tumors were measured every other day. The tumor volumes (Vs) were calculated by the equation: V = L × W2/2. On day 30, the mice without obvious tumors were rechallenged with 1 × 106 B16F10-OVA cells by IV injection. A control group (n = 5) was also treated with 1 × 106 B16F10-OVA cells by IV injection. On day 48, all the mice were killed and the lungs were collected for photograph. For regular B16F10 tumor model, all the protocols were the same as those of B16F10-OVA model except for the does, which was equivalent to 30 μg active lipid and 3 μg TRP2180–188 mRNA.
Statistics.
Statistical analysis was performed using Tukey’s multiple comparisons test in GraphPad Prism version 9.0.0 (GraphPad Software). *P < 0.05 was considered statistically significant. **P < 0.01 and ***P < 0.001 were considered highly significant.
Supplementary Material
Acknowledgments
We acknowledge support from NIH Grant R01 EB027170-01. We acknowledge Jennifer M. Khirallah for reviewing the manuscript.
Footnotes
Competing interest statement: Q.X. and J.C. are inventors on a pending patent related to this work filed by Tufts University. The pending patent was licensed to Hopewell Therapeutics, of which Q.X. is a founder and interim CEO and J.C. is a consultant.
This article is a PNAS Direct Submission. D.A. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207841119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Creech C. B., Walker S. C., Samuels R. J., SARS-CoV-2 vaccines. JAMA 325, 1318–1320 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Zhao Z., et al. , New insights from chemical biology: Molecular basis of transmission, diagnosis, and therapy of SARS-CoV-2. CCS Chemistry 3, 1501–1528 (2021). [Google Scholar]
- 3.Pardi N., Hogan M. J., Weissman D., Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 65, 14–20 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Jahanafrooz Z., et al. , Comparison of DNA and mRNA vaccines against cancer. Drug Discov. Today 25, 552–560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan C., et al. , Cancer/testis antigens: From serology to mRNA cancer vaccine. Semin. Cancer Biol. 76, 218–231 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J., Chen J., Xu Q., Current developments and challenges of mRNA vaccines. Annu. Rev. Biomed. Eng. 24, 85–109 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Karikó K., et al. , Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wittrup A., Lieberman J., Knocking down disease: A progress report on siRNA therapeutics. Nat. Rev. Genet. 16, 543–552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moss K. H., Popova P., Hadrup S. R., Astakhova K., Taskova M., Lipid nanoparticles for delivery of therapeutic RNA oligonucleotides. Mol. Pharm. 16, 2265–2277 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Maier M. A., et al. , Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 21, 1570–1578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassett K. J., et al. , Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids 15, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhi D., et al. , Transfection efficiency of cationic lipids with different hydrophobic domains in gene delivery. Bioconjug. Chem. 21, 563–577 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald K., et al. , Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: A randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 383, 60–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou X., Zaks T., Langer R., Dong Y., Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldén M., et al. , Intracellular reverse transcription of Pfizer BioNTech COVID-19 mRNA vaccine BNT162b2 in vitro in human liver cell line. Curr. Issues Mol. Biol. 44, 1115–1126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boettler T., et al. , SARS-CoV-2 vaccination can elicit a CD8 T-cell dominant hepatitis. J. Hepatol. 10.1016/j.jhep.2022.03.040. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loughrey D., Dahlman J. E., Non-liver mRNA delivery. Acc. Chem. Res. 55, 13–23 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Jiang H., Wang Q., Sun X., Lymph node targeting strategies to improve vaccination efficacy. J. Control. Release 267, 47–56 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Thapa Magar K., Boafo G. F., Li X., Chen Z., He W., Liposome-based delivery of biological drugs. Chin. Chem. Lett. 33, 587–596 (2022). [Google Scholar]
- 21.Yin D., Zhang M., Chen J., Huang Y., Liang D., Shear-responsive peptide/siRNA complexes as lung-targeting gene vectors. Chin. Chem. Lett. 32, 1731–1736 (2021). [Google Scholar]
- 22.Qiu M., et al. , Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. U.S.A. 118, e2020401118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N.-N., et al. , A thermostable mRNA vaccine against COVID-19. Cell 182, 1271–1283.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M., et al. , Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 113, 2868–2873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu M., et al. , Lung-selective mRNA delivery of synthetic lipid nanoparticles for the treatment of pulmonary lymphangioleiomyomatosis. Proc. Natl. Acad. Sci. U.S.A. 119, e2116271119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dienz O., Rincon M., The effects of IL-6 on CD4 T cell responses. Clin. Immunol. 130, 27–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida K., et al. , Anti-PD-1 antibody decreases tumour-infiltrating regulatory T cells. BMC Cancer 20, 25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yunna C., Mengru H., Lei W., Weidong C., Macrophage M1/M2 polarization. Eur. J. Pharmacol. 877, 173090 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Yamano T., Kaneda Y., Huang S., Hiramatsu S. H., Hoon D. S. B., Enhancement of immunity by a DNA melanoma vaccine against TRP2 with CCL21 as an adjuvant. Mol. Ther. 13, 194–202 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Vasievich E. A., Ramishetti S., Zhang Y., Huang L., Trp2 peptide vaccine adjuvanted with (R)-DOTAP inhibits tumor growth in an advanced melanoma model. Mol. Pharm. 9, 261–268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., et al. , Multifunctional oncolytic nanoparticles deliver self-replicating IL-12 RNA to eliminate established tumors and prime systemic immunity. Nat. Can. 1, 882–893 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauffman K. J., et al. , Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 109, 78–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linares-Fernández S., Lacroix C., Exposito J.-Y., Verrier B., Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 26, 311–323 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Majzner R. G., Mackall C. L., Tumor antigen escape from CAR T-cell therapy. Cancer Discov. 8, 1219–1226 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.