Abstract
The por genes of the predominant serovars of Neisseria gonorrhoeae circulating in a high-frequency transmitter core group located in Nairobi, Kenya, were examined for nucleotide sequence polymorphism. The level of por gene diversity did not differ significantly between core group-derived gonococcal strains and gonococcal strains originating elsewhere. However, por mosaicism appeared to be more frequent among core group-derived strains, suggesting that recombination of different por sequences may be a important strategy by which N. gonorrhoeae generates por gene diversity within core group populations. Despite extensive sequence variability, por expressed by gonococcal isolates of different geographic origin exhibited conserved patterns of nucleotide change, suggesting that diversity among por alleles may also be finite.
For many obligate human pathogens, including Neisseria gonorrhoeae, ecological success depends on the ability to evade human host defenses (38). Variation of surface-exposed molecules appears to be one common theme that has evolved among microbial pathogens for immune evasion, and N. gonorrhoeae represents one of the most thoroughly characterized bacteria in this respect (34). A number of outer membrane proteins that are involved in gonococcal pathogenesis (e.g., Pil and Opa) are extremely polymorphic, a consequence of both mutation (5, 43) and rearrangement (16, 22, 35, 42, 44, 55) between alleles of their multigene families (2, 24).
The porin protein (Por) is the major outer membrane protein of N. gonorrhoeae (28) that is encoded by an essential, single-copy gene. Although Por is antigenically diverse among strains, its expression is thought to be stable within a given strain. Thus, it exhibits allelic variability. These properties make the gonococcal porin an excellent marker for strain classification and epidemiologic studies. Immunological and biochemical data have determined that there are two structural variants of the porin protein, IA and IB, that are further subclassified into serovars based on reactivity to a panel of Por-specific monoclonal antibodies (MAbs). This hierarchy forms the basis of a serologic typing system for the gonococcus (29, 48).
Porins are expressed ubiquitously among gram-negative bacteria. The structures and functions of several gram-negative porins have been thoroughly characterized by analyses including X-ray crystallographic studies (19, 27, 53). Identical polypeptide monomers assemble into a stable trimer that forms a channel through the outer membrane of the bacterial cell. Porins characterized thus far are composed of antiparallel beta strands that fold into a cylinder-like molecule. Extensive studies suggest that the gonococcal porin conforms to this basic model (4, 32). Topological data show that the mature gonococcal porin has eight surface-exposed regions (loops) that vary in length (49). More limited sequence data indicate that variation is largely confined to these regions of the mature protein (17, 33). MAbs that bind Por surface loops mediate complement-mediated bacterial lysis in vitro (21, 30, 51, 52), suggesting that loop-specific antibodies may be protective in vivo. MAb specificity for some Por epitopes may be abolished by a single amino acid change (11). Epidemiologic data suggest that the immune response directed against Por confers partial, serovar-specific immunity against gonococcal cervicitis (40) and gonococcal salpingitis (9). Thus, there is interest in Por as a potential gonococcal vaccine candidate.
One hypothesis which would explain the extent and nature of Por heterogeneity is that it is a result of selection by protective immunological responses in the host populations. In human populations, endemic gonococcal infection is sustained by high-frequency transmitter core groups who have high rates of sex partner change, frequent sexually transmitted infections, and a high risk of transmitting infection to others (54). Since these individuals are frequently infected with different gonococcal strains, they have greater opportunity to develop protective immune responses to extant strains. If the majority of core group members develop protective immunity to an individual gonococcal strain, herd immunity for the entire population would ensue, resulting in strain extinction. Continuing survival of N. gonorrhoeae in a human host population would therefore depend on the production of new Por antigenic variants. If Por is an important target of protective immunity, diversification of the por gene would be necessary for the ecological success of N. gonorrhoeae in the face of serovar-specific herd immunity (6).
If this hypothesis is correct, Por heterogeneity may be greatest in gonococcal isolates from a core group within which these events are occurring. Since 1985, we have longitudinally studied gonococcal infections (40) and other sexually transmitted diseases (7) in a cohort of sex workers in Nairobi, Kenya. Genetic studies of variation in the chlamydial major outer membrane protein have shown that chlamydial strains from this core group (designated KEN) exhibit greater heterogeneity than isolates from non-core group populations (6). Epidemiologic studies from this population have provided some evidence for acquired immunity to gonococcal infection (infection rates decline with increasing duration of prostitution) and suggested that there is strain specific protection against homologous strain reinfection (40). To determine how surviving in a core group, where any potential host immunologic defenses are likely to be maximal, affects genetic diversity at the por locus, we examined por gene variability in gonococci isolated from this core group population.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Isolates of N. gonorrhoeae were cultured from endocervical swabs as part of a longitudinal study of 302 commercial sex workers in the lower socioeconomic district of Pumwani in Nairobi, Kenya, from July 1991 to December 1995. Swabs were streaked onto Thayer-Martin medium and incubated at 37°C and 5% CO2 for 48 h before further characterization. N. gonorrhoeae was identified by colony morphology, oxidase test, and Gram stain reaction. Isolates were subcultured once after multiple colonies were picked, stocked in skim milk containing 10% glycerol, and frozen at −70°C. At the time of analysis, gonococcal isolates were again cultured on Thayer-Martin medium and a single colony was cloned for further analysis. Isolates were serotyped by using a panel of MAbs according to the method of Knapp et al. (29).
Amplification and sequencing of por.
One representative gonococcal isolate from each of the most prevalent gonococcal serovars and four serologically untypeable serovars were selected for analysis of por. Primers Ngopor1sense and Ngopor2antisense (hereafter referred to as primers 1 and 2) were used for initial amplification of por. All primers used for amplification and sequencing were synthesized on an Oligo 1000 DNA synthesizer (Beckman Instruments Canada, Mississauga, Ontario, Canada) and purified by high-pressure liquid chromatography. The amplification reaction mixture consisted of approximately 100 ng of genomic DNA, 5 μM each primer, 250 μM dATP, dCTP, dTTP, and dGTP, 10 mM Tris-HCl (pH 8.85), 25 mM KCl, 5 mM (NH4)2SO4, 2 mM MgSO4, 2.5 U of Pwo polymerase (Boehringer-Mannheim Canada, Laval, Quebec, Ontario, Canada), and dimethyl sulfoxide-H2O (10%, vol/vol). Reactions were performed on a Geneamp 9600 (Perkin-Elmer Cetus, Mississauga, Ontario, Canada), using the following protocol: 35 cycles of 95°C for 60 s, 57°C for 60 s, and 72°C for 10 s, followed by a single cycle of 72°C for 7 min. Amplification products were resolved on a 1% agarose gel (Tris-borate-EDTA buffer [pH 8.3]) and visualized by staining the gel in an ethidium bromide solution (1 mg/ml). Bands corresponding to the known molecular weight of the por gene were excised, and the DNA was purified with the Prep-a-Gene DNA purification kit (Bio-Rad Laboratories, Mississauga, Ontario, Canada) according to the manufacturer’s instructions. Sequences of purified amplification products were determined with a dye terminator cycle sequencing kit with Amplitaq DNA polymerase FS (Applied Biosystems, Mississauga, Ontario, Canada) according to the manufacturer’s specifications. Primers used in the sequencing reactions were constructed from highly conserved regions of the por open reading frame (Table 1). Sequencing reaction mixtures contained approximately 250 ng of DNA and 1 μM primer in a 20-μl volume. Reactions were performed with the following protocol: 25 cycles of denaturation at 95°C for 15 s, primer annealing at 55°C for 10 s, and elongation at 72°C for 4 min. Unincorporated nucleotides were removed with Centri-sep spin columns (Princeton Separations, Adelphia, N.J.). Products were dried in a vacuum at room temperature, resuspended in 4 μl of formamide–50 mM EDTA (5:1, vol/vol), and resolved on an model 373A automated DNA sequencer (Applied Biosystems).
TABLE 1.
Nucleotide sequences and locations of primers used for amplification and dideoxy sequencing of por
| Primer | Sequence (5′ → 3′) | Locationa | Specificity |
|---|---|---|---|
| Ngopor1sense | CAATGAAAAAATCCCTGATTG | −2 to 19 (P9) | 1A and 1B |
| Ngopor2antisense | TTTGCAGATAGAATTTCTGG | 1000 to 1018 (P9) | 1A and 1B |
| Ngor3sense | CTGATTGCCCTGACTTTGGCAG | 13 to 34 (P9) | 1A and 1B |
| Ngopor4antisense | AGAAGTGCGTTTGGAGAAGTCG | 933 to 954 (P9) | 1A and 1B |
| Ngopor5sense | CAAGAAGACCTCGGCAAGG | 194 to 212 (P9) | 1A and 1B |
| Ngopor6antisense | TCGTATTCCGCACCGACAACCACT | 846 to 888 (P9) | 1A and 1B |
| Ngopor7sense | ACGCTACGATTCTCCCG | 441 to 457 (P9) | 1A and 1B |
| Ngopor8Asense | GCTTCTTCGTGCAATATGCCG | 557 to 577 (FA19) | 1A |
| Ngopor8Bsense | GCACAATACGCCGGCTTGG | 580 to 598 (P9) | 1B |
| Ngopor9antisense | TCGTTAGGCACGTATTG | 481 to 497 (FA19) | 1A |
| Ngopor9Bantisense | AGCCTGCATGGTAAGATTC | 532 to 550 (P9) | 1B |
| Ngopor10Aantisense | CTGCATGGTAAGATTCGCTG | 516 to 535 (FA19) | 1A |
| Ngopor10Bantisense | GCACGAAGAAGCCGCTG | 564 to 580 (P9) | 1B |
| Ngopor11Asense | CCGTACGCTACGATTCTTCCC | 437 to 456 (FA19) | 1A |
| Ngopor11Bsense | GCGCCGGTAGCCTGAACAGC | 323 to 342 (P9) | 1B |
Sequence alignment and computer analyses.
Additional por gene sequences were obtained from the GenBank database (Table 1). Nucleotide sequences were aligned by using the CLUSTALW algorithm (25). The maximum chi-squared test (45) was used to assess the significance of mosaic gene structures, using the MAXCHI computer program (35a). Nucleotide sequence statistics were calculated with the MEGA package of sequence analysis programs (31).
RESULTS
Molecular cloning and sequencing of por.
Gonococcal strains representing the predominant serovars circulating in the core group were selected for por gene sequence analysis. Using primers 1 and 2 (Table 1), a DNA fragment of approximately 1 kb was amplified from 33 gonococcal serovars (17 IA and 16 IB) corresponding to the correct size of published por sequence data. Sequencing primers were selected from highly conserved regions of published por sequences.
Nucleotide diversity at the por locus.
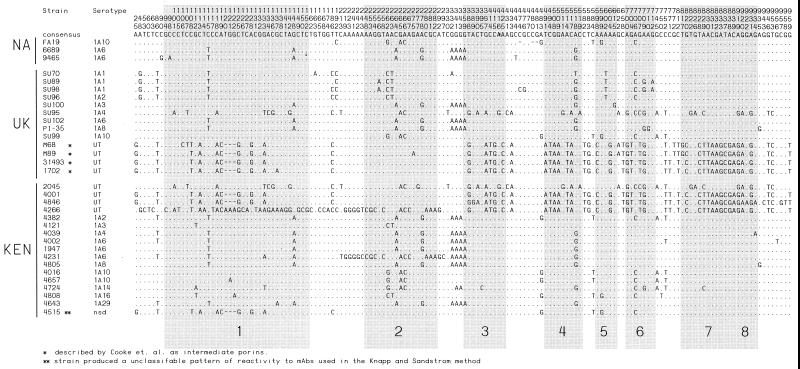
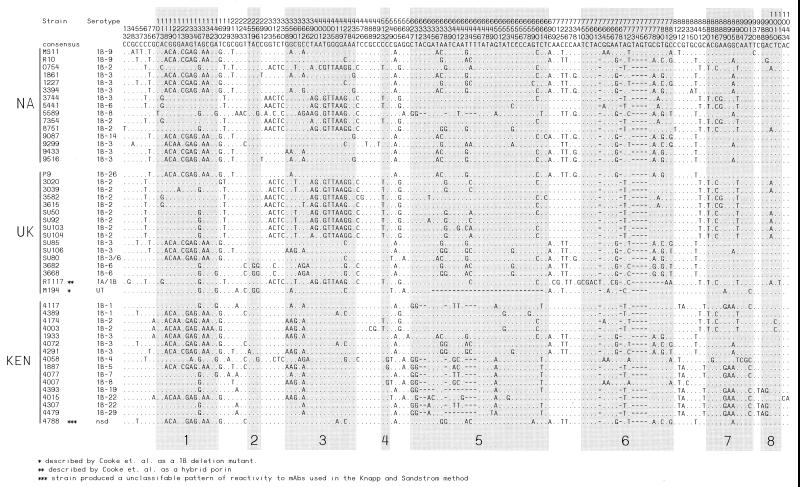
The por genes of 17 IA and 16 IB Kenyan gonococcal strains that were sequenced as part of this study were compared with 47 other por gene sequences that have been deposited in genetic databases (Table 2). Overall, 33 IA por genes and 47 IB por genes were analyzed. Among the 33 IA por genes, there were 30 unique IA por alleles. A total of 151 (15.4%) polymorphic sites were identified for the aligned IA por sequences (Fig. 1). Pairwise comparison revealed that diversity among the IA por sequences from the 17 gonococcal strains originating in the core group (KEN) was not significantly different than that of the 16 IA por from gonococcal strains originating outside the group (UK [United Kingdom] and NA [North America] groups) (4.1 and 3.5% of nucleotide sites, respectively). Variation among IB por genes was comparable to that observed for IA por. Of the 47 IB por genes, 45 distinct IB por alleles were observed. One hundred and forty-three (13.6%) polymorphic sites (Fig. 2) were identified along the aligned IB por sequences. As with IA por, the diversity of IB por from the 16 core group-derived gonococcal strains was not significantly greater than that of IB por sequences from the 37 non-core group-derived strains (3.8 and 3.6% of nucleotide sites, respectively).
TABLE 2.
Sources of por nucleotide sequence data referred to in this study
| Strain(s) | GenBank accesion no. | Reference(s) |
|---|---|---|
| MS11, FA19 | M21289, J03029 | 14, 15 |
| R10 | J03017 | 23 |
| 0754, 1861, 1227, 3394, 5441, 5589, 7354, 8751, 9087, 9299, 9433, 9516, 6689, 9465, 3744 | AF044787, AF044790, AF044789, AF044791, AF044795, AF044794, AF044786, AF044784, AF044796, AF044788, AF044792, AF044793, AF044782, AF044783, AF044785 | 26 |
| 3020, 3039, 3582, 3615, 3682, 3668, SU50, SU85, SU92, SU103, SU104, SU106, SU80 | U75631, U75632, U75633, U75634, U75636, U75635, U75640, U75642, U75643, U75637, U75638, U75639, U75641 | 17 |
| RT117, M194, M68, M89, 31493, 1702 | AF015117, AF015121, AF015119, AF015120, AF015118, AF015122, | 18 |
| P9 | X52823 | 10 |
| SU70, SU89, SU98, SU96, SU100, SU95, SU102, PI-35, SU99 | L19959, L19958, L19966, L19960, L19964, L19963, L19962, L19965, L19961 | 33 |
| 4117, 4389, 4174, 4003, 1933, 4072, 4291, 4058, 1887, 4077, 4007, 4393, 4015, 4307, 4479, 4788, 2045, 4001, 4846, 4266, 4382, 4121, 4039, 4002, 1947, 4231, 4805, 4016, 4657, 4724, 4808, 4643, 4515 | AF090794, AF090796, AF090801, AF090802, AF090800, AF090803, AF090793, AF090797, AF090804, AF090795, AF090805, AF090798, AF090806, AF090792, AF090799, AF090807, AF090822, AF090823, AF090824, AF090808, AF090811, AF090813, AF090816, AF090817, AF090818, AF090821, AF090819, AF090810, AF090809, AF090812, AF090814, AF090820, AF090815 | This study |
FIG. 1.
Locations of polymorphic sites (including gaps) along the IA por alignment. Vertical numbers indicate the position of each site in the corresponding alignments where at least one sequence differs from the consensus sequence at the top. Sites that are identical to the consensus are represented as dots. Shaded boxes indicate segments of the gene that encode surface-exposed loops as predicted by a topological model of the IA gonococcal porin (44). Sequences are grouped according to the geographic areas in which the corresponding gonococcal strains were first isolated.
FIG. 2.
Locations of polymorphic sites in the IB por nucleotide sequence alignments. The format used is the same as for the IA alignment in Fig. 1.
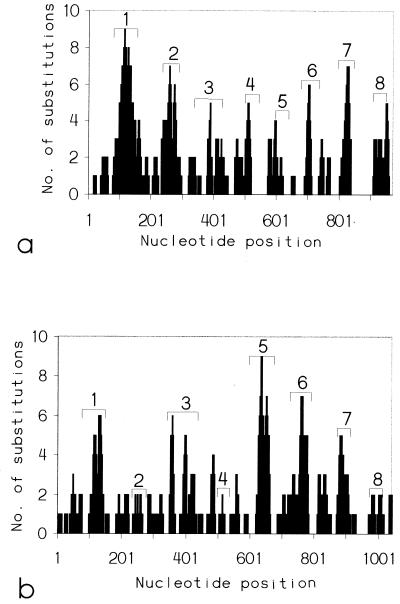
For both IA and IB, the majority of nucleotide substitutions (76 and 72%, respectively) were located within segments of the open reading frame that encode surface-exposed loops as predicted by a putative, topological model of the gonococcal porin (49) (Fig. 3).
FIG. 3.
Distribution of nucleotide substitutions (including gaps) along the IA por (a) and IB por (b) open reading frames calculated by using a sliding window of 20 nucleotides. Numbers with bracketed areas indicate the relative nucleotide positions that encode the surface-exposed loops, predicted by topological models of the IA and IB gonococcal porins (44).
Rates of synonymous and nonsynonymous substitution.
Comparison of synonymous and nonsynonymous substitution rates for a given gene can provide an estimate of selection exerted on the expressed protein. The average proportion of synonymous and nonsynonymous substitutions were calculated for different regions of por that encode surface-exposed and non-surface-exposed regions of the protein (39). For the entire coding sequence of IA and IB por, the mean rates of nonsynonymous substitution (4.07 ± 0.41 and 3.28 ± 0.35, respectively) did not differ significantly from the mean rates of synonymous substitution (2.65 ± 0.52 and 4.58 ± 0.35, respectively). For gene segments that encode the surface-exposed loops, the rates of nonsynonymous substitutions (8.23 ± 0.93 and 8.55 ± 0.98, respectively) were significantly higher (P < 0.01) than the rates of synonymous substitutions (3.10 ± 0.96 and 3.24 ± 0.1, respectively), suggesting the presence of selection for amino acid replacement in these regions. For segments of the IA por gene that encode the non-surface-exposed domains, the synonymous substitution rate (2.37 ± 0.59) was not significantly greater than the rate for nonsynonymous substitutions (1.45 ± 0.3), while for segments of the IB por gene that encode non-surface-exposed domains, the rate of synonymous substitutions (4.41 ± 0.96) was significantly higher (P < 0.001) than the rate of nonsynonymous substitutions (1.15 ± 0.28), suggesting the presence of selection against amino acid replacement.
Comparison of the rates of synonymous and nonsynonymous change in por from core group-derived (KEN) and non-core group-derived (UK and NA) gonococcal strains revealed no significant differences for the entire gene or for the different coding segments (e.g., surface exposed versus non-surface exposed) (data not shown).
Evidence for horizontal exchange between por sequences.
Examination of the polymorphic sites within IA and IB por sequences revealed a mosaic organization to several IA and IB por genes where groups of nucleotide sequence were very similar or identical in one region of the gene but highly variable elsewhere (Fig. 1 and 2). For example, por from strains 4266 (UT) and 1933 (IB) were identical in the first 295 nucleotide positions except for a single nucleotide substitution at position 75 (0.3% divergence), while in the remaining 775 nucleotide positions, the two sequences differed at 136 sites (18% divergence). Furthermore, the remaining 755 sites of the 4266 porin were identical to another por gene from the IA strain 4001. To assess the significance of mosaic gene structures, the maximum chi-squared test (45) was applied to the IA and IB por nucleotide sequences. The test involves comparison of two hypothetical, parental sequences with the potential, recombinant sequence. Crossover sites are identified as those nucleotide positions that maximize the differences between the proportion of sites occupied by identical or by different nucleotides, both before and after the putative crossover site. The significance of the putative crossover site is then assessed by comparison of observed data against T trial pairs. The observed mosaic structure is significant at the level of P < 1/T. Since several of the por sequences were identical or very similar, a subset of the most divergent por sequences (14 IA and 24 IB; >0.8% divergence) were chosen for maximum chi-squared analysis. Applying the test to the 38 por sequences (Fig. 4) identified 19 por genes as having significant (P < 10−4) mosaic structure. Of these, 16 alleles (3 IA and 13 IB) were identified as having a unique mosaic gene structure. Exchange involving members of the same subtype appeared to be more prevalent among IB por than IA por. Three por genes (strains 4266 [KEN], 4231 [KEN], and RT117 [UK]) were identified as having a significant mosaic structure that involved exchange between IA and IB por sequences.
FIG. 4.
Significant mosaic IA and IB por gene structures identified with the maximum chi-squared test (40) and approximate positions of crossover points. All crossover points were significant to P < 0.0001.
Overall, mosaicism appeared to be more prevalent among por sequences from core group-derived gonococcal strains than non-core group-derived strains. Nine of the 24 IB por sequences analyzed for mosaicism were derived from core group-derived strains, and of these, 8 IB sequences were identified as having a significant mosaic structure. Of the 15 por sequences from non-core group gonococcal strains, only 6 were identified as having a mosaic gene structure. For the IA sequences analyzed, nine were from core group-derived strains, and five of these were identified as having mosaic gene structure. None of the IA por sequences from non-core group-derived strains appeared to have a mosaic organization. Thus, overall 13 of 19 strains from the core group population were mosaics, compared to 6 of 19 strains from non-core group populations (P < 0.023, two-tailed χ2).
DISCUSSION
In human populations, endemic gonococcal infection is sustained by core groups whose members are highly sexually active and connected to many sexual networks (54). Core group members not only disproportionately transmit infection but are repeatedly exposed to a high proportion of gonococcal strains infecting the total population. Thus, these individuals have an opportunity to develop protective immune responses against most gonococcal variants circulating in a population. To determine if the extent of por heterogeneity differs in gonococcal isolates from core group and non-core group populations, we compared gonococcal por DNA sequence variation among gonococcal strains isolated from a high-frequency-transmitter core group to published sequences. The results indicate that point mutations in por are not more frequent in organisms isolated from the core group population. However, mosaic por molecules were detected at a high frequency in isolates from the core group population, indicating the recombination may be an important mechanism through which the gonococcus generates heterogeneity in por within core group populations.
Current knowledge of the extent of por gene diversity has been limited primarily to gonococcal strains that are epidemiologically linked (10, 17, 23, 26, 33). The present study is based on a relatively large number of gonococcal isolates isolated from members of a well-characterized high-frequency transmitter core group. Women in this study population have an average gonococcal infection prevalence per visit of 28% ± 19% and an incidence of 18 ± 16 infections per 100 person-months and thus a very extensive immunologic experience with the gonococcus. Smith et al. (47) have shown that relative to housekeeping genes, IA por genes have a significantly higher rate of nonsynonymous than synonymous substitutions, indicating that mutations are nonrandom and that selection for the mutations is occurring. Data from the present study extend these findings to IB por alleles. The accumulation of nonsynonymous codon changes is most dramatic in sequences that encode the surface-exposed loops of the protein, implying that some force acting in the extracellular milieu is driving the process. One hypothesis is that host immune selection is driving this process, as has been seen with Chlamydia trachomatis strains isolated from a high-frequency transmitter core group (6). However, in this study, for both IA and IB por, the level of variation among por from core group-derived gonococcal strains was not significantly greater than that of por from non-core group-derived strains. This suggests that immune selection of por point mutations is unlikely to be a major mechanism in the generation of por diversity within core groups and that other forces are selecting for mutation in the surface exposed loops of porin.
The similarity of por genes from diverse geographic regions was remarkable. In several instances, gonococcal strains of different geographic origins appear to have acquired identical por genes or segments of the same gene. The identification of common patterns of mutation in these surface-exposed segments from gonococcal strains of different geographic origin suggests that diversity of por may be finite, which may be a reflection of functional constraints that are imposed on the porin molecule. Recent studies suggest that the gonococcal porin may play a active role in gonococcal pathogenesis (1, 3, 36, 37, 41, 50). Structural constraints may therefore limit loop peptide motifs to a finite number of sequence variants that retain functionality.
Comparison of IA and IB por indicates that molecular events that have contributed to nucleotide sequence diversity are fundamentally different. Codon changes in gene segments that encode surface-exposed loops were equally frequent in IA and IB por; however, in non-surface-exposed regions, selection against amino acid replacement appears to be significantly greater for transmembrane segments of IB por than of IA por. Nucleotide deletions and insertions, which are largely absent in IA, appear to be very common in IB por. In general, these differences may be a reflection of the different biological properties associated with each porin subclass, such as resistance to killing by normal human serum (41), local versus disseminated infection (8, 13), and antibiotic resistance (12).
Isogenic gonococcal strains expressing hybrid porins have been engineered in vitro (14, 15). However, recombination of por appears to be much rarer in nature (18, 29). In this study we identified por recombinants by comparative sequence analysis, using the approach of Smith et al. (46), who have argued that a mosaic gene structure where regions of extensive nucleotide sequence diversity are interspersed with segments of highly conserved sequence indicates that localized recombination has occurred. In the present study, mosaicism appeared to be more prevalent in por from core group-derived gonococcal strains than non-core group-derived strains, suggesting that recombination of por is more frequent among core group-derived strains. Since the gonococcal chromosome carries only one copy of the por gene, coinfection of an individual with different gonococcal strains would seem a prerequisite for por recombination to occur. Since gonococcal infections are more frequent in core group than in non-core group individuals, the likelihood of coinfection is greater in core group populations. Genetic rearrangement of the chromosomal por locus with any por sequences acquired by transformation with DNA from a coinfecting gonococcal strain could then occur, perhaps resulting in the creation of unique loop combinations and immunologically distinct porins to which the infected individual is naive. Strains bearing the new porin would then be selected for by the immune response directed against the parent porins. Recombination might also help to maintain gonococcal strain fitness and virulence, through competition between coinfecting strains and recombinant strains. In an environment in which several strains are competing, the most biologically fit and virulent of the parental strains and any recombinants will be more likely to be transmitted. Because of the greater opportunity for genetic exchange within a core group, evolutionary forces are continually selecting for augmented virulence of gonococcus (20).
Although there is extensive variation in the repertoire of por alleles, it may be finite given the conserved patterns of variation observed in por from gonococcal strains of different geographic origins. These data have important implications for the development of Por-based gonococcal vaccines.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada (GR13301) and the National Institutes of Health. F. A. Plummer is a Medical Research Council of Canada Senior Scientist. J. N. Simonsen is an American Foundation for AIDS Research scholar.
REFERENCES
- 1.Bauer F J, Rudel T, Stein M, Meyer T F. Mutagenesis of the Neisseria gonorrhoeae porin reduces invasion in epithelial cells and enhances phagocyte responsiveness. Mol Microbiol. 1999;31:903–913. doi: 10.1046/j.1365-2958.1999.01230.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhat K S, Gibbs C P, Barrera O, Morrison S G, Jahnig F, Stern A, Kupsch E M, Meyer T F, Swanson J. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol Microbiol. 1991;5:1889–1901. doi: 10.1111/j.1365-2958.1991.tb00813.x. . Erratum, 6:1073–1076, 1992.) [DOI] [PubMed] [Google Scholar]
- 3.Bjerknes R, Guttormsen H K, Solberg C O, Wetzler L M. Neisserial porins inhibit human neutrophil actin polymerization, degranulation, opsonin receptor expression, and phagocytosis but prime the neutrophils to increase their oxidative burst. Infect Immun. 1995;63:160–167. doi: 10.1128/iai.63.1.160-167.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake M S, Gotschlich E C. Purification and partial characterization of the major outer membrane protein of Neisseria gonorrhoeae. Infect Immun. 1982;36:277–283. doi: 10.1128/iai.36.1.277-283.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks G F, Olinger L, Lammel C J, Bhat K S, Calvello C A, Palmer M L, Knapp J S, Stephens R S. Prevalence of gene sequences coding for hypervariable regions of Opa (protein II) in Neisseria gonorrhoeae. Mol Microbiol. 1991;5:3063–3072. doi: 10.1111/j.1365-2958.1991.tb01866.x. [DOI] [PubMed] [Google Scholar]
- 6.Brunham R, Yang C, Maclean I, Kimani J, Maitha G, Plummer F. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J Clin Investig. 1994;94:458–463. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunham R C, Kimani J, Bwayo J, Maitha G, Maclean I W, Yang C, Shen C, Roman S, Nagelkerke N J, Cheang M, Plummer F A. The epidemiology of Chlamyida trachomatis within a sexually transmitted disease core group. J Infect Dis. 1996;17:950–956. doi: 10.1093/infdis/173.4.950. [DOI] [PubMed] [Google Scholar]
- 8.Brunham R C, Plummer F, Slaney L, Rand F, DeWitt W. Correlation of auxotype and protein I type with expression of disease due to Neisseria gonorrhoeae. J Infect Dis. 1985;152:339–343. doi: 10.1093/infdis/152.2.339. [DOI] [PubMed] [Google Scholar]
- 9.Buchanan T M, Eschenbach D A, Knapp J S, Holmes K K. Gonococcal salpingitis is less likely to recur with Neisseria gonorrhoeae of the same principal outer membrane protein antigenic type. Am J Obstet Gynecol. 1980;138:978–980. doi: 10.1016/0002-9378(80)91091-1. [DOI] [PubMed] [Google Scholar]
- 10.Butt N J, Lambden P R, Heckels J E. The nucleotide sequence of the por gene from Neisseria gonorrhoeae strain P9 encoding outer membrane protein PIB. Nucleic Acids Res. 1990;18:4258. doi: 10.1093/nar/18.14.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butt N J, Virji M, Vayreda F, Lambden P R, Heckels J E. Gonococcal outer-membrane protein PIB: comparative sequence analysis and localization of epitopes which are recognized by type-specific and cross-reacting monoclonal antibodies. J Gen Microbiol. 1990;136:2165–2172. doi: 10.1099/00221287-136-11-2165. [DOI] [PubMed] [Google Scholar]
- 12.Bygdeman S M, Mardh P A, Sandstrom E G. Susceptibility of Neisseria gonorrhoeae to rifampicin and thiamphenicol: correlation with protein I antigenic determinants. Sex Transm Dis. 1984;11:366–370. doi: 10.1097/00007435-198410001-00012. [DOI] [PubMed] [Google Scholar]
- 13.Cannon J G, Buchanan T M, Sparling P F. Confirmation of association of protein I serotype of Neisseria gonorrhoeae with ability to cause disseminated infection. Infect Immun. 1983;40:816–819. doi: 10.1128/iai.40.2.816-819.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbonetti N, Simnad V, Elkins C, Sparling P F. Construction of isogenic gonococci with variable porin structure: effects on susceptibility to human serum and antibiotics. Mol Microbiol. 1990;4:1009–1018. doi: 10.1111/j.1365-2958.1990.tb00673.x. [DOI] [PubMed] [Google Scholar]
- 15.Carbonetti N H, Simnad V I, Seifert H S, So M, Sparling P F. Genetics of protein I of Neisseria gonorrhoeae: construction of hybrid porins. Proc Natl Acad Sci USA. 1988;85:6841–6845. doi: 10.1073/pnas.85.18.6841. . (Erratum, 86:1317, 1989.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connell T D, Black W J, Kawula T H, Barritt D S, Dempsey J A, Kverneland K, Jr, Stephenson A, Schepart B S, Murphy G L, Cannon J G. Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol Microbiol. 1988;2:227–236. doi: 10.1111/j.1365-2958.1988.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 17.Cooke S J, de la Paz H, La Poh C, Ison C A, Heckels J E. Variation within serovars of Neisseria gonorrhoeae detected by structural analysis of outer-membrane protein PIB and by pulsed-field gel electrophoresis. Microbiology. 1997;143:1415–1422. doi: 10.1099/00221287-143-4-1415. [DOI] [PubMed] [Google Scholar]
- 18.Cooke S J, Jolley K, Ison C A, Young H, Heckels J E. Naturally occurring isolates of Neisseria gonorrhoeae, which display anomalous serovar properties, express PIA/PIB hybrid porins, deletions in PIB or novel PIA molecules. FEMS Microbiol Lett. 1998;162:75–82. doi: 10.1111/j.1574-6968.1998.tb12981.x. [DOI] [PubMed] [Google Scholar]
- 19.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 20.Ebert D. Experimental evolution of parasites. Science. 1998;282:1432–1435. doi: 10.1126/science.282.5393.1432. [DOI] [PubMed] [Google Scholar]
- 21.Elkins C, Carbonetti N H, Varela V A, Stirewalt D, Klapper D G, Sparling P F. Antibodies to N-terminal peptides of gonococcal porin are bactericidal when gonococcal lipopolysaccharide is not sialylated. Mol Microbiol. 1992;6:2617–2628. doi: 10.1111/j.1365-2958.1992.tb01439.x. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs C P, Reimann B Y, Schultz E, Kaufmann A, Haas R, Meyer T F. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature. 1989;338:651–652. doi: 10.1038/338651a0. [DOI] [PubMed] [Google Scholar]
- 23.Gotschlich E C, Seiff M E, Blake M S, Koomey M. Porin protein of Neisseria gonorrhoeae: cloning and gene structure. Proc Natl Acad Sci USA. 1987;84:8135–8139. doi: 10.1073/pnas.84.22.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas R, Meyer T F. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44:107–115. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- 25.Higgins D G, Sharp PM. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 26.Hobbs M M, Alcorn T M, Davis R H, Fischer W, Thomas J C, Martin I, Ison C, Sparling P F, Cohen M S. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J Infect Dis. 1999;179:371–381. doi: 10.1086/314608. [DOI] [PubMed] [Google Scholar]
- 27.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 28.Johnston K H, Holmes K K, Gotschlich E C. The serological classification of Neisseria gonorrhoeae. I. Isolation of the outer membrane complex responsible for serotypic specificity. J Exp Med. 1976;143:741–758. doi: 10.1084/jem.143.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knapp J S, Tam M R, Nowinski R C, Holmes K K. Serological classification of Neisseria gonorrhoeae with use of monoclonal antibodies to gonococcal outer membrane I. J Infect Dis. 1984;150:44–48. doi: 10.1093/infdis/150.1.44. [DOI] [PubMed] [Google Scholar]
- 30.Kohl P K, Olsen D A, Buchanan T M. Monoclonal antibodies to protein I for serotyping of Neisseria gonrrhoeae: correlation of serotype with bactericidal activity. Zentbl Bakteriol Mikrobiol Hyg A. 1989;270:517–526. doi: 10.1016/s0176-6724(89)80023-9. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 32.Mauro A, Blake M, Labarca P. Voltage gating of conductance in lipid bilayers induced by porin from outer membrane of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:1071–1075. doi: 10.1073/pnas.85.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mee B J, Thomas H, Cooke S J, Lambden P R, Heckels J E. Structural comparison and epitope analysis of outer-membrane protein PIA from strains of Neisseria gonorrhoeae with differing serovar specificities. J Gen Microbiol. 1993;139:2613–2620. doi: 10.1099/00221287-139-11-2613. [DOI] [PubMed] [Google Scholar]
- 34.Meyer T F. Evasion mechanisms of pathogenic Neisseriae. Behring Inst Mitt. 1991;88:194–199. [PubMed] [Google Scholar]
- 35.Meyer T F, Mlawer N, So M. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30:45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- 35a.Molecular Biology Group, University of Sussex. 30 July 1999, revision date. MAXCHI program [Online.] http://epunix.biols.susx.ac.uk/ [30 July 1999, last date accessed.]
- 36.Mosleh I M, Huber L A, Steinlein P, Pasquali C, Gunther D, Meyer T F. Neisseria gonorrhoeae porin modulates phagosome maturation. J Biol Chem. 1998;273:35332–35338. doi: 10.1074/jbc.273.52.35332. [DOI] [PubMed] [Google Scholar]
- 37.Muller A, Gunther D, Dux F, Naumann M, Meyer T F, Rudel T. Neisserial porin (PorB) causes rapid calcium influx in target cells and induces apoptosis by the activation of cysteine proteases. EMBO J. 1999;18:339–352. doi: 10.1093/emboj/18.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy T F. Antigenic variation of surface proteins as a survival strategy for bacterial pathogens. Trends Microbiol. 1994;2:427–428. doi: 10.1016/0966-842x(94)90797-8. [DOI] [PubMed] [Google Scholar]
- 39.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 40.Plummer F A, Simonsen J N, Chubb H, Slaney L, Kimata J, Bosire M, Ndinya-Achola J O, Ngugi E N. Epidemiologic evidence for the development of serovar-specific immunity after gonococcal infection. J Clin Investig. 1989;83:1472–1476. doi: 10.1172/JCI114040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ram S, McQuillen D P, Gulati S, Elkins C, Pangburn M K, Rice P A. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae. J Exp Med. 1998;188:671–680. doi: 10.1084/jem.188.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segal E, Billyard E, So M, Storzbach S, Meyer T F. Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell. 1985;40:293–300. doi: 10.1016/0092-8674(85)90143-6. [DOI] [PubMed] [Google Scholar]
- 43.Segal E, Hagblom P, Seifert H S, So M. Antigenic variation of gonococcal pilus involves assembly of separated silent gene segments. Proc Natl Acad Sci USA. 1986;83:2177–2181. doi: 10.1073/pnas.83.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seifert H S, Ajioka R S, Marchal C, Sparling P F, So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988;336:392–395. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- 45.Smith J M. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 46.Smith J M, Dowson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 47.Smith N H, Maynard Smith J, Spratt B G. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: evidence of positive Darwinian selection. Mol Biol Evol. 1995;12:363–370. doi: 10.1093/oxfordjournals.molbev.a040212. [DOI] [PubMed] [Google Scholar]
- 48.Tam M R, Buchanan T M, Sandstrom E G, Holmes K K, Knapp J S, Siadak A W, Nowinski R C. Serological classification of Neisseria gonorrhoeae with monoclonal antibodies. Infect Immun. 1982;36:1042–1053. doi: 10.1128/iai.36.3.1042-1053.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Ley P, Virji H J E M, Hoogerhout P, Poolman J T. Topology of outer membrane porins in pathogenic Neisseria spp. Infect Immun. 1991;59:2963–2971. doi: 10.1128/iai.59.9.2963-2971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Putten J P, Duensing T D, Carlson J. Gonococcal invasion of epithelial cells driven by P.IA, a bacterial ion channel with GTP binding properties. J Exp Med. 1998;188:941–952. doi: 10.1084/jem.188.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Virji M, Fletcher J N, Zak K, Heckels J E. The potential protective effect of monoclonal antibodies to gonococcal outer membrane protein IA. J Gen Microbiol. 1987;133:2639–2646. doi: 10.1099/00221287-133-9-2639. [DOI] [PubMed] [Google Scholar]
- 52.Virji M, Zak K, Heckels J E. Monoclonal antibodies to gonococcal outer membrane protein IB: use in the investigation of the potential protective effect of antibodies directed against conserved and type-specific epitopes. J Gen Microbiol. 1986;133:1621–1629. doi: 10.1099/00221287-132-6-1621. [DOI] [PubMed] [Google Scholar]
- 53.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- 54.Yorke J A, Hethcote H W, Nold A. Dynamics and control of the transmission of gonorrhea. Sex Transm Dis. 1978;5:51–56. doi: 10.1097/00007435-197804000-00003. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q Y, DeRyckere D, Lauer P, Koomey M. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc Natl Acad Sci USA. 1992;89:5366–5370. doi: 10.1073/pnas.89.12.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]