Abstract
Screening of derivatives of Rhizobium etli KIM5s randomly mutagenized with mTn5SSgusA30 resulted in the identification of strain KIM-G1. Its rough colony appearance, flocculation in liquid culture, and Ndv− Fix− phenotype were indicative of a lipopolysaccharide (LPS) defect. Electrophoretic analysis of cell-associated polysaccharides showed that KIM-G1 produces only rough LPS. Composition analysis of purified LPS oligosaccharides from KIM-G1 indicated that it produces an intact LPS core trisaccharide (α-d-GalA-1→4[α-d-GalA-1→5]-Kdo) and tetrasaccharide (α-d-Gal-1→6[α-d-GalA-1→4]-α-d-Man-1→5Kdo), strongly suggesting that the transposon insertion disrupted a locus involved in O-antigen biosynthesis. Five monosaccharides (Glc, Man, GalA, 3-O-Me-6-deoxytalose, and Kdo) were identified as the components of the repeating O unit of the smooth parent strain, KIM5s. Strain KIM-G1 was complemented with a 7.2-kb DNA fragment from KIM5s that, when provided in trans on a broad-host-range vector, restored the smooth LPS and the full capacity of nodulation and fixation on its host Phaseolus vulgaris. The mTn5 insertion in KIM-G1 was located at the N terminus of a putative α-glycosyltransferase, which most likely had a polar effect on a putative β-glycosyltransferase located downstream. A third open reading frame with strong homology to sugar epimerases and dehydratases was located upstream of the insertion site. The two glycosyltransferases are strain specific, as suggested by Southern hybridization analysis, and are involved in the synthesis of the variable portion of the LPS, i.e., the O antigen. This newly identified LPS locus was mapped to a 680-kb plasmid and is linked to the lpsβ2 gene recently reported for R. etli CFN42.
Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium strains, collectively known as rhizobia (59), are capable of inducing the formation of specialized symbiotic organs called nodules on the roots or stems of particular leguminous host plants. Specific diffusible plant and bacterial metabolites (i.e., flavonoids and lipo-chitooligosaccharides, respectively) trigger the first steps of the nodulation process (20, 21). The rhizobia colonize the central nodular tissues by means of a complex infection process, leading to the formation of a highly efficient N2-fixing association (7, 60). Surface polysaccharides, including lipopolysaccharides (LPSs), are involved in the normal infection process of all rhizobial associations studied so far (28, 35). Rhizobia nodulating hosts that form determinate nodules, such as beans (Phaseolus vulgaris) and soybeans (Glycine max), require an intact LPS structure for complete nodule development and N2 fixation (29, 38). All transposon insertion mutants of Rhizobium etli, Bradyrhizobium japonicum, and B. elkanii isolated so far that are affected in LPS synthesis fail to form normal infection threads. Their growth is blocked either in the root hairs or in the underlying cortical cells. These mutants have never been observed to be released into symbiosomes (37, 55), eliciting incompletely developed nodules that do not fix N2 (Ndv− Fix− phenotypes).
LPS is a major and distinctive molecular component of the outer membranes of gram-negative bacteria, and lipid A, the hydrophobic moiety of LPS, forms much of the outer leaflet of the outer membrane. It is composed of an acylated disaccharide that is differently substituted in rhizobia and enteric bacteria. Attached to this disaccharide is a nonrepeating core oligosaccharide (core OS), the lipid A-proximal portion of which is conserved in structure among related bacterial species (52). The LPS core region is also highly conserved between R. etli and R. leguminosarum strains, and both contain two core OS components that are released by mild acid hydrolysis. These are generally referred to as the core trisaccharide (α-d-GalA-1→4[α-d-GalA-1→5]-Kdo) and core tetrasaccharide (and α-d-Gal-1→6[α-d-GalA-1→4]-α-d-Man-1→5Kdo) (4, 23), and they are easily separated by high-performance anion-exchange chromatography (HPAEC) (14). The lipid A moiety linked to the core OS constitutes the rough LPS (R-LPS), because mutant strains producing only R-LPS display a characteristically rough (less glossy) colony appearance. The R-LPS is often capped by a hydrophilic O-antigen polysaccharide (O-PS), forming a mature LPS molecule termed smooth LPS (S-LPS). The O-PS is usually the most variable component of the LPS, being strain specific in Rhizobium and Bradyrhizobium strains, although not in Sinorhizobium spp. (22, 40). The variability results primarily from differences in the sugar composition of the repeating units, as is the case for the members of the family Enterobacteriaceae (65). Other variable factors include the repeat unit size, the linkages between the monosaccharide components, and the presence of noncarbohydrate substituents.
The genetics of LPS biosynthesis in rhizobia has been only partially investigated, in contrast to the abundant information on pathogenic Enterobacteriaceae. In these and many other bacteria, the genes involved in O-chain and core biosynthesis are clustered on the chromosome, forming the wb* (formerly rfb) and wa* (formerly rfa) regions, respectively (40, 42). This is not the case for R. etli and R. leguminosarum strains; at least three major genetic regions encoding enzymes involved in LPS biosynthesis have been identified in R. etli CFN42T (=CE3) (38). Two of these lps regions, termed α and γ regions, are located at distinct loci on the chromosome (15, 16, 38), and a third region, called β-lps, is plasmid borne (8, 16, 38). The large α-lps region is involved both in O-PS and core OS biosynthesis (12, 15), whereas the β-lps region encodes enzymes thought to be involved in core OS synthesis (15, 24). Poole et al. (39) and Allaway et al. (1) reported on the presence of a distinct locus in Rhizobium leguminosarum bv. viciae involved in LPS core OS synthesis that is not complemented by cosmid DNA from any of the α, β, γ regions of R. etli CFN42; it was, however, complemented with cosmid gene libraries derived from R. leguminosarum bv. viciae and R. leguminosarum bv. phaseoli. This R. leguminosarum-specific LPS locus was named δ-lps. Currently no sequence or enzymatic data are available for Rhizobium spp. functions involved in O-PS synthesis. Here we report on a novel plasmid-borne and strain-specific LPS locus of R. etli KIM5s involved in the synthesis of the variable O-PS of the LPS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains used in this work are listed in Table 1. Rhizobial strains were grown in PY (36) at 28°C or on the minimal medium (MM) described by Kingsley and Bohlool (30). Escherichia coli strains were grown in antibiotic medium no. 3 (PA; Oxoid) at 37°C. Antibiotics were added at the following concentrations: ampicillin (Ap), 75 μg ml−1; kanamycin (Km), 50 and 75 μg ml−1 for E. coli and Rhizobium strains, respectively; spectinomycin (Sp) and streptomycin (Sm), 50 and 75 μg ml−1, respectively, for rhizobial strains and 50 μg ml−1 for E. coli strains. Plasmids used in this work are listed in Table 2.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype or host/origin | Source or referencea |
|---|---|---|
| Escherichia coli | ||
| DH5α | recA1 ΔlacU169 φ80dlacZΔM15 | Stratagene |
| S17-1 | thi pro hsdR hsdM+ recA, RP4 integrated in the chromosome, 2-Tc::Mu-Km::Tn7(Tpr Smr) | 53 |
| S17-1 λ-pir | λ-pir lysogen of S17-1 | 19 |
| GM2163 | F− ara-14 leuB6 thi-1 fhuA31 lacY1 tsx-78 galK2 galT22 supE44 hisG4 rpsL136 (Strr) xyl-5 mtl-1 dam13::Tn9(Camr) dcm-6 mcrB1 hsdR2(rK−mK+) mcrA | New England Biolabs |
| Rhizobium etli KIM5s derivatives | ||
| KIM-G1 | mTn5SSgusA30 insertion mutant, LPS− Ndv− Fix− Smr Spr | This study |
| KIM-G1/pRePV7.2CMCS-21 | KIM-G1 complemented with pRePV7.2CMCS-21, Kmr Smr Spr | This study |
| Wild-type rhizobiaa | ||
| R. etli KIM5s (Spr) | Phaseolus vulgaris/Mexico | J. Handelsman, University of Wisconsin, Madison |
| R. etli CFN42T | P. vulgaris/Mexico | E. Martínez, CFN |
| R. etli CIAT7014 | P. vulgaris/Colombia | CIAT |
| R. tropici CIAT899T | P. vulgaris/Colombia | CIAT |
| Sinorhizobium meliloti GRT3 | Medicago sativa/Spain | J. Sanjuan, CSIC |
CIAT, Centro International de Agricultura Tropical, Cali, Colombia; CIFN, Centro de Investigación sobre Fijación de Nitrógeno, Cuernavaca, morelos, Mexico; CSIC, Consejo Superior the Investigaciones Cientificas, Granada, Spain.
Classification at the species level of strains KIM5s and CIAT7014 was performed by PCR-restriction fragment length polymorphism analysis of rrs and rrl genes as described elsewhere (61a).
TABLE 2.
Constructs used in this study
| Construct | Relevant features | Source or reference |
|---|---|---|
| pBluescript II SK+ | Standard cloning and sequencing vector; Apr, lacZ+, fl(+) ori, ColE1 ori; high copy number; 2,964 bp | Stratagene |
| pBBR1MCS-2 | Mobilizable, replicative broad-host-range vector; Kmr; 5,144 bp | 31 |
| pAM130 | Plasmid used for mTn5gusA transposon mutagenesis. Sm/Sp, Ap; mTn5SSgusA30 (R. etli nifH-gusA-trpA ter translational fusion in pUT/mini-Tn5 Sm/Sp | 66 |
| pCAM140 | Promoterless mTn5gusA transposon for promoter probing mutagenesis; Ap; Sm/Sp; loaded on pUT | 66 |
| pPVgusAESK | Promoterless gusA gene from pCAM140 cloned as an EcoRI fragment into pSK | This study |
| pRePV-KG1 | mTn5 insertion of strain KIM-G1 cloned as a 8.5-kb ClaI fragment in pSK | This study |
| pRePV1 | pRePV-KG1 EcoRI subclone in pSK containing the I border of the mTn5 and 3.8 kb of flanking genomic DNA | This study |
| pRePV2 | pRePV1 XbaI subclone containing 1.3 kb of flanking genomic DNA linked to the I border of the mTn5 | This study |
| pRePV7.2CSK | 7.2-kb KIM5s wild-type DNA fragment cloned in pSK, spanning the insertion site of the mTn5 | This study |
| pRePV7.2CMCS-21 | 7.2-kb insert of pRePV7.2CSK subcloned in broad-host-range vector pBBRMCS-2; complements strain KIM-G1, ORF2, -3, and -44 being in opposite transcriptional orientation to the lacZ promoter of the vector | This study |
Transposon mutagenesis and conjugal transfer of plasmids.
Derivatives of R. etli KIM5s carrying mTn5SSgusA30 insertions were obtained after biparental matings using E. coli S17-1 λ-pir (19) transformed with pCAM130 (66) as the donor strain. Matings were carried out on nitrocellulose filters for 16 h on PY plates at 30°C. Selection of mTn5 insertion mutants was achieved by plating appropriate dilutions on MM containing Sp and Sm. The conjugal transfer of pRePV7.2CMCS-21 into strain KIM-G1 was performed as described above, using E. coli S17-1 (53) as the donor strain, selecting the transconjugants on MM containing Km and Sm.
Cloning of the mTn5 insertion of strain KIM-G1.
Genomic DNA of KIM-G1 was extracted from broth cultures in late exponential phase by using a standard cetyltrimethylammonium bromide protocol (3). Purified DNA was digested with ClaI and XhoI and then vacuum transferred onto nylon membranes by the method of Southern (54). The Southern blots were probed with a gusA-specific, digoxigenin (DIG)-labelled PCR probe derived from pPVgusAESK, using the T3/T7 primers and a DIG-PCR labelling kit (Boehringer GmbH, Mannheim, Germany) as recommended by the manufacturer. Hybridization and posthybridization washings were performed under stringent conditions (68°C and 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) and signal detection was performed by using anti-DIG Fab fragments coupled to alkaline phosphatase with colorimetric detection as instructed by the manufacturer (Boehringer). To clone the hybridizing fragments, about 20 μg of genomic DNA from strain KIM-G1 was restricted with ClaI and XhoI and fractionated on a low-melting-point agarose gel (Biozym, Hess. Oldendorf, Germany), purifying the DNA from appropriate gel fractions by enzymatic digestion of the agarose with an agarase (GELase; Epicentre Technologies). The purified DNA was ligated to pBluescript SK+ with T4 ligase (USB-Amersham) and used to transform CaCl2-competent E. coli DH5α cells (49), selecting transformants resistant to Ap, Sp, and Sm.
Generation and screening of a partial genomic library of R. etli KIM5s size-fractionated DNA fragments.
Plasmids pRePV1 and pRePV2 (see Fig. 3B) were chosen to generate DIG-labelled PCR probes (KIM-P#1 and KIM-P#2, respectively), using the T3/T7 primers, as described above. Probe KIM-P#1 was used to screen Southern blots of genomic DNA from wild-type KIM5s restricted with ClaI. To clone the hybridizing fragment, about 20 μg of genomic DNA of KIM5s was restricted with ClaI and fractionated on a low-melting-point agarose gel, and the DNA of the size fraction corresponding to the hybridization signal was purified as described above. This pool of DNA fragments was ligated into dephosphorylated pBluescript SK+ and used to transform E. coli DH5α. All transformants were streaked out in an ordered manner for colony lifting, using nylon membranes as recommended by the manufacturer (Boehringer). The membranes were screened with the internal 850-bp HindIII fragment from KIM-P#1, which had been gel purified. This purification step was required to remove the flanking vector sequences. Stringent hybridization and detection were performed as described above.
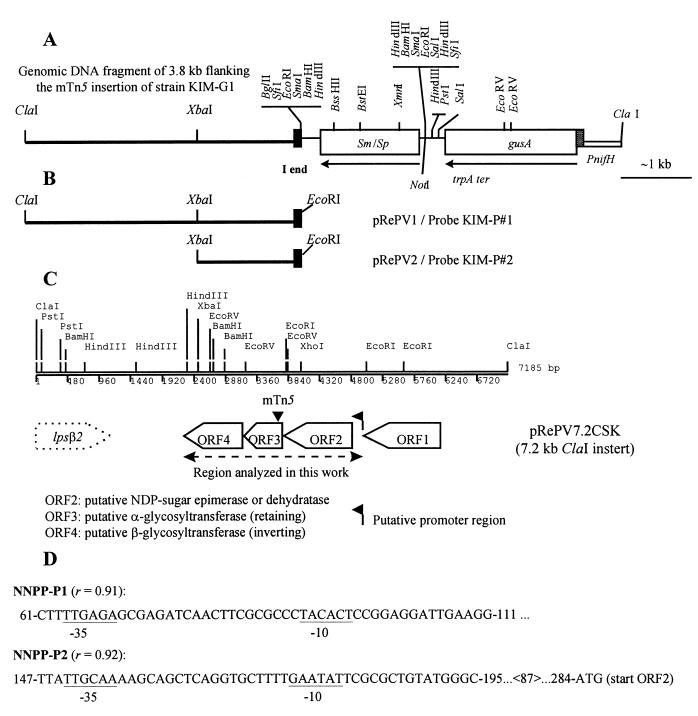
FIG. 3.
(A) Physical map of pRePV-KG1 carrying the cloned insertion of strain KIM-G1. (B) Physical maps of the subclones pRePV1 and pRePV2, carrying the I border of the mTn5 insertion along with the flanking genomic DNA fragments. These subclones were used for the generation of the DIG-labelled DNA probes KIM-P#1 and KIM-P#2. (C) Genetic and physical maps of the insert of pRePV7.2CSK, which spans over the mTn5 insertion site of KIM-G1, shown as a filled triangle. Transcriptional orientations of the ORFs are indicated by arrowheads. The approximate location of two potential promoter sequences detected by the NNPP is indicated by a flag. Only 1,682 nt of the 3′ end of the lpsβ2 gene are present on pRePV7.2CSK. (D) Nucleotide sequences of the two putative promoters predicted by the NNPP to be located in the region between ORF1 and ORF2, showing the presumed −10 and −35 sequences (underlined). The indicated residue positions correspond to those in GenBank accession no. AF127522.
Filter blot hybridization of rhizobial plasmid profiles.
Plasmid profiles of selected rhizobial strains were visualized by the method of Hynes and MacGregor (26), using 0.6% horizontal agarose gels. Plasmids were blotted onto nylon membranes by capillary transfer (49). Hybridization with probe KIM-P#1 was performed as described above, using a chemiluminescence detection system with CDP-Star (Boehringer).
DNA sequencing.
Subclones of pRePV7.2CSK were made in pBluescript SK, and both strands of the resulting plasmids were subjected to cycle sequencing in a Gene Amp 2400 thermocycler (Perkin-Elmer), using IRD-labelled T3/T7 primers (MWG-Biotech, Ebersberg, Germany) and a Thermosequenase fluorescent labelled primer cycle sequencing kit with 7-deaza-dGTP (Amersham International, Braunschweig, Germany), based on the dideoxy-chain termination method of Sanger et al. (50), as instructed by of the manufacturer. The cycle sequencing products were resolved, and the sequences were automatically read by using a model 4000 Li-COR automatic DNA sequencer and the corresponding sequence reading and editing software (MWG-Biotech).
Analysis of nucleotide and amino acid sequences.
Contig assembly, sequence editing, identification of open reading frames (ORFs), and determination of potential coding sequences and deduced amino acid sequences were carried out with GeneCompar version 2.0 (Applied Maths, Kortrijk, Belgium). A remote search for potential promoter regions was performed at the Neural Network for Promoter Prediction (NNPP) server (41). Remote searches for sequence similarities were performed at the National Center for Biotechnology Information server, using the different BLAST programs (2). Multiple alignments of amino acid sequences were performed with GeneCompar. Hydrophobic cluster analysis (HCA) plots of protein sequences were obtained from the HCA server, using DRAWHCA (10).
DOC-PAGE analysis of crude LPS extracts.
Crude LPS extracts were prepared from late-log-phase bacterial cultures by extraction of cell pellets with hot phenol-water as described elsewhere (11). The extracts were fractionated by electrophoresis on 18% polyacrylamide gels (PAGE) assembled in the Bio-Rad (Richmond, Calif.) minigel casting system, using deoxycholic acid (DOC) as the detergent for dissociating hydrophobic LPS aggregates, as described by Krauss et al. (32). Gels were silver stained by the procedure of Tsai and Frasch (58). To visualize capsular polysaccharides (KPS), an alcian blue prestain (18) is required prior to silver staining (43).
HPAEC-PAD of the LPS core OS.
The extracted polysaccharides were subjected to mild acid hydrolysis in 1% acetic acid for 1 h at 105°C (12). The lipid A precipitate was removed by centrifugation, and the supernatant containing the water-soluble polysaccharides and oligosaccharides, including the core fragments and O-PS, was analyzed by HPAEC on a Carbo Pac PA-1 column (Dionex) equipped with pulsed amperometric detection (PAD), as described elsewhere (12).
Purification and analysis of LPS oligosaccharides.
The extraction and purification protocols used are presented in previous reports (43, 44). The cell pellets from 2-liter cultures were extracted with hot phenol-water. The aqueous phase was fractionated by size exclusion chromatography over Sephadex G-150 superfine (Pharmacia, Uppsala, Sweden), and the column was eluted with 0.2 M NaCl–1 mM EDTA–10 mM Tris base–0.25% deoxycholic acid (pH 9.25). The fractions (2 ml) were assayed colorimetrically for Kdo by the thiobarbituric acid (TBA) assay (63) and for uronic acid by the hydroxybiphenyl assay (5); LPS-containing fractions were positively identified by PAGE analysis. The LPS-containing fractions were then pooled, dialyzed, and freeze-dried (43), and the LPS pools were subjected to mild acid hydrolysis (1% acetic acid, 100°C, 90 min) followed by centrifugation to remove the insoluble lipid A. The oligosaccharides were separated by gel filtration on Bio-Gel P-2 (Bio-Rad), using 50 mM ammonium formate (pH 5) as an eluent. The fractions were assayed for Kdo as described above, and the pools were freeze-dried several times to remove the ammonium formate.
Glycosyl composition and linkage analyses.
Glycosyl residue compositions were determined by gas chromatography-mass spectrometry (GC-MS) analysis of the trimethylsilyl methyl glycoside derivatives (68), using a 30-m DB-1 fused silica column (J&W Scientific, Folsum, Calif.) on a 5890A GC-MSD apparatus (Hewlett-Packard, Palo Alto, Calif.). The location of endogenous O-Me groups was determined by GC-MS analysis of the alditol acetate derivatives, using a 30-m SP-2330 column (Supelco). Inositol was used as an internal standard, and retention times were compared to authentic monosaccharide standards.
Plant nodulation assays.
P. vulgaris cv. Saxa seeds (Kornhaus Cölbe, Germany) were surface sterilized and germinated as described elsewhere (61a). Inoculum was derived from exponentially growing batch cultures that were diluted in N-free LN leguminous nutrient solution (64) to reach about 5 × 106 CFU ml−1. Seedlings were dipped into this suspension and transplanted to growth pouches filled with N-free LN nutrient solution. Cultivation took place in a controlled-environment chamber (15 h of light and 9 h of darkness at 25 and 18°C, respectively, and 75% relative humidity) for 21 days.
Nucleotide sequence accession number.
The 2,709-bp sequence described in this work has been deposited at GenBank nucleotide sequence database under accession no. AF127522.
RESULTS
R. etli KIM-G1 is an R-LPS mutant.
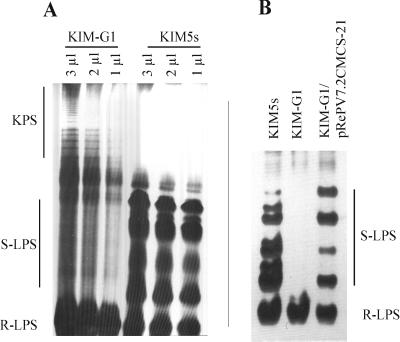
Strain KIM-G1 is a prototrophic mTn5 insertion derivative of wild-type strain KIM5s. It was identified as a putative LPS-defective mutant on PY or 20E plates (36, 64) due to its rough colony appearance and tendency to flocculate (autoagglutinate) in liquid culture. It elicited few small, round, white nodules on P. vulgaris cv. Saxa that did not reduce acetylene (data not shown). In addition, no bacteroids could be recovered from these structures, suggesting that strain KIM-G1 did not infect them. These phenotypic traits of R. etli mutants are commonly associated with defective LPS production (38). This was confirmed by comparative DOC-PAGE analysis of hot phenol-water extracts of cell-associated polysaccharides from R. etli KIM5s and its derivative KIM-G1. Figure 1A shows an alcian blue/silver-stained gel loaded with these extracts. The wild-type strain yielded a disperse band that represents R-LPS and several forms of S-LPS containing sequential degrees of polymerization (DPs) of the O antigen. KIM5s seems to display a regulated (modal) distribution of O-antigen chain lengths since the LPS molecules with two attached O units are consistently produced in lower quantity than molecules containing one, three, or four repeating units, as deduced from the relative band intensities revealed by DOC-PAGE (Fig. 1). In contrast, strain KIM-G1 displayed only the R-LPS band, which had the same electrophoretic mobility as the parental strain. Interestingly, strain KIM-G1, but not KIM5s, appears to produce high-molecular-weight KPS which is visible only with alcian blue prestaining (43), migrating slower than the S-LPS bands of the wild type and displaying a characteristic ladder pattern with multiple bands (Fig. 1A). This product was not examined further.
FIG. 1.
DOC-PAGE analysis of crude hot phenol-water extracts of cell-associated polysaccharides from wild-type R. etli KIM5s and its mTn5 insertion derivative KIM-G1. (A) Alcian blue/silver-stained gel loaded with the indicated volumes of extract, revealing the S-LPS phenotype of KIM5s and several DPs of the O unit (S-LPS). KIM-G1 exhibits only R-LPS, lacking the O-PS. KIM-G1, but not its parental strain, produces a high-molecular-weight polysaccharide that stains specifically with alcian blue, migrating as a ladder pattern. This product most probably corresponds to acidic Kdo-rich KPS. (B) Silver-stained LPSs produced by strains KIM5s, KIM-G1, and the complemented strain KIM-G1/pRePV7.2CMCS-21. Synthesis of an intact O antigen is restored in the complemented strain.
HPAEC and composition analysis of purified LPS from KIM-G1 indicate that it has an intact core but lacks the O antigen.
To determine if the defect associated with strain KIM-G1 was in the biosynthesis of the LPS core or the O antigen, a polysaccharide preparation was obtained from 5 liters of cultured cells. The oligosaccharides from the LPS of R. etli KIM5s and KIM-G1 were released from the lipid A moiety by mild acid hydrolysis and analyzed by HPAEC-PAD. The core fingerprints of both strains were similar to each other and to those of other R. etli and R. leguminosarum strains (data not shown), yielding both the core α-d-GalA-1→4[α-d-GalA-1→5]-Kdo trisaccharide and the α-d-Gal-1→6[α-d-GalA-1→4]-α-d-Man-1→Kdo tetrasaccharide that are highly conserved in these bacteria (12, 13, 27, 29). In addition, composition analysis of the Sephadex G-150-purified R-LPS indicated that the mutant produced all of the glycosyl residues associated with the core oligosaccharides. This demonstrates that KIM-G1 is competent to synthesize an intact LPS core.
Purification and analysis of the O antigen from strain KIM5s.
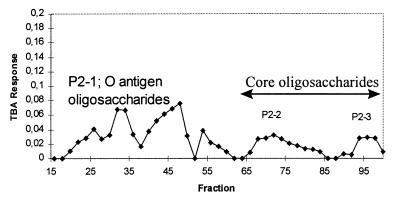
As the mutant did not produce any detectable O antigen (by PAGE), subsequent studies focused on the O antigen of the wild-type strain. The S-LPS of strain KIM5s was isolated by size exclusion chromatography on Sephadex G-150 (data not shown) and subjected to mild acid hydrolysis. The insoluble lipid A was removed by centrifugation, and the released oligosaccharides, both core and O antigen, were then separated on Bio-Gel P2 (Fig. 2). The TBA assay (for Kdo) yielded three major peaks; peaks P2-2 and P2-3 were shown by HPAEC and composition analysis to contain the tetrasaccharide and trisaccharide from the LPS core and were not studied further. P2-1 (fractions 17 to 60) contained the higher-molecular-weight oligosaccharides, and the broad elution suggested a significant degree of heterogeneity in size; PAGE analysis also showed that the O antigen is heterogeneous in size (Fig. 1). An aliquot of pool P2-1 (fractions 17 to 60) was examined by HPAEC, which confirmed that there was no detectable core oligosaccharides in that preparation (data not shown). Therefore, pool P2-1 contained only O-antigen oligosaccharides.
FIG. 2.
Elution profile of purified and hydrolyzed LPS core and O-antigen regions from strain KIM5s separated on a Bio-Gel P2 column. Peaks P2-2 and P2-3 were shown by HPAEC and composition analysis to contain the tetrasaccharide and trisaccharide from the LPS core. The P2-1 pool (fractions 17 to 60) contained the high-molecular-weight oligosaccharides corresponding to the O antigen. Its broad elution suggests that the O polysaccharide is heterogeneous in size.
Composition analysis of pool P2-1 showed a complex array of glycosyl components; however, five sugars accounted for 81% of the detected carbohydrate. This fraction included Glc, Man, GalA, 3-O-Me-6-deoxytalose, and Kdo in approximately a 2:1:1:1:1 molar ratio, and this most likely represents the repeating unit that yields the ladder pattern on the polyacrylamide gel (Fig. 1). There were lesser amounts of five other sugars: GlcNAc, Fuc, Xyl, Gal, and quinovosamine constituted the remaining 19% of the detected carbohydrates. This indicates that these glycosyl residues are not part of the repeating unit but are present in an inner O-antigen oligosaccharide or an oligosaccharide cap at the nonreducing terminus of the O antigen, as has been found in the LPSs of other Rhizobium spp. strains (10a, 23, 28).
Cloning of the mTn5SSgusA30 insertion of strain KIM-G1 and of a wild-type fragment spanning the insertion site.
Enzymatic digestion of the genomic DNA of KIM-G1 with ClaI and XhoI followed by Southern hybridization analysis with a gusA-specific probe indicated that a single transposon insertion had taken place, yielding single hybridization signals of about 8.5 and 11.5 kb, respectively (data not shown). The ClaI fragment was cloned in pBluescript SK, yielding pRePV-KG1, which confers Ap, Sm, and Sp resistance (Fig. 3A). Restriction mapping of this plasmid together with Southern hybridization analysis indicated that about 3.8 kb of genomic DNA from KIM-G1 adjacent to the I border (19) of the mTn5 insertion were cloned in pRePV-KG1 (data not shown). The 3.8-kb fragment was subcloned as an EcoRI fragment in pBluescript SK to yield pRePV1. This plasmid was the source for pRePV2, which has the 1.3-kb XbaI fragment adjacent to the I border cloned as an EcoRI-XbaI fragment (Fig. 3B). The cloned fragments were DIG labelled, and the resulting probes were named KIM-P#1 and KIM-P#2, respectively (Fig. 3B). A partial genomic DNA library of ClaI size-fractionated DNA fragments from R. etli KIM5s around 7 kb in size was screened with probe KIM-P#2, yielding clone pRePV7.2CSK (Fig. 3C).
Complementation of KIM-G1 with pRePV7.2CMCS-21.
The recombinant plasmid pRePV7.2CSK carries a 7.2-kb DNA fragment from KIM5s spanning the insertion site of KIM-G1, as determined from sequence analysis (Fig. 3C). This wild-type DNA fragment was subcloned into the broad-host-range cloning vector pBBR1MCS-2 (31), yielding pRePV7.2CMCS-21 and pRePV7.2CMCS-22, respectively, depending on the insert orientation. E. coli S17-1 was transformed with pRePV7.2CMCS-21, which has the gene disrupted by the mTn5 insertion oriented in the opposite direction to the lacZ promoter of the vector and was conjugally transferred into KIM-G1. Strain KIM-G1/pRePV7.2CMCS-21 recovered the glossy colony morphology of the parental strain and did not flocculate when grown in liquid culture (data not shown), suggesting that the wild-type LPS had been restored in the complemented strain. This inference was confirmed by DOC-PAGE analysis of LPS extracts of KIM-G1/pRePV7.2CMCS-21, as shown in Fig. 1B. Finally, strain KIM-G1/pRePV7.2CMCS-21 was capable of full and effective nodulation on P. vulgaris cv. Saxa (data not shown). Sections of the nodules formed by the complemented strain presented the reddish color characteristic of leghemoglobin-containing N2-fixing nodules. These nodules had specific nitrogenase activities comparable to that of nodules occupied by the wild-type strain, as measured by the acetylene reduction assay (data not shown).
The mTn5 inserted in a glycosyltransferase (GT) locus.
The 2,709-nucleotide (nt) sequence spanning the site of insertion in strain KIM-G1 (Fig. 3C) encodes three ORFs, ORF2, ORF3, and ORF4, which most likely form a single transcriptional unit. The mTn5 insertion was found in ORF3.
The start codon ATG of ORF2 is located 279 nt downstream of the stop codon of ORF1 (not discussed in this report). NNPP (41) located two putative promoter sequences (Fig. 3D) with high correlation coefficients (r = 0.9) within this intergenic region. ORF2 is 816 nt long, encoding a 29.7-kDa protein of 272 amino acids (aa). A potential 5′-AAG-3′ ribosomal binding site (RBS) was located 7 nt upstream of the putative ATG start codon. The highest similarities of ORF2 were displayed to (Mycobacterium tuberculosis EpiB [Z8034-3]) (83% identity) dTDP-glucose 4,6-dehydratase B. japonicum and Azorhizobium brasiliense and UDP-4-glucose-epimerases (AF039306 and Z25478). Sequence similarity was particularly high at the N termini of these proteins, where the signature motif GxxGxxG (34) and other conserved residues characteristic of bacterial NAD(P)-binding domains were identified in all aligned proteins, located between positions 8 and 14 of the KIM5s ORF2 product. Notable is that the product encoded by ORF2 is some 50 residues shorter in its C terminus than the related proteins.
The ATG start codon of ORF3 is located 14 bp downstream of the TAG stop codon of ORF2 and is preceded by a potential 5′-AGGA-3′ RBS located 12 bp upstream of the presumptive ATG start codon. ORF3 is 705 bp long, potentially encoding a 235-aa protein of 26.6 kDa. It showed weak but significant similarities to sugar transferases, with the highest matches corresponding to several α-GTs, such as WbpY, an α-1,3-d-rhamnosyltransferase involved in the synthesis of the A-band d-rhamnan O chain from Pseudomonas aeruginosa (48), and the α-mannosyltransferases MtfB and WbaW from E. coli and Salmonella choleraesuis, respectively, which are also involved in O-chain biosynthesis and are part of the rfb operons of these enteric bacteria (8, 57). It is noteworthy that the product encoded by ORF3 is shorter than these proteins, having a truncated N terminus. The KIM5s ORF3 product and related proteins display the signature sequence Ex7E (positions 144 to 152 in ORF3) previously found in some α-GTs (6, 25). Interestingly, even though E. coli WbdP (62) was the protein with the highest homology score in the BLASTP search (32% identity over 150 residues), it does not have the conserved Ex7E motif and therefore cannot be properly aligned in this region (data not shown).
The site of insertion of the mTn5 in strain KIM-G1 was precisely mapped by sequencing over the I border cloned in pRePV2 (Fig. 3B). The insertion disrupted codon 16 (P) of ORF3 and therefore is located at the 5′ end of the coding sequence.
The putative ATG start codon of ORF4 is located 54 nt downstream of the TAA stop codon of ORF3 and is preceded by a potential 5′-AAG-3′ RBS located 5 nt upstream of the ATG codon. ORF4 is 828 nt long, encoding a putative 276-aa protein of 31.1 kDa. Since no apparent termination or promoter sequences could be located between ORF3 and ORF4, the mTn5 insertion in ORF3 most likely has also a polar effect on ORF4. The product encoded by ORF4 displays significant sequence homology in its N-terminal part with many bacterial proteins, most of which are predicted to be GTs. The KIM5s ORF4 product is similar over almost the entire length with a few proteins, particularly a hypothetical protein from Helicobacter pylori HP0102 (AE000532) (∼45% overall sequence identity). Among the proteins of known function, we found enzymes belonging to different classes of β-GTs, i.e., β3-GlcT (WaaV [AF019746]) from E. coli, β3-GlcNAcT (Cps14I [X85787]) and β4-GalT (Cps14J [X85787]) from Streptococcus pneumoniae, and β6-GlcT (ExoO [L20758]) from Sinorhizobium meliloti. All of this information strongly suggests that ORF4 encodes a β-GT. A comparison of peptide sequences revealed two conserved motifs, Ex4(D/N)x2SxD and Gx5Nx Gx5Gx9D (positions 35 to 45 and 68 to 92, respectively, of ORF4), which constitute a signature for this group of GTs. These peptide motifs comprise three invariant acidic residues which could play an important role in catalysis, since these residues possess appropriate side chain reactivity for glycosyl transfer (51). The three proteins HP0102, WbdO, and WcaE all share with ORF4 a high degree of conservation within the second motif (ExDxGYDYMNKGx5G), which could be characteristic of a subfamily of β-GTs.
The presence of many stop codons located at short intervals in all three reading frames in the downstream sequence (data not shown) strongly suggests that ORF4 is the last one in the presumed transcriptional unit.
The mutated DNA region of strain KIM-G1 is located on a plasmid.
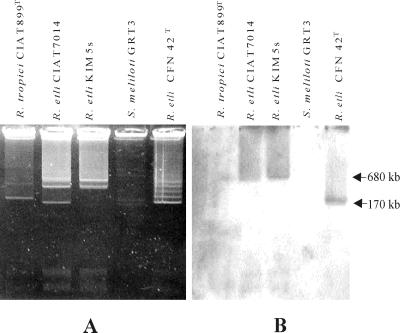
Plasmid profiles of several Rhizobium spp. and Sinorhizobium spp. strains were displayed by using a modified Eckhardt procedure (Fig. 4A). The plasmids were blotted onto nylon membranes for probing with the DNA probe KIM-P#1 (Fig. 3B). The Southern hybridization results presented in Fig. 4B indicate that the hybridizing DNA region is plasmid encoded. The plasmid profiles of R. etli KIM5s and CIAT7014 had not been previously reported. The Eckhardt plasmid profiling experiments suggest that the two plasmid DNA bands of KIM5s might actually correspond each to two plasmids of similar size, as judged from the band intensities and visual inspection at high magnification of the profiles of replicate samples (data not shown). Probe KIM-P#1 hybridized with the upper band (Fig. 4B), which was estimated to have a size of about 680 kb, based on the known sizes of the plasmids present in CFN42 (34a). Importantly, strain CIAT7014 displays a plasmid profile similar to that of KIM5s, although it has an additional small plasmid of about 150 kb not present in KIM5s. Comparison of the plasmid profiles of 19 R. etli and R. tropici strains indicated that KIM5s and CIAT7014 displayed the most similar plasmid profiles, suggesting that these strains have closely related plasmid backgrounds (61). The lowest plasmid band displayed by strain CFN42 actually comprises plasmids pCFN42a and pCFN42b (8). The latter carries the lpsβ2 region (24), which is recognized by probe KIM-P#1 (Fig. 3). The lpsβ region was reported to be conserved among R. etli and R. leguminosarum bv. phaseoli strains (24).
FIG. 4.
Eckhardt gel displaying plasmid profiles (A) and the corresponding Southern hybridization signals (B) obtained with probe KIM-P#1 (Fig. 3B). The two plasmid DNA bands displayed by strain KIM5s could each comprise two plasmids of similar size, as might be also the case for CIAT7014, which has an additional smaller plasmid of about 150 kb. The probe hybridizes only with plasmids from R. etli strains, those from KIM5s and CIAT7014 being about 680 kb in size. The signal yielded by strain CFN42 is due to the lpsβ2 region located on pCFN42b (of ca. 170 kb).
The DNA region encoding GTs has a strain-specific distribution.
pRePV2 was chosen as template to synthesize DNA probe KIM-P#2, which is specific for the locus mutated by the mTn5 insertion of KIM-G1, spanning the sequences of ORF3 and ORF4 (Fig. 3B).
Southern blots of EcoRI-digested genomic DNAs of 18 bean microsymbionts were probed with KIM-P#2. The hybridization and the posthybridization washings were performed at high stringency (0.5× SSC and 68°C). Under these conditions, only strains KIM5s and CIAT7014 yielded strong hybridization signals (data not shown). When the ionic strength of the washing solution was reduced to 0.1× SSC, only these two strains hybridized with the probe (data not shown). These results suggest that the DNA region mutated in KIM-G1 has a strain-specific distribution, further supporting the notion that the putative GTs encoded by ORF3 and ORF4 are involved in the synthesis of the variable O antigen and demonstrate that the hybridization signal yielded by the plasmid profile of strain CFN42 with probe KIM-P#1 (Fig. 4) is due to the lpsβ2 gene located on pCFN42b.
DISCUSSION
In this paper, we report on the mutagenesis, cloning, complementation, and sequence analysis of a novel DNA region from R. etli KIM5s involved in LPS O-antigen biosynthesis. The mTn5 insertion derivative KIM-G1 exhibited a rough colony appearance, tended to flocculate in liquid culture, and when inoculated on P. vulgaris cv. Saxa elicited pseudonodules devoid of leghemoglobin or bacteria. These phenotypic traits had been previously shown to be characteristic for R. etli strains defective in LPS production (29, 36–38).
Comparative DOC-PAGE analysis of the LPS from wild-type strain KIM5s and KIM-G1 confirmed that the mutation affected LPS production, as KIM-G1 appeared to produce only R-LPS, which had the same relative mobility as the R-LPS of the parental strain (Fig. 1). Subsequent HPAEC and composition analyses showed that the mutant strain produced an intact core, with the core tri- and tetrasaccharides that are common in R. etli and R. leguminosarum (4, 12, 13, 29). No O-chain residues were detected in LPS preparations from strain KIM-G1. From these results, we concluded that the mutation in KIM-G1 specifically affects O-PS synthesis, leaving the core OSs unaffected. Thus, the mutated locus is involved in LPS O-chain biosynthesis.
Although the structures of LPS molecules are not as well established for rhizobia as they are for enteric bacteria, for clarity we have designated the tetrasaccharide and trisaccharide components of the R. etli (and R. leguminosarum) LPS as the core, as has been done in other recent studies (23, 27). This designation is based on the fact that purified R-LPS of R. etli contains only these moieties and an additional Kdo residue (10a, 47). The other carbohydrate residues, such as the GlcNAc, Fuc, Xyl, Gal, and quinovosamine residues (i.e., the minor components), identified in this study are found only in association with S-LPS and therefore may be defined as O-antigen components. In addition, the tetrasaccharide and trisaccharide components have been found in every strain of R. etli and R. leguminosarum studied to date (29), showing that they are conserved in these species. In contrast, the other components are strain specific, and this variable OS portion defines the O antigens of many species, including R. etli and R. leguminosarum (29).
The O chains of R. etli and R. leguminosarum strains that have been characterized, such as that from strain CFN42, contain complex repeating units and nonrepeating oligosaccharides (10a, 23, 29). Strain KIM5s also produces a complex O antigen that appears to contain sequential DPS of a repeating unit, as well as nonrepeating oligosaccharides. The apparently modal (regulated) distribution of O-chain lengths displayed by KIM5s (Fig. 1) is reminiscent of the situation found in the heteropolymeric O antigens produced, for example, by Salmonella enterica strains of serogroups B and E and could indicate the action of a control system for O-chain polymerization such as the Salmonella Rol protein (52, 65). However, there is no experimental evidence indicating the existence of such a system in the rhizobia, and control of the O-chain length in these bacteria may involve completely different mechanisms, such as O-methylation of the capping glycosidic residue(s) (29, 65).
The monosaccharide composition of the O-PS from KIM5s is clearly different from that of CFN42 (23), demonstrating that it is a strain-specific antigen. The structural specificity of the O antigen indicates that each strain must carry unique genes involved in LPS biosynthesis. This notion is supported by genetic evidence presented in this report based on Southern hybridization data for genomic DNAs from 18 bean microsymbionts and probe KIM-P#2, which hybridized only with the homologous strain KIM5s and one other strain, CIAT7014. This result suggests that the mutated region, which encodes two GTs, appears to have a strain-specific distribution among R. etli strains.
In this regard, Rhizobium spp. are quite different from Sinorhizobium spp., in which the LPS is a highly conserved antigen throughout the genus (22, 46). The K antigens are the strain-specific antigens in the sinorhizobia (22, 43, 45, 46). The production of K antigens by cultured cells of wild-type strains of Rhizobium spp. has not been observed, which was also true of R. etli KIM5s. In contrast, its mutant derivative KIM-G1 was found to produce a non-LPS, acidic polysaccharide that migrated as a low-mobility ladder pattern on polyacrylamide gels (Fig. 1A). This polysaccharide required an alcian blue prestain to appear on the gel, similar to the K antigens of Sinorhizobium spp. (43). This phenomenon has been observed in other LPS mutants of rhizobia (47) and in a pb− derivative from R. etli CFN42 (61). One possible explanation for the production of a specific component by the mutant but not by the wild-type strain is that by this means the cell can compensate in part for the loss of the O antigen. It is not clear, however, why laboratory-cultured cells of wild-type R. etli do not produce this class of polysaccharide. Conceivably the unique polysaccharide produced by R. etli KIM-G1 is, in fact, a K antigen, as genetic analysis of R. etli has shown that it carries the K-antigen-specific rkpABCDEF operon (47).
Three ORFs were identified in this report as the likely components of the putative transcriptional unit disrupted by the mTn5 insertion in strain KIM-G1. ORF2, located upstream of the insertion site, probably encodes a sugar nucleotide epimerase or dehydratase, containing the strictly conserved motif GxxGxxG at its N terminus. This motif is characteristic of prokaryotic NAD(P)-binding domains (34, 67). The highest homology was found to EpiB, a putative dTDP-glucose-4,6 dehydratase from M. tuberculosis (17). Such an enzyme may be involved in the synthesis of 6-deoxy and dideoxy sugars (33), which are present in the O antigen of KIM5s.
ORF3 was disrupted at its 5′ end by the mTn5 insertion in strain KIM-G1, and complementation of KIM-G1 with pRePV7.2CMCS-2 unequivocally demonstrated that the insertion is the sole cause for the mutant phenotype. When provided in trans, this plasmid restores the S-LPS, full nodulation and N2 fixation capabilities lost by the mutant. ORF3 could encode for an α-GT, as it exhibits the ExE motif which is highly conserved in the family of prokaryotic α-ManTs (25). This motif has also been found in other prokaryotic GTs, including α3-GalTs, α6-GalTs, and α2-GlcNAcTs (6). Geremia et al. (25) have proposed that all prokaryotic α-mannosyltransferases share the signature peptide ExFGxxxxE, a more specific version of the ExE motif. This signature peptide is also present in the R. leguminosarum core OS α-mannosyltransferase LpcC (27). The putative α-1,3-d-rhamnosyltransferase WbpY from P. aeruginosa (48) also shares this more specific motif, indicating that it is not absolutely specific for α-mannosyltransferases. Even though ORF3 displays significant homology to several α-mannosyltransferases, and mannose was identified as one of the components of the O-PS from KIM5s, it does not contain the FG residues of the diagnostic ExFGxxxxE signature peptide. For these reasons, and as supported by HCA (data not shown), ORF3 is tentatively considered to encode an α-GT of unknown substrate specificity.
The mTn5 insertion in ORF3 probably has a polar effect on the downstream ORF4 (Fig. 3C). ORF4 most likely encodes a β-GT, since it displays significant homology in its N terminus with a number of known β-GTs. Two peptide motifs (Ex4 (D/N)x2SxD and Gx5NxGx5Gx9D) similar to those found in some β-GTs (6, 51) were found to be conserved among numerous proteins. However, the position of the conserved acidic residues is slightly different in the KIM5s ORF4 product and related proteins.
Upon HCA analysis, the KIM5s ORF4 product displays similarities over the entire sequence (data not shown) to putative GTs WcaE and WbdO (56, 62) from E. coli. It also contains a more specific version (ExDxGIYDAMNKGx5Gx9D) of the second peptide motif described above, which could be a signature for a subfamily of β-GTs.
The lpsβ2 gene, recently described by García de los Santos and Brom for R. etli CFN42 and located on plasmid pCFN42b (24), is found in KIM5s downstream of ORF4, in an opposite transcriptional orientation (Fig. 3C). The complete lpsβ2 gene and two other ORFs located further upstream were cloned in an overlapping 8.0-kb EcoRI fragment, which was found by using the same strategy as for pRePV7.2CSK (61). The complete KIM5s Lpsβ2 gene product shows 96% identity with that of CNF42, has the same length, and complements strain CE168 (lpsβ2::Tn5 derivative of CFN42) (15); consequently, these genes are considered to be allelic forms (61), and thus probe KIM-P#1 hybridizes with pCFN42 (Fig. 4). Interestingly, the KIM5s lpsβ2 allele is present as a monocistronic transcript, lacking the lpsβ1 gene found in CFN42 (24), indicating a difference in gene organization around the lpsβ2 locus in these two Mexican R. etli strains. Furthermore, since lpsβ1 and lpsβ2 appear to be the only LPS biosynthesis genes on pCFN42b (24), and since probe KIM-P#2 hybridizes with genomic DNAs from KIM5s and CIAT7014, but not with genomic DNA from CFN42, the locus that we identified in strain KIM-G1 actually represents a novel plasmid-encoded lps region, mapped to a 680-kb plasmid present in strains KIM5s and CIAT7014, but not in other R. etli strains tested. The nucleotide sequence of this region is the first one available for Rhizobium spp. encoding for gene products involved in LPS O-chain synthesis.
ACKNOWLEDGMENTS
We thank J. A. Herrera and J. Sanjuan for help with the plasmid profiling experiments, and we thank H. Thierfelder and E.-M. Kurz for excellent technical assistance.
B. L. Reuhs was supported by National Science Foundation grant MCB-9728564 and by the U.S. Department of Energy-funded Center for Plant and Microbial Complex Carbohydrate Research under grant DE-FG02-93ER-20097. P. Vinuesa was the recipient of a TMR-Network grant from the EU. This work was supported by the Deutsche Forschungsgemeinshaft through the SFB 395.
REFERENCES
- 1.Allaway D, Jeyretnam B, Carlson R W, Poole P S. Genetic and chemical characterization of a mutant that disrupts synthesis of the lipopolysaccharide core tetrasaccharide in Rhizobium leguminosarum. J Bacteriol. 1996;178:6403–6406. doi: 10.1128/jb.178.21.6403-6406.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Warren G, Miller W, Myers E U, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J C, Struhl K S, editors. Current protocols in molecular biology, section 2.4. New York, N.Y: John Wiley, Inc.; 1990. [Google Scholar]
- 4.Bhat U R, Bhagyalakshmi S K, Carlson R W. Re-examination of the structures of the lipopolysaccharide core oligosaccharides from Rhizobium leguminosarum biovar phaseoli. Carbohydr Res. 1991;220:219–227. doi: 10.1016/0008-6215(91)80020-n. [DOI] [PubMed] [Google Scholar]
- 5.Blumenkrantz N J, Asboe-Hansen B. A new method for the quantitative determination of uronic acid. Anal Biochem. 1973;54:484–489. doi: 10.1016/0003-2697(73)90377-1. [DOI] [PubMed] [Google Scholar]
- 6.Breton C, Bettler E, Joziasse D H, Geremia R A, Imberty A. Sequence-function relationships of prokaryotic and eukaryotic galactosyltransferases. J Biochem. 1998;123:1000–1009. doi: 10.1093/oxfordjournals.jbchem.a022035. [DOI] [PubMed] [Google Scholar]
- 7.Brewin N J. Tissue and cell invasion by Rhizobium: the structure and development of infection threads and symbiosomes. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 417–429. [Google Scholar]
- 8.Brom S, García de los Santos A, Stepkowsky T, Flores M, Dávila G, Romero D, Palacios R. Different plasmids of Rhizobium leguminosarum bv. phaseoli are required for optimal symbiotic performance. J Bacteriol. 1992;174:5183–5189. doi: 10.1128/jb.174.16.5183-5189.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown P K, Romana L K, Reeves P R. Molecular analysis of the rfb gene cluster of Salmonella muenchen (strain M67): the genetic basis of the polymorphism between groups C2 and B. Mol Microbiol. 1992;6:1385–1394. doi: 10.1111/j.1365-2958.1992.tb00859.x. [DOI] [PubMed] [Google Scholar]
- 10.Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon J P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Carlson, R. Personal communication.
- 11.Carlson R W, Kalembasa S, Turowski D, Pachori P, Noel K D. Characterization of the lipopolysaccharide from a Rhizobium phaseoli mutant that is defective in infection thread development. J Bacteriol. 1987;169:4923–4928. doi: 10.1128/jb.169.11.4923-4928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson R W, Garcia F, Noel K D, Hollingsworth R I. The structures of the lipopolysaccharide core components from Rhizobium leguminosarum biovar phaseoli CE3 and two of its symbiotic mutants, CE109 and CE309. Carbohydr Res. 1990;195:101–110. doi: 10.1016/0008-6215(89)85092-x. [DOI] [PubMed] [Google Scholar]
- 13.Carlson R W, Reuhs B L, Chen T-B, Bhat U R, Noel K D. Lipopolysaccharide core structures in Rhizobium etli mutants deficient in O antigen. J Biol Chem. 1995;270:11783–11788. doi: 10.1074/jbc.270.20.11783. [DOI] [PubMed] [Google Scholar]
- 14.Carlson R W, Kannenberg E L, Forsberg L S, Xie S. Rhizobium etli lipopolysaccharide structure: comparison with the LPSs from Rhizobium leguminosarum and related Rhizobium strains. In: Martinez E, Hernández G, editors. Highlights of nitrogen fixation research. New York, N.Y: Plenum Publishing Corporation; 1999. [Google Scholar]
- 15.Cava J R, Elias P M, Turowski D A, Noel K D. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J Bacteriol. 1989;171:8–15. doi: 10.1128/jb.171.1.8-15.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cava J R, Tao H, Noel K D. Mapping of complementation groups within a Rhizobium leguminosarum CFN42 chromosomal region required for lipopolysaccharide synthesis. Mol Gen Genet. 1990;221:125–128. [Google Scholar]
- 17.Cole S T, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 18.Corzo J, Pérez-Galdona R, León-Barrios M, Gutiérrez-Navarro M A. Alcian blue fixation allows silver staining of the isolated polysaccharide component of bacterial lipopolysaccharides in polyacrylamide gels. Electrophoresis. 1991;12:439–441. doi: 10.1002/elps.1150120611. [DOI] [PubMed] [Google Scholar]
- 19.de Lorenzo V, Herrera M, Jakubzik U, Timmis K. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative bacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dénarié J, Debellé F, Promé J C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 21.Fisher R F, Long S R. Rhizobium-plant signal exchange. Nature. 1992;357:655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- 22.Forsberg L S, Reuhs B L. Structural characterization of the K antigens from Rhizobium fredii USDA257: evidence for a common structural motif, with strain-specific variation, in the capsular polysaccharides of Rhizobium spp. J Bacteriol. 1997;179:5366–5371. doi: 10.1128/jb.179.17.5366-5371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsberg L S, Carlson R W. The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359. J Biol Chem. 1998;273:2747–2757. doi: 10.1074/jbc.273.5.2747. [DOI] [PubMed] [Google Scholar]
- 24.García de los Santos A, Brom S. Characterization of two plasmid-borne lpsβ loci of Rhizobium etli required for lipopolysaccharide synthesis and for optimal interaction with plants. Mol Plant-Microbe Interact. 1997;10:891–902. doi: 10.1094/MPMI.1997.10.7.891. [DOI] [PubMed] [Google Scholar]
- 25.Geremia R A, Petroni E A, Ielpi L, Henrissat B. Towards a classification of glycosyltransferases based on amino acid sequence similarities: prokaryotic α-mannosyltransferases. Biochem J. 1996;318:133–138. doi: 10.1042/bj3180133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hynes M F, MacGregor N F. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen fixing nodules of Rhizobium leguminosarum. Mol Microbiol. 1990;4:567–574. doi: 10.1111/j.1365-2958.1990.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 27.Kadrmas J L, Allaway D, Studholme R E, Sullivan J T, Ronson C W, Poole P S, Raetz C R H. Cloning and overexpression of glycosyltransferases that generate the lipopolysaccharide core of Rhizobium leguminosarum. J Biol Chem. 1998;273:26432–26440. doi: 10.1074/jbc.273.41.26432. [DOI] [PubMed] [Google Scholar]
- 28.Kannenberg E L, Brewin N J. Host-plant invasion by Rhizobium: the role of cell-surface components. Trends Microbiol. 1994;2:277–283. doi: 10.1016/0966-842x(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 29.Kannenberg E L, Reuhs B L, Forsberg L S, Carlson R W. Lipopolysaccharides and K antigens: their structures, biosynthesis and functions. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 119–154. [Google Scholar]
- 30.Kingsley M T, Bohlool B B. Extracellular polysaccharide is not responsible for aluminum tolerance of Rhizobium leguminosarum bv. phaseoli CIAT899. Appl Environ Microbiol. 1992;58:1095–1101. doi: 10.1128/aem.58.4.1095-1101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 32.Krauss J H, Weckesser J, Mayer H. Electrophoretic analysis of lipopolysaccharides of purple nonsulfur bacteria. Int J Syst Bacteriol. 1988;38:157–163. [Google Scholar]
- 33.Liu H W, Thorson J S. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu Rev Microbiol. 1994;48:233–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 34.Macpherson D F, Manning P A, Morona R. Characterization of the dTDP-rhamnose biosynthetic genes encoded in the rfb locus of Shigella flexneri. Mol Microbiol. 1994;11:281–292. doi: 10.1111/j.1365-2958.1994.tb00308.x. [DOI] [PubMed] [Google Scholar]
- 34a.Martínez-Romero, E. Personal communication.
- 35.Niehaus K, Becker A. The role of microbial surface polysaccharides in the Rhizobium-legume interaction. Plant-Microbe Interact. 1998;29:73–116. doi: 10.1007/978-1-4899-1707-2_3. [DOI] [PubMed] [Google Scholar]
- 36.Noel K D, Sánchez A, Fernández L, Leermans J, Cevallos M. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noel K D, Vandenbosh K A, Kulpaca B. Mutations in Rhizobium phaseoli that lead to arrested development of infection threads. J Bacteriol. 1986;168:1392–1401. doi: 10.1128/jb.168.3.1392-1401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noel K D. Rhizobial polysaccharides required in symbiosis with legumes. In: Verma D P S, editor. Molecular signals in plant-microbe communications. Boca Raton, Fla: CRC Press; 1992. pp. 341–357. [Google Scholar]
- 39.Poole P S, Schofield N A, Reid C J, Drew E M, Walshaw D L. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology. 1994;140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- 40.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1035–1063. [Google Scholar]
- 41.Reese M G, Harris N L, Eeckman F H. Large scale sequencing specific neural networks for promoter and splice site recognition. In: Hunter L, Klein T, editors. Pacific Symposium on Biocomputing ’96. Singapore, Singapore: World Scientific Publications; 1995. pp. 737–738. [Google Scholar]
- 42.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 43.Reuhs B L, Carlson R W, Kim J S. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J Bacteriol. 1993;175:3570–3580. doi: 10.1128/jb.175.11.3570-3580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuhs B L, Kim J S, Badgett A, Carlson R W. Production of cell-associated polysaccharides of Rhizobium fredii USDA205 is modulated by apigenin and host root extract. Mol Plant-Microbe Interact. 1994;7:240–247. doi: 10.1094/mpmi-7-0240. [DOI] [PubMed] [Google Scholar]
- 45.Reuhs B L. Acidic capsular polysaccharides (K antigens) of Rhizobium, p. 331–336. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: IS-MPMI; 1996. [Google Scholar]
- 46.Reuhs B L, Geller D P, Kim J S, Fox J E, Kolli V S K, Pueppke S G. Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl Environ Microbiol. 1998;64:4930–4938. doi: 10.1128/aem.64.12.4930-4938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reuhs, B. L. Unpublished data.
- 48.Roccheta H L, Burrows L L, Pacan J C, Lam J S. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol Microbiol. 1998;28:1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T A. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saxena I M, Brown M R, Fevre M, Geremia R A, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 54.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 55.Stacey G, So J-S, Roth L E, Lakshmi S K, Carlson R W. A lipo-polysaccharide mutant of Bradyrhizobium japonicum that uncouples plant from bacterial differentiation. Mol Plant-Microbe Interact. 1991;4:332–340. doi: 10.1094/mpmi-4-332. [DOI] [PubMed] [Google Scholar]
- 56.Stevenson G, Andrianopoulus K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugiyama T, Kido N, Komatsu T, Ohta M, Jann K, Jann B, Saeki A, Kato A. Genetic analysis of Escherichia coli O:9 rfb: identification and DNA sequence of phosphomannomutase and GDP-mannose pyrophophorylase genes. Microbiology. 1994;140:59–71. doi: 10.1099/13500872-140-1-59. [DOI] [PubMed] [Google Scholar]
- 58.Tsai C, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 59.van Berkum P, Eardly B D. Molecular evolutionary systematics of the Rhizobiaceae. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–24. [Google Scholar]
- 60.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vinuesa, P. Unpublished data.
- 61a.Vinuesa P, Rademaker J L W, de Bruijn F J, Werner D. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl Environ Microbiol. 1998;64:2096–2104. doi: 10.1128/aem.64.6.2096-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Reeves P R. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weissbach A, Hurwitz J. The formation of 2-keto-3-deoxyheptanoic acid in extracts of Escherichia coli B. J Biol Chem. 1958;234:705–709. [PubMed] [Google Scholar]
- 64.Werner D, Wilcockson J, Zimmermann E. Adsorption and selection of rhizobia by ion exchange papers. Arch Microbiol. 1975;105:27–32. doi: 10.1007/BF00447108. [DOI] [PubMed] [Google Scholar]
- 65.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 66.Wilson K J, Sessich A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 67.Wyk P, Reeves P R. Identification and sequence of a gene for abequose synthase, which confers antigenic specificity on group B salmonellae: homology with galactose epimerase. J Bacteriol. 1989;171:5687–5693. doi: 10.1128/jb.171.10.5687-5693.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.York W S, Darvill A G, McNeil M, Stevenson T T, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]