Abstract
We previously identified the prpBCDE operon, which encodes catabolic functions required for propionate catabolism in Salmonella typhimurium. Results from 13C-labeling experiments have identified the route of propionate breakdown and determined the biochemical role of each Prp enzyme in this pathway. The identification of catabolites accumulating in wild-type and mutant strains was consistent with propionate breakdown through the 2-methylcitric acid cycle. Our experiments demonstrate that the α-carbon of propionate is oxidized to yield pyruvate. The reactions are catalyzed by propionyl coenzyme A (propionyl-CoA) synthetase (PrpE), 2-methylcitrate synthase (PrpC), 2-methylcitrate dehydratase (probably PrpD), 2-methylisocitrate hydratase (probably PrpD), and 2-methylisocitrate lyase (PrpB). In support of this conclusion, the PrpC enzyme was purified to homogeneity and shown to have 2-methylcitrate synthase activity in vitro. 1H nuclear magnetic resonance spectroscopy and negative-ion electrospray ionization mass spectrometry identified 2-methylcitrate as the product of the PrpC reaction. Although PrpC could use acetyl-CoA as a substrate to synthesize citrate, kinetic analysis demonstrated that propionyl-CoA is the preferred substrate.
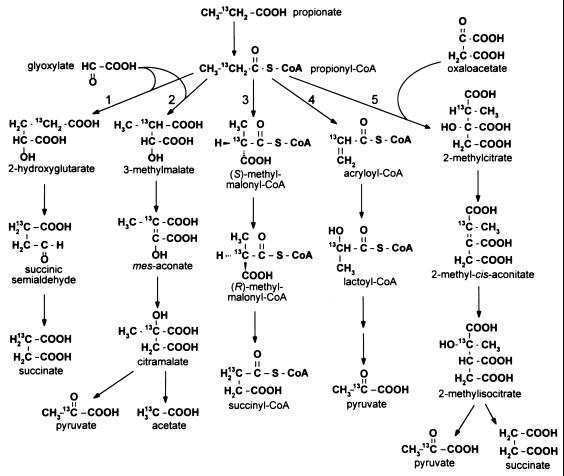
Fatty acids are important sources of carbon and energy for procaryotes. Medium-chain (C6 to C12) and long-chain (>C12) fatty acids are common breakdown products of cell membrane components. The catabolism of these compounds is known to proceed by β-oxidation to acetyl coenzyme A (acetyl-CoA), which can enter the tricarboxylic acid cycle. Short-chain fatty acids (<C6) are common fermentation by-products of microorganisms and are catabolized by procaryotes through different routes. The most complicated of these oxidation pathways is the breakdown of propionate. Pathways for propionate catabolism in procaryotes were recently reviewed by Textor et al. (33), and those proposed to exist in Escherichia coli and Salmonella typhimurium are shown in Fig. 1.
FIG. 1.
Proposed propionate breakdown pathways in E. coli and S. typhimurium. Pathways: 1, α-hydroxyglutarate; 2, citramalate; 3, methylmalonyl-CoA; 4, acryloyl-CoA; 5, 2-methylcitric acid cycle.
In E. coli, 14CO2 evolution experiments and 13C nuclear magnetic resonance spectroscopy (13C-NMR) studies led researchers to conclude that propionate was oxidized to pyruvate through acryloyl-CoA and lactoyl-CoA intermediates (14, 38). Furthermore, 13C-NMR studies and crude enzyme assays indicated E. coli could also catabolize propionate through the 2-hydroxyglutarate, citramalate, and methylmalonyl-CoA pathways (7, 39). Crude enzyme assays by Fernandez-Briera and Garrido-Pertierra suggested that S. typhimurium also possessed the acrylate pathway for propionate breakdown (8). Unfortunately, experiments to fully characterize the genes and/or enzymes for any of these oxidation pathways in E. coli or S. typhimurium were never performed.
Recently, we identified a locus on the S. typhimurium chromosome that was required for growth on propionate (11). Genetic and molecular characterization of this locus identified five genes, prpRBCDE, all of which are involved in propionate oxidation (12). Surprisingly, sequence comparisons indicated that the Prp enzymes were not similar to propionyl-CoA dehydrogenase or lactoyl-CoA dehydratase, which catalyze steps of the acryloyl-CoA pathway. The only propionate catabolism pathway consistent with the proposed functions encoded by the prpBCDE operon was the 2-methylcitric acid cycle of fungi (31, 32). This pathway was recently identified in E. coli as the route of propionate breakdown (19, 33). A chromosomal region of E. coli, nearly identical to the prpRBCDE locus of S. typhimurium, was identified by the E. coli genome-sequencing project (4). Recently, the PrpC protein from this locus in E. coli was demonstrated to catalyze the condensation of propionyl-CoA with oxaloacetate to form 2-methylcitrate, and it was inferred to yield 2-methylcitrate. However, no spectroscopic evidence was reported to document that 2-methylcitrate was the product of the reaction (9, 33).
Studies with E. coli showed evidence for the existence of the 2-methylcitric acid cycle (19, 33). The similarities between the prp loci in E. coli and S. typhimurium suggested that this pathway was also present in S. typhimurium. However, the intermediates of the 2-methylcitric acid cycle in E. coli were not identified, and the prp locus was not shown to be required for growth on propionate. To identify the propionate oxidation pathway in S. typhimurium, we focused our efforts on the identification of intermediates in the pathway. In all proposed catabolic pathways (Fig. 1), the oxidation state of the methylene carbon of propionate changes significantly throughout the pathway. These changes were monitored by 13C-NMR. We used a combination of in vivo and in vitro 13C-NMR experiments to identify the propionate breakdown intermediates in S. typhimurium. We purified and kinetically characterized the PrpC enzyme and demonstrated by spectroscopic means that PrpC catalyzes the synthesis of 2-methylcitrate from propionyl-CoA and oxaloacetate.
MATERIALS AND METHODS
Culture media and growth conditions.
Nutrient broth at 0.8% (wt/vol) containing 85 mM NaCl (6) was routinely used as rich medium. No-carbon E (NCE) medium was supplemented with 1 mM MgSO4 and used as minimal medium (6). Final concentrations of compounds in the culture medium were as follows: methionine, 0.5 mM; acetate 50 mM; propionate, 30 mM; and pyruvate, 30 mM. Antibiotic concentrations in rich medium were as follows: ampicillin, 50 μg/ml (100 μg/ml for plasmids); kanamycin, 25 μg/ml (50 μg/ml for plasmids); tetracycline, 20 μg/ml. All chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise stated. A list of strains and plasmids used and their genotypes is provided in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α/F′ | F′/endA1 hsdR17 (rk− mk+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | New England Biolabs |
| BL21(λDE3) | F−ompT hsdSB(rB− mB+) dcm gal λ(DE3) | Novagen |
| JE4570 | BL21(λDE3)/pPRP62 (T7 rpo+ prpC+ bla+) | This work |
| S. typhimurium | ||
| JR501 | hsdSA29 hsdSB121 hsdL6 metA22 metE551 trpC2 ilv-452 rpsL120 xyl-404 galE719 H1-b H2-en,n,x (Fels2−) fla-66 nml | 23 |
| MST226 | aceA101::Tn10 | S. Maloy |
| MST229 | aceB102::Tn10 | S. Maloy |
| MST2026 | aceK::Tn10 | S. Maloy |
| RT906 | pps-81::MudI1734(kan+) | R. Jeter |
| RT910 | pps-85::Tn10Δ16Δ17 | R. Jeter |
| TR6583 | metE205 ara-9 | K. Sanderson via J. Roth |
| Derivatives of TR6583 | ||
| JE3907 | prpC167 zai-6386::Tn10Δ16Δ17 | 12 |
| JE3909 | prpD169 zai-6386::Tn10Δ16Δ17 | 12 |
| JE3913 | prpC173 zai-6386::Tn10Δ16Δ17 | 12 |
| JE3914 | prpD174 zai-6386::Tn10Δ16Δ17 | 12 |
| JE3946 | prpB195 zai-6386::Tn10Δ16Δ17 | 12 |
| JE3961 | prpB210 zai-6386::Tn10Δ16Δ17 | 12 |
| JE4184 | pGP1-2 (T7 rpo+ kan+), pPRP38 (prpE+ bla+) | 13 |
| JE4287 | pGP1-2 (T7 rpo+ kan+), pT7-6 (bla+) | 13 |
| JE4330 | pGP1-2 (T7 rpo+ kan+), pPRP22 (prpC+ bla+) | 12 |
The TR6583 (metE205 ara-9) genetic background was used for determination of all growth phenotypes. Isolated colonies of strains were patched to nutrient broth, grown overnight at 37°C, and printed to agar plates of NCE medium with appropriate supplements. TR6583 was used as a positive control for these experiments.
Recombinant DNA and genetic techniques.
Restriction and modification enzymes were purchased from Promega (Madison, Wis.) and used as specified by the manufacturer. All DNA manipulations were performed in E. coli DH5α/F′. Plasmids were transformed into E. coli or S. typhimurium by CaCl2 heat shock as described previously (2). Plasmids transferred to S. typhimurium were first transformed into recombination-deficient S. typhimurium JR501 (34). Plasmids from strain JR501 were quick-transformed into other S. typhimurium strains as described previously (23).
Construction of plasmid pPRP62.
Plasmid pPRP22 was digested with NdeI and BamHI to remove a 2.2-kb fragment containing prpC+. This fragment was cloned into vector pET15b (Novagen, Madison, Wis.) cut with the same enzymes. The resulting plasmid was 7.9 kb long, encoded ampicillin resistance, and was referred to as pPRP62.
Genetic crosses.
Transductions involving phage P22 HT105/1 int201 (27, 28) were performed as described previously (5, 6).
13C-NMR. (i) Accumulation of [13C-2]propionate breakdown intermediates.
Overnight cultures (5 ml) of each strain were prepared in NCE medium supplemented with 50 mM glycerol and 1 mM propionate. The overnight cultures were subcultured into 500 ml of fresh medium in 1-liter flasks and grown aerobically at 37°C. After 24 h of growth, cells were harvested by centrifugation at 10,500 × g at 4°C for 10 min and resuspended in 50 ml of NCE medium supplemented with 20 mM glycerol and 5 mM [2-13C]propionate. The cultures were grown aerobically at 37°C in 125-ml flasks for an additional 24 h. Culture growth was stopped with 3.3 ml of 60% perchloric acid, and the mixture was stored at −80°C. To obtain 13C-NMR spectra, samples were thawed at room temperature and cell debris was removed by centrifugation at 4°C for 10 min at 10,500 × g. The supernatant was neutralized with 10 M KOH, and any insoluble material was removed by centrifugation. Neutralized samples were lyophylized and resuspended in 2.5 ml of 10% D2O, and insoluble material in the sample was removed with a syringe filter. The suspension was transferred to a 10-mm (inner diameter [i.d.]) NMR tube (Wilmad Glass, Buena, N.J.), into which a sealed tetramethylsilane (TMS) capillary was introduced to serve as external reference. Samples were stored for no longer than 24 h at 4°C before a 13C-NMR spectrum was obtained.
(ii) Preparation of PrpE product for 13C-NMR.
The following were combined in a 2.5-ml reaction mixture: CoA, 2.5 μmol; ATP, 12.5 μmol; MgCl2, 25 μmol; PrpE dialyzed cell extract, 200 μg; and phosphate buffer, 125 μmol (pH 7.5). The mixture was preincubated at 37°C for 10 min, and the reaction was started with 5 μmol of [2-13C]propionate. The reaction mixture was incubated for 1 h at 37°C and was stopped by heating at 95°C for 2 min. The mixture was centrifuged for 2 min in 1.5-ml microcentrifuge tubes, and a 2.25-ml sample was transferred into a 10-mm (i.d.) NMR tube. The volume was brought to 2.5 ml with 100% D2O to make a 10% final D2O concentration. As above, TMS was used as external reference. The prpE gene was overexpressed in strain JE4184, and dialyzed cell extract of this strain was prepared as described previously (13).
(iii) In vitro synthesis and spectroscopic characterization of 2-methylcitrate.
The following were combined in a 1-ml reaction mixture: propionyl-CoA, 5.0 μmol; oxaloacetate, 5.0 μmol; phosphate buffer, 50 μmol (pH 7.5); and PrpC protein, 20 μg. The reaction mixture was incubated for 1 h at 37°C, and the reaction was stopped by adding HCl to a final pH of <2. The sample was extracted four times with 5 ml of diethyl ether, and the organic phase was combined and dried in a SpeedVac concentrator (Savant Instruments, Farmingdale, N.Y.). The remaining residue was resuspended in 600 μl of 100% D2O and placed in a 5-mm (i.d.) NMR tube (Wilmad Glass, Buena, N.J.) for 1H-NMR analysis. Negative-ion electrospray ionization (ESI) mass spectrometry was performed on a sample of the reaction product. Negative-ion ESI mass spectra were obtained with a Perkin-Elmer Sciex API 365 triple-quadrupole spectrometer equipped with an ion spray source on samples resuspended in 50% acetonitrile.
(iv) In vitro synthesis of [13C-2]methylcitrate.
The following were combined in a 1.5-ml reaction mixture: CoA, 2 μmol; ATP, 2 μmol; MgCl2, 10 μmol; PrpE dialyzed cell extract, 100 μg; and phosphate buffer, 75 μmol (pH 7.5). The mixture was preincubated at 37°C for 10 min, and the reaction was started with 10 μmol of [2-13C]propionate. After 30 min, a 0.5-ml mixture of 5 μmol of oxalacetate, 25 μmol of phosphate buffer (pH 7.5), and dialyzed cell extract (50 μg of protein) of prpC-overexpressing strain JE4330 was added to the reaction mixture. The mixture was incubated for 1 h at 37°C and was stopped by heating at 95°C for 2 min. To chelate excess Mg2+ ions, 10 μmol of EDTA was added to the mixture and the sample was prepared for 13C-NMR as described above. The prpC gene was overexpressed in strain JE4330, and dialyzed cell extract of this strain was prepared in the same manner as for PrpE, as described previously (13).
(v) Acquisition of NMR spectra.
13C-NMR spectra were acquired at 100.6 MHz with a deuterium lock on a Bruker Instruments DMX-400 Avance console with a 9.4-T wide-bore magnet (Nuclear Magnetic Resonance Facility at the University of Wisconsin—Madison). 13C-NMR proton-decoupled and proton-coupled spectra were obtained with a 90° pulse angle and relaxation time of 5 s, and the spectra were Fourier transformed with 5-Hz line broadening. 1H-NMR spectra were acquired at 401.13 MHz with a deuterium lock and water suppression on the same Bruker DMX-400 instrument. 1H-NMR spectra were obtained with a 90° pulse angle and relaxation of 1 s, and the spectra were Fourier transformed with 0.5-Hz line broadening. Homonuclear decoupling was performed by standard procedures. Chemical shifts presented in this work were relative to that of TMS, which was set to 0.0 ppm. [13C]propionate, D2O, and TMS were purchased from Cambridge Isotope Labs (Andover, Mass.).
Biochemical characterization of PrpC. (i) Overexpression of PrpC and preparation of cell extracts.
Strain JE4570 was used for overexpression and purification of PrpC. One liter of strain JE4570 was grown in Luria-Bertani broth plus ampicillin at 37°C with shaking to a cell density of 0.5 at 600 nm. At this point, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 0.3 mM to induce expression, and the culture was grown for an additional 3 h at 37°C. Cells were harvested by centrifugation for 10 min at 10,500 × g and 4°C in an RC-5B refrigerated superspeed centrifuge (DuPont, Wilmington, Del.). The cell pellet was resuspended in 4 ml of cold 20 mM Tris-HCl buffer (pH 7.9) containing 0.5 M NaCl and 5 mM imidazole. Cells were broken by sonication (10 min, 50% duty, setting 3) with a model 550 Sonic Dismembrator (Fisher Scientific, Itasca, Ill.). Cell debris was removed by centrifugation in 50 ml of Nalgene polypropylene copolymer Oakridge tubes (Fisher Scientific) at 31,000 × g for 30 min at 4°C. The supernatant was removed and kept at 4°C until the purification step.
(ii) Purification of PrpC.
PrpC was purified from cell extracts by using His · Bind resin (Novagen) as specified by the manufacturer. Following purification, the enzyme was placed into a Slide-A-Lyzer cassette (Pierce Chemical Co.) and dialyzed at 4°C into 1 liter of 50 mM HEPES buffer (pH 7.5) containing 0.2 M KCl and 5 mM EDTA. After 3 h, the dialysis buffer was switched to 1 liter of 50 mM HEPES buffer (pH 7.5) containing 0.2 M KCl, 5 mM EDTA, and 0.54 M glycerol. The enzyme was stored at −80°C in this buffer, and no detectable loss in enzyme activity was observed after 6 months.
(iii) Enzyme assay conditions.
Acyl-CoA consumption was quantitated spectrophotometrically as described by Srere et al. (29). This assay detects the level of free CoA in the reaction mixture by using 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). CoA reacts with DTNB to form a TNB− anion and a CoA-TNB adduct; the anion absorbs at 412 nm with an extinction coefficient of 13,600 M−1 cm−1. Standard citrate synthase assay mixtures (0.8 ml) contained 50 mM HEPES buffer (pH 7.5), 0.1 M KCl, 0.54 M glycerol, 0.15 mM DTNB, 0.05 mM oxaloacetate, and 1 mM acetyl-CoA. For 2-methylcitrate synthase assays, the same conditions were used except that 0.1 mM propionyl-CoA was used instead of acetyl-CoA. For these assays, buffer and substrates were preincubated in 1.5-ml methacrylate cuvettes (Fisher Scientific) at 37°C in a Lambda 6 spectrophotometer (Perkin-Elmer, Norwalk, Conn.) equipped with a circulating water jacket. After 5 min, the assays were started by adding PrpC enzyme. The progress of the reaction was monitored for 10 min at 412 nm.
(iv) Kinetic analysis of PrpC.
Approximate Km values were initially determined for citrate and methylcitrate synthase activity by varying one substrate at a fixed concentration of the other substrate. These Km values were used to set up bisubstrate kinetic experiments to identify the true Km values. Assays were carried out by varying both substrates at four different concentrations ranging from 5 to 0.5 Km (16 assays). Experiment sets were performed at least four times for both citrate synthase and methylcitrate synthase assays. True Km and Vmax values were determined by fitting the reaction rates to the ordered bisubstrate rate equation, using the program GraFit 4.0 (18). For the data fitting, simple weighting was used and estimates were made from the apparent Km and Vmax values for each data set.
Other procedures.
Protein concentrations were determined, by the method of Kunitz (16), from a standard curve generated with bovine serum albumin. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (17) with 12% polyacrylamide gels and were visualized with Coomassie blue (25). Mid-range standards (14 to 97.4 kDa) were used for SDS-PAGE (Promega).
RESULTS
Accumulation of propionate catabolites in a prpBCDE+ strain.
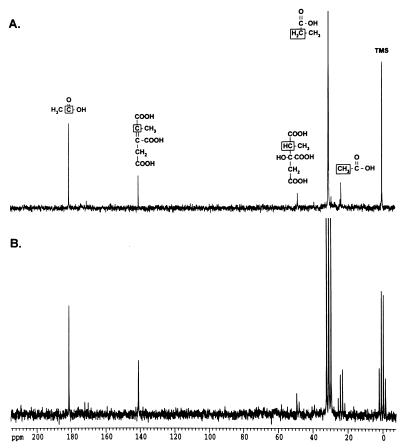
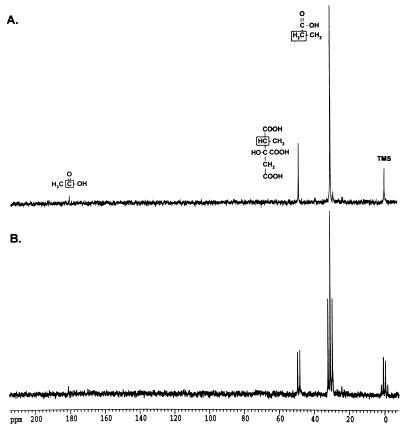
To identify breakdown intermediates in a strain capable of catabolizing propionate, strain TR6583 (prpBCDE+) was grown in NCE medium containing glycerol (20 mM) and [2-13C]propionate (5 mM) as carbon and energy sources. Glycerol was needed to slow the catabolism of propionate enough to allow the identification of pathway intermediates by 13C-NMR. Figure 2A shows the proton-decoupled spectrum of two intermediates whose chemical shifts correlated with those expected for a methine carbon at 48.8 ppm and for an alkene carbon at 140.5 ppm. To help identify these compounds, proton-coupled (off-resonance) spectra were obtained. The carbon signal at 48.8 ppm was shown to have one bound hydrogen (JCH = 132 Hz), while the carbon signal at 140.5 ppm did not have any attached hydrogens (Fig. 2B). This pattern of signals was consistent with intermediates of the 2-methylcitric acid cycle or the citramalate pathway (Fig. 1). The methine carbon signal would correspond to 2-methylcitrate or 3-methylmalate, whereas the alkene carbon signal would be consistent with 2-methyl-cis-aconitate or mes-aconate. Natural-abundance 13C-NMR showed that the corresponding alkene carbon in mes-aconate was located at 138.9 ppm, suggesting that the unknown signal at 140.5 ppm was probably due to an intermediate of the 2-methylcitric acid cycle (data not shown). Additionally, natural-abundance 13C-NMR spectra of citrate and cis-aconitate were obtained, and the corresponding carbons in these compounds showed similar peak positions and C-H coupling (45.8 ppm [JCH = 128 Hz] and 143.7 ppm, respectively) to those of the unknown intermediates. From these experiments, we assigned the 48.8-ppm methine carbon to 2-methylcitrate and the 140.5-ppm alkene carbon to 2-methyl-cis-aconitate.
FIG. 2.
Propionate catabolites visualized by 13C-NMR spectroscopy. Strain TR6583 (prpBCDE+) was grown in minimal medium supplemented with [2-13C]propionate to accumulate 13C-labeled propionate breakdown intermediates. (A) Proton-decoupled 13C-NMR spectrum of an extract from this culture. (B) Proton-coupled 13C-NMR spectrum of this extract. The boxed carbon atoms in the chemical structures above the peaks indicate the source of the carbon signals.
Signals for acetate (carboxylic carbon, 183 ppm; methyl carbon, 23 ppm) were also evident in the spectrum. The assignment of these signals was verified by direct addition of acetate to the sample. The acetate signals were due to the natural-abundance 13C levels, which were verified by preparing samples from cultures that were not grown in the presence of [2-13C]propionate (data not shown).
The above results obtained with the prpBCDE+ strain TR6583 strongly suggested that in S. typhimurium, propionate was catabolized via the 2-methylcitric acid cycle. To obtain further support for this hypothesis, we used mutant strains of S. typhimurium carrying mutations in prpB, prpC, or prpD to block propionate oxidation.
Ordering the propionate catabolic pathway through mutant analysis.
Each 13C-NMR experiment described below was performed at least twice, and experiments with additional point mutations in each prp gene yielded identical 13C-NMR peak profiles.
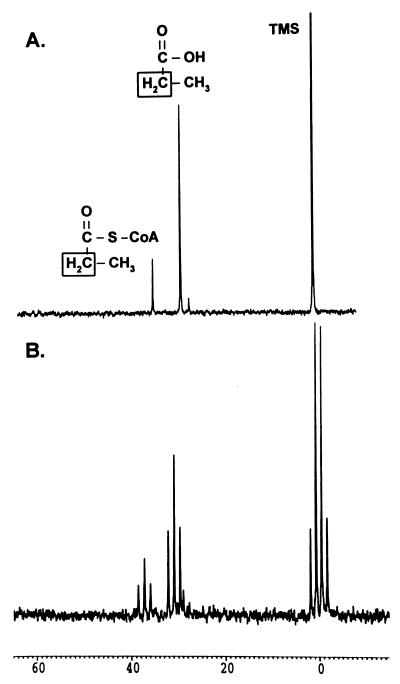
(i) The first step of the pathway: conversion of propionate to propionyl-CoA by PrpE.
To properly interpret the results of the 13C-NMR experiments reported herein, we determined the chemical shift of [2-13C]propionyl-CoA in the spectra. Crude cell extracts of strain JE4184 containing high levels of propionyl-CoA synthetase (PrpE) (13) were used to synthesize [2-13C]propionyl-CoA from propionate, ATP, and CoA. Carbon signals at 28.9 and 37.2 ppm were observed upon incubation of the substrates with PrpE (Fig. 3). These signals were not observed in reaction mixtures containing control cell extract of strain JE4287 (data not shown). The 37.2-ppm signal was present in the 13C natural-abundance spectrum of authentic propionyl-CoA (stored in 50 mM phosphate buffer [pH 7.5]). The chemical shift of the C-2 methylene carbon of the PrpE product was consistent with the shift reported for this carbon (37.5 ppm) in synthetic propionyl-CoA (22). As expected, the proton-coupled spectrum of [2-13C]propionyl-CoA showed the methylene oxidation state of the C-2 carbon of propionate was retained. The peak at 28.9 ppm was not identified.
FIG. 3.
In vitro conversion of [2-13C]propionate to [2-13C]propionyl-CoA by PrpE. Dialyzed cell extracts of strain JE4184 were used in these experiments. (A) Proton-decoupled 13C-NMR spectrum of the reaction products. (B) Proton-coupled 13C-NMR spectrum of the reaction products.
(ii) Evidence for a functional 2-methylcitric acid cycle in Salmonella typhimurium.
To allow for the accumulation of 13C-labeled intermediates, a mixture of [2-13C]propionate and glycerol was fed to mutant strains carrying lesions in prpB, prpC, or prpD. After a period of incubation, the cells were processed as described in Materials and Methods, and the 13C-NMR spectrum of each sample was obtained.
(a) Intermediates accumulating in prpC mutants.
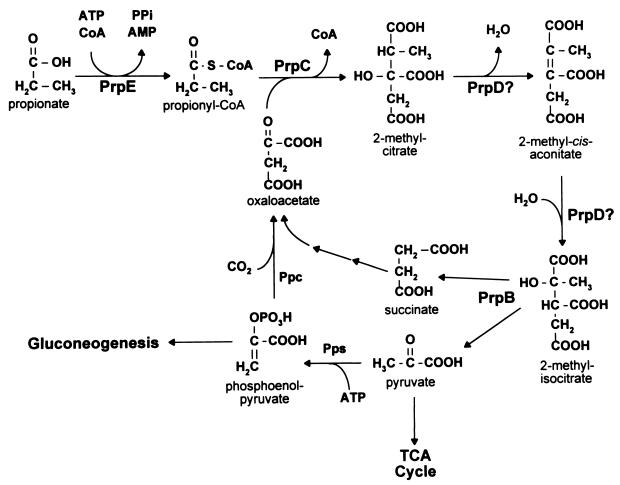
The 13C-NMR spectrum of samples of prpC mutants showed the expected signal for [2-13C]propionate at 30.8 ppm. Interestingly, no signal corresponding to propionyl-CoA was detected. We concluded that PrpC was responsible for the synthesis of 2-methylcitrate from propionyl-CoA and oxaloacetate (see Fig. 8), which was supported by 2-methylcitrate synthase assays with pure PrpC enzyme (see below). The lack of a signal for propionyl-CoA is discussed below.
FIG. 8.
Pathway for propionate catabolism in S. typhimurium. The results presented in this paper support the breakdown of propionate via 2-methylcitrate. The reactions catalyzed by enzymes encoded in the prpBCDE operon are indicated. A possible route for oxaloacetate regeneration and gluconeogenesis is shown (pps, PEP synthetase; Ppc, PEP carboxylase).
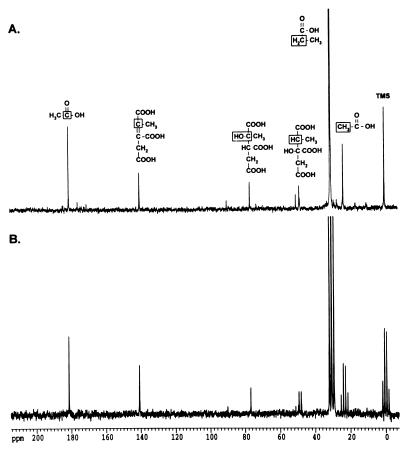
(b) Intermediates accumulating in prpD mutants.
Figure 4A shows that the 48.8-ppm carbon signal observed in the spectrum of the prpBCDE+ strain accumulated in the prpD mutant strain. Unlike in the prpBCDE+ strain, the intermediate with a chemical shift of 140.5 ppm did not accumulate in the mutant. The proton-coupled spectrum of the sample (Fig. 4B) shows that the 48.8-ppm carbon was a methine carbon with same JCH coupling (JCH = 132 Hz) as the signal observed in the spectrum of the prpBCDE+ strain. Propionyl-CoA did not accumulate in prpD mutants. In sample preparations, acetate (carboxylic carbon, 183 ppm; methyl carbon, 23 ppm) and formate (170 ppm) signals were present at different levels in the spectrum (data not shown). We determined that the acetate and formate signals were from natural-abundance 13C levels by preparing prpD mutant samples without [2-13C]propionate. We concluded that mutants lacking prpD function accumulate 2-methylcitrate. Hence, either PrpD was involved in the conversion of 2-methylcitrate to 2-methylisocitrate, or it was catalyzing the conversion of the latter to pyruvate and succinate. Results of experiments with prpB mutants (see below) indicate that PrpD was needed to convert 2-methylcitrate to 2-methylisocitrate (see Fig. 8).
FIG. 4.
Propionate catabolites that accumulate in a prpD mutant. Strain JE3914 (prpD174) was grown in minimal medium supplemented with [2-13C]propionate to accumulate 13C-labeled propionate breakdown intermediates. (A) Proton-decoupled spectrum of an extract from this culture. (B) Proton-coupled spectrum of this extract. The boxed carbon atoms in the chemical structures above the peaks indicate the source of the carbon signals.
(c) Intermediates accumulating in prpB mutants.
Figure 5A shows the proton-decoupled spectrum of propionate catabolites that accumulate in prpB point mutants. Signals corresponding to 2-methylcitrate (48.8 ppm) and 2-methyl-cis-aconitate (140.5 ppm) were observed, with an additional signal at 76.8 ppm in the region expected for an alcohol. Data from the proton-coupled spectrum (Fig. 5B) were consistent with 2-methylcitrate (48.8 ppm) and 2-methyl-cis-aconitate (140.5 ppm). This spectrum also showed that no hydrogens were attached to the carbon with a chemical shift of 76.8 ppm.
FIG. 5.
Propionate catabolites that accumulate in a prpB mutant. Strain JE3946 (prpB195) was grown in minimal medium supplemented with [2-13C]propionate to accumulate 13C-labeled propionate breakdown intermediates. (A) Proton-decoupled spectrum of an extract from this culture. (B) Proton-coupled spectrum of this extract. The boxed carbon atoms in the chemical structures above the peaks indicate the source of the carbon signals.
A control experiment with natural-abundance citramalate demonstrated that the signal at 76.8 ppm was not citramalate (74.5 ppm). The intermediate with a 76.8-ppm chemical shift was proposed to be 2-methylisocitrate. As with the wild-type strain, acetate signals in the spectra of prpB mutants were derived primarily from 13C natural abundance (see above). We concluded that prpB mutants lack 2-methylisocitrate lyase activity. Lack of this activity resulted in the accumulation of 2-methylcitrate and 2-methyl-cis-aconitate. As with the prpC and prpD mutants, propionyl-CoA did not accumulate in prpB mutants.
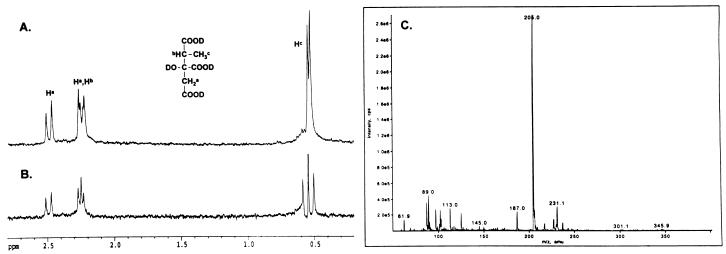
Analysis of the PrpC reaction product.
The in vivo 13C-NMR experiments suggested that PrpC had 2-methylcitrate synthase activity. To confirm this conclusion, His-tagged PrpC was incubated with propionyl-CoA and oxaloacetate, and the reaction product was analyzed by 1H-NMR spectroscopy. The 1H-NMR spectrum (Fig. 6A) showed four unique signals at 0.5 ppm (d, J = 7.2 Hz), 2.2 ppm (d, J = 16.3 Hz), 2.2 ppm (q, undetermined coupling), and 2.4 ppm (d, J = 16.3 Hz). The signals were consistent with the predicted 1H-NMR spectrum of 2-methylcitrate. To verify that the methine and methyl protons are coupled, homonuclear decoupling was used to remove the doublet at 2.2 ppm. As expected for 2-methylcitrate, removing this doublet decoupled the methine proton (Fig. 6B). The identity of the PrpC product was also confirmed by ESI mass spectrometry (Fig. 6C). In negative-ion mode, the mass of the primary ion was m/z = (205)− which corresponds to the mass of 2-methylcitrate (206 Da). The other ions in the mass spectrum were primarily contaminants from the in vitro PrpC reaction. The combined in vivo and in vitro NMR results and the mass spectrum demonstrated that the PrpC reaction product is 2-methylcitrate.
FIG. 6.
Spectroscopic analysis of the PrpC reaction product. (A) 1H-NMR spectrum; (B) homodecoupling 1H-NMR spectrum with irradiation at the methyl doublet; (C) ESI mass spectrum. The structure shown is 2-methylcitrate.
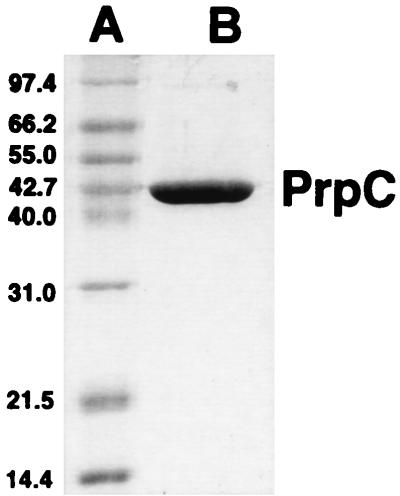
Purification of PrpC.
PrpC was purified to homogeneity by using an N-terminal His tag (see Materials and Methods). SDS-PAGE analysis (Fig. 7) indicated that His-tagged PrpC was an approximately 45-kDa monomer, which was consistent with the molecular mass of PrpC (43-kDa monomer) without the N-terminal His tag. By using the DTNB assay of Srere et al. (29), PrpC was tested for 2-methylcitrate synthase, citrate synthase, and glyoxylate condensation activity. In agreement with our previous results (see above), PrpC readily catalyzed the synthesis of 2-methylcitrate from propionyl-CoA and oxaloacetate. PrpC also catalyzed the synthesis of citrate from acetyl-CoA and oxaloacetate, demonstrating that the enzyme has citrate synthase activity as well. Glyoxylate did not substitute for oxaloacetate in the reaction. To identify the preferred substrate for the enzyme, bisubstrate kinetic experiments were performed with propionyl-CoA or acetyl-CoA as the substrate (Table 2). The kinetic data supported a sequential reaction mechanism for PrpC (data not shown). The kcat/Km specificity values were about 30:1 in favor of propionyl-CoA, which supports our conclusion that PrpC is the 2-methylcitrate synthase enzyme of S. typhimurium. The kinetic data were in good agreement with those reported for E. coli PrpC and for the 2-methylcitrate synthase from the Antarctic bacterium DS2-3R (9). These comparisons suggest that the His tag on PrpC has little, if any, deleterious effect on enzyme activity.
FIG. 7.
SDS-PAGE analysis of homogeneous PrpC. Lanes: A, molecular mass standards (in decreasing mass order): phosphorylase b, bovine serum albumin, glutamate dehydrogenase, ovalbumin, aldolase, carbonic anhydrase, lysozyme; B, His-tagged PrpC (3 μg) (approximately 45-kDa monomer).
TABLE 2.
Kinetic parameters for PrpC citrate synthase and 2-methylcitrate synthase activity
| Parameter | Valuea |
|---|---|
| Citrate synthase activity | |
| Km acetyl-CoA | 285 ± 20 μM |
| Km oxaloacetate | 14 ± 4 μM |
| Vmax | 2 ± 0.1 μmol/min/mg |
| kcat | 1.4 ± 0.1b s−1 |
| kcat/Km acetyl-CoA | 5 × 103 M−1 s−1 |
| 2-Methylcitrate synthase activity | |
| Km propionyl-CoA | 48 ± 8 μM |
| Km oxaloacetate | 12 ± 2 μM |
| Vmax | 10 ± 0.3 μmol/min/mg |
| kcat | 7.4 ± 0.2b s−1 |
| kcat/Km propionyl-CoA | 150 × 103 M−1 s−1 |
Values reported as mean ± standard deviation.
kcat values were calculated per subunit of His-tagged PrpC.
In vitro coupling of PrpE and PrpC yields 2-methylcitrate.
2-[2-13C]methylcitrate was synthesized in vitro (see Materials and Methods) to verify that the 13C-labeled compound accumulating in prpD mutants was 2-methylcitrate. The proton-decoupled 13C-NMR spectra of the reaction mixture obtained after incubation of the substrates with the PrpE and PrpC enzymes showed a signal with a chemical shift of the methylene carbon for [2-13C]propionate (30.8 ppm) and a signal at 48.8 ppm (data not shown). The latter signal had a chemical shift identical to that of the intermediate that accumulated in prpD mutants (Fig. 4A). Proton-coupled 13C-NMR spectra confirmed the identity of the carbon signal by demonstrating that one hydrogen was attached with JCH = 132 Hz (data not shown). Control crude cell extract of a strain containing the overexpression plasmid (pT7-7) lacking prpC showed only signals for propionate (30.8 ppm) and propionyl-CoA (37.2 ppm). On the basis of these results, we concluded that the signal at 48.8 ppm was due to PrpC activity. This experiment was later repeated with pure PrpC enzyme and purified preparations of PrpE (data not shown), and the same result was obtained.
The glyoxylate shunt is not required for the catabolism of propionate in S. typhimurium.
A functional glyoxylate shunt was reported to be required for growth of E. coli on propionate as the carbon and energy source (33). We tested the ability of glyoxylate shunt mutants of S. typhimurium carrying null alleles of aceB (encoding isocitrate lyase), aceA (encoding malate synthase), or aceK (encoding isocitrate dehydrogenase phosphatase/kinase) to grow on various carbon sources. As expected, these strains failed to grow on acetate. However, they did grow on propionate and pyruvate as carbon sources. These results demonstrated that, unlike in E. coli, the glyoxylate shunt was not required for the breakdown of propionate in S. typhimurium. To identify the anaplerotic pathway required in S. typhimurium, we tested pps (encoding phosphoenolpyruvate synthetase) mutants for growth on pyruvate and propionate. These mutants failed to grow on either carbon source, indicating that Pps is essential for growth of S. typhimurium on propionate.
DISCUSSION
It was previously reported that S. typhimurium catabolizes propionate via the acryloyl-CoA pathway (8). The data presented in this paper clearly eliminate this pathway. Our work indicates that S. typhimurium catabolizes propionate via the 2-methylcitric acid cycle. We propose that the Prp enzymes encoded by the prpBCDE operon catalyze the reactions of this pathway, as shown in Fig. 8. In this pathway, PrpE is the propionyl-CoA synthetase (13) and PrpC is the 2-methylcitrate synthase. PrpD is needed for the synthesis of 2-methylisocitrate, but it is unclear whether PrpD is responsible only for the dehydration of 2-methylcitrate to 2-methyl-cis-aconitate or whether it can also convert the latter to 2-methylisocitrate. PrpB is the 2-methylisocitrate lyase that cleaves the 2-methylisocitrate into pyruvate and succinate (10) (Fig. 8).
Because of a previous report (21), we considered the possibility that S. typhimurium catabolizes propionate via the closely related citramalate pathway (Fig. 1). However, natural-abundance 13C-NMR data obtained with authentic mes-aconate and citramalate did not support the existence of this pathway in S. typhimurium. In addition, PrpC failed to catalyze the synthesis of citramalate from glyoxylate and propionyl-CoA in vitro (data not shown).
Flow of propionate through the 2-methylcitrate cycle. (i) prp mutants do not accumulate propionyl-CoA.
It is of interest that propionyl-CoA did not accumulate in any of the prp mutants tested, particularly in prpC mutants. Although this observation may be of physiological significance, it could also be explained as an artifact of the use of crude enzyme preparations. It is possible that phosphotransacetylase (Pta) and acetate kinase (AckA) are degrading propionyl-CoA to propionate in the presence of phosphate buffer. Genetic experiments by Van Dyk and LaRossa with S. typhimurium suggest that the Pta and AckA enzymes can catalyze these reactions (36). Cleaner preparations of enzymes and a detailed analysis of their catalytic activities are needed to properly address this result.
(ii) Conversion of 2-methylcitrate to 2-methylisocitrate.
The similarities of the TCA cycle and the 2-methylcitric cycle would suggest that the conversion of 2-methylcitrate to 2-methylisocitrate is catalyzed by an aconitase. However, the 13C-NMR results indicate that PrpD is required for this conversion, and this enzyme shows no sequence similarities to aconitases or other related enzymes (12). It is possible that PrpD activity is not sufficient for the conversion of 2-methylcitrate to 2-methylisocitrate; the existence of two separate enzymes catalyzing this conversion is not unprecedented. Two separate dehydratases were identified for the 2-methylcitric acid cycle in Yallowia lipolytica, one specific for 2-methylcitrate and the other specific for 2-methylisocitrate (1, 30). Unfortunately, the genes encoding these dehydratase activities have not been identified. If PrpD only converts 2-methylcitrate to 2-methyl-cis-aconitate, the gene encoding 2-methylisocitrate dehydratase must lie elsewhere on the S. typhimurium chromosome. Beef liver and Saccharomycopsis lipolytica aconitases use 2-methyl-cis-aconitate and 2-methylisocitrate as substrates (26), which lends support to the notion that the TCA cycle aconitase of S. typhimurium catalyzes this half-reaction. It is also possible that other hydratases such as fumarase are catalyzing this reaction. To complete the assignment of functions in our model (Fig. 8), the enzyme(s) catalyzing the conversion of 2-methylcitrate to 2-methylisocitrate need to be identified.
(iii) The last step in the pathway.
In prpB point mutants, 2-methylcitrate and 2-methyl-cis-aconitate appear to accumulate to higher levels than 2-methylisocitrate (Fig. 5). This result may reflect the equilibrium of the methylcitrate-methylisocitrate interconversion, which may favor 2-methylcitrate formation. For mammalian aconitases, it is known that citrate formation is preferred over isocitrate in the TCA cycle (3, 15). In the 2-methylcitric acid cycle, PrpB activity (2-methylisocitrate lyase) is probably necessary to drive propionate breakdown to completion.
Regeneration of oxaloacetate.
The source of oxaloacetate for the 2-methylcitric acid cycle is an important issue in propionate breakdown. Recent isotope labeling studies with E. coli indicate that oxaloacetate is regenerated mainly through the glyoxylate shunt and, not surprisingly, glyoxylate shunt mutants do not grow on propionate (33). However, these results contrast with results of previous studies of E. coli indicating that the requirement for the shunt was strain dependent. Some strains regenerated oxaloacetate through phosphoenolpyruvate (PEP) synthetase and PEP carboxylase or through the glyoxylate shunt (14). S. typhimurium glyoxylate shunt mutants grow well on propionate, which suggests that oxaloacetate can be generated from pyruvate via the PEP synthetase and PEP carboxylase reactions (Fig. 8). In support of this proposal, pps (encoding PEP synthetase) mutants failed to utilize propionate as a substrate. It is noteworthy that the Cra (FruR) regulatory protein is required for expression of pps and ppc (encoding PEP carboxylase) (24). The inability of cra mutants to grow on propionate (11) may be the result of insufficient levels of oxaloacetate rather than a direct effect on the transcription of the prpBCDE operon. Low levels of oxaloacetate would result in lower levels of 2-methylcitrate, the proposed coactivator of prpBCDE expression (35). This PEP synthetase and carboxylase pathway of oxaloacetate synthesis for propionate catabolism appears more plausible than the glyoxylate shunt, especially since mutations that eliminate aceBAK functions do not affect the ability of S. typhimurium to grow on propionate (see above). The regulation of these anaplerotic pathways may be the source of differences among E. coli strains and S. typhimurium.
The difficulties of E. coli to grow on propionate may be due to evolution.
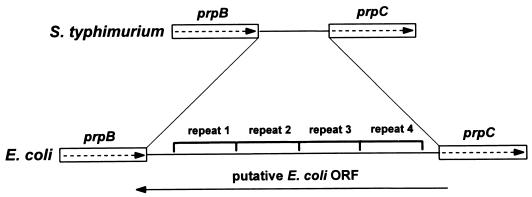
An interesting difference between E. coli and S. typhimurium prpBCDE operons is the gap between the prpB and prpC genes (Fig. 9). Sequence data from the E. coli genome project were compared to the prp operon sequence previously obtained for S. typhimurium (12). The gap distance is 439 bp in E. coli and only 122 bp in S. typhimurium. Analysis of this gap shows that the first 82 bp are 34% identical between these bacteria while the last 40 bp are 73% identical. The missing 318 bp in S. typhimurium are actually part of four 91-bp repeats in the E. coli sequence. In E. coli, this region was proposed to contain an open reading frame (ORF) that is transcribed in the opposite direction to the prp operon (4). However, the presence of these repeats suggests that this region may have undergone some rearrangements, and in fact, it does not contain an ORF. It is possible that these repeats negatively affect the expression of the E. coli prpCDE operon. Interestingly, some wild-type E. coli strains are unable to grow on propionate as the sole carbon and energy source (14, 33). However, these strains were reported to spontaneously revert to growth on propionate at a frequency of 10−7 (14). It would be of interest to determine the sequence of the region between prpB and prpC in these “revertants.” No defects in the ability of wild-type S. typhimurium to grow on propionate have ever been noted.
FIG. 9.
Comparison of the prpBC intergenic region between S. typhimurium and E. coli. The 91-bp repeats and proposed ORF in E. coli are indicated.
Distribution of the 2-methylcitric acid cycle among procaryotes.
Previous studies on the 2-methylcitric acid cycle demonstrated that it was widely distributed among fungi (20). Considering that the 2-methylcitric acid cycle has now been identified in both S. typhimurium and E. coli (33), perhaps it is time to investigate the prevalence of this pathway among procaryotes. With the recent sequencing of many procaryotic genomes, homologues of the Prp enzymes can be easily identified. For example, sequence comparisons indicate that Bacillus subtilis has a cluster of four genes that encode proteins homologous to the PrpC, PrpD, PrpB, and PrpR enzymes (in that order) (20a). Homologues of the PrpC and PrpD enzymes can also be found in Mycobacterium tuberculosis (5a). The only other biochemical evidence for the pathway was a report of 2-methylcitrate synthase activity in extracts Pseudomonas aeruginosa (37). As more genome sequences become available and procaryotes are tested for activities of the 2-methylcitric acid cycle, we may find that this pathway is as widely distributed among procaryotes as among fungi.
ACKNOWLEDGMENTS
This work was supported by NSF grant MCB 9724924 to J.C.E.-S. A.R.H. was supported by NIH biotechnology training grant GM08349 and an NSF predoctoral fellowship. NMR studies were performed at the National Magnetic Resonance Facility at Madison, which is supported by the NIH Biomedical Technology Program (RR02301) with additional equipment funding from the University of Wisconsin, NSF Academic Infrastructure Program (BIR-9214394), NIH Shared Instrumentation Program (RR02781, RR08438), NSF Biological Instrumentation Program (DMB-8415048), and the U.S. Department of Agriculture.
We thank S. Maloy and R. Jeter for strains. We thank M. Anderson for technical assistance with the NMR experiments. A. Harms obtained the ESI mass spectra at the University of Wisconsin—Madison Biotechnology Center.
REFERENCES
- 1.Aoki H, Uchiyama H, Umetsu H, Tabuchi T. Isolation of 2-methylcitrate dehydratase, a new enzyme serving in the methylcitric acid cycle for propionate metabolism, from Yallowia lipolytica. Biosci Biotechnol Biochem. 1995;59:1825–1828. [Google Scholar]
- 2.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1989. [Google Scholar]
- 3.Blair J M. Magnesium and aconitase equilibrium: determination of apparent stability constants of magnesium substrate complexes from equilibrium data. Eur J Biochem. 1969;8:287–291. doi: 10.1111/j.1432-1033.1969.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Chan R K, Botstein D, Watanabe T, Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high transducing lysate. Virology. 1972;50:883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 5a.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Sqares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 6.Davis R W, Botstein D, Roth J R. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 7.Evans C T, Sumegi B, Srere P A, Sherry A D, Malloy C R. [13C]propionate oxidation in wild-type and citrate synthase mutant Escherichia coli: evidence for multiple pathways of propionate utilization. Biochem J. 1993;291:927–932. doi: 10.1042/bj2910927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Briera A, Garrido-Pertierra A. A degradation pathway of propionate in Salmonella typhimurium LT-2. Biochimie. 1988;70:757–768. doi: 10.1016/0300-9084(88)90105-8. [DOI] [PubMed] [Google Scholar]
- 9.Gerike U, Hough D W, Russell N J, Dyall-Smith M L, Danson M J. Citrate synthase and 2-methylcitrate synthase: structural, functional and evolutionary relationships. Microbiology. 1998;144:929–935. doi: 10.1099/00221287-144-4-929. [DOI] [PubMed] [Google Scholar]
- 10.Grimek, T. L., and J. C. Escalante-Semerena. 1999. Unpublished results.
- 11.Hammelman T A, O’Toole G A, Trzebiatowski J R, Tsang A W, Rank D, Escalante-Semerena J C. Identification of a new prp locus required for propionate catabolism in Salmonella typhimurium LT2. FEMS Microbiol Lett. 1996;137:233–239. doi: 10.1111/j.1574-6968.1996.tb08111.x. [DOI] [PubMed] [Google Scholar]
- 12.Horswill A R, Escalante-Semerena J C. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horswill A R, Escalante-Semerena J C. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology. 1999;145:1381–1388. doi: 10.1099/13500872-145-6-1381. [DOI] [PubMed] [Google Scholar]
- 14.Kay W W. Genetic control of the metabolism of propionate by Escherichia coli K12. Biochim Biophys Acta. 1972;264:508–521. doi: 10.1016/0304-4165(72)90014-1. [DOI] [PubMed] [Google Scholar]
- 15.Krebs H A. The equilibrium constants of the fumarase and aconitase systems. Biochem J. 1953;54:78–82. doi: 10.1042/bj0540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunitz M J. Crystalline inorganic pyrophosphatase isolated from baker’s yeast. J Gen Physiol. 1952;35:423–450. doi: 10.1085/jgp.35.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage and structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Leatherbarrow R J. GraFit version 4.0. Staines, United Kingdom: Erithacus Software Ltd.; 1998. [Google Scholar]
- 19.London R E, Allen D L, Gabel S A, DeRose E F. Carbon-13 nuclear magnetic resonance study of metabolism of propionate by Escherichia coli. J Bacteriol. 1999;181:3562–3570. doi: 10.1128/jb.181.11.3562-3570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyakoshi S, Uchiyama H, Someya T, Satoh T, Tabuchi T. Distribution of the methylcitric acid cycle and β-oxidation for propionate catabolism in fungi. Agric Biol Chem. 1987;51:2381–2387. [Google Scholar]
- 20a.Mizuno M, Masuda S, Takemaru K, Hosono S, Sato T, Takeuchi M, Kobayashi Y. Systematic sequencing of the 283 kb 210 degrees-232 degrees region of the Bacillus subtilis genome containing the skin element and many sporulation genes. Microbiology. 1996;142:3103–3111. doi: 10.1099/13500872-142-11-3103. [DOI] [PubMed] [Google Scholar]
- 21.Rabin R, Reeves H C, Wegener W S, Megraw R E, Ajl S J. Glyoxylate in fatty acid metabolism. Science. 1965;150:1548–1558. doi: 10.1126/science.150.3703.1548. [DOI] [PubMed] [Google Scholar]
- 22.Robbins J E, Ball M S, Williams S A. Synthesis and NMR spectra of 13C-labeled coenzyme A esters. Anal Biochem. 1986;157:84–88. doi: 10.1016/0003-2697(86)90199-5. [DOI] [PubMed] [Google Scholar]
- 23.Ryu J-I, Hartin R J. Quick transformation of Salmonella typhimurium LT2. BioTechniques. 1990;8:43–45. [PubMed] [Google Scholar]
- 24.Saier M H, Jr, Ramseier T M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasse J. Detection of proteins. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1991. pp. 10.6.1–10.6.8. [Google Scholar]
- 26.Schloss J V, Emptage M H, Cleland W W. pH profiles and isotope effects for aconitase from Saccharomycopsis lipolytica, beef heart, and beef liver. α-Methyl-cis-aconitate and threo-ds-α-methylisocitrate as substrates. Biochemistry. 1984;23:4572–4580. doi: 10.1021/bi00315a010. [DOI] [PubMed] [Google Scholar]
- 27.Schmieger H. A method for detection of phage mutants with altered transduction ability. Mol Gen Genet. 1971;100:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 28.Schmieger H, Bakhaus H. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants) Mol Gen Genet. 1973;120:181–190. doi: 10.1007/BF00267246. [DOI] [PubMed] [Google Scholar]
- 29.Srere P A, Brazil H, Gonen L. The citrate condensing enzyme of pigeon breast muscle and moth flight muscle. Acta Chem Scand. 1963;17:S129–S134. [Google Scholar]
- 30.Tabuchi T, Aoki H, Uchiyama H, Nakahara T. 2-Methylcitrate dehydratase, a new enzyme functioning at the methylcitric acid cycle of propionate metabolism. Agric Biol Chem. 1981;45:2823–2829. [Google Scholar]
- 31.Tabuchi T, Hara S. Production of 2-methylcitric acid from n-paraffins by mutants of Candida lipolytica. Agric Biol Chem. 1974;38:1105–1106. [Google Scholar]
- 32.Tabuchi T, Serizawa N. A hypothetical cyclic pathway for the metabolism of odd-carbon n-alkanes or propionyl-CoA via seven-carbon tricarboxylic acids in yeasts. Agric Biol Chem. 1975;39:1055–1061. [Google Scholar]
- 33.Textor S, Wendisch V F, De Graaf A A, Müller U, Linder M I, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- 34.Tsai S P, Hartin R J, Ryu J. Transformation in restriction-deficient Salmonella typhimurium LT2. J Gen Microbiol. 1989;135:2561–2567. doi: 10.1099/00221287-135-9-2561. [DOI] [PubMed] [Google Scholar]
- 35.Tsang A W, Horswill A R, Escalante-Semerena J C. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J Bacteriol. 1998;180:6511–6518. doi: 10.1128/jb.180.24.6511-6518.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Dyk T K, LaRossa R A. Involvement of ack-pta operon products in α-ketobutyrate metabolism in Salmonella typhimurium. Mol Gen Genet. 1987;207:435–440. doi: 10.1007/BF00331612. [DOI] [PubMed] [Google Scholar]
- 37.Watson D, Lindel D L, Fall R. Pseudomonas aeruginosa contains an inducible methylcitrate synthase. Curr Microbiol. 1983;8:17–21. [Google Scholar]
- 38.Wegener W S, Reeves H C, Ajl S J. Propionate oxidation in Escherichia coli. Arch Biochem Biophys. 1967;121:440–442. doi: 10.1016/0003-9861(67)90098-7. [DOI] [PubMed] [Google Scholar]
- 39.Wegener W S, Reeves H C, Rabin R, Ajl S J. Alternate pathways of metabolism of short-chain fatty acids. Bacteriol Rev. 1968;32:1–26. doi: 10.1128/br.32.1.1-26.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]