Abstract
Cryptococcus neoformans is a pathogenic fungus which most commonly affects the central nervous system and causes fatal meningoencephalitis primarily in patients with AIDS. This fungus produces a thick extracellular polysaccharide capsule which is well recognized as a virulence factor. Here, we describe the isolation and characterization of a novel gene, CAP10, which is required for capsule formation. Complementation of the acapsular cap10 mutant produced an encapsulated strain and the deletion of CAP10 from a wild strain resulted in an acapsular phenotype. The molecular mass of the hemagglutinin epitope-tagged Cap10p is about 73 kDa, which is similar to the size predicted from sequence analysis. When CAP10 was fused with a hybrid green fluorescent protein construct, the fluorescence signals appeared as patches in the cytoplasm. Using a reporter gene construct, we found that CAP10 was expressed at high levels in late-stationary-phase cells. In addition, we found that the expression levels of CAP10 are modulated by the transcriptional factor STE12α. Deletion of STE12α downregulated the expression levels of CAP10 while overexpression of STE12α upregulated the expression levels of CAP10. Animal model studies indicate that deletion of the CAP10 gene results in the loss of virulence, and complementation of the acapsular phenotype of cap10 restores virulence. Thus, CAP10 is required for capsule formation and virulence.
Cryptococcus neoformans is a pathogenic fungus which most commonly affects the central nervous system and causes fatal meningoencephalitis in AIDS patients (20). This fungus produces a thick extracellular polysaccharide capsule which is a well-recognized virulence factor (15, 22). The predominant capsular polysaccharide of C. neoformans is glucuronoxylomannan (GXM), which consists of an O-acetylated, α-1,3-mannose backbone with xylosyl and glucuronosyl side chains. The extent of O-acetylation and xylosyl substitution varies with serotype. The biochemical pathway for synthesis of the polysaccharide capsule, however, is not clear.
Several acapsular mutants have been isolated by classic mutational approaches (3, 27). Molecular cloning by direct complementation of acapsular mutants has resulted in the isolation of three genes, CAP59, CAP60, and CAP64, which are required for capsule formation (2–4). While these genes are not essential for growth in vitro, each has been shown to be required for virulence in the murine model. DNA sequence analysis was not indicative of their biochemical function except that Cap59p and Cap60p contained a putative transmembrane domain and shared sequence similarity at the center of their coding regions. Functional analysis indicated that the transmembrane domain of Cap59p was required for its ability to complement the cap59 acapsular mutant (2). Immunogold electron microscopy revealed the location of Cap60p to be around the nuclear membrane (3).
Difficulties in using histochemical methods to localize gene products in C. neoformans (3, 29) have been encountered. The green fluorescent protein (GFP) from the bioluminescent jellyfish Aequorea victoriae (24) has emerged as a useful marker in studying protein localization in a variety of organisms. The formation of fluorophore appears to be cell autonomous besides the requirement for molecular oxygen (9, 18, 25). Direct visualization of gene expression in individual cells is therefore possible without distortion caused by fixation, sectioning, and staining.
Although the molecular mechanisms of regulation in capsule synthesis are not clear, it has been shown that a homolog of the GPA1 gene encoding the G-protein alpha subunit in the signal transduction pathway influences capsule production in response to iron limitation (1). Recently, another gene involved in the pheromone response signal transduction cascade, STE12α, was also found to modulate the expression of several capsule-associated genes, including CAP59, CAP60, and CAP64 (5). The STE12α gene of C. neoformans shares sequence similarity with the Saccharomyces cerevisiae STE12 gene and its homologs, but STE12α exists only in MATα strains of C. neoformans (31). The STE12α gene is required for haploid fruiting on filamentous agar but not for mating. Experimental infections in the murine model suggested that the STE12α gene is important for virulence in C. neoformans (5).
We isolated and characterized a novel capsular gene, CAP10, which may also encode a protein containing a transmembrane domain. CAP10 is required for capsule formation and its deletion abolishes the ability of the fungus to cause fatal infection in mice. Cellular location of the CAP10 gene product was determined by tagging the Cap10p with a new hybrid GFP specific for C. neoformans. In addition, the Escherichia coli β-glucuronidase (GUS) gene was used as a reporter to monitor the levels of expression of CAP10 during different stages of growth. The importance of STE12α in regulating CAP10 expression was also demonstrated.
MATERIALS AND METHODS
Strains and media.
The cDNA library was constructed by J. C. Edman from mRNA of log-phase cells. Table 1 summarizes the strains used in this study. C. neoformans var. neoformans strains B-3501 (MATα) and B-3502 (MATa) have been described previously (19). B-4500 is a wild-type congenic strain of B-4476 (21). B-4500FO2 is a ura5 auxotroph and LP1 is an ade2 ura5 strain derived from B-4500 (3). Strain cap10F2 is an F2 progeny obtained from a cross between the acapsular mutant, cap10C (3), and a MATa strain. Strain cap10F2FO is a ura5 auxotroph of cap10F2 and was isolated according to the methods described previously (23). TYCC245F1FO is a ura5 auxotroph of Δste12α and TYCC259 is a ura5 auxotroph containing GAL7(p)::STE12α (5). YEPD contains 1% yeast extract, 2% Bacto Peptone, and 2% dextrose. Minimal medium (YNB) contains 6.7 g of yeast nitrogen base without amino acids (Difco) and 20 g of glucose per liter. 5-Fluoroorotic acid (5-FOA) medium contains 6.7 g of yeast nitrogen base (Difco), 1 g of 5-FOA, 50 mg of uracil, and 20 g of glucose per liter.
TABLE 1.
List of strains relevant to this study
| Strain | Genotype | Source or reference |
|---|---|---|
| B-3501 | MATα | 19 |
| B-3502 | MATa | 19 |
| B-4500 | MATα | 21 |
| B-4476 | MATa | 21 |
| B-4500FO2 | MATα ura5 | 3 |
| cap10C | MATα cap10 | This study |
| cap10F2 | MATα cap10 | This study |
| cap10F2FO | MATα cap10 ura5 | This study |
| CIP3 | MATα ura5 cap10 pCIP3(URA5) | This study |
| LP1 | MATα ura5 ade2 | 3 |
| TYCC133 | MATα ura5 cap10 pYCC133(URA5 CAP10) | This study |
| TYCC150 | MATα ura5 ade2 Δcap10::ADE2 | This study |
| TYCC259 | MATα ura5 ade2 ADE2::GAL7(p)::STE12α | 5 |
| TYCC245F1FO | MATα ura5 Δste12::ADE2 | 5 |
Transformation of C. neoformans.
The electroporation method described by Edman and Kwon-Chung (13) was used to transform C. neoformans. TYCC133 is a stable encapsulated transformant of cap10F2FO containing pYCC133 and CIP3 is a stable acapsular transformant of cap10F2FO containing pCIP3. The stable transformants were uracil prototrophs obtained after three transfers on YEPD medium.
Preparation and analysis of nucleic acid and proteins.
Genomic DNA isolation and analysis were as described previously (2). Random hexamer priming was used to label the DNA probes to specific activities of >108 dpm/μg (14). Total RNA was isolated by using the FastRNA kit (Bio 101, Vista, Calif.) and poly(A)+ RNA was isolated by using the Oligotex mRNA kit (Qiagen, Valencia, Calif.). Northern blot analysis was performed as described previously (6). Following each hybridization, the blot was exposed to PhosphorImager Screen and the CAP10 specific signal, normalized to that of the actin gene, was quantified with ImageQuant 1.1 (Molecular Dynamics). DNA sequencing was performed by the dideoxy-mediated chain-termination method using a Sequenase version 2.0 kit (U.S. Biochemicals, Cleveland, Ohio). Programs of the University of Wisconsin Genetics Computer Group (Madison, Wis.) were used for analysis of nucleic acid sequences (11).
Total protein isolation, polyacrylamide gel electrophoresis, and Western blot analyses were as described previously (3). The membrane was incubated with anti-hemagglutinin (HA) monoclonal antibody (BAbCO, Richmond, Calif.) followed by secondary antibody obtained from the Western-Star chemiluminescence detection system (Tropix, Bedford, Mass.) and was used as suggested by the manufacturers.
Construction of plasmids.
Table 2 summarizes the plasmids used in this study. The URA5-containing plasmid, pCIP3, was obtained from J. C. Edman. The BamHI-EcoRI fragment of pYCC76 (3), which contained the functional ADE2 gene, was cloned into the BamHI-EcoRI site of pBC(KS+) to yield pYCC123. To recover free plasmids from C. neoformans, genomic DNA from encapsulated transformants was digested with NotI, ligated, and transformed into E. coli. Plasmid pYCC125 was one of several plasmids recovered from E. coli which were able to complement the mutation of cap10F2FO. Plasmids pYCC130, pYCC131, and pYCC132 were subclones of pYCC125 in pCIP3 (Fig. 1A).
TABLE 2.
List of plasmids relevant to this study
| Plasmid | Description | Source |
|---|---|---|
| pBIN 35S-mgfp5-ER | GFP designed for plant | J. Haseloff |
| pCIP3 | URA5 in pBluescript | J. Edman |
| pYGFP3 | GFP designed for C. albicans | B. Cormack |
| pYCC123 | ADE2 in pBC KS | This study |
| pYCC125 | CAP10 subclone; see Fig. 1 | This study |
| pYCC130 | CAP10 subclone; see Fig. 1 | This study |
| pYCC131 | CAP10 subclone; see Fig. 1 | This study |
| pYCC132 | CAP10 subclone; see Fig. 1 | This study |
| pYCC133 | CAP10 subclone; see Fig. 1 | This study |
| pYCC147 | 1.6-kb XbaI fragment at 3′ end of CAP10 | This study |
| pYCC150 | Deletion construct of CAP10; see Fig. 2A | This study |
| pYCC152 | HA epitope-tagged CAP10 | This study |
| pYCC203 | Three-HA epitope-tagged CAP10 | This study |
| pYCC330 | CAP10(p)::GUS in pPM8 | This study |
| pYCC331 | Promoterless GUS in pPM8 | This study |
| pYCC352 | Hybrid GFP-tagged CAP10 | This study |
FIG. 1.
Genomic structure of CAP10. (A) Restriction map of CAP10. Overlapping subclones of pYCC125 are depicted. Plasmids were transformed into the acapsular strain cap10F2FO. The capsular phenotypes of the resulting transformants were scored as indicated. Arrow represents transcriptional direction of CAP10 and triangles represent introns. Filled box represents the coding region of CAP10. B, BamHI; Bg, BglII; D, HindIII; M, MscI; N, NsiI; Xb, XbaI. (B) Southern blot analysis. Genomic DNA of B-4500 was digested with different restriction enzymes and fractionated on an 0.8% agarose gel. The DNA blot was hybridized with the 3.3-kb fragment of pYCC133. (C) Chromosomal location of CAP10. The chromosomal DNA was separated by CHEF gel electrophoresis and stained with ethidium bromide (I). The gel-separated chromosomal DNA was transferred to a nylon membrane and hybridized with the CAP10 probe (II). B-3501 (MATα) and B-3502 (MATa) are wild-type strains.
To construct a partial library, genomic DNA of B-4500 was digested with XbaI and fractionated on a 1.0% agarose gel. The region from 1.5 to 2.5 kb was gel isolated and cloned into pBluescript vector. The library was screened with the 1.2-kb BamHI-NotI fragment of pYCC125. One of the positive clones, pYCC147, containing a 1.6-kb insert was isolated. The deletion construct, pYCC150 (Fig. 2A), was constructed as follows. The 1.2-kb MscI-XbaI fragment of pYCC133 (Fig. 1A) was replaced with the 3.0-kb EcoRI-XbaI fragment of the ADE2 gene from pYCC123 to give pYCC149. The 1.2-kb NsiI-NotI fragment of pYCC147 was cloned into the BamHI-NotI site of pYCC149 to give pYCC150. The 5′ rapid amplification of cDNA ends (RACE) method was performed in accordance with the protocol accompanying the Marathon cDNA amplification kit (Clontech, Palo Alto, Calif.).
FIG. 2.
Deletion of the CAP10 gene. (A) CAP10 locus and gene replacement vector. B, BamHI; Bg, BglII; M, MscI; N, NsiI; S, SmaI; X, XhoI; Xb, XbaI. (B) Southern blot analysis. Genomic DNAs were prepared from the wild-type strain B-4500 and an acapsular strain, TYCC150. DNAs were digested with XhoI and separated in a 0.8% agarose gel. The blot was hybridized with a probe of the 6.0-kb SmaI-NsiI fragment of pYCC150 (I) or the 1.7-kb MscI-NsiI fragment which was deleted from pYCC150 (II). Arrows at 2.0 and 0.8 kb (B-4500; I) indicate the faint signals corresponding to ADE2 hybridization signals.
The HA epitope (YPYDYPDYA) was inserted in frame at the carboxyl terminus of Cap10p by PCR amplification of pYCC147 as described previously (3). The resulting plasmid (pYCC151) was sequenced to confirm that no errors had been introduced during amplification. The 3′ end of CAP10 in pYCC133 was replaced with the tagged fragment in pYCC151 to generate pYCC152. GETP1 containing three tandem copies of HA was developed by M. Tyers and B. Futcher. The BstXI-XbaI fragment of GETP1 was cloned into pYCC151 to give pYCC199 and the 3′ end of CAP10 in pYCC133 was replaced with the three-HA-tagged fragment in pYCC199 to generate pYCC203.
To create the CAP10(p)::GUS fusion, an NdeI site was created at the first ATG site of the CAP10 coding region by PCR and the resulting promoter was cloned into the NdeI site created at the first ATG of the GUS gene. Plasmid pYCC330 contained the CAP10(p)::GUS reporter gene construct in pPM8 which contained URA5, telomeres, and a 1.08-kb STAB sequence (a gift from P. Mondon). The telomeres increase the transformation frequency (12) and the 1.08-kb STAB sequence confers stability to the episomal plasmid (28). Plasmid pYCC331 contained a promoterless GUS gene in pPM8.
Construction of the GFP-containing plasmid, pYCC352, was briefly as follows. The first 77 amino acids of GFP, which includes the chromophore region of GFP, were from the 0.6-kb SalI-NdeI fragment of pYGFP3 developed for Candida species (7). The rest of the GFP was from the 0.5-kb NdeI-SacI fragment of pBIN 35S-mgfp5-ER (a gift from J. Haseloff), in which a cryptic intron between the 127th and 155th amino acids was removed from a thermotolerant mutant of GFP. This hybrid GFP was inserted into the C terminus of Cap10p and the entire fusion construct was placed in the plasmid pCIP3 to yield pYCC352.
GUS activity assay.
To monitor the GUS activity in different growing stages, transformants of B-4500FOF2 containing pYCC330 were inoculated in 10 ml of minimal medium containing 2% raffinose in 50-ml Falcon tubes and incubated at 30°C with shaking at 200 rpm for 24 h. One milliliter of this culture was then diluted in 9 ml of fresh minimal medium containing 2% glucose and reincubated as described above. Cells were harvested at 5, 10, 15, 25, 45, and 75 h after transfer and GUS activity was assayed as previously described (30). GUS activity was expressed as picomoles of 4-methylumbelliferone produced per minute per 200 μg of protein. To study the effect of overexpression of STE12α on CAP10 expression, TYCC259 was transformed with pYCC330. For galactose induction, cells were inoculated in 10 ml of minimal medium containing 2% raffinose in a 50-ml Falcon tube and incubated at 30°C with shaking at 200 rpm for 24 h. One milliliter of the culture was inoculated into 9 ml of fresh minimal medium containing either 2% glucose or 2% galactose as a carbon source. Cells were harvested after an additional 20-h incubation. To assay GUS activity in cultures of stationary phase, B-4500FO2 and TYCC245F1FO were transformed with pYCC330 separately. Cells were grown in 10 ml of minimal medium containing 2% raffinose for 24 h and 1 ml of this culture was diluted into 9 ml of fresh minimal medium containing 2% glucose. This culture was then grown for an additional 45 h under the same conditions. Cells were harvested and GUS activity was assayed. Six independent transformants from each strain were assayed for GUS activity.
Protein localization.
The immunofluorescence and immunogold localization methods were as described previously (3). GFP was visualized by using an Axiovert 100TV microscope (Carl Zeiss, Jena, Germany), and images were recorded using a C5810 color chilled 3CCD camera (Hamamatsu Corporation, Bridgewater, N.J.) and processed using Adobe Photoshop.
Virulence study.
Female BALB/c mice (body weight, 20 g) were injected via the lateral tail vein with each yeast strain as described previously (2) and mortality was monitored.
Nucleotide sequence accession number.
The GenBank nucleotide accession number for CAP10 is AF144574.
RESULTS
Cloning of the CAP10 gene.
We have generated 16 acapsular strains by mutagenesis (3). Among the 16 acapsular strains, four were complemented by CAP59, four were complemented by CAP60, and three were complemented by CAP64 (3). From the remaining five strains, one (cap10C) was randomly chosen in an attempt to complement its acapsular phenotype with a library of genomic DNA from B-4500. Several encapsulated transformants of cap10F2FO were obtained after enriching with a two-polymer aqueous-phase system (2). The plasmid responsible for the complementation was recovered and transformed into E. coli. The plasmid, pYCC125, containing a 5.0-kb insert, complemented the acapsular mutation of cap10F2FO but not any other acapsular mutants in our collection (Fig. 1A). The 5-kb insert was subcloned to minimize the region required for complementation. Plasmid pYCC133 containing a 3.3-kb insert was the smallest clone obtained (Fig. 1A). We designated the newly isolated gene CAP10.
Sequence analysis and genomic structure of CAP10.
DNA sequences of the genomic and cDNA clones of CAP10 were determined. Because the full-length cDNA was absent in our cDNA library, the 5′ portion of the cDNA was obtained by the 5′ RACE method. Comparisons of the genomic and cDNA sequences revealed the presence of three introns in CAP10. The presence of multiple introns in genes is a commonly observed feature in C. neoformans. Unlike the other three CAP genes (2–4), no other transcript was detected in close proximity to the CAP10 locus when pYCC125 was used as a probe. The CAP10 gene encodes a putative protein containing 640 amino acids with a calculated molecular mass of 73 kDa. Database searches did not reveal any gene sharing significant similarity with CAP10. However, a putative type II transmembrane region was detected close to the N terminus.
Southern blot analysis of CAP10 suggested the existence of a single copy of the CAP10 gene in the genome of B-4500 (Fig. 1B). Two of the previously isolated capsule-related genes, CAP59 and CAP60, are both on chromosome I and CAP64 is on chromosome III. To determine the chromosomal location of CAP10, chromosomal DNA was separated by contour-clamped homogeneous electric field (CHEF) electrophoresis and the resulting blot was hybridized with a probe of pYCC133. The result showed that CAP10 was on a chromosome which is different from the other three capsule-related genes (Fig. 1C).
The importance of CAP10 in capsule formation and virulence.
A positive-negative selection method was used to delete CAP10 from a wild-type strain (2). This method required a double crossover at the flanking region of the gene. However, the largest clone, pYCC125, which complemented the cap10 mutation contained only 90 bp beyond the stop codon of CAP10 (Fig. 1A). To construct the CAP10 deletion construct, a longer 3′ flanking region of CAP10 was obtained by screening an XbaI-digested partial genomic library. The deletion construct, pYCC150, which contained 1.1 kb flanking both 5′ and 3′ regions of CAP10 is shown in Fig. 2A. This plasmid was used to transform an ade2 ura5 strain, LP1, and the yeast cells were plated on 5-FOA plates. The DNA of acapsular transformants was isolated and analyzed by Southern blotting. Figure 2B shows that the >12-kb band in B-4500 was replaced by an 8.0-kb and a 2.0-kb band in TYCC150, which indicated the replacement of the wild-type CAP10 with the deletion construct. The replacement event was further supported by the lack of hybridization signal in TYCC150 when the blot was hybridized with a probe of the deleted DNA fragment of CAP10 (Fig. 2BII). These results indicated that the acapsular phenotype of the transformant was a result of the CAP10 disruption.
Previous studies have shown that each of the other three capsule-associated genes in C. neoformans is required to produce fatal infections in mice. Two sets of animal experiments were conducted to study the importance of CAP10 in virulence. First, four strains of C. neoformans were used to infect groups of mice, including a stable Cap+ transformant of cap10F2FO (TYCC133), an acapsular transformant of cap10F2FO containing vector only (CIP3), an acapsular mutant (cap10F2), and a wild-type congenic strain (B-4500). Both TYCC133 and B-4500 produced fatal infection in all eight mice within 65 days (Fig. 3A). In contrast, CIP3 and the acapsular mutant (cap10F2) remained healthy when the experiment was terminated at 100 days postinfection. The mortality rate in mice infected with TYCC133 was higher than that in mice infected with B-4500. It was due to the slightly larger size of inoculant in mice receiving TYCC133. However, the size of inoculum among mice that received TYCC133, CIP3, and cap10F2 was similar.
FIG. 3.
Virulence test. Groups of eight mice each were injected with about 106 viable cells and monitored for 100 days to determine mortality. (A) B-4500, a wild-type strain; TYCC133, a stable Cap+ transformant of cap10F2FO; CIP3, a stable Cap− transformant of cap10F2FO harboring only the vector sequence; cap10F2, a cap10 mutant. (B) B-4500FO2, a CAP10 ura5 auxotroph; TYCC150, a ura5 auxotrophic cap10 disruptant.
In the second set of experiments, virulence of an acapsular strain produced by deletion of CAP10 (TYCC150) and a congenic encapsulated strain (B-4500FO2) was compared. B-4500FO2 produced fatal infection in all eight mice within 60 days whereas mice injected with TYCC150 remained healthy over 100 days (Fig. 3B). The slightly faster killing by B-4500FO2 compared to B-4500 may have been due to differences between batches of mice used in these experiments (Fig. 3A and B). These results corroborated the hypothesis that capsule is required for the virulence of C. neoformans.
The influence of different stages of growth on the expression of CAP10.
The GUS reporter gene has been successfully used to study the expression of several genes in C. neoformans (5, 30). We constructed a plasmid, pYCC330, in which the coding region of GUS was placed under the control of the CAP10 promoter. This construct was transformed into B4500FO2. GUS activity was measured by using protein extracts from transformants of different stages of growth (see Materials and Methods). Noticeable GUS activity was observed from overnight cultures using raffinose as a carbon source (Fig. 4). When cultures were transferred from raffinose to glucose medium, GUS activity decreased initially and its activity stayed at low levels for 25 h. However, GUS activity increased significantly after prolonged incubation in glucose medium (>45 h). At 3 days after transfer, GUS activity increased about sixfold compared to the activity of 5-h cultures. These results indicated that the expression of the CAP10(p)::GUS was influenced by different growth stages.
FIG. 4.
GUS activity of cultures from different growth stages. The GUS reporter construct, pYCC330, was transformed into the wild-type strain B-4500FO2. Protein extracts from three independent transformants were isolated at different time points and the GUS activity was determined. Error bars represent the sample standard deviations.
CAP10 expression is regulated by STE12α.
Because STE12α of C. neoformans regulates the expression of several virulence-associated genes (5), it was of interest to test whether STE12α regulates the expression of CAP10. Poly(A)+ RNA was isolated from 45-h glucose-grown cultures of the ste12α disruptant and the wild-type strain. The mRNA levels of CAP10 were 1.7-fold higher in the wild-type strain than in the ste12α disruptant (Fig. 5A). To test whether the disruption of STE12α affects the CAP10(p)::GUS reporter activity, pYCC330 was transformed into an STE12α strain (B-4500FO2) and ste12α disruptant (TYCC245F1FO). GUS activity was determined from the same stationary-phase culture of both sets of transformants. A significant decrease in GUS activity was observed in transformants of the ste12α disruptant compared to transformants of the wild-type strain (Fig. 5B). Furthermore, to study the effect of overexpression of STE12α on CAP10 expression, pYCC330 was transformed into TYCC259. TYCC259 contains a GAL7(p)::STE12α construct, which can overexpress STE12α when galactose is used as a sole carbon source. GUS activity was measured from protein extracts of glucose- and galactose-grown cultures. GUS activity in galactose-grown culture was slightly higher than that in glucose-grown culture (Fig. 5C). Because the GUS activity in the transformants containing just the vector was also higher in galactose-grown cells than in glucose-grown cells (Fig. 5C), it was possible that the observed differences in GUS activity were caused by differences in culture medium and were not induced by overexpression of STE12α. To evaluate this possibility, GUS activity from transformants of a wild-type strain (B-4500FO) containing pYCC330 grown in both culture media was determined. No significant differences in GUS activity were observed between glucose- and galactose-grown cultures for transformants of B-4500FO containing pYCC330 (glucose, 8.00 ± 1.56, versus galactose, 7.37 ± 2.16). Thus, the differences of GUS activity in transformants of TYCC259 were caused by overexpression of STE12α, which induced the CAP10(p)::GUS reporter gene activity. Therefore, the observed changes of GUS activity in transformants containing the vector may be due to the existence of a cryptic promoter in the vector. These data indicated that STE12α modulates the expression of CAP10.
FIG. 5.
STE12α modulates CAP10 expression. (A) Northern blot analysis. Poly(A)+ RNA isolated from the wild-type strain (B-4500) and the ste12α disruptant (TYCC245F1) was fractionated in a 2.2 M formaldehyde–1% agarose gel and transferred to a nylon membrane. The blot was hybridized with a probe of CAP10 cDNA and a probe of actin cDNA. The phosphorimaging results were used to determine the relative signal intensities. (B) The effect of the deletion of STE12α on GUS reporter activity. (C) The effect of STE12α overexpression on GUS reporter activity. CAP10 denotes the transformants containing pYCC330 and vector denotes the transformants containing the promoterless GUS (pYCC331). Data are averages of GUS activity from six independent transformants. Error bars represent the standard deviations.
Localization of Cap10p.
To understand the function of CAP10, we attempted to determine the cellular location of the CAP10 gene product by peptide epitope-tagging methods. Nine amino acids of the influenza virus HA protein were inserted into the C terminus of CAP10, and the resulting construct was transformed into a cap10 strain. The resulting transformants produced capsule, indicating that insertion of the HA tag at the C terminus of Cap10p did not interfere with its function. Total proteins were extracted from these capsule-containing transformants and analyzed by Western blotting using an anti-HA antibody. The size of the protein detected by Western blotting corroborated the predicted molecular weight (Fig. 6I). Immunofluorescence and immunoelectron microscopy were used to determine the cellular location of Cap10p. However, we did not obtain satisfactory results due to technical difficulties, such as suboptimal levels of fluorescence and poor preservation of organelles in immunogold electron microscopy studies (data not shown). Similar negative results were obtained even when three tandem copies of HA were used to tag Cap10p.
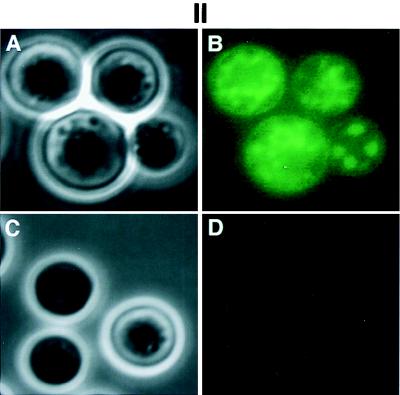
FIG. 6.

Detection and localization of Cap10p. (I) Immunoblot analysis. Yeast cells were grown in YNB and total protein extracts were analyzed by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis, incubated with anti-HA antibody, and analyzed using the Western-Star chemiluminescence detection system. Lane 1, a wild-type strain, B-4500; lane 2, an encapsulated transformant of TYCC150 containing pYCC152. (II) Localization of Cap10p-GFP fusion protein. Yeast cells were grown on YNB medium for no more than 24 h at 30°C. Panels A and B show the transformants of TYCC150 containing Cap10p-GFP fusion construct, pYCC352. Panels C and D show the transformants of B-4500FO2 containing pCIP3. Shown are phase-contrast micrographs (A and C) and fluorescence micrographs (B and D).
A modified GFP, yGFP3, has been engineered and successfully expressed in Candida albicans (7). Two modifications have been introduced in the coding region of GFP in yGFP3. First, several mutations surrounding the chromophore of GFP were introduced in the amino acid sequence between residues 64 and 72. These alterations increased the fluorescent intensity of GFP 100-fold compared to the wild-type GFP construct (8). Secondly, many codons were modified for optimal translation in C. albicans. The yGFP3-tagged Cap10p, however, failed to produce satisfactory fluorescence of GFP in C. neoformans. Recently, a different version of GFP, mgfp5, in which a cryptic intron present between amino acids 127 and 155 was removed from a thermotolerant mutant of GFP was successfully expressed in a plant system (17, 26). Experiments suggested that it is possible to increase fluorescence intensity by further modification of the chromophore region of this thermotolerant mutant GFP (26). In order to express GFP successfully in C. neoformans, we constructed a hybrid protein. The first 77 amino acids of the hybrid GFP, which contains the chromophore region, were derived from yGFP3 and the remainder were derived from the C-terminal portion of mgfp5. This hybrid GFP was inserted at the C terminus of Cap10p and the resulting plasmid, pYCC352, was transformed into the cap10 disruptant, TYCC150. The resulting transformants produced abundant capsule. When these encapsulated transformants were viewed by fluorescence microscopy, green fluorescent signals appeared as patches within the cytoplasm (Fig. 6IIB). The green fluorescence could be detected only in transformants containing hybrid GFP and not in the controls.
DISCUSSION
Cloning and characterization of CAP10 revealed that the gene contains three introns and encodes a novel protein. CAP10 was not contiguous with another transcript in close proximity. This observation was different from those obtained with the other three CAP genes which are tightly linked with other genes: CAP59 with L27, CAP60 with CEL1, and CAP64 with PRE1 (2–4). Animal studies demonstrated that cap10 mutants constructed by deletion or mutagenesis were unable to produce fatal infection in mice, as demonstrated with other acapsular strains of cap59, cap60, and cap64 (2–4). Complementation of the cap10 mutation restored capsule and virulence. Thus, CAP10 is the fourth characterized gene required for capsule formation and virulence in C. neoformans.
The GFP-tagged Cap10p appeared as patches within the cytoplasm of yeast cells. Because of the presence of a putative type II transmembrane region close to the N terminus, we speculate that Cap10p may be associated with certain types of organelles, although insertion of GFP may have affected the location. We used HA epitope-tagging and immunoelectron microscopy to further define the location of Cap10p without satisfactory results. Similar difficulties have been encountered using histochemical methods to localize gene products in C. neoformans (3, 29). Raising high-titer antibodies against Cap10p may increase the sensitivity of detecting the protein and reveal the definite location of Cap10p.
We have previously used several versions of modified GFP, including yGFP3, to tag Cap10p, but without success. The yGFP3 was used as a reporter by fusing it to different promoters of C. neoformans (10). It is not clear why the CAP10-yGFP3 fusion construct failed to produce strong fluorescence. The only GFP construct that yielded satisfactory results was a hybrid GFP, pYCC352. This hybrid protein contained a portion of yGFP3 engineered for C. albicans at the N terminus and a portion of GFP designed for the plant system at the C terminus. The success of expressing this hybrid GFP in C. neoformans may be due to combinative effects: the removal of the cryptic intron from a thermotolerant GFP mutant (17, 26) and introducing modified chromophore region to increase the fluorescence intensity (7, 8). This hybrid GFP was also successfully used to localize the Cap60p (data not shown) which, by immunogold electron microscopy, has been localized to the nuclear membrane (3). Several factors influenced the results of our hybrid GFP expression. When the promoter of CAP10 in the GFP fusion construct was replaced with a strong inducible GAL7 promoter of C. neoformans (30), the resulting construct was able to complement the acapsular mutation of TYCC150 on galactose medium. However, no GFP signal was detected in these encapsulated transformants from galactose-grown culture (data not shown). Thus, overexpression of fusion GFP showed an adverse effect on the GFP fluorescence signal. Physiological conditions of yeast cells also affected the level of GFP signals. GFP fluorescence was reliably detected only when cultures were grown on agar for no more than 24 h. When older cultures were used, not only did the GFP signals fade but also many yeast cells showed copious autofluorescence. Therefore, it may be important to have appropriate expression levels of the fusion construct for detection of GFP signals in C. neoformans. The expression levels of the fusion construct, however, appeared to have no effect on its function to complement the acapsular mutation.
Using the GUS gene as a reporter system, we found that CAP10 expression is influenced by different stages of growth; the CAP10 gene was expressed at much higher levels during the late stationary phase. It appears that there is a basal level of expression of CAP10 in young cultures and the expression of CAP10 increases when the nutrient of the medium is depleted. These data appear to corroborate the observations that yeast cells produce abundant capsule in late stationary phase (16), although it is not clear how this process is regulated. Interestingly, GUS activity and accumulation of CAP10 mRNA decreased in a strain containing a deletion of a well-conserved transcriptional factor, STE12α. In addition, the CAP10(p)::GUS reporter activity was induced by overexpression of STE12α. Thus, CAP10 expression is modulated by STE12α. Since STE12α is present only in MATα cells, it would be of interest to know what transcriptional factor(s) controls the expression of CAP10 in MATa strains. STE12α is a global regulator, which also controls the expression of several genes involved in virulence, such as capsule and phenol oxidase production (5). Although four capsule-associated genes have been isolated, the regulation of expression of these genes is not well defined. Further investigation on the mechanisms of regulating age-dependent CAP10 expression and how STE12α modulates CAP10 expression may lead to further understanding of the regulation of capsule synthesis.
ACKNOWLEDGMENTS
We thank L. Penoyer for technical assistance, A. Varma for a critical reading of the manuscript, B. Cormack and J. Haseloff for GFP plasmids, and J. Hanover and H. Edskes for help with fluorescence microscopy.
REFERENCES
- 1.Alspaugh J A, Perfect J R, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y C, Kwon-Chung K J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang Y C, Kwon-Chung K J. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Y C, Penoyer L A, Kwon-Chung K J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang Y C, Wickes B L, Kwon-Chung K J. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. The STE12α gene of Cryptococcus neoformans, abstr. F-63; p. 263. [Google Scholar]
- 6.Chang Y C, Wickes B L, Kwon-Chung K J. Further analysis of the CAP59 locus of Cryptococcus neoformans: structure defined by forced expression and description of a new ribosomal protein-encoding gene. Gene. 1996;167:179–183. doi: 10.1016/0378-1119(95)00640-0. [DOI] [PubMed] [Google Scholar]
- 7.Cormack B P, Bertram G, Egerton M, Gow N A, Falkow S, Brown A J. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 8.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 9.Cubitt A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 10.del Poeta M, Toffaletti D L, Rude T H, Sparks S D, Heitman J, Perfect J R. Cryptococcus neoformans differential gene expression detected in vitro and in vivo with green fluorescent protein. Infect Immun. 1999;67:1812–1820. doi: 10.1128/iai.67.4.1812-1820.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edman J C. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol Cell Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edman J C, Kwon-Chung K J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 15.Fromptling R A, Shadomy H K, Jacobson E S. Decreased virulence in stable acapsular mutants of Cryptococcus neoformans. Mycopathologia. 1982;79:23–29. doi: 10.1007/BF00636177. [DOI] [PubMed] [Google Scholar]
- 16.Granger D L, Perfect J R, Durack D T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Investig. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haseloff J, Siemering K R, Prasher D C, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heim R, Prasher D C, Tsien R Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon-Chung K J. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- 20.Kwon-Chung K J, Bennett J E. Medical mycology. Philadelphia, Pa: Lea & Febiger; 1992. pp. 397–446. [Google Scholar]
- 21.Kwon-Chung K J, Edman J C, Wickes B L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon-Chung K J, Varma A, Edman J C, Bennett J E. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J Med Vet Mycol. 1992;30:61–69. [PubMed] [Google Scholar]
- 24.Morise H, Shimomura O, Johnson F H, Winant J. Intermolecular energy transfer in the bioluminescent system of Aequorea. Biochemistry. 1974;13:2656–2662. doi: 10.1021/bi00709a028. [DOI] [PubMed] [Google Scholar]
- 25.Prasher D C, Eckenrode V K, Ward W W, Prendergast F G, Cormier M J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 26.Siemering K R, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 27.Still C N, Jacobson E S. Recombinational mapping of capsule mutations in Cryptococcus neoformans. J Bacteriol. 1983;156:460–462. doi: 10.1128/jb.156.1.460-462.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varma A, Kwon-Chung K J. Construction of stable episomes in Cryptococcus neoformans. Curr Genet. 1998;34:60–66. doi: 10.1007/s002940050366. [DOI] [PubMed] [Google Scholar]
- 29.Vartivarian S E, Reyes G H, Jacobson E S, James P G, Cherniak R, Mumaw V R, Tingler M J. Localization of mannoprotein in Cryptococcus neoformans. J Bacteriol. 1989;171:6850–6852. doi: 10.1128/jb.171.12.6850-6852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickes B L, Edman J C. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- 31.Wickes B L, Edman U, Edman J C. The Cryptococcus neoformans STE12alpha gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]