Abstract
Myxococcus xanthus is a gram-negative soil bacterium that produces the polyketide antibiotic TA. In this study, we describe the analysis of an M. xanthus gene which encodes a homologue of the prolipoprotein signal peptidase II (SPase II; lsp). Overexpression of the M. xanthus SPase II in Escherichia coli confers high levels of globomycin resistance, confirming its function as an SPase II. The M. xanthus gene encoding the lsp homologue is nonessential for growth, as determined by specific gene disruption. It has been mapped to the antibiotic TA gene cluster, and the disrupted mutants do not produce the antibiotic, indicating a probable involvement in TA production. These results suggest the existence of more than one SPase II protein in M. xanthus, where one is a system-specific SPase II (for TA biosynthesis).
The secretion of most proteins, in both prokaryotes and eukaryotes, is targeted through amino-terminal signal (or leader) peptides which are specifically cleaved by signal peptidases. Three types of signal peptidases have been described in bacteria: type I, LepB, which is responsible for the majority of exported preproteins; type II, Lsp, which is involved in the processing of lipoproteins; and type III, which is implicated in the processing of pili subunits. Lipoproteins, processed by signal peptidase II (SPase II), are characterized by the modification of a cysteine residue, located within a conserved consensus signal peptide sequence, by glyceryltransferase and O-acyltransferase. In all bacteria so far analyzed, SPase II was found to be an essential membrane protein anchored to the cytoplasmic membrane (37). The enzyme recognizes the conserved amino acid motif in the prolipoprotein and cleaves it in front of the cysteine residue. An additional acyl group is then attached to the NH2-terminal cysteine by N-acyltransferase (48).
Several lsp genes encoding SPase II have been cloned and sequenced from both gram-positive bacteria (Staphylococcus aureus [51], Staphylococcus carnosus [47], Bacillus subtilis [28], and Mycoplasma genitalium [6]) and gram-negative bacteria (Escherichia coli [12], Enterobacter aerogenes [13], Pseudomonas fluorescens [14], and Haemophilus influenzae [5]). In all of these gram-negative bacteria, the lsp gene is located within the ilaS-lsp operon, consisting of ilaS and three open reading frames (ORFs), probably with unrelated functions: orfX, orf149, and orf316 (20).
Myxococcus xanthus is a gram-negative, gliding, soil bacterium that feeds on proteins and peptides and undergoes a complex life cycle that includes cell-to-cell interactions, signaling, and fruiting body formation (30, 40). In the last process, tens of thousands of cells glide, upon starvation, toward an aggregation center to form a multicellular fruiting body (40), where individual rod-shaped cells differentiate into spherical, dormant, environmentally resistant myxospores. Genetic and biochemical evidence indicates that essential intercellular signaling occurs at multiple stages during the developmental process and that cell-to-cell interactions are required for proper transcriptional regulation of developmental genes. M. xanthus produces the polyketide antibiotic TA (33, 34), which inhibits cell wall synthesis by interfering with the polymerization of the lipid-disaccharide-pentapeptide (50). The transposon Tn5lac (16) was used as a promoter probe to study the expression and transcriptional regulation of genes required for the antibiotic TA biosynthesis (42, 43, 45). A series of transposition mutants blocked in antibiotic production were obtained and assigned the TA operon to a chromosomal segment of at least 36 kb (44). These genes are organized in type I polyketide synthetase modules (25), and the region contains several other genes, all involved in postmodification steps of polyketide biosynthesis (23, 24). Among these genes, we identified a homologue of an SPase II protein.
Information about lipoproteins and their biosynthesis in M. xanthus is very scarce. Only a few lipoproteins have been identified; these include MlpA, Tgl, CelA, and two other proteases, PrtA and PrtB. MlpA is a 33-kDa lipoprotein required for normal development of M. xanthus and contains the conserved lipo-box sequence at the putative cleavage site of SPase II. When MlpA is expressed in E. coli in the presence of globomycin (Glm; a specific inhibitor of SPase II), it behaves as a 35-kDa protein rather than the 33-kDa protein seen with no antibiotic, suggesting that the antibiotic prevents its processing by the E. coli SPase II (9). Tgl is a membrane protein required for the production of the type IV pili, which are essential for the social motility involved in the development process of M. xanthus. Tgl contains an SPase II recognition sequence at its N terminus, predicting the removal of 19 amino acid residues by SPase II (31, 32). The third lipoprotein, CelA, is an extracellular endoglucanase with an unusual signal peptide which contains three prolines in its N terminus (8a) and is thought to be processed by the type II secretion system. The secretion of extracellular proteases, such as PrtA and PrtB, is required for external protease activities involved in the provision of nutrients to M. xanthus. Both proteins contain a lipoprotein signal peptide which is presumably processed by the type II secretion system (29).
In this study, we report the genetic analysis of an SPase II gene encoding a homologue from M. xanthus, designated taG. We provide evidence for its possible involvement in the biosynthesis of the antibiotic TA, as well as for the existence of more than one SPase II in this bacterium.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The M. xanthus strain used in this study was the previously described wild-type strain ER-15 (42). E. coli TG1, DH10β, and XL1-Blue MR were used for cloning and DNA manipulations. E. coli Y815 was used for complementation analyses. This strain is temperature sensitive for growth due to a mutation in the SPase II gene. It also carries an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lpp gene (coding for the major outer membrane lipoprotein, also known as Braun’s lipoprotein [4]) on a low-copy-number plasmid pHY001 that encodes for tetracycline resistance (49). The plasmids pKS [negative control, pBluescript KS(+)] and pKSA11 (positive control, contains the complete ileS-lsp-pyrR operon from B. subtilis) were used in the complementation assays and are described elsewhere (28). A conjugative-tagged Tn1000 transposition system was used for the sequencing, as described elsewhere (26), using E. coli MH1578 and MH1599 (39). E. coli vectors used for cloning and sequencing were pUC18, pUC19 (22), and SUPERCOS-1 (Stratagene, La Jolla, Calif.). Plasmid pQE9 (Qiagen, Chatsworth, Calif.) was used for the expression of the six-His–SPase II fusion proteins.
Media and growth conditions.
E. coli was grown at 30, 37, or 42°C, as required, in Luria broth (LB) or on LB agar with the appropriate antibiotics as described previously (35). M. xanthus was grown at 32°C on 1 CT–0.5 CTS–CTK medium (44) or CTT medium (10) or on media solidified by 1.5% Bacto agar (Difco) and maintained as described previously (42). LB or AB3 agar medium (Difco) was used for the lsp complementation assays as described previously (28).
General DNA procedures.
Standard genetic techniques, Southern blot analysis, plasmid preparations, hybridization, and in vitro DNA manipulations were as described earlier (35). Isolation of total DNA from M. xanthus was as described elsewhere (3). Cosmid DNA and DNA templates for sequencing reactions and M. xanthus electroporation were purified by Qiagen columns. Electroporation of M. xanthus was performed as described before (15). Conjugative transposition of Tn1000 for sequencing was done as previously described (26).
DNA sequencing and analysis.
Automated DNA sequencing was performed on double-stranded DNA templates by the dideoxynucleotide chain termination method (36), using an Applied Biosystems (Foster City, Calif.) model 373A sequencer. Several cosmids, originating from the chromosomal regions that were found through gene disruption to be essential for TA production, were further analyzed, and the cosmid clone pPYCA111 was subcloned into pUC18 and pUC19 (22) plasmid vectors. A 1.051-kb fragment was sequenced by using the conjugative transposon Tn1000 mutagenesis walking system as described previously (26). Analysis and assembly of the primary DNA sequence data were completed by using the software provided by MacVectora 3.5 (International Biotechnologies Inc.) and Sequence Navigator (Applied Biosystems). The searches for DNA and protein sequence homologies in databases were achieved through the use of the BLAST (2) and FASTA (27) programs.
Overexpression of TaG in E. coli and complementation tests.
In order to construct a recombinant plasmid which would overexpress a six-His–taG fusion protein, a 515-bp fragment containing the entire taG gene was amplified from the cosmid pPYCA111 by PCR. The forward primer was 5′CCGGATCCATGAACACTCCTTCC3′, into which a BamHI site (underlined) was incorporated in order to form an in-frame fusion immediately upstream of the first ATG initiation codon (in bold). The reverse primer, 5′CCCAAGCTTCTATCCGGGAGACAAC3′, incorporated a HindIII site (underlined). The same procedure was applied to clone and overexpress an E. coli six-His–lsp fusion protein. A 521-bp fragment, containing the E. coli lsp gene, was amplified from E. coli chromosomal DNA by PCR. The forward primer was 5′CCGGATCCAGTCAATCGATCTGTTCAACAGGG3′, into which a BamHI site (underlined) was incorporated immediately upstream of the second codon of the gene (AGT; in bold). A HindIII site (underlined) was incorporated into the reverse primer, 5′CCCAAGCTTGCGTCAGCATCGCATCCGGCAGGG3′. The PCR products were digested with BamHI and HindIII and cloned into the vector pQE9. In the resulting plasmids, pPYMXSP-II (M. xanthus taG) and pPYECSP-II (E. coli lsp), the cloned genes were fused in frame to the six-His sequence at their 5′ ends and were expressed under the control of a T5 promoter and two lac operator sequences. The overexpression of both proteins in E. coli was determined by their ability to increase the levels of Glm (a generous gift from M. Inukai, Sankyo Co. Ltd, Tokyo, Japan) resistance in E. coli TG1 and Y815, carrying pPYMXSP-II or pPYECSP-II.
The biochemical activity of the M. xanthus SPase II in E. coli was assayed in vivo by its ability to complement the growth of the temperature-sensitive SPase II mutant E. coli Y815, which is sensitive to IPTG at 42°C, due to the accumulation of lipid-modified prolipoprotein (49). Dilutions of E. coli Y815 carrying the plasmids pKSA11 and pPYECSP-II (both as positive controls), pQE9 and pKS (both as negative controls), and pPYMXSP-II (taG) were plated in two series of LB or AB3 agar medium (Difco) supplemented with 100 μg of ampicillin ml−1, 15 μg of tetracycline ml−1, and 0 to 6 mM IPTG. The plates were incubated for 1 day at the permissive temperature (30°C) and then for 7 days at either the permissive (30°C) or the nonpermissive (42°C) temperature. In addition, we monitored the growth of E. coli Y815 transformed with plasmids pQE9, pKSA11, pPYMXSP-II, and pPYECSP-II at 42°C in LB medium supplemented with 100 μg of ampicillin ml−1, 15 μg of tetracycline ml−1, and 0 to 6 mM IPTG.
General protein procedures.
The procedure for protein extraction from M. xanthus strains was similar to that described previously (35). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (19). Western blot analysis was performed as described elsewhere (35).
Endoglucanase activity.
Extracellular CelA activity was determined by the degradation of carboxymethylcellulose on plates. Twenty colonies from each mutant strain (ER-taGH and ER-taG) and twenty colonies from the wild-type strain ER-15 were grown on CTT plates containing 1% carboxymethylcellulose (Sigma). After 4 days at 30°C, the utilization of carboxymethylcellulose was determined by the addition of 0.1% Congo red for 10 min, followed by a rinse with 10 ml of 1 M NaCl solution. The colonies that produced active endoglucanase were surrounded by a clear yellow halo on a red background.
General extracellular protease activity.
The general extracellular proteolytic activity of M. xanthus was examined by proteolysis of skim milk on plates. Twenty colonies from each mutant strain (ER-taGH and ER-taG) and twenty colonies from the wild-type strain ER-15 were grown on CTT plates containing 2% skim milk. The halo zones surrounding the colonies, as an indication of extracellular proteolytic activity, were measured at different growth times.
Developmental analysis.
Fruiting body development was examined on TPM (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, 8 mM MgSO4 [final pH 7.6]) agar (1.5%) plates as described previously (17).
Antibiotic TA production assay.
The production of the antibiotic TA was determined by the disc assay, as described previously (44).
Nucleotide sequence accession number.
The DNA sequence reported here was submitted to the EMBL database and will appear in data libraries under accession no. AJ223309.
RESULTS
Isolation and analysis of the M. xanthus SPase II homologue.
A cosmid library of M. xanthus ER-15 DNA was screened with TA-specific probes (23–25). The recombinant cosmid pPYCA111, which hybridized to these probes, was further characterized through restriction enzyme analysis and subcloned into pUC19. The two contiguous SphI inserts in pPY12 and pPY13, 0.303 and 0.754 kb, respectively, were further analyzed. High-stringency Southern blotting, using the SphI inserts as probes on SphI-restricted M. xanthus ER-15 chromosomal DNA, indicated that they are present as single copies in M. xanthus. In order to characterize further the cloned region and its deduced function, the nucleotide sequence of the 1.051-kb fragment was determined. A computer homology search of this sequence with available databases revealed significant homology to several SPases II (Lsp) and to several cytochrome P450 mono-oxygenases.
The two ORFs within this fragment were identified through the use of the University of Wisconsin Genetic Computer Group (7) and MacVectora 3.5 (International Biotechnologies Inc.) programs and were found to be transcribed in the same direction. ORF1, designated taG, begins at nucleotide 84 (ATG) and stops at position 596 (TAG), encoding a protein of 171 amino acids (Mr = 19,489.5). The frequency of G+C base pairs at the third position of each codon in this frame was found to be 83.2%, a value which is typical of ORFs found in organisms (such as M. xanthus) with a high percentage of G+C (total G+C content of this sequence is 63.3%). ORF2, designated taH, encodes a cytochrome P450 mono-oxygenase and was found to be involved in TA production. The structure and function of this ORF, which starts at position 648 (ATG) and ends outside the insert of pPY13, is discussed elsewhere (24).
Homology with other lipoprotein SPase II sequences.
The deduced amino acid sequence of ORF1 (taG) was compared to known SPase II sequences in the EMBL database and was found to display the highest homology to E. coli (42% identity and 51.2% similarity) (12) and P. fluorescens (31.5% identity and 44.3% similarity) (14) SPase II proteins. These homologies identify ORF1 as an SPase II homologue of M. xanthus. The homology between the M. xanthus ORF and those of other SPase II proteins, including that of E. coli, were analyzed by the program BESTFIT and is displayed in Fig. 1. The sequence alignment reveals several areas of high identity and particularly three generally conserved regions, which are designated I, II, and III (Fig. 1) (28).
FIG. 1.
Multiple alignment of the deduced amino acid sequences of SPases II. Shaded boxed letters indicate residues identical in more than four of the six organisms. The predicted membrane-spanning segments of the SPases II (A to D) are indicated by black lines, and the conserved regions (I to III) are indicated by dashed lines.
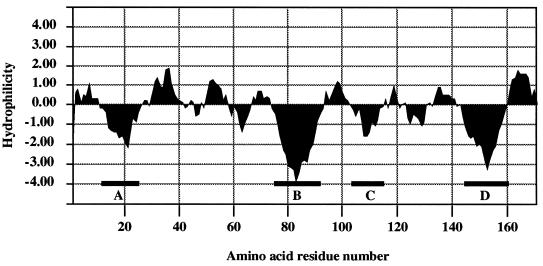
Hydrophobicity profile.
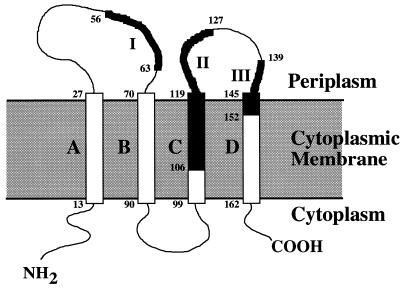
The hydrophobicity profile (18) of the M. xanthus SPase II homologue TaG contains four putative hydrophobic domains (Fig. 1 and 2, segments A to D), which could represent transmembrane domains similar to those proposed for other SPase II proteins (21, 28). Following the proposed positive inside rule (46), a model for the transmembrane structure of TaG is outlined in Fig. 3. In this model, the conserved regions I, II, and III are assigned to specific domains of the proposed structure. Region I is located in the carboxy-terminal part of the first periplasmic loop; region II is located in transmembrane segment C and the amino-terminal part of the second periplasmic loop; and region III is assigned to the carboxy-terminal moiety of the second periplasmic loop, as well as to transmembrane segment D (Fig. 3).
FIG. 2.
Hydrophilicity profile of the M. xanthus SPase II protein according to the algorithm developed by Kyte and Doolittle (18). The four major hydrophobic segments are indicated (A to D).
FIG. 3.
A proposed model for the membrane topology of M. xanthus SPase II. The four predicted hydrophobic transmembrane segments (A to D) are indicated. The conserved regions (I to III) are indicated in bold. The numbers represent amino acid residues.
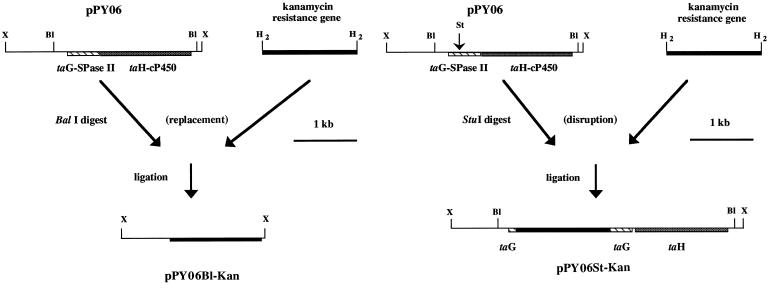
Specific deletion and disruption of the taG gene.
The involvement of TaG in TA biosynthesis and its role in cellular growth were assessed in mutant strains with two different types of gene deletion and disruption. First, a kanamycin resistance gene was inserted into pPY06, replacing a 2.33-kb BalI fragment (Fig. 4A), thus deleting the entire taG gene and the neighboring gene, taH. Second, a StuI restriction site was inserted into pPY06 (using PCR) to replace a 44-bp fragment, starting at position 133 (133 to 177) of pPY12, and a kanamycin resistance gene was inserted into pPY06 at this position, thus disrupting taG (Fig. 4B). The resulting plasmids (pPY06B1-Kan and pPY06St-Kan, respectively) (Fig. 4A and B) were linearized and electroporated into M. xanthus ER-15. The facts that pUC19 cannot replicate in M. xanthus and that the electroporated DNA was linear meant that any kanamycin-resistant colony resulted from an integration of the linearized DNA into the chromosome via homologous recombination. Kanamycin-resistant transformants were obtained and shown by Southern blot analysis to have the expected disruptions (see below). This result indicated that this gene encoding the lsp homologue is not required for growth and is the first demonstration of a nonessential SPase II protein. The transformants were plated on CTK medium for 5 days, and twenty colonies of each strain were examined for TA production on 0.5 CTS agar. The results clearly indicate that all kanamycin-resistant colonies (20 of 20 colonies) were blocked in TA production, strongly suggesting that taG is involved in the production of an active TA molecule but is not essential for normal growth. Two mutants with deletions of taG (SPase II), one of each strain, were picked up for further analysis (see below) and were designated ER-taGH (carrying a kanamycin gene replacing all taG and taH genes) and ER-taG (carrying a kanamycin gene insertion in taG).
FIG. 4.
Specific gene deletion and disruption of taG and taH by a kanamycin resistance gene insertion into pPY06 replacing a 2.3-kb BalI site, forming the plasmid pPY06B1-Kan (A), and a 44-bp fragment at the beginning of taG (at a specific StuI site formed by PCR), forming the plasmid pPY06St-Kan (B). The resulting plasmids were linearized and electroporated into M. xanthus ER-15. Restriction enzymes: St, StuI; X, XhoI; Bl, BalI; H2, HindII.
Southern blot analysis of mutant strains.
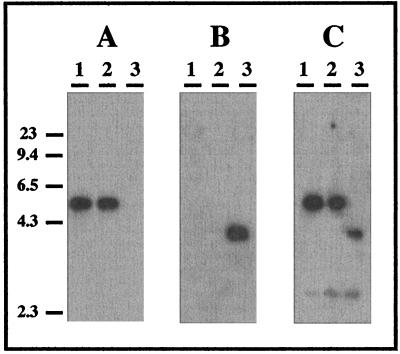
Chromosomal DNAs from M. xanthus ER-15 and ER-taGH were digested with SalI and analyzed through Southern hybridization with three different probes: a kanamycin probe, a specific taG probe (the fragment cloned in pPY12), and an insert from the plasmid pPY06 which contains both taG-taH and the flanking regions of the deleted 44-bp fragment. The results, presented in Fig. 5, demonstrate that the kanamycin probe hybridized to ER-taGH but not to ER-15, whereas the specific taG probe hybridized to ER-15 but not to ER-taGH, indicating that the kanamycin resistance gene indeed replaced the taG-taH fragment in ER-15. Similar results, obtained from strain ER-taG, confirmed the insertion of the kanamycin resistance gene into the designated site of the taG gene (data not shown).
FIG. 5.
Southern hybridization analysis of cosmid pPYCA111 DNA (lanes 1) and chromosomal DNA from M. xanthus ER-15 (wild type, lanes 2) and ER-taGH (deleted mutant, lanes 3) probed with three different 32P-labeled probes: a specific taG probe (the fragment cloned in pPY12) (A); a kanamycin resistance gene (B); and the insert from the plasmid pPY06, which contains both taG-taH and their flanking regions (C). The cosmid and chromosomal DNA were digested with SalI and separated on a 1% gel prior to Southern blotting. Hybridization was carried out in 50% formamide at 42°C. Filters were washed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate)–0.1% sodium dodecyl sulfate at 42 and 50°C (second wash). Marker positions, indicated at the left, are those of HindIII-digested λ DNA.
Overexpression and activity of the M. xanthus TaG protein in E. coli.
The overexpression of lsp genes from either gram-negative or gram-positive bacteria was reported to increase the levels of Glm resistance in E. coli (28) and was used previously to clone several lsp genes (13, 14, 47, 51). We used this finding to determine if M. xanthus taG can function as an SPase II in E. coli. Since M. xanthus genes frequently fail to be expressed in E. coli under their own regulatory elements, the taG gene was also subcloned into the E. coli six-His-tagged expression vector pQE9. The resulting plasmid, pPYMXSP-II, carries the taG gene, fused to a 5′ six-His sequence, under the control of elements made up of a phage T5 promoter and two lac operator sequences. As a control, we cloned the E. coli lsp gene in the same way into the same vector (pQE9), forming the plasmid pPYECSP-II.
E. coli TG1 was transformed with various plasmids (pUC19, pQE9, SUPERCOS-1, pPY06, pPYCA111, pPYECSP-II, and pPYMXSP-II; see Materials and Methods), and the resulting strains were grown in LB medium containing Glm (0 to 200 μg ml−1) at 30, 37, and 42°C, with the addition of IPTG when required. The results, summarized in Table 1, clearly demonstrate that the M. xanthus SPase II homologue (TaG) expressed from the plasmid pPYMXSP-II and the E. coli SPase II (Lsp) expressed from the plasmid pPYECSP-II increased the level of Glm resistance from 25 μg ml−1 to more than 200 μg ml−1 at all temperatures tested. E. coli strains carrying any of the other plasmids failed to grow at Glm concentrations higher than 25 μg ml−1, the MIC of Glm for E. coli TG1. Since two of the plasmids (pPY06 and cosmid pPYCA111) carry the entire native taG gene, it can be concluded that this gene fails to be expressed in E. coli when controlled by its own regulatory elements. In a similar experiment, the expression of taG and the E. coli lsp gene in E. coli Y815 at 30°C (by plasmids pPYMXSP-II and pPYECSP-II) led to the increase of Glm resistance from 25 μg ml−1 to more than 200 μg ml−1. As with E. coli TG1, all other plasmid-containing strains failed to grow at Glm concentrations higher than 25 μg ml−1.
TABLE 1.
Biological activity of SPase II genes from various bacteria in E. colia
| Plasmid | Gene | Growth at the following Glm concn (μg ml−1)
|
Complementation growth of E. coli Y815b | ||||
|---|---|---|---|---|---|---|---|
| 0 | 10 | 25 | 50 | 200 | |||
| None | + | + | + | − | − | − | |
| pUC19 | + | + | + | − | − | NT | |
| pQE9 | + | + | + | − | − | − | |
| SUPERCOS-1 | + | + | + | − | − | NT | |
| pPY06c | taG | + | + | + | − | − | NT |
| pPYCA111c | taG | + | + | + | − | − | NT |
| pPYMXSP-IId | 6 His-taG | + | + | + | + | + | ± |
| pPYECSP-IIe | 6 His-lsp | + | + | + | + | + | + |
| pKS | NT | NT | NT | NT | NT | − | |
| pKSA11f | B. subtilis lsp | NT | NT | NT | NT | NT | + |
Bacterial cultures (E. coli TG1 and its transformants) were grown in LB containing 0 to 200 μg of Glm ml−1 and 100 μg of ampicillin ml−1 (if required) at 30, 37, and 42°C. The results (+, growth; ±, limited growth; and −, no growth) were the same for all temperatures tested, and therefore, the temperature is not indicated. The same experiment was also performed with E. coli Y815 and its transformants, except that Glm resistance was determined only at 30°C. NT, not tested.
Complementation growth on LB or AB3 plates.
taG was cloned with its own regulatory elements.
taG was cloned under pQE9 regulatory elements.
E. coli lsp was cloned under pQE9 regulatory elements.
B. subtilis ileS-lsp-pyrR operon was cloned in pBluescript KS(+) as described elsewhere (28).
The in vivo biochemical activity of the M. xanthus SPase II in E. coli was further examined by determining its ability to complement the growth of E. coli Y815, which is temperature sensitive in the presence of IPTG due to the accumulation of lipid-modified prolipoprotein (49). This strain was transformed with plasmids pPYMXSP-II, pPYECSP-II, and pQE9 (negative control), and the resulting transformants were tested for their ability to grow at 42°C (nonpermissive temperature) in the presence of IPTG (0 to 6 mM). We additionally used, in the same experiment, E. coli Y815 transformed with the plasmids pKS (negative control) and pKSA11, which contains the B. subtilis lsp operon, previously shown to complement this mutation (28). The results are summarized in Table 1 and indicate that the strain transformed with the plasmid pKSA11 (B. subtilis lsp) formed visible colonies after 4 to 5 days, complementing the temperature-sensitive mutation of E. coli Y815 at 42°C in the presence of IPTG. The strain transformed with pPYECSP-II (E. coli lsp) formed visible colonies after 5 days, the strain transformed with pPYMXSP-II (M. xanthus taG) formed only a few small visible colonies after 6 to 7 days, and all of the rest of the strains did not form any colonies at all, even after 2 weeks. The results indicate that the M. xanthus taG gene, expressed from plasmid pPYMXSP-II, confers only limited complementation of the E. coli SPase II temperature-sensitive mutation. These results were confirmed by quantitative determination of growth rates in liquid medium of E. coli Y815 transformed with plasmids pQE9, pKSA11, pPYMXSP-II, and pPYECSP-II at 42°C in the presence of IPTG (0 to 6 mM). The growth of E. coli Y815 transformed with plasmid pQE9 (negative control) was strongly inhibited by the addition of IPTG, while the growth of E. coli Y815 transformed with plasmids pPYECSP-II and pKSA11 was IPTG independent, as expected from the complementation of the temperature-sensitive mutation. E. coli Y815 transformed with pPYMXSP-II (expressing taG) displayed very limited growth in the presence of IPTG, in agreement with the results obtained on solid media, indicating limited or no complementation of the E. coli system by TaG. These results imply that TaG is not fully compatible with the E. coli lipoprotein processing system, possibly as a result of its more restricted substrate specificity. The findings support the proposal that M. xanthus has more than one SPase II protein, one (TaG) of which represents a nonessential, system-specific, prolipoprotein peptidase.
Normal growth and developmental analysis of ER-taGH and ER-taG.
The growth rates of ER-taGH and ER-taG were indistinguishable from that of ER-15, indicating that TaG and TaH or any protein processed through their activities is not essential for normal growth. Moreover, all strains (ER-15, ER-taGH, and ER-taG) developed to mature fruiting bodies when examined on TPM agar plates at 30°C, suggesting that the protein encoded by taG or any protein processed by this gene product is not essential for the development process of M. xanthus. These results suggest that Tgl, which is required for the social motility of M. xanthus (31, 32) and consequently for normal development, is not likely to be processed by TaG and hence support the existence of at least two SPase II proteins in M. xanthus.
MlpA processing in ER-15, ER-taGH, and ER-taG.
The existence of more than one SPase II protein in M. xanthus was examined through Southern hybridization analysis, performed on SalI-digested chromosomal DNA extracted from the wild-type strain ER-15. For this purpose, we used the taG gene as a probe at different hybridization stringencies. At high stringency, only one fragment could be observed, whereas no specific hybridization could be detected at low stringency (data not shown), underlining the need for a different approach to verify the existence of other SPase II proteins in M. xanthus. We consequently studied the processing of the lipoprotein MlpA in the mutant strains ER-taGH and ER-taG as well as in the wild-type strain ER-15. The MlpA proteins were purified from cultures grown in 1 CT medium (normal growth) and in 0.5 CTS medium (promoting TA biosynthesis) and analyzed by Western blots, using anti-MlpA polyclonal antibodies (a generous gift from S. Inouye) (9). The data indicate that the size of MlpA is the same in all strains (about 33 kDa), in both 1 CT and 0.5 CTS media. It is therefore likely that MlpA is not processed by TaG, supporting our conclusion that there are at least two SPase II proteins in M. xanthus.
Endoglucanase activity (CelA processing).
The lipoprotein CelA is an extracellular endoglucanase, containing a type II signal peptide at its N terminus, presumably processed by the type II secretion system. The activity of the extracellular CelA can be determined through the degradation of carboxymethyl cellulose on plates. Twenty colonies from each mutant strain (ER-taGH and ER-taG) and twenty colonies from the wild-type strain ER-15 were grown on CTT agar plates containing 1% carboxymethylcellulose. The degradation of carboxymethylcellulose was determined by the addition of 0.1% Congo red to the plates after 4 days of incubation at 30°C. The formation of similar clear halos around the colonies of all strains clearly demonstrated that both mutant and wild-type strains can degrade carboxymethylcellulose, presumably due to the secretion of an active CelA protein. If the lipoprotein CelA is processed by the type II secretion system, as assumed from its properties and signal peptide, it has to be processed and secreted by an SPase II protein different from TaG.
General extracellular protease activity.
M. xanthus feeds on proteins and peptides and secretes many proteolytic enzymes to the medium (8). At least two lipoprotein proteases, PrtA and PrtB (29), which contain a type II signal peptide at their N termini and are thought to be processed by SPase II, were identified and characterized in M. xanthus. In an attempt to detect extracellular, proteolytic activity in this bacterium, we determined the proteolysis of skim milk on plates, assuming that the loss of PrtA and PrtB activities (due to the absence of an SPase II activity) would be detected. Twenty colonies from each mutant strain (ER-taGH and ER-taG) and from the wild-type strain ER-15 were grown on CTT plates containing 2% skim milk. No differences in the sizes of halos around the colonies of all strains could be detected, demonstrating that both mutant and wild-type strains possess the same extracellular proteolytic activities. Although the results are indirect, they are compatible with the suggestion that the secreted lipoprotein proteases, including PrtA and PrtB, are likely to be processed by an SPase II protein different from TaG.
DISCUSSION
We determined and analyzed the complete nucleotide sequence of a novel gene (lsp) encoding an M. xanthus SPase II. The behavior of SPase II mutant strains suggests that the product of the lsp gene is required for the production of the antibiotic TA but not for vegetative growth or for the developmental process of the bacterium. As all known SPase II proteins are essential, these results strongly indicate, for the first time, the existence of more than one SPase II protein in a bacterium. One of these proteins has to be a system-specific SPase II, since it is involved in TA production. These conclusions are further reinforced by the observations that the processing of at least three M. xanthus lipoproteins (MlpA, CelA, and Tgl), thought to be processed by SPase II, is not impaired in the absence of TaG. The localization of taG in the TA gene cluster and its limited complementation of an E. coli lsp temperature-sensitive mutant, although confirming Glm resistance, underlines our suggestion for the existence of a system-specific SPase II in M. xanthus.
The DNA sequence data of TaG predict a protein of 171 amino acid residues with a calculated molecular mass of 19,489.5 Da, which is consistent with the size of other known SPase II proteins. No ribosomal binding site motif (Shine-Dalgarno) was found 5′ to taG. In E. coli, the SPase II (lsp) gene is organized in an operon consisting of ileS, lsp, and three ORFs: orfX, orf149, and orf316 (20). It was therefore suggested that the lsp gene in gram-negative bacteria is part of an operon, associated with ileS, which encodes the isoleucyl-tRNA synthetase. All gram-negative bacterial lsp genes analyzed so far are organized like the E. coli cluster. In the gram-positive bacteria, B. subtilis, S. aureus, S. carnosus, and M. genitalium, the lsp and ileS genes are not assigned to one operon, although the lsp and orf4 genes seem to be part of one operon (6, 28, 47, 51). In contrast to other gram-negative bacteria, M. xanthus taG is not located near ileS but rather is part of the antibiotic TA gene cluster and is flanked by genes essential for TA biosynthesis (24). The gene 5′ to taG is an acyl enzyme which is a homologue of the B. subtilis pksG gene (1, 38), whereas the gene 3′ to taG is a cytochrome P450 mono-oxygenase (24). All three genes (acyl enzyme, cytochrome P450, and taG) are presumed to be part of one operon involved in TA production, similar to other polyketide synthase systems (11).
The sequence alignment of taG with other homologous genes revealed three conserved regions (Fig. 1). The alignment data suggest that regions II and III are more likely to be involved in the catalytic site, which is probably located in the second periplasmic loop. Region I seems to be the least conserved of the three (in M. xanthus). This observation is consistent with the suggestion of Sankaran and Wu (37) that SPase II belongs to a novel class of aspartic proteases, as SPase II contains three conserved aspartic acid residues, two of which are located in region II and one that is in region III.
The overexpression of taG or the lsp gene in E. coli confers high Glm resistance, indicating that both proteins are properly synthesized in E. coli. Yet, while the E. coli lsp gene product complements the growth of E. coli Y815 at the nonpermissive temperature (42°C), TaG (under the same control elements) conferred a very limited complementation. This result differs from that obtained with the B. subtilis SPase II enzyme (28) which complements the same E. coli strain (Table 1). The result could mean that the physiological activity of the M. xanthus protein is not fully compatible with that of the E. coli prolipoprotein processing system. Alternatively, it is possible that although the M. xanthus SPase II confers Glm resistance at 42°C, it is not physiologically functional at this temperature, as this organism does not grow at temperatures above 37°C. A more interesting possibility is that taG encodes a specific SPase II involved in the biosynthesis of antibiotic TA (as suggested by its gene organization in the TA gene cluster and by the data presented in this study) that has a more restricted substrate specificity and therefore cannot fully complement the E. coli SPase II mutation. The observation that the expressed M. xanthus protein confers resistance to Glm at 30, 37, and 42°C but confers only limited complementation to the temperature sensitivity of E. coli Y815 at 42°C supports the latter possibility, i.e., that taG encodes a TA-specific SPase II. Consequently, it is very likely that Glm binding and the processing of lipid-modified prolipoproteins represent two independent activities.
The disruption of taG in ER-taG and the deletion of taG and taH in ER-taGH indicate a possible involvement in TA biosynthesis and suggest that TaG may be required for the production of an active TA molecule. However, it is possible that the block in TA production is the result of a polar effect on taH (cytochrome P450) or other genes located 3′ to taG (in ER-taG). This possibility is difficult to exclude since there are no self-replicating plasmids in M. xanthus which could be used for trans complementation of the disrupted and deleted gene(s). A specific nonpolar disruption of taG should confirm its involvement in TA biosynthesis. At the moment, the exact function of taG in TA biosynthesis and in the processing of lipoproteins is an enigma.
In bacteria, lipoproteins play important roles in nutrient uptake, antibiotic resistance, adhesion mechanisms (targeting bacteria to specific substrates), protein secretion, sporulation, and germination (41). The complete pathway determining the biosynthesis and functions of the M. xanthus lipoprotein is as yet unknown, but like gram-positive bacilli and streptomyces, M. xanthus undergoes a complex life cycle, including cell-to-cell interactions, signaling, secretion of proteases, and fruiting body formation (40). Lipoproteins may play a part in one or more of these processes.
In summary, this work describes, for the first time, a gram-negative SPase II homologue which is found to be in a gene cluster organized differently from that of other known gram-negative lsp genes. In contrast to other bacteria, this gene is not essential for growth, suggesting the existence of more than one SPase II protein in bacteria. The future analysis of all the genes required for TA biosynthesis may provide clues as to the substrate of this SPase II. The observation that TaG appears not to process lipoproteins such as MlpA, CelA, and Tgl and its failure to complement the temperature-sensitive mutation of E. coli Y815 strengthen this suggestion. Identification and analysis of other lsp genes should clarify the mechanism of type II secretion in M. xanthus.
ACKNOWLEDGMENTS
We are very grateful to Sierd Bron and Harold Tjalsma from the University of Groningen, The Netherlands, for generously providing E. coli Y815 and plasmids pKS and pKSA11 and for the helpful suggestions in constructing the complementation assay. We are likewise grateful to M. Inukai from Sankyo Co. Ltd., Tokyo, Japan, for his generous gift of globomycin and to S. Inouye for generously providing polyclonal antibodies raised against the MlpA protein.
This work was supported in part by the Pasha Gol Chair for Applied Microbiology (E.R.), the Morris and Manja Leigh Chair for Biophysics and Biotechnology (E.Z.R.), a British Council Clore Foundation Scholarship awarded to Y.P., and grants from the Cancer Research Campaign (no. SP1937/0301) and the Welcome Trust (no. 038060/Z/93/Z) to E.O.
REFERENCES
- 1.Albertini A M, Caramori T, Scoffone F, Scotti C, Galizzi A. Sequence around the 159° region of the Bacillus subtilis genome: the pksX locus spans 33.6-kb. Microbiology. 1995;141:299–309. doi: 10.1099/13500872-141-2-299. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;251:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Avery L, Kaiser D. In situ transposon replacement and isolation of spontaneous tandem duplication. Mol Gen Genet. 1983;191:99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- 4.Braun V, Rehn K. Chemical characterization, spatial distribution and function of lipoproteins (murein lipoprotein) of E. coli cell wall. The specific effect of trypsin on membrane structure. Eur J Biochem. 1969;10:423–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 5.Fleischmann R D, Adams M D, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 6.Fraser C M, Gocayne J D, White O, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 7.Genetics Computer Group. Sequence analysis software package, version 7. Madison: University of Wisconsin Genetics Computer Group; 1991. [Google Scholar]
- 8.Guespin-Michel J, Letouvet-Pawlak B, Petit F. Protein secretion in myxobacteria. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 235–255. [Google Scholar]
- 8a.Guespin-Michel, J. Personal communication.
- 9.Hanlon W A, Martinez-Canamero M, Inouye M, Inouye S. MlpA, a lipoprotein required for normal development of Myxococcus xanthus. J Bacteriol. 1995;177:7150–7154. doi: 10.1128/jb.177.24.7150-7154.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgkin J, Kaiser D. Cell-to-cell stimulation of motility in non-motile mutants of Myxococcus xanthus. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 12.Innis M A, Tokunmaga M, Williams M E, Loranger J M, Chang S Y, Chang S, Wu H C. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci USA. 1984;81:3708–3712. doi: 10.1073/pnas.81.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaki L, Kawakami M, Beers R, Hom R, Wu H C. Cloning and nucleotide sequence of the Enterobacter aerogenes signal peptidase II (lsp) and flanking genes. J Bacteriol. 1990;172:469–472. doi: 10.1128/jb.172.1.469-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isaki L, Beers R, Wu H C. Nucleotide sequence of the Pseudomonas fluorescens signal peptidase II (lsp) gene. J Bacteriol. 1990;172:6512–6517. doi: 10.1128/jb.172.11.6512-6517.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashefi A, Hartzell P L. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF-defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 16.Kroos L, Kaiser D. Construction of Tn5lac, a transposon that fuses lacZ expression to exogenous promoters and its introduction into Myxococcus xanthus. Proc Natl Acad Sci USA. 1984;81:5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Miller K W, Bouvier J, Stragier P, Wu H C. Identification of the genes in Escherichia coli iles-lsp operon. Analysis of multiple polycistronic mRNA made in vivo. J Biol Chem. 1987;262:7391–7397. [PubMed] [Google Scholar]
- 21.Muñoa F J, Miller K, Beers R, Graham M, Wu H C. Membrane topology of Escherichia coli prolipoprotein signal peptidase (signal peptidase II) J Biol Chem. 1991;266:17667–17672. [PubMed] [Google Scholar]
- 22.Norrander J, Kempe T, Messing J. Construction of improved M13 vector. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 23.Paitan Y, Orr E, Ron E Z, Rosenberg E. A NusG-like transcription anti-terminator is involved in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus. FEMS Microbiol Lett. 1999;170:221–227. doi: 10.1111/j.1574-6968.1999.tb13377.x. [DOI] [PubMed] [Google Scholar]
- 24.Paitan Y, Orr E, Ron E Z, Rosenberg E. Cloning and characterization of a Myxococcus xanthus cytochrome P-450 hydroxylase required for biosynthesis of the polyketide antibiotic TA. Gene. 1999;228:147–153. doi: 10.1016/s0378-1119(98)00609-x. [DOI] [PubMed] [Google Scholar]
- 25.Paitan Y, Alon G, Orr E, Ron E Z, Rosenberg E. The first gene in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus codes for a unique PKS module coupled to a peptide synthetase. J Mol Biol. 1999;286:465–474. doi: 10.1006/jmbi.1998.2478. [DOI] [PubMed] [Google Scholar]
- 26.Paitan Y, Boulton N, Ron E Z, Rosenberg E, Orr E. Molecular analysis of the DNA gyrB gene from Myxococcus xanthus. Microbiology. 1998;144:1641–1647. doi: 10.1099/00221287-144-6-1641. [DOI] [PubMed] [Google Scholar]
- 27.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 28.Prágai Z, Tjalsma H, Bolhuis A, van Dijl J, Venema G, Bron S. The signal peptidase II (lsp) gene of Bacillus subtilis. Microbiology. 1997;143:1327–1333. doi: 10.1099/00221287-143-4-1327. [DOI] [PubMed] [Google Scholar]
- 29.Quillet L, Bensmail L, Barray S, Guespin-Michel J. Cloning and sequencing of two genes, prtA and prtB, from Myxococcus xanthus, encoding PrtA and PrtB proteases, both of which are required for the protease activity. Gene. 1997;198:135–140. doi: 10.1016/s0378-1119(97)00303-x. [DOI] [PubMed] [Google Scholar]
- 30.Reichenbach H. Biology of the myxobacteria: ecology and taxonomy. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 13–62. [Google Scholar]
- 31.Rodriguez-Soto J P, Kaiser D. The tgl gene: social motility and stimulation in Myxococcus xanthus. J Bacteriol. 1997;179:4361–4371. doi: 10.1128/jb.179.13.4361-4371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Soto J P, Kaiser D. Identification and localization of Tgl protein, which is required for Myxococcus xanthus social motility. J Bacteriol. 1997;179:4372–4381. doi: 10.1128/jb.179.13.4372-4381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg E, Vaks B, Zuckerberg A. Bactericidal action of an antibiotic produced by Myxococcus xanthus. Antimicrob Agents Chemother. 1973;4:507–513. doi: 10.1128/aac.4.5.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenberg E, Varon M. Antibiotics and lytic enzymes. In: Rosenberg E, editor. Myxobacteria, development and cell interaction. New York, N.Y: Springer-Verlag; 1984. pp. 109–125. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sankaran K, Wu H C. Signal peptidase II—specific signal peptidase for bacterial lipoproteins. In: von Heijne G, editor. Signal peptidases. R. G. Austin, Tex: Landes Company; 1994. pp. 17–29. [Google Scholar]
- 38.Scotti C, Piatti M, Cuzzoni A, Perani P, Tognoni A, Grandi G, Galizzi A, Albertini A M. A Bacillus subtilis large ORF coding for a polypeptide highly similar to polyketide synthases. Gene. 1993;130:65–71. doi: 10.1016/0378-1119(93)90347-6. [DOI] [PubMed] [Google Scholar]
- 39.Sedgwick S G, Morgan B A. Locating, DNA sequencing and disrupting yeast genes using tagged Tn1000. Methods Mol Genet. 1994;3:131–140. [Google Scholar]
- 40.Shimkets L J. Social and developmental biology of myxobacteria. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolchinsky S, Fuchs N, Varon M, Rosenberg E. Use of Tn5lac to study expression of genes required for antibiotic TA production. Antimicrob Agents Chemother. 1992;36:2322–2327. doi: 10.1128/aac.36.10.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varon M, Rosenberg E. Transcriptional regulation of genes required for antibiotic TA synthesis in Myxococcus xanthus. FEMS Microbiol Lett. 1996;136:203–208. [Google Scholar]
- 44.Varon M, Fuchs N, Monosov M, Tolchinsky S, Rosenberg E. Mutation and mapping of genes involved in antibiotic TA production in Myxococcus xanthus. Antimicrob Agents Chemother. 1992;36:2316–2321. doi: 10.1128/aac.36.10.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varon M, Paitan Y, Rosenberg E. Trans-acting regulation of antibiotic TA genes in Myxococcus xanthus. FEMS Microbiol Lett. 1997;155:141–146. doi: 10.1016/s0378-1097(97)00378-9. [DOI] [PubMed] [Google Scholar]
- 46.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1994;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 47.Witke C, Götz F. Cloning and nucleotide sequence of the signal peptidase II from Staphylococcus carnosus. FEMS Microbiol Lett. 1995;126:233–240. doi: 10.1111/j.1574-6968.1995.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 48.Wu H C, Tokunaga M. Biogenesis of lipoproteins in bacteria. Curr Top Microbiol Immunol. 1986;125:127–157. doi: 10.1007/978-3-642-71251-7_9. [DOI] [PubMed] [Google Scholar]
- 49.Yamagata H, Ippolito C, Inukai M, Inouye M. Temperature- sensitive processing of outer membrane lipoprotein in an Escherichia coli mutant. J Bacteriol. 1982;152:1163–1168. doi: 10.1128/jb.152.3.1163-1168.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zafriri D, Rosenberg E, Mirelman D. Mode of action of Myxococcus xanthus antibiotic TA. Antimicrob Agents Chemother. 1981;19:349–351. doi: 10.1128/aac.19.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X J, Wu H C. Nucleotide sequence of the Staphylococcus aureus signal peptidase II (lsp) gene. FEBS Lett. 1992;299:80–84. doi: 10.1016/0014-5793(92)80105-p. [DOI] [PubMed] [Google Scholar]