Abstract
Anaerobic mineralization of ethylbenzene by the denitrifying bacterium Azoarcus sp. strain EB1 was recently shown to be initiated by dehydrogenation of ethylbenzene to 1-phenylethanol. 1-Phenylethanol is converted to benzoate (benzoyl coenzyme A) via acetophenone as transient intermediate. We developed in vitro assays to examine ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities in cell extracts of this strain. With p-benzoquinone as the electron acceptor, cell extracts of Azoarcus sp. strain EB1 catalyzed ethylbenzene oxidation at a specific rate of 10 nmol min−1 [mg of protein]−1 and an apparent Km for ethylbenzene of approximately 60 μM. The membrane-associated ethylbenzene dehydrogenase activity was found to oxidize 4-fluoroethylbenzene and propylbenzene but was unable to transform 4-chloro-ethylbenzene, the ethyltoluenes, and styrene. Enzymatic ethylbenzene oxidation was stereospecific, with (S)-(−)-1-phenylethanol being the only enantiomer detected by chiral high-pressure liquid chromatography analysis. Moreover, cell extracts catalyzed the oxidation of (S)-(−)-1-phenylethanol but not of (R)-(+)-1-phenylethanol to acetophenone. When cell extracts were dialyzed, (S)-(−)-1-phenylethanol oxidation occurred only in the presence of NAD+, suggesting that NAD+ is the physiological electron acceptor of 1-phenylethanol dehydrogenase. Both ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities were present in Azoarcus sp. strain EB1 cells that were grown anaerobically on ethylbenzene, 1-phenylethanol, and acetophenone, but these activities were absent in benzoate-grown cells.
Gasoline spills and leaking underground fuel storage vessels have resulted in the release of BTEX compounds (benzene, toluene, ethylbenzene, and the xylene isomers) to the natural environment. The toxic properties (8, 21) of these compounds, in combination with their relatively high water solubility (7), have resulted in compromised water resources, which are often rendered anaerobic by oxygen-consuming microorganisms. Aerobic degradation of alkylbenzenes is initiated by the well-characterized oxygenases, which use molecular oxygen as a cosubstrate (12). However, recent studies with anaerobic microorganisms have shown that novel enzymatic reactions can be used as a means of initiating mineralization of alkylbenzenes in the absence of molecular oxygen. In vitro studies on anaerobic toluene oxidation in denitrifying and sulfate-reducing bacteria have shown that anaerobic toluene degradation is initiated by the addition of toluene to fumarate to form benzylsuccinate (3–5, 19). It has also been shown that in Azoarcus sp. strain T, m-xylene mineralization (16) is initiated by a homologous activation to form (3-methylbenzyl)succinate from m-xylene and fumarate. Benzylsuccinate and (3-methylbenzyl)succinate are mineralized via benzoate (or benzoyl coenzyme A [benzoyl-CoA]) and 3-methylbenzoate (or 3-methyl benzoyl-CoA), respectively, as transient intermediates.
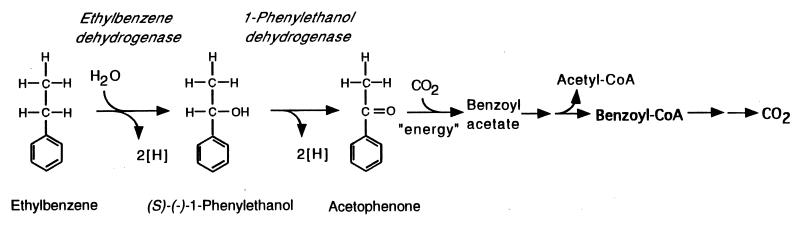
In contrast to anaerobic metabolism of methylbenzenes, ethylbenzene mineralization is initiated by dehydrogenation at the methylene carbon of the alkyl side chain to form 1-phenylethanol (2) (Fig. 1). 1-Phenylethanol is further oxidized to acetophenone (Fig. 1). It has been hypothesized (20) and preliminary evidence supports (2) that acetophenone is then carboxylated to form benzoyl acetate (benzoyl acetyl-CoA). Benzoyl acetyl-CoA is proposed to be further degraded via a thiolytic cleavage to form benzoyl-CoA, a common intermediate of anaerobic aromatic metabolism (11), and acetyl-CoA. We now report the first in vitro characterization of ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase in cell extracts of strain EB1.
FIG. 1.
Proposed initial reactions in anaerobic ethylbenzene mineralization (2, 21). Ethylbenzene is dehydrogenated at the methylene carbon to form 1-phenylethanol, and the hydroxyl group of 1-phenylethanol is derived from water. 1-Phenylethanol is further oxidized via acetophenone to benzoyl-CoA.
Because strain EB1 shows 99% identity to Azoarcus sp. based upon the previously determined 16S rDNA sequence (2), we refer to this bacterium as Azoarcus sp. strain EB1.
MATERIALS AND METHODS
Growth of Azoarcus sp. strain EB1.
Unless otherwise noted, cells of Azoarcus sp. strain EB1 were grown anaerobically in 1- or 2-liter batch cultures on mineral medium supplemented with ethylbenzene and nitrate as described previously (2).
Preparation of cell extract.
Cells (1 to 10 liters) growing exponentially, with a doubling time of 11 h, to an optical density at 400 nm of 0.3 were harvested under anaerobic conditions by centrifugation (10,000 × g for 20 min at 4°C). The cell pellet was washed in 100 ml of anoxic 20 mM Tris-HCl buffer (pH 7.5), recentrifuged, and resuspended in approximately 3 ml of the same buffer. The cells were then ruptured anaerobically by three passages through a French pressure cell at 138 MPa. Unbroken cells and cellular debris were removed by anaerobic centrifugation (12,000 × g for 15 min at 4°C). The supernatant was defined as cell extract. The cell extract (8 to 20 mg of protein/ml) was used immediately or stored in 1-ml aliquots at −20°C.
Dialyzed cell extract was prepared by dialyzing 1 to 2 ml of cell extract overnight against 1 liter of 20 mM Tris-HCl buffer (pH 7.5) at 4°C. The dialysis tubing had a molecular mass cutoff of 12,000 to 14,000 Da.
Fractionation of cell extract.
Cell extract was separated into membrane and cytoplasmic fractions by ultracentrifugation (150,000 × g for 2 h at 4°C). The supernatant was defined as the cytoplasmic fraction. The solid pellet was resuspended in anoxic 20 mM Tris-HCl (pH 7.5) buffer to a volume equal to the precentrifugation volume and defined as the membrane fraction.
Ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase assays.
Anaerobic in vitro assays of ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities were performed at room temperature (23°C) in 5-ml vials fitted with Teflon-lined Mininert (Alltech Associates, Inc., Deerfield, Ill.) caps. The 1-ml assay mixture contained 20 mM Tris-HCl buffer (pH 7.5) and 1 μmol of p-benzoquinone (added from a 20 mM stock solution in 70% ethanol) as an electron acceptor. Approximately 800 nmol of ethylbenzene was added for the ethylbenzene dehydrogenase standard assay, and approximately 800 nmol of 1-phenylethanol was added for the 1-phenylethanol dehydrogenase assay. Addition of 0.25 to 1 mg of protein as cell extract initiated the reactions. The assay mixture was incubated for 1 to 20 min in an anaerobic chamber (10% hydrogen, 10% carbon dioxide, 80% nitrogen [Coy Laboratory Products, Grass Lake, Mich.]) while shaking (approximately 100 rpm). Addition of 100 μl of concentrated hydrochloric acid was used to stop the reaction at selected time intervals. The transformation products, 1-phenylethanol or acetophenone or both, were extracted into 1 ml of hexane (with an efficiency of 40 and 70%, respectively) and quantified by gas chromatography with flame ionization detection (GC-FID) or gas chromatography-mass spectroscopy (GC-MS) or both. Product yields of duplicate assays were typically within ca. 10%. Rates for ethylbenzene dehydrogenase activity were calculated by dividing the sum of the amount of 1-phenylethanol and acetophenone formed by the elapsed time. The rate for 1-phenylethanol dehydrogenase was calculated by dividing the amount of acetophenone produced by the elapsed time.
Aerobic assays were performed by purging air into the assay mixture for 2 min before adding the volatile substrate. The reaction was initiated by addition of ethylbenzene or 1-phenylethanol.
For substrate specificity assays, 800 nmol of the substrate (o-, m-, or p-ethyltoluene, 4-fluoroethylbenzene, 4-chloroethylbenzene, propylbenzene, or styrene) was added. Oxidation products (alcohols and ketones homologous to 1-phenylethanol and acetophenone) of these substrates were identified and quantified by GC-FID or GC-MS analysis following extraction into 1 ml of hexane.
The assay buffer used for optimum pH determination was an anaerobic mixture of constant ionic strength containing 100 mM Tris, 50 mM 2-(N-morpholino)ethanesulfonic acid (MES), and 50 mM acetic acid (10).
Km values were calculated from a nonlinear least-squares fit of experimental data to the equation v = Vmax × S/(Km + S) by using the computer program SCIENTIST (MicroMath Scientific Software, Salt Lake City, Utah). The range of substrate concentrations used for the determination of Km values was 10 to 800 μM ethylbenzene for ethylbenzene dehydrogenase activity and 10 to 600 μM 1-phenylethanol for 1-phenylethanol dehydrogenase activity.
Other enzyme assays.
Malate dehydrogenase (22) and NADH:p-benzoquinone oxidoreductase activities were determined in photometric assays by monitoring the absorbance at 340 nm. The 1-ml NADH:p-benzoquinone oxidoreductase assay mixture contained 20 mM Tris-HCl buffer (pH 7.5), 1 mM p-benzoquinone, 0.25 mM NADH, and approximately 0.02 mg of protein as cell extract. The reaction was started by the addition of p-benzoquinone. The progress of the reaction was monitored by measuring the decrease in absorbance at 340 nm. Under our assay conditions, NADH oxidation by p-benzoquinone occurred both chemically and enzymatically. Therefore, the abiotic rate (assay mixture without cell extract; approximately 100 nmol/min) was subtracted from the total rate to determine the rate of the enzyme-catalyzed reaction.
Cell suspension experiment.
Cell suspension experiments were performed as previously described (2) with ethylbenzene and nitrate as substrates. The cell suspension reactions were halted by centrifugation at 10,000 × g (20 min at 4°C) after 2 h of anaerobic incubation while shaking (approximately 100 rpm). The supernatant was immediately extracted with 20 ml of hexane. The hexane extract was concentrated approximately fourfold and analyzed by chiral high-pressure liquid chromatography (HPLC).
Chemical analysis.
Optical density and absorbance measurements were made with an HP model 8451A diode array spectrophotometer (Hewlett-Packard).
1-Phenylethanol, acetophenone, 1-phenyl-1-propanol, propiophenone, 4-fluoro-1-phenylethanol, 4-fluoroacetophenone, 4-chloro-1-phenylethanol, and 4-chloroacetophenone were analyzed with an Hewlett-Packard 5890 Series II gas chromatograph-flame ionization detector (Hewlett-Packard) equipped with a DB 1701 fused-silica capillary column (length, 30 m; inside diameter, 0.32 mm; J&W Scientific) with a flow rate of 1 ml/min. Compounds were identified by comparison of retention times to authentic standards, and their identities were confirmed by GC-MS. Detection limits were approximately 1 nmol. Analyses of 1-phenylethanol, acetophenone, 1-phenyl-1-propanol, and propiophenone were isothermal at 90°C. The retention times of 1-phenylethanol, acetophenone, 1-phenyl-1-propanol, and propiophenone were 11.5, 10.4, 17.5, and 16.6 min, respectively. 4-Fluoro-1-phenylethanol and 4-fluoroacetophenone were analyzed by GC-FID with an 80°C isocratic temperature setting. The retention times for 4-fluoro-1-phenylethanol and 4-fluoroacetophenone were 22.4 and 16.1 min, respectively. 4-Chloro-1-phenylethanol and 4-chloroacetophenone were analyzed by GC-FID with a temperature program of isocratic 90°C for 15 min followed by a temperature gradient of 10°C per min up to 200°C. The retention times for 4-chloro-1-phenylethanol and 4-chloroacetophenone were 22.2 and 20.6 min, respectively.
Products of the ethylbenzene dehydrogenase assays with 2-, 3-, or 4-ethyltoluene as substrate were analyzed by GC-MS. No authentic standards were commercially available for comparison; therefore, products could be identified only by peak detection and an analysis of the molecular mass. The detection limit was estimated to be approximately 10 nmol based on the detection limit of similar compounds such as 1-phenylethanol and acetophenone.
GC-MS analyses were performed as described previously (2).
Chiral high-performance liquid chromatography (HPLC) was performed with an HP 1050 series pump system (Hewlett-Packard) equipped with a 25-cm Chiralpak AD (Chiral Technologies Inc., Exton, Penn.) column and 100-μl sample loop. Chromatographic conditions were as described previously (18). (R)-(+)-1-phenylethanol and (S)-(−)-1-phenylethanol were identified by comparison of retention times to those of authentic chiral standards and by coinjection with optically pure (R)-(+)-1-phenylethanol and (S)-(−)-1-phenylethanol. Retention times varied by several minutes per day but were approximately 48 min for (S)-(−)-1-phenylethanol and 45 min for (R)-(+)-1-phenylethanol. The response also varied by day, with a detection limit of approximately 10 nmol.
Protein was determined by the method of Bradford (6) by using a commercially available dye-binding assay (Bio-Rad Laboratories, Hercules, Calif.). Bovine serum albumin was used as the standard.
Chemicals and solutions.
Chemicals were reagent grade and were purchased from Sigma or Aldrich (Milwaukee, Wis.). 4-Fluoroethylbenzene and 4-chloroethylbenzene were purchased from Lancaster Synthesis, Inc. (Windam, N.H.). All solutions except for p-benzoquinone (20 mM) and menaquinone (10 mM) were prepared with distilled water; p-benzoquinone and menaquinone solutions were prepared with 70% ethanol in distilled water.
RESULTS
Ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities in cell extracts.
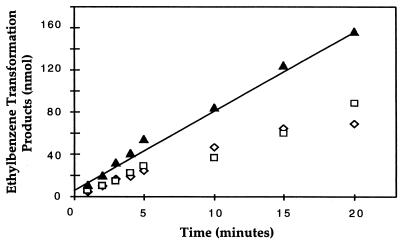
The denitrifying bacterium Azoarcus sp. strain EB1 was previously shown to completely mineralize ethylbenzene to carbon dioxide via the initial metabolic intermediates 1-phenylethanol and acetophenone (2). We developed assays to characterize ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities in vitro. When establishing assays for these redox reactions, we examined physiological and artificial electron acceptors with a standard-state redox potential (E0′) that was more positive than that of the redox pair, 1-phenylethanol/ethylbenzene (E0′ = −12 mV) (calculated from data in references 9 and 13) and of acetophenone/1-phenylethanol (E0′ = −265 mV) (calculated from data in references 1 and 13). Initial experiments revealed that when 1 mM p-benzoquinone was used as the electron acceptor, ethylbenzene was oxidized to both 1-phenylethanol and acetophenone. We did not expect ethylbenzene to be transformed beyond 1-phenylethanol and acetophenone, since our in vitro assay mixture did not contain an energy source (e.g., ATP), which was expected to be required for carboxylation of acetophenone to benzoyl acetate (Fig. 1). As is evident from Table 1, the highest specific activity was detected with p-benzoquinone. Among all electron acceptors tested, none was identified that served only for ethylbenzene oxidation but not for 1-phenylethanol oxidation. Therefore, we assayed ethylbenzene dehydrogenase activity with p-benzoquinone and defined the activity as the sum of 1-phenylethanol and acetophenone formed per unit time. Under standard assay conditions, ethylbenzene dehydrogenase activity was found in cell extracts of Azoarcus sp. strain EB1 at a specific rate of 5 to 10 nmol min−1 [mg of protein]−1. The amounts of reaction products increased linearly over time for at least 20 min, as shown in Fig. 2. Ethylbenzene dehydrogenase activity was linearly dependent upon the protein concentration, and heat-treated cell extracts did not display any activity (data not shown). The optimum pH for the assay was 7.5, with constant-ionic-strength buffers between pH 4 and 10 tested (data not shown). The ethylbenzene dehydrogenase activity was not oxygen sensitive, since addition of the reducing agent titanium(III) citrate (up to 0.2 mM) to the standard assay mixture did not increase activity (data not shown). The assay could also be performed under aerobic conditions without a loss of activity.
TABLE 1.
Electron acceptors for ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities in cell extracts
| Electron acceptor tested | Activitya of ethylbenzene dehydrogenase (%) | Activitya of 1-phenylethanol dehydrogenase* (%) |
|---|---|---|
| p-Benzoquinone (E0′ = +293 mV) | 100 | 100 |
| Oxygen (E0′ = +820 mV) | 40 | 30 |
| Nitrate (E0′, nitrate/nitrite = +430 mV) | 11 | 17 |
| Dichlorophenol indophenol (E0′ = +217 mV) | 58 | 29 |
| 1,4-Napthoquinone (E0′ = +36 mV) | <1 | 22 |
| Methylene blue (E0′ = +11 mV) | 2 | 6 |
| Menaquinone (E0′ = −80 mV) | 6 | ND |
| NAD+ (E0′ = −320 mV) | NDb | 25 |
| NADP+ (E0′ = −320 mV) | ND | 10 |
| None | 10 | 10 |
Activity measured as percentage of activity in cell extracts with p-benzoquinone as the electron acceptor (approximately 10 nmol min−1 [mg of protein]−1 for ethylbenzene dehydrogenase and approximately 15 nmol min−1 [mg of protein]−1 for 1-phenylethanol dehydrogenase). Assay conditions were as described in Materials and Methods. All electron acceptors tested were at 1 mM in the in vitro assay (except for oxygen).
ND, not determined.
FIG. 2.
Time course of ethylbenzene dehydrogenase activity. The amount of ethylbenzene transformed in cell extracts increased linearly with time up to 20 min. The amount of ethylbenzene transformed (▴) was equal to the sum of 1-phenylethanol (◊) and acetophenone (□) formed. The assay mixtures contained approximately 1 mg of protein as cell extract. Values are means of duplicate assays.
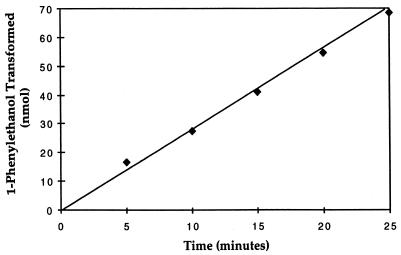
Because acetophenone was a significant product of the standard ethylbenzene dehydrogenase assay, we also examined 1-phenylethanol dehydrogenase activity. As shown in Table 1, the use of p-benzoquinone as an electron acceptor resulted in the highest 1-phenylethanol dehydrogenase activity with cell extracts. The amount of acetophenone formed from 1-phenylethanol increased linearly over time (Fig. 3) at a specific rate of 10 to 15 nmol min−1 [mg of protein]−1. Unfrozen cell extract had a specific activity of approximately 50 nmol min−1 [mg of protein]−1. The rate of 1-phenylethanol dehydrogenase activity was protein dependent, and heat-treated cell extracts were inactive (data not shown). The 1-phenylethanol dehydrogenase activity did not appear to be oxygen sensitive, since acetophenone was formed in the ethylbenzene dehydrogenase assay under aerobic conditions and oxygen could serve as an electron acceptor for 1-phenylethanol dehydrogenase activity (Table 1).
FIG. 3.
Time course of 1-phenylethanol dehydrogenase activity. The amount of 1-phenylethanol transformed by cell extracts increased linearly with time up to 25 min. The assay mixtures contained approximately 0.25 mg of protein as cell extract. Values are means of duplicate assays.
Ethylbenzene and 1-phenylethanol oxidation rates were dependent on the substrate concentration as predicted by standard enzyme kinetics. For ethylbenzene dehydrogenase activity, the apparent Km for ethylbenzene was approximately 62 μM (s.d. = 11). For 1-phenylethanol dehydrogenase activity, the apparent Km for 1-phenylethanol was approximately 25 μM (s.d. = 4) (data not shown).
Localization of the ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities.
The cytoplasmic and membrane fractions were tested for ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities. As shown in Table 2, the membrane fraction contained approximately half of the recovered ethylbenzene dehydrogenase activity whereas 70% of the 1-phenylethanol dehydrogenase activity was found in the cytoplasmic fraction. Malate dehydrogenase (22) and NADH:p-benzoquinone oxidoreductase activities were used as cytoplasmic and membrane marker activities, respectively. The distribution of ethylbenzene dehydrogenase activity between the cytoplasmic and membrane fractions suggests that ethylbenzene dehydrogenase activity is membrane associated. Preliminary experiments show that ethylbenzene dehydrogenase activity can be solubilized by a high sodium chloride concentration (data not shown).
TABLE 2.
Localization of ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities in cell extracts of Azoarcus sp. strain EB1
| Sample | Activity (nmol/min) (%) of:
|
|||
|---|---|---|---|---|
| Ethylbenzene dehydrogenase | 1-Phenylethanol Dehydrogenase | Malate Dehydrogenase | NADH:p-benzoquinone oxidoreductasea | |
| Cell extract | 3 (100) | 25 (100) | 43 (100) | 2,760 (100) |
| Membrane fraction | 1 (33) | 2 (8) | 6 (14) | 992 (36) |
| Cytoplasmic fraction | 1 (33) | 17 (68) | 42 (98) | 816 (30) |
Enzyme-catalyzed activity only. See Materials and Methods for more details.
Substrate specificity of ethylbenzene dehydrogenase activity.
We tested whether several ethylbenzene derivatives could be transformed by cell extracts of ethylbenzene-grown Azoarcus sp. strain EB1. After the incubation period, reaction products were extracted and analyzed by GC-FID and GC-MS. In assay mixtures that contained o-, m-, or p-ethyltoluene as the substrate, no transformation products could be detected following 10 min of incubation (the detection limit was approximately 10 nmol). Moreover, addition of 700 nmol of these compounds to the standard ethylbenzene dehydrogenase assay mixture did not inhibit ethylbenzene oxidation (data not shown). Oxidation products of the halogenated ethylbenzene, 4-chloroethylbenzene, were not detected by GC-FID or GC-MS analysis (detection limit, 1 nmol) following 10 min or 1 h of incubation, but oxidation products of 4-fluoroethylbenzene were detected. After 10 min of incubation, approximately 200 nmol of 4-fluoroacetophenone were detected. We also tested propylbenzene as a substrate and detected small amounts of 1-phenylpropanol (3 nmol) and propiophenone (0.5 nmol) by GC-FID and GC-MS analysis following 20 min of incubation.
As we showed previously, ethylbenzene oxidation to 1-phenylethanol is a dehydrogenation reaction where the hydroxyl group of 1-phenylethanol is derived from water. One possible mechanism for this enzymatic reaction is the initial desaturation of the ethyl side chain to form styrene, followed by hydration of styrene to form 1-phenylethanol. To test the hypothesis that styrene is a free intermediate of enzymatic ethylbenzene oxidation, styrene was used as a substrate for ethylbenzene dehydrogenase or as an inhibitor of ethylbenzene oxidation or both. No products were detected by GC-FID or GC-MS analysis following a 30-min incubation of the ethylbenzene dehydrogenase assay mixture with styrene as the sole substrate, nor did addition of styrene (800 nmol) to the standard ethylbenzene dehydrogenase assay mixture inhibit ethylbenzene dehydrogenase activity. The specific oxidation rate of ethylbenzene in the presence of styrene was 9.6 nmol min−1 [mg of protein]−1, compared to 10 nmol min−1 [mg of protein]−1 in the absence of styrene. This observation suggests that styrene is not a free intermediate of ethylbenzene oxidation to 1-phenylethanol.
Stereospecificity of ethylbenzene dehydrogenase and stereoselectivity of 1-phenylethanol dehydrogenase.
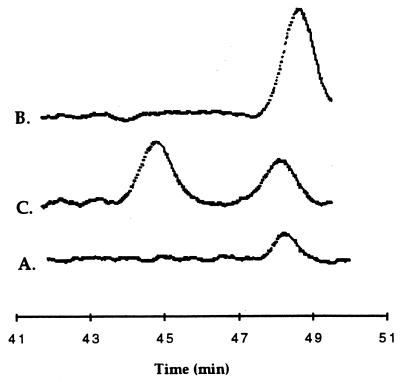
Because the C-1 carbon of 1-phenylethanol is a chiral center, we investigated whether (S)-(−)-1-phenylethanol, (R)-(+)-1-phenylethanol, or both enantiomers were formed from ethylbenzene. The concentration of 1-phenylethanol in the in vitro assay mixture was too low to be analyzed by chiral HPLC. Therefore, we isolated 1-phenylethanol from ethylbenzene-metabolizing cell suspensions of Azoarcus sp. strain EB1. When the metabolically produced 1-phenylethanol, coinjected with (S)-(−)-1-phenylethanol, was applied to the chiral HPLC column, the 1-phenylethanol coeluted with the (S)-(−)-1-phenylethanol standard (as evidenced by a single larger peak [Fig. 4]). However, when metabolically produced 1-phenylethanol, coinjected with (R)-(+)-1-phenylethanol, was analyzed, an additional peak eluting at the time of (R)-(+)-1-phenylethanol elution was detected which was not present in the metabolic sample. These chromatograms revealed that only the (S) enantiomer of 1-phenylethanol was formed from ethylbenzene by Azoarcus sp. strain EB1.
FIG. 4.
Chiral HPLC analysis of 1-phenylethanol formed from ethylbenzene in cell suspension. (A) Chiral HPLC chromatograph of the sample alone, approximately 100 μM of 1-phenylethanol. (B) Chiral HPLC chromatograph of the sample coinjected with 100 μM (S)-(−)-1-phenylethanol. (C) Chiral HPLC chromatograph of the sample coinjected with 100 μM (R)-(+)-1-phenylethanol. Retention times are normalized to standards. Sample A was analyzed on a different day. See Materials and Methods.
We also investigated the stereoselectivity of the 1-phenylethanol dehydrogenase activity in cell extract assays. We found that (S)-(−)-1-phenylethanol was oxidized to acetophenone but that (R)-(+)-1-phenylethanol was not transformed (data not shown). In addition, 100 μM (R)-(+)-1-phenylethanol did not inhibit the oxidation of (S)-(−)-1-phenylethanol (at 20 to 200 μM) to acetophenone. These results provide experimental evidence that 1-phenylethanol dehydrogenase is a stereoselective enzyme, oxidizing only the (S) enantiomer of 1-phenylethanol.
1-Phenylethanol dehydrogenase activity in dialyzed cell extracts.
In the standard 1-phenylethanol dehydrogenase assay, we observed 1-phenylethanol oxidation with p-benzoquinone as the electron acceptor. Of this activity, 70% was found in the cytoplasmic fraction (Table 2). To investigate this unusual coupling of a quinone to the cytoplasmic 1-phenylethanol dehydrogenase, we examined this activity more closely. Using the standard 1-phenylethanol dehydrogenase assay, we found that 1-phenylethanol dehydrogenase activity increased linearly with p-benzoquinone concentration up to 5 mM (data not shown). These findings led us to reason that an electron carrier that coupled 1-phenylethanol oxidation to p-benzoquinone reduction may be present in the cell extract. Hence, we dialyzed the cell extract overnight and reexamined the in vitro assay. Following dialysis, the rate of 1-phenylethanol oxidation with p-benzoquinone as electron acceptor was reduced to 2 nmol min−1 [mg of protein]−1. In contrast, when NAD+, a common electron acceptor for cytoplasmic alcohol dehydrogenases, was used as the sole electron acceptor in the assay, the specific activity before and after dialysis was similar, i.e., 5 (Table 1) and 7 nmol min−1 [mg of protein]−1, respectively. To test whether NAD+ may be involved as an electron carrier between 1-phenylethanol dehydrogenase and p-benzoquinone reductase, we attempted to reconstitute the 1-phenylethanol dehydrogenase activity observed in our standard cell extract assay by using dialyzed cell extract. When these assay mixtures were amended with 10 μM NAD+ plus 1 mM p-benzoquinone, a specific activity for 1-phenylethanol oxidation of 30 nmol min−1 [mg of protein]−1 was observed. This result suggests that the 1-phenylethanol dehydrogenase activity detected in undialyzed cell extracts may involve a coupling of 1-phenylethanol dehydrogenase activity, which presumably used residual NAD+ as the electron acceptor, and an NADH:p-benzoquinone oxidoreductase activity, which oxidized NADH. Enzymatic NADH oxidation by p-benzoquinone occurred very rapidly (0.7 μmol min−1 [mg of protein]−1). Therefore, this activity would not be the rate-limiting step in the assay with dialyzed cell extract or in the standard 1-phenylethanol dehydrogenase assay (Fig. 3; Table 1).
We further examined the effects of NAD+ and p-benzoquinone concentrations on 1-phenylethanol dehydrogenase activity in dialyzed cell extracts. Increasing the concentration of NAD+ from 10 μM to 1 mM with the p-benzoquinone concentration kept constant at 1 mM did not increase the activity (data not shown). Similar results were obtained with 2.5 mM p-benzoquinone. These results suggest that the apparent Km of 1-phenylethanol dehydrogenase for NAD+ may be less than 10 μM. When the concentration of NAD+ was kept constant at 10 μM, increasing the p-benzoquinone concentration to up to 10 mM resulted in a linear increase in activity up to 0.2 μmol min−1 [mg of protein]−1, which would suggest a high Km for p-benzoquinone in the NADH:p-benzoquinone oxidoreductase activity. This observation is consistent with our earlier observation with undialyzed cell extracts.
The 1-phenylethanol dehydrogenase activity was much lower with NAD+ as the sole electron acceptor than with small amounts of NAD+ plus 1 mM p-benzoquinone. In addition, a spectrophotometric assay monitoring the absorbance at 340 nm for 1-phenylethanol dehydrogenase showed that the rate of NADH formation decreased rapidly (data not shown). One possible explanation is an inhibition of 1-phenylethanol dehydrogenase activity by NADH. We also found that addition of 1 mM NADH to the 1-phenylethanol dehydrogenase assay mixture resulted in an 86% decrease in activity (data not shown). To circumvent this apparent inhibition by NADH, we investigated the reverse reaction of 1-phenylethanol dehydrogenase, the reduction of acetophenone to 1-phenylethanol. In this new assay, enzymatic reduction of acetophenone to 1-phenylethanol was monitored spectrophotometrically (NADH depletion at 340 nm) or by GC (1-phenylethanol formation). We discovered that the specific activity as measured by the decrease in NADH and/or the formation of 1-phenylethanol consisted of two rates, an initial high rate (5 to 10 nmol min−1 [mg of protein]−1) followed by a much lower rate (0.5 to 2 nmol min−1) after 1 to 3 min. Only the initial high rate was dependent upon protein concentration (data not shown).
The 1-phenylethanol dehydrogenase assays with dialyzed cell extracts suggest that NAD+ is a likely physiological electron acceptor for 1-phenylethanol dehydrogenase. Furthermore, 1-phenylethanol dehydrogenase activity may be tightly regulated by the concentration of NADH.
Induction of enzyme activities.
We investigated which anaerobic growth substrates would lead to the expression of ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities in Azoarcus sp. strain EB1. We found that when Azoarcus sp. strain EB1 was grown anaerobically on ethylbenzene, 1-phenylethanol, or acetophenone, both ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities were found at high specific rates (Table 3). Conversely, if cells were grown anaerobically on benzoate, neither 1-phenylethanol dehydrogenase nor ethylbenzene dehydrogenase activity was found. This suggests that the initial anaerobic ethylbenzene degradation intermediates were able to induce these activities but benzoate was not.
TABLE 3.
Induction of enzyme activities involved in anaerobic ethylbenzene oxidation in Azoarcus sp. strain EB1
| Growth substratea | Activityb (nmol min−1 [mg of protein]−1) (%) of
|
|
|---|---|---|
| Ethylbenzene dehydrogenase | 1-Phenylethanol dehydrogenase | |
| Ethylbenzene | 7 (100) | 12 (100) |
| 1-Phenylethanol | 6 (85) | 89 (740) |
| Acetophenone | 5 (71) | 47 (390) |
| Benzoate | <DLc | <DL |
Azoarcus sp. strain EB1 was grown under denitrifying conditions with the growth substrates indicated as the sole carbon and electron source.
Activities were measured in vitro.
DL, detection limit.
DISCUSSION
Anaerobic ethylbenzene mineralization is initiated by a novel enzymatic reaction, the oxidation of ethylbenzene to 1-phenylethanol catalyzed by an ethylbenzene dehydrogenase activity (2, 20) (Fig. 1). This activity and a 1-phenylethanol dehydrogenase activity represent the first reactions in the anaerobic ethylbenzene degradation pathway (Fig. 1). We examined the ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities in cell extracts of ethylbenzene grown cells of Azoarcus sp. strain EB1. By using p-benzoquinone as an electron acceptor, the ethylbenzene dehydrogenase activity of approximately 10 nmol min−1 [mg of protein]−1 detected in vitro accounts for approximately 10% of the observed rate of ethylbenzene consumption in cell suspension—0.1 μmol min−1 [mg of protein]−1 (2) (Fig. 2). The highest specific activity in vitro was observed when p-benzoquinone served as the electron acceptor (Table 1), and this activity was membrane associated (Table 2). These findings suggest that a quinone is the physiological electron acceptor for ethylbenzene dehydrogenase. Recently, Rabus and Heider (19) reported ethylbenzene dehydrogenase activity in cell extracts of ethylbenzene-grown denitrifying strain EbN1 at a specific rate of 10 to 30 pmol min−1 [mg of protein]−1. In these studies, nitrate was used as the electron acceptor. The 1,000-fold-higher specific activity observed in our cell extract may be due to the selection of p-benzoquinone as the electron acceptor and to differences in experimental handling.
We explored the substrate specificity of the ethylbenzene dehydrogenase activity. When ring-substituted ethylbenzenes were tested as substrates for ethylbenzene dehydrogenase activity, only 4-fluoroethylbenzene was oxidized. After 10 min of incubation in the standard ethylbenzene dehydrogenase assay, 4-fluoroethylbenzene was transformed to an extent similar to that of ethylbenzene (approximately 0.2 μmol compared to 0.1 μmol), suggesting that activation of ethylbenzene at the para position may not be essential for the enzymatic dehydrogenation reaction. The methyl- and 4-chloro-substituted ethylbenzenes may not have been oxidized due to steric hindrance by the ring substituents.
The effect of the alkyl side chain on ethylbenzene dehydrogenase activity was examined with propylbenzene as substrate. Propylbenzene, which is not a growth substrate for Azoarcus sp. strain EB1 (15a), was transformed to 1-phenylpropanol and propiophenone at a low rate (approximately 2 nmol was detected after 10 min), suggesting that the methyl group in the alpha position with respect to the methylene carbon of ethylbenzene may not be crucial for catalysis. The low activity observed with propylbenzene as the substrate may be due to suboptimal binding of the substrate at the active site. The propylbenzene- and ethylbenzene-mineralizing, denitrifying bacterium PbN1 was found to utilize 1-phenyl-1-propanol and propiophenone, but not 1-phenyl-2-propanol, as growth substrates under denitrifying conditions (20). It is therefore conceivable that an enzyme activity related to the ethylbenzene dehydrogenase activity described here is involved in the first step of anaerobic propylbenzene mineralization.
In one possible reaction mechanism of ethylbenzene dehydrogenase, ethylbenzene may be transformed to 1-phenylethanol in a two-step process consisting of a dehydrogenation followed by a hydration. A two-step dehydrogenation-hydration reaction has been proposed for p-cresol methylhydroxylase (14). This enzyme catalyzes the oxidation of p-cresol under aerobic (14) and denitrifying (15) conditions by dehydrogenation of p-cresol to a quinone methide, which is hydrated to form p-hydroxybenzyl alcohol. However, for p-cresol methylhydroxylase activity, the hydroxyl group para to the alkyl group is required for activity (17), suggesting that the para hydroxyl group may assist in the removal of a hydride. Oxidation of ethylbenzene could also involve a dehydrogenation-hydration mechanism, which would form styrene as an intermediate. When we assayed styrene as the substrate in the standard ethylbenzene dehydrogenase assay, we did not detect any 1-phenylethanol or acetophenone. Furthermore, styrene did not inhibit the rate of ethylbenzene oxidation. These observations are consistent with the notion that styrene is not a free intermediate in ethylbenzene oxidation to 1-phenylethanol.
Similar to p-cresol methylhydroxylase, the ethylbenzene dehydrogenase activity is insensitive to molecular oxygen. This is interesting since Azoarcus sp. strain EB1 can grow on ethylbenzene only under denitrifying conditions (2). However, this strain is able to grow aerobically with 1-phenylethanol. This may suggest that the expression of ethylbenzene dehydrogenase or ethylbenzene uptake may be coregulated by molecular oxygen.
The second enzyme of anaerobic ethylbenzene mineralization is 1-phenylethanol dehydrogenase (Fig. 1). 1-Phenylethanol dehydrogenase activity was found in the cytoplasmic fraction of Azoarcus sp. strain EB1 (Table 2). Examination of the 1-phenylethanol dehydrogenase activity with dialyzed cell extract suggests that NAD+ is the most likely electron acceptor in this reaction. Consistent with the stereospecific formation of (S)-(−)-1-phenylethanol by ethylbenzene dehydrogenase (Fig. 4) is the finding that in cell extracts, only this enantiomer was oxidized to acetophenone.
Under denitrifying conditions, both enzyme activities, ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase, were present in cells grown with 1-phenylethanol and acetophenone as the substrate, whereas in benzoate-grown cells these activities were not detected. These findings are consistent with the different protein banding patterns detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cell extracts from strain EbN1 grown on ethylbenzene or benzoate (19).
The ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities described here support the proposed pathway of anaerobic ethylbenzene and 1-phenylethanol degradation (Fig. 1) (2). The information concerning these activities and the assays developed to measure them can now be used for purification of the environmentally relevant ethylbenzene dehydrogenase and 1-phenylethanol dehydrogenase activities.
ACKNOWLEDGMENTS
Funding for this project was provided by a grant from the U.S. Environmental Protection Agency through the Western Region Hazardous Substance Research Center and National Science Foundation grant MCB 9733535.
We thank Bettina Rosner for technical advice.
REFERENCES
- 1.American Institute of Chemical Engineers, Department of Chemical Engineering, Penn State University. Physical and thermodynamic properties of pure chemicals. Data compilation. Design Institute for Physical Property Data. Pa: University Park; 1991. [Google Scholar]
- 2.Ball H A, Johnson H A, Reinhard M, Spormann A M. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J Bacteriol. 1996;178:5755–5761. doi: 10.1128/jb.178.19.5755-5761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beller H R, Spormann A M. Anaerobic activation of toluene and o-xylene by addition to fumarate in denitrifying strain T. J Bacteriol. 1997;179:670–676. doi: 10.1128/jb.179.3.670-676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beller H R, Spormann A M. Benzylsuccinate formation as a means of anaerobic toluene activation by sulfate-reducing strain PRTOL1. Appl Environ Microbiol. 1997;63:3729–3731. doi: 10.1128/aem.63.9.3729-3731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biegert T, Fuchs G, Heider J. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem. 1996;238:661–668. doi: 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Cline P V, Delfino J J, Rao P S C. Partitioning of aromatic constituents into water from gasoline and other complex solvent mixtures. Environ Sci Technol. 1991;25:914–920. [Google Scholar]
- 8.Dean B J. Recent findings on the genetic toxicology of benzene, toluene, xylenes, and phenols. Mutat Res. 1985;154:153–181. doi: 10.1016/0165-1110(85)90016-8. [DOI] [PubMed] [Google Scholar]
- 9.Dean J A, editor. Lange’s handbook of chemistry. 14th ed. New York, N.Y: McGraw-Hill Publishers; 1992. [Google Scholar]
- 10.Ellis K J, Morrison J F. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs G, Mohamed M E S, Altenschmidt U, Kock J, Lack A, Brackmann R, Lochmeyer C, Oswald B. Biochemistry of anaerobic biodegradation of aromatic compounds. In: Ratledge C, editor. Biochemistry of microbial degradation. Boston, Mass: Kluwer Academic Publishers; 1994. pp. 513–553. [Google Scholar]
- 12.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 181–252. [Google Scholar]
- 13.Guthrie P. Equilibrium constants for a series of simple aldol condensations and linear free energy relations with other carbonyl addition reactions. Can J Chem. 1978;56:962–973. [Google Scholar]
- 14.Hopper D J. The hydroxylation of p-cresol and its conversion to p-hydroxybenzaldehyde in Pseudomonas putida. Biochem Biophys Res Commun. 1976;69:462–468. doi: 10.1016/0006-291x(76)90544-1. [DOI] [PubMed] [Google Scholar]
- 15.Hopper D J, Bossert I D, Rhodes-Roberts M E. p-Cresol methylhydroxylase from a denitrifying bacterium involved in anaerobic degradation of p-cresol. J Bacteriol. 1991;173:1298–1301. doi: 10.1128/jb.173.3.1298-1301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Johnson, H., and A. Spormann. Unpublished data.
- 16.Krieger, C. K., H. R. Beller, M. Reinhard, and A. M. Spormann. Initial reactions in anaerobic oxidation of m-xylene by the denitrifying bacterium Azoarcus sp. strain T. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 17.McIntire W, Hopper D J, Singer T J. p-Cresol methylhydroxylase: assay and general principles. Biochem J. 1985;228:325–335. doi: 10.1042/bj2280325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naemura K, Murata M, Tanaka R, Yano M, Hirose K, Tobe Y. Enantioselective acylation of primary and secondary alcohols catalyzed by lipase QL from Alcaligenes sp.: a predictive active-site model for lipase QL to identify which enantiomer of an alcohol reacts faster in this acylation. Tetrahedron Asymmetry. 1996;7:3285–3294. [Google Scholar]
- 19.Rabus R, Heider J. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch Microbiol. 1998;170:377–384. [Google Scholar]
- 20.Rabus R, Widdel F. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol. 1995;163:96–103. doi: 10.1007/BF00381782. [DOI] [PubMed] [Google Scholar]
- 21.Sittig M. Handbook of toxic and hazardous chemicals and carcinogens. 2nd ed. Park Ridge, N.J: Noyes Publications; 1985. [Google Scholar]
- 22.Stams A J M, Kremer D R, Nicolay K, Weenk G H, Hansen T A. Pathway of propionate formation in Desulfobulbus propionicus. Arch Microbiol. 1984;139:167–173. [Google Scholar]