Abstract
The protein HoxA is the central regulator of the Alcaligenes eutrophus H16 hox regulon, which encodes two hydrogenases, a nickel permease and several accessory proteins required for hydrogenase biosynthesis. Expression of the regulatory gene hoxA was analyzed. Screening of an 8-kb region upstream of hoxA with a promoter probe vector localized four promoter activities. One of these was found in the region immediately 5′ of hoxA; the others were correlated with the nickel metabolism genes hypA1, hypB1, and hypX. All four activities were independent of HoxA and of the minor transcription factor ς54. Translational fusions revealed that hoxA is expressed constitutively at low levels. In contrast to these findings, immunoblotting studies revealed a clear fluctuation in the HoxA pool in response to conditions which induce the hox regulon. Quantitative transcript assays indicated elevated levels of hyp mRNA under hydrogenase-derepressing conditions. Using interposon mutagenesis, we showed that the activity of a remote promoter is required for hydrogenase expression and autotrophic growth. Site-directed mutagenesis revealed that PMBH, which directs transcription of the structural genes of the membrane-bound hydrogenase, contributes to the expression of hoxA under hydrogenase-derepressing conditions. Thus, expression of the hox regulon is governed by a positive feedback loop mediating amplification of the regulator HoxA. These results imply the existence of an unusually large (ca. 17,000-nucleotide) transcript.
Alcaligenes eutrophus H16 is a facultative lithoautotroph that can utilize hydrogen as its sole source of energy. Two biochemically and physiologically distinct [NiFe] metalloenzymes catalyze the oxidation of H2: a membrane-bound hydrogenase (MBH), which couples H2 oxidation to electron transport phosphorylation, and a cytoplasmic hydrogenase (SH), which catalyzes H2-dependent reduction of NAD+ (reviewed in references 14 and 15). The hydrogenases supply the organism with energy during lithoautotrophic growth on CO2. They are also synthesized in the presence of poor organic substrates, permitting the organism to utilize H2 as a supplemental energy source (16).
Both A. eutrophus hydrogenases are nickel metalloenzymes. A. eutrophus possesses a high-affinity Ni permease which ensures a supply of Ni for their synthesis (12). Both enzymes undergo a complex maturation process, which converts the inactive precursor forms to catalytically active enzymes. The quintessential steps in the two maturation pathways lead to the assembly of the nickel-containing active sites. These sites contain a special coordination structure containing one nickel atom, one iron atom, and three diatomic ligands. The architecture of this metallocenter appears to be conserved in the [NiFe] hydrogenases (1, 13). The assembly of the hydrogenase [NiFe] metallocenter in A. eutrophus and in other bacteria is mediated by a set of specialized proteins encoded by the hyp genes. One of the functions of the Hyp proteins is to donate Ni to the hydrogenase apoprotein (7, 8). HypX may be instrumental in inserting the diatomic ligands into the nascent metallocenter (6). The underlying mechanism and the specific contributions of the Hyp proteins are fascinating but so far have thwarted analysis (28). Metallocenter assembly seems to be intimately connected to C-terminal proteolytic processing of the Ni-containing large subunit of the hydrogenase enzyme. Both the MBH and the SH undergo C-terminal proteolytic processing, and each enzyme has its own specific protease (4, 22, 29, 43).
Detailed studies on the expression of the hydrogenase structural genes have been carried out in our laboratory (39). Both the MBH and SH genes are transcribed from ς54-dependent promoters. Expression of these genes is controlled at the transcriptional level. The central regulatory agent governing the hydrogenase regulon is a transcriptional activator encoded by the gene hoxA (10). HoxA triggers the activation of the promoters in response to energy limitation, e.g., during growth on poor organic substrates such as glycerol or during autotrophic growth. The actual physiological cue is unknown. The deduced amino acid sequence of HoxA reveals several features typical of response regulators of the NtrC family. The N-terminal part of the protein is homologous to the receiver domains of the response regulators and has an aspartate residue at the usual position. The mode of action of this protein is, however, unconventional. HoxA mediates activation of the cognate promoters in the absence of a sensory kinase. Furthermore, mutations altering the conserved phosphoryl acceptor residue do not abolish transcriptional activation by HoxA (24). Recently, a hydrogen-sensing system which mediates H2-dependent control of hydrogenase expression was discovered in strains of Alcaligenes (24). Remarkably, this system, consisting of a dimeric H2 receptor and a histidine protein kinase, is cryptic in the wild-type strain but can be activated by a mutation leading to a single-amino-acid exchange. This mutation seems to occur at a high frequency and may represent a genetic switch allowing the organism to shift between two modes of regulation.
The main goal of the study reported here was to investigate the expression of the hydrogenase regulator, HoxA, itself. We used a promoter probe vector to search for and quantify promoters directing transcription of hoxA. Additional information on the transcription of the hoxA region was obtained from RNase protection experiments. Plasmid-borne translational fusions were used to monitor expression of hoxA under different growth conditions and to compare the expression of hoxA and other hydrogenase genes. Finally, the cellular levels of regulator were assayed via Western immunoblotting.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. A. eutrophus H16 is the wild-type strain harboring the endogenous megaplasmid pHG1. Strains HF09 (37) and HF18 (17) are derivatives of H16. Escherichia coli S17-1 (41) served as a donor in conjugative transfers. Strains of A. eutrophus were grown in a modified Luria broth containing 0.25% sodium chloride and 0.4% fructose (LBF medium) or in mineral salts medium as described previously (39). Synthetic media for heterotrophic growth contained 0.4% fructose (FN medium), 0.4% succinate (SN medium), or 0.2% fructose and 0.2% glycerol (FGN medium). Lithoautotrophic cultures were grown in mineral salts medium under an atmosphere of hydrogen, carbon dioxide, and oxygen of 8:1:1 (vol/vol/vol). For assays of hydrogenase activity, cells were cultivated in the above media containing 1 μM NiCl2 in place of the standard trace elements mixture SL6. Strains of E. coli were grown in LB medium or in M9 medium containing glycerol (30). Solid media contained 1.5% agar. Antibiotics were added where appropriate (for A. eutrophus, kanamycin [350 μg/ml] and tetracycline [15 μg/ml]; for E. coli, kanamycin [25 μg/ml], tetracycline [15 μg/ml], and ampicillin [50 μg/ml]).
TABLE 1.
Strains, plasmids, and synthetic oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristicsa or sequence | Source or reference |
|---|---|---|
| Strains | ||
| A. eutrophus | ||
| H16 | MBH+ SH+ | DSM428, ATCC 17699 |
| HF18 | H16 hoxA18, MBH− SH− | 17 |
| HF09 | H16 rpoN09, MBH− SH− | 37 |
| HF457 | H16 hoxTΩR-B2 | This study |
| HF491 | H16 hoxKΔ171-R4 | This study |
| E. coli | ||
| S17-1 | recA pro thi hsdR chr::RP4-2 Tra+ | 41 |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) e14−(F′ lacIqlacZΔM15 proAB traD36) | 48 |
| Plasmids: | ||
| pEDY305 | Tcr RK2 ori Mob+, promoterless lacZ gene | 39 |
| pPHU234 | Tcr RK2 ori Mob+, ′lacZ translational fusion vector | 20 |
| pPHU278 | Φ(aph-lacZ), Paph deleted | 20 |
| pGMΩ1 | Derivative of pUC18 carrying a polar Ω cassette | 40 |
| pLO2 | sacB oriTRP4 ColE1 ori, Kmr Mob+, counterselectable suicide vector | 25 |
| pRZ10 | Φ(acoR-lacZ) | 23 |
| pCH103 | Derivative of pSUP202 carrying the hyp locus | 11 |
| pCH104 | Derivative of pSUP202 carrying the hyp locus | 11 |
| pCH300 | 2.2-kb SalI-BamHI fragment of pGE43 in pBluescript KS+ | This study |
| pCH301 | 2.2-kb PstI-KpnI fragment of pGE43 in pBluescript KS+ | This study |
| pCH303 | 2.6-kb XhoI-SalI fragment (Klenow enzyme treated) of pGE43 in pBluescript SK+ | This study |
| pCH304 | 2.7-kb SalI-XhoI fragment of pGE43 in pBluescript KS+ | This study |
| pCH427 | hoxT subcloned in pLO1, KmrsacB oriTRP4 ColE1 ori | 4 |
| pCH679 | Ω cassette of pGMΩ1 inserted as a SmaI fragment into the EcoRV site of pCH427 | This study |
| pCH680 | Deletion derivative lacking 171 bp of the hoxK upstream region introduced into pLO2 as 686-bp PstI-XhoI fragment | This study |
| pGE43 | Derivative of pVK101 carrying the hyp region | 10 |
| pGE281 | 8.5-kb EcoRI (Klenow enzyme-treated)-HpaI fragment of pCH104 in pPHU234, Φ(hoxA-lacZ) | This study |
| pGE282 | 6.1-kb BstEII (Klenow enzyme-treated)-HpaI fragment of pCH104 in pPHU234, Φ(hoxA-lacZ) | This study |
| pGE283 | 1.9-kb BspEI (Klenow enzyme-treated)-HpaI fragment of pCH104 in pPHU234, Φ(hoxA-lacZ) | This study |
| pGE319 | Derivative of pEDY305 with PMBH upstream of lacZ | 39 |
| pGE320 | Derivative of pEDY305 with PSH upstream of lacZ | 39 |
| pGE322 | 667-bp XhoI-PvuII fragment of pCH104 in pEDY305 | O. Lenz and B. Friedrich |
| pGE323 | 516-bp PvuII fragment of pCH303 in pEDY305 | O. Lenz and B. Friedrich |
| pGE324 | 711-bp BamHI-NheI fragment pCH104 in pEDY305 | This study |
| pGE325 | 1739-bp NheI-HpaI fragment of pCH104 in pEDY305 | O. Lenz and B. Friedrich |
| pGE326 | 463-bp SnaBI fragment of pCH301 in pEDY305 | This study |
| pGE327 | 721-bp FspI-Eco47III fragment of pCH303 in pEDY305 | This study |
| pGE328 | 233-bp segment of pRZ10 subcloned as a PCR product in pEDY305 | This study |
| pGE413 | 2.1-kb EcoRI-PvuII fragment of pCH103 in pPHU234, Φ(hypB1-lacZ) | This study |
| pGE417 | 1-kb AscI-NheI fragment of pCH301 in pEDY305 | This study |
| pGE418 | 350-bp segment of pCH301 subcloned as PCR product in pEDY305 | This study |
| pGE420 | 393-bp segment of pCH103 subcloned as a PCR product in pEDY305 | This study |
| Oligonucleotides | ||
| BF213 | 5′-gcgGATCCATGACCACCAGG-3′ (M90471, 150–166) | |
| BF214 | 5′-tggccCGGGCGCGTGCTGCGG-3′ (M90471, 383–368) | |
| BF250 | 5′-ccagCTAGCGATTCCCGCACG-3′ (X70183, 2466–2482) | |
| BF251 | 5′-ccgCGCGCCTCAGCGCGAACG-3′ (X70183, 2802–2785) | |
| BF360 | 5′-acagagatcTCGCGGAGTTGGTGTTGC-3′ (M96433, 8579–8596) | |
| BF361 | 5′-gcaacaGCTGAGCTCATGCATGGACAG-3′ (M96433, 8971–8951) |
MBH, membrane-bound hydrogenase activity; SH, NAD-reducing hydrogenase activity; Tra, transfer of mobilizable plasmids; ori, origin of replication; chr::RP4-2, chromosomally integrated copy of RP4-2; Mob, mobilizability. For oligonucleotides, foreign or altered nucleotides are shown in lowercase, and accession numbers of DDBJ/EMBL/GenBank entries and coordinates are given in parentheses.
Plasmid and strain construction.
For the generation of an interposon mutant, an Ω cassette was excised from plasmid pGMΩ1 (kindly supplied by H. P. Schweitzer, Calgary, Alberta, Canada) by digestion with SmaI and inserted into the EcoRV site of pCH427. The resulting plasmid (pCH679) was used to transfer the allele hoxTΩR-B2 into A. eutrophus H16 via an allelic exchange procedure (25), yielding strain HF457. A deletion derivative with a lesion in the MBH promoter region was isolated by a similar strategy. Plasmid pCH128 was cut with BamHI and BglII and religated. A 686-bp PstI-XhoI fragment spanning the site of the deletion was transferred to the suicide vector pLO1, resulting in plasmid pCH680. The latter plasmid was used to generate the mutant HF491. Promoter test constructs were obtained by inserting the fragments listed in Table 1 directly into pEDY305. Oligonucleotides BF213 and BF214 were used to amplify a 233-bp segment of the acoR promoter region on plasmid pRZ10. The PCR product was cut with BamHI and SmaI and inserted into pEDY305. Similarly, amplified segments of the hypD and hypA1 upstream regions were obtained using the primer pairs BF250-BF251 and BF360-BF361, respectively. After cutting with NheI and BssHII (hypD fragment) and BglII and PvuII (hypA1 fragment), the products were likewise inserted into pEDY305.
Conjugative plasmid transfer.
Mobilizable plasmids were transferred from E. coli S17-1 to A. eutrophus by a spot mating technique (41). Donor and recipient strains were grown on LB and LBF media, respectively. Transconjugants were selected on FN plates containing the appropriate antibiotics.
Recombinant DNA techniques.
Standard DNA techniques were used in this study (2). Large-scale isolation of plasmid DNA was carried out by the alkaline lysis procedure followed by ethidium bromide-cesium chloride gradient centrifugation. Smaller amounts of pure plasmid DNA were isolated by using Qiagen Tip-20 columns (Qiagen GmbH) according to the manufacturer’s instructions. DNA fragments used in plasmid constructions were isolated from agarose gels by using the Qiaex II system (Qiagen GmbH).
Western immunoblot analysis.
Strains of A. eutrophus were grown under standard conditions as described above. Mid-log-phase cells were harvested, resuspended in 50 mM potassium phosphate buffer (pH 7.0), and homogenized by three passages through a French pressure cell. Soluble and membrane fractions were separated by spinning for 30 min at 140,000 × g; 20-μg samples were separated by polyacrylamide gel electrophoresis (PAGE) through sodium dodecyl sulfate (SDS)–10% polyacrylamide gels. BenchMark Prestained Protein Ladder (Life Technologies Inc.) was used as a molecular mass standard. Following SDS-PAGE, proteins were transferred to Protran BA85 membranes (Schleicher & Schuell) (45). Blots were treated with rabbit polyclonal antisera (diluted 1:1,000) raised against HypD, HypX, or HoxA and developed with alkaline phosphatase conjugate (Dianova).
RNase protection assays.
Riboprobes were synthesized by using a MAXIscript kit (Ambion, Inc.) and 32P-labeled UTP (800 Ci/mmol; Dupont NEN). XmnI-linearized plasmid pCH300 and NarI-linearized plasmid pCH304 served as templates for the generation of the riboprobes E and F1C, respectively. The riboprobes were 354 and 91 nucleotides (nt) long, respectively. The in vitro transcripts were purified by two rounds of ethanol precipitation. Total RNA was prepared by a hot-phenol procedure (19). Total RNA (5 to 20 μg) was added to 30 μl of hybridization buffer (40 mM PIPES [pH 6.4], 0.4 M NaCl, and 1 mM EDTA in a 1:4 (vol/vol) mixture of water-deionized formamide) containing 105 to 106 cpm of the appropriate riboprobe. After an initial denaturation step (5 min at 85°C), hybridization proceeded for at least 8 h at 45°C. Then 350 μl of RNase digestion cocktail (10 mM Tris-HCl, [pH 7.5], 300 mM NaCl, 5 mM EDTA, 40 μg of RNase A per ml, 2 μg of RNase T1 per ml) was added, and the mixture was incubated for 30 min at 30°C. Treatment with proteinase K (10 μl of 20% [wt/vol] SDS, 2.5 μl of proteinase K [20 mg/ml]; 37°C for 15 min) was followed by phenol extraction and precipitation in the presence of 10 μg of yeast tRNA. The pellet was dissolved in 3 to 5 μl of formamide loading buffer and applied to a 6% sequencing gel. In vitro transcripts of known length served as size standards. Quantitation of the protected fragments was done by analysis of scanned images obtained in a Molecular Dynamics PhosphorImager 445 SI, using IP-Labgel software (Scanalytics, Inc.).
Enzyme assays.
Independent single colonies of the strains to be tested were picked from plates and inoculated in liquid media. Precultures were incubated for 15 to 20 h at 35°C. Since the hydrogenase system is repressed at temperatures above 33°C, this step ensures that the cells are uniformly devoid of hydrogenase at the beginning of an experiment. SH (hydrogen:NAD+ oxidoreductase; EC 1.12.1.2) activity was assayed by spectrophotometric determination of H2-dependent NAD reduction in detergent-treated cells. MBH (ferredoxin:H+ oxidoreductase; EC 1.18.99.1) activity was determined by measuring H2-dependent methylene blue reduction in isolated membranes. One unit of hydrogenase activity is the amount of enzyme which catalyzes the formation of 1 μmol of product per min. β-Galactosidase was assayed by the standard method (30), and the activity (in units) was calculated according to Miller except that cell density was measured at 436 nm. Unless otherwise indicated, enzyme activities were assayed in mid-log-phase cells, i.e., cells grown to optical densities of 3 in SN medium, 5 in FN medium, 8 in FGN medium, and 4 under lithoautotrophic conditions. Protein determinations were done according to the method of Lowry et al. (26).
RESULTS
Mapping promoters upstream of hoxA.
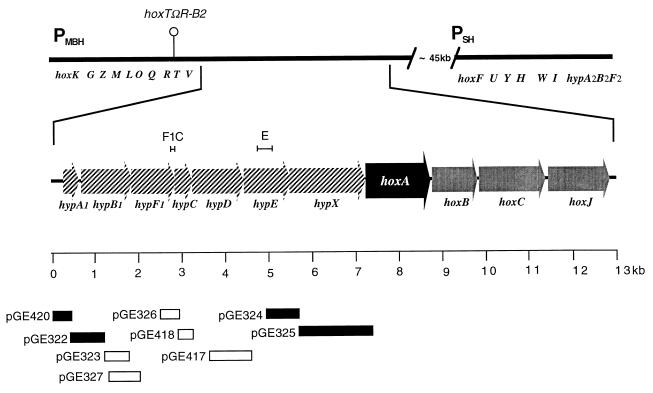
As a first step in the analysis of the expression of the hydrogenase regulator HoxA, we screened the region upstream of hoxA (Fig. 1) by using a promoter assay vector. Plasmid pEDY305, a broad-host-range vector designed in our laboratory for promoter assays in gram-negative bacteria (39), was chosen for this study. DNA fragments from the 8-kb region containing genes hypA1 through hoxA were inserted into the multiple cloning site of pEDY305. The resulting recombinant plasmids were introduced into A. eutrophus H16 via conjugal transfer from the E. coli donor strain S17-1. Transconjugants were streaked onto FN plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and scored for β-galactosidase activity. Four of the transconjugants tested gave a positive reaction in the plate test. These strains contained the plasmids pGE420, pGE322, pGE324, and pGE325, harboring DNA segments upstream of the genes hypA1, hypB1, hypX, and hoxA, respectively (Fig. 1). This indicated that the inserted DNA contained functional promoters which directed transcription of the vector-borne reporter gene. The four plasmid-harboring strains were cultivated in liquid media for quantitative β-galactosidase assays. Cultures were grown under hydrogenase-derepressing conditions (FGN medium). All four strains produced low but significant levels of β-galactosidase activity indicating moderate promoter activities (Table 2). The strongest activity (829 U for pGE420) resided in the hypA1 upstream region; the weakest (173 U for pGE324) was in the hypX upstream region. For comparison, plasmids pGE319 and pGE320, containing the structural gene promoters PMBH and PSH, respectively, were included in the experiment. These constructs produced 14,445 and 12,961 U, respectively (Table 2). We also measured the β-galactosidase activity in liquid cultures of selected transconjugants testing negative in the plate test. The activity of these strains was not more than double the background activity, i.e., the level of activity in the wild-type strain harboring vector plasmid pEDY305.
FIG. 1.
The A. eutrophus hydrogenase genes. The upper part of the diagram represents the two hydrogenase gene clusters on the A. eutrophus megaplasmid pHG1. The genes of the MBH region (hoxKGZMLOQRTV) and SH region (hoxFUYHWIhypA2B2F2) are labeled below the bar. The promoters PMBH and PSH are indicated. The site of an interposon in the polar mutation in strain H16 hoxTΩR-B2 is marked by a hairpin. The segment containing the regulatory genes hoxA, hoxB, hoxC, and hoxJ and the upstream hyp genes (hatched arrows) is enlarged. The scale bar pertains to the enlarged part of the drawing. The inserts of the various promoter test constructs are represented by bars. Open bars represent plasmids testing negative on X-Gal indicator plates; solid bars denote positive constructs. The brackets above the genetic map indicate the sequences used to generate the riboprobes F1C and E.
TABLE 2.
Activities of promoter-test plasmids in wild-type and mutant strains of A. eutrophus
| Strain | β-Galactosidase activity (U)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| pGE420 | pGE322 | pGE324 | pGE325 | pGE328 (PacoR) | pGE320 (PSH) | pGE319 (PMBH) | pEDY305 | |
| H16 | 829 ± 13 | 219 ± 3 | 173 ± 4 | 319 ± 13 | 450 ± 20 | 12,961 ± 960 | 14,445 ± 773 | 23 ± 1 |
| HF09 (H16 rpoN09) | 499 ± 54 | 227 ± 5 | 194 ± 3 | 1,052 ± 22 | 184 ± 22 | 146 ± 3 | 89 ± 5 | 18 ± 1 |
| HF18 (H16 hoxA18) | 579 ± 66 | 232 ± 7 | 233 ± 10 | 1,711 ± 56 | 346 ± 64 | 78 ± 2 | 62 ± 1 | 22 ± 0 |
FGN cultures; mean values ± standard errors; n = 5.
Cultures of the four transconjugants grown under hydrogenase-repressing conditions contained similar levels of β-galactosidase activity as under derepressing conditions (data not shown). In contrast, the activity of the structural gene promoters is negligible in repressed cells (39). This suggested that the promoters present on the four test plasmids are not subject to the same regulation as PMBH and PSH. The latter promoters are controlled by the transcriptional activator HoxA (39). To test the dependence of the promoters contained in pGE420, pGE322, pGE324, and pGE325 on HoxA, we introduced these plasmids into the hoxA mutant HF18 and assayed the β-galactosidase activity in cultures grown in FGN medium (Table 2). All four plasmids produced significant levels of β-galactosidase in the HoxA− background. In the case of pGE325, the activity was even higher in the mutant than in the wild type. In contrast, the activity of the structural gene promoters was dramatically higher in the HoxA+ background. We also tested the four new constructs in a ς54-deficient strain (HF09). Again the activities produced by the four plasmids were comparable to or higher than the wild-type background (Table 2). The ς54-dependent promoters PMBH and PSH gave only basal levels of β-galactosidase in the mutant. Thus, the promoters cloned in the four test plasmids are dependent neither on HoxA nor on the alternative transcription factor ς54. For comparison, we included a control plasmid with a constitutive promoter in the experiment. We chose the promoter of the A. eutrophus acoR gene, which controls the acetoin catabolism operon, since expression of this gene has been shown to be constant under different growth conditions (23). The test plasmid containing the acoR promoter (pGE328) showed significant promoter activity in both the RpoN− and HoxA− backgrounds, albeit at somewhat lower levels.
Expression of hoxA is independent of hydrogenase expression.
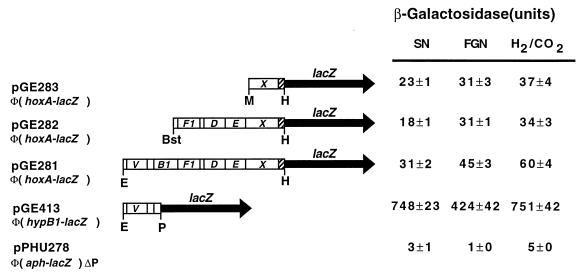
One or more of the promoters identified in our screening could contribute to the expression of hoxA. To monitor the expression of this regulatory gene under different physiological conditions, we constructed in-frame fusions to the lacZ gene in a mobilizable, broad-host-range vector. Three Φ(hoxA-lacZ) fusions with identical fusion joints (codon 95 of hoxA) but different amounts of upstream sequence were assembled. A derivative of the same vector containing a promoterless kanamycin resistance gene fused in frame to lacZ (20) served as a control. An A. eutrophus transconjugant harboring the smallest fusion plasmid (pGE283) produced low levels of β-galactosidase activity (Fig. 2). The activities were, nevertheless, significantly higher than the background level assayed in the negative control, indicating that hoxA is in fact expressed under the control of a proximal promoter. Similar amounts of activity were assayed in both repressed (SN medium) and derepressed cells (FGN medium and lithoautotrophic cultures). Thus, HoxA is expressed constitutively at a low level. The second construct (pGE282), which carried additional upstream sequences and the promoter activity associated with the hypX upstream region, gave similar β-galactosidase values. Apparently the distal promoter does not contribute significantly to the expression of hoxA. The third fusion plasmid (pGE413) carried 8.5 kb of the region upstream of hoxA and, thus, a contiguous sequence spanning the segments contained in the promoter test plasmids pGE420, pGE322, pGE324, and pGE325. The β-galactosidase activities associated with this plasmid were higher than those determined for the smaller Φ(hoxA-lacZ) fusions. This indicates that distal promoters contribute to the transcription of hoxA, but the effect is not simply additive.
FIG. 2.
Expression of hoxA and hyp genes. Translational fusions for hoxA and hypB1 were generated by inserting restriction fragments into vector plasmid pPHU234. The resulting recombinants were mobilized into A. eutrophus H16, and reporter gene activity was monitored under hydrogenase-repressing (SN medium) and -derepressing (FGN medium and lithoautotrophic cultures) conditions. The relevant segment of each plasmid is shown schematically at the left; the corresponding β-galactosidase activities are shown at the right. The solid arrows denote the lacZ gene of the vector (not to scale). Bars represent the inserted A. eutrophus sequences. The shaded segment indicates the 5′ part of hoxA. Some of the genes are labeled for clarity (see Fig. 1). The restriction enzymes used to generate the insert are given below the bar. Bst, BstEII; E, EcoRI; H, HpaI; M, MroI; P, PvuII. pPHU278 is a control plasmid containing a Φ(aph-lacZ) fusion deleted for the aph promoter (20). Values are means of five independent measurements ± standard errors.
To compare the expression of hoxA and the upstream hyp genes, we constructed a comparable plasmid-borne fusion for the hypB1 gene. The Φ(hypB1-lacZ) also expressed β-galactosidase independent of the growth regimen (Fig. 2). The activities assayed were 10- to 20-fold higher than those produced by the Φ(hoxA-lacZ) fusions.
Immunoreactive HoxA is present only in derepressed cells.
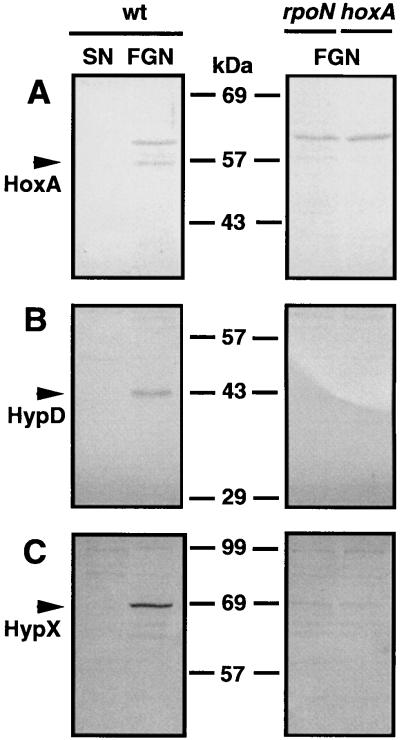
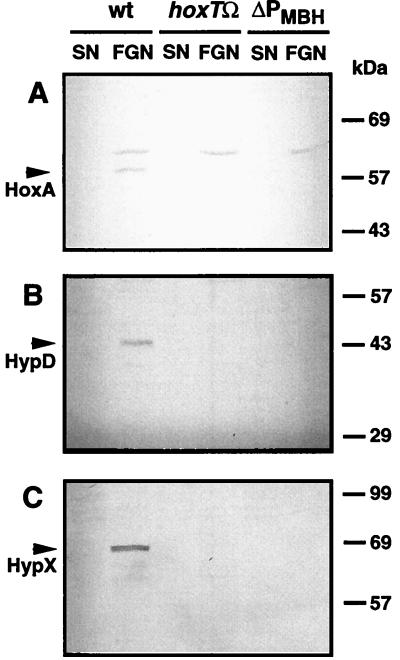
The results described above show that expression of the hoxA gene is constitutive within the scope of the experimental conditions tested. This finding agrees with the promoter activity data, which indicate that transcription from the promoters in the hyp region is not significantly modulated under the relevant growth conditions. Together the two sets of data suggest that the concentration of regulator in the cell is more or less constant. To assay the cellular levels of HoxA directly, we carried out immunoblotting experiments using anti-HoxA serum. Soluble extracts from cells of A. eutrophus grown under hydrogenase-repressing and -derepressing conditions were separated by SDS-PAGE and immobilized on nitrocellulose membranes. Surprisingly, significant amounts of immunoreactive HoxA were present in cells grown in FGN medium but not in succinate-grown cells (Fig. 3). Cells grown on other carbon sources known to mediate repression of the hydrogenase system were likewise devoid of immunologically detectable HoxA (data not shown). In some cases, an additional band of cross-reacting material was present in the immunoblots. However, control experiments with hoxA mutants and overproducing strains left no doubt that the 57-kDa species was HoxA (data not shown).
FIG. 3.
Cellular levels of HoxA, HypD, and HypX in strains of A. eutrophus under different growth conditions. Cultures of A. eutrophus H16 (wild type [wt]) and of the mutant strains HF09 (rpoN) and HF18 (hoxA) were grown to mid-log phase under hydrogenase-repressing (SN) and -derepressing (FGN) conditions. Cells were homogenized, and 20-μg samples of the soluble proteins were separated by SDS-PAGE on 10% gels. Proteins were subsequently transferred to nitrocellulose membranes and stained with antisera directed against HoxA (A), HypD (B), and HypX (C). Arrows indicate the immunoreactive species. Positions of the molecular mass standards are given between the panels.
The paradoxical finding described above might be due to differential stability of protein HoxA. However, assays of the same extracts using anti-HypD and anti-HypX sera revealed a similar pattern for the two Hyp proteins: antigenic material was detectable in the glycerol-grown cells but not in the succinate-grown cells (Fig. 3). The appearance of immunologically detectable amounts of the three unrelated proteins following derepression on glycerol argues against differential stability as the basis of the variations in cellular HoxA concentration. Western analysis of soluble proteins from two mutant strains provided additional clues on the fluctuations of the cellular HoxA pool. A. eutrophus HF09 (RpoN−) and HF18 (HoxA−) were cultivated in FGN medium, and soluble extracts were tested along with extracts from the wild-type cells. Mutant and wild-type extracts were screened on the same membrane to permit direct comparison (Fig. 3). Staining with anti-HoxA gave a scarcely visible band for the RpoN− extract. Thus, the amount of HoxA in the RpoN− background was significantly reduced. Staining with anti-HypD or anti-HypX gave scarcely visible bands, indicating radically lower levels of antigenic material in both cases (Fig. 3). Comparable findings were obtained for the HoxA− extracts. Only traces of HypD and HypX were visible. As expected, a band with an apparent molecular mass of 57 kDa corresponding to HoxA was absent. These data show that the presence of immunologically detectable quantities of HoxA, HypD, and HypX in glycerol-grown cells requires ς54-dependent transcription. Furthermore, the elevated levels of HypD and HypX require the hydrogenase regulator HoxA.
hyp mRNA is more abundant in derepressed cells.
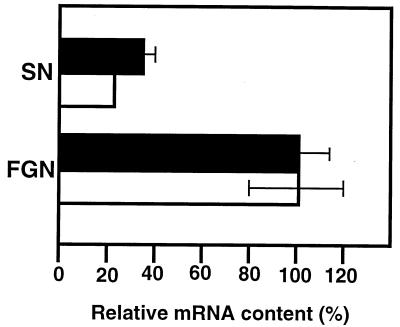
The data described above led us to conclude that the fluctuation in the HoxA pool is not a protein-stability-related phenomenon. We hypothesized that an additional promoter or promoters direct transcription of the hoxA gene under hydrogenase-derepressing conditions. This promoter might have escaped detection in our screening procedure, or it might be located outside of the region screened. To investigate this, we used an RNase protection assay to quantitate transcripts from the hyp region in cells growing under repressing (SN medium) and derepressing (FGN medium) conditions. Since the immunological data showed that the expression of the genes hypD, hypX, and hoxA was affected in a similar fashion, we chose two different segments within this stretch of DNA to use as templates for generating riboprobes. The riboprobes were complementary to the hypF1-hypC border (probe F1C) and to an internal segment of hypE (probe E) (Fig. 1). The two riboprobes consistently yielded similar results: approximately three- to fourfold more hyp mRNA was present in derepressed cells (Fig. 4). This increase in transcript concentration, although not dramatic, could account for the observed fluctuation in the cellular levels of the three gene products.
FIG. 4.
Relative abundance of A. eutrophus hyp mRNA in repressed and derepressed cells. Total RNA was isolated from A. eutrophus cells grown to mid-log phase on SN or FGN medium; 5 to 20 μg of each sample was hybridized to 32P-labeled riboprobe (105 to 106 cpm) for 8 h at 45°C. Following RNase treatment, the protected hybrids were separated in 6% sequencing gels. Labeled species were quantitated by densitometric analysis of scanned images. The bars (solid, probe E; open, probe F1C) represent radioactivities of protected fragments, taking the value for the FGN culture as 100% in each case. Each bar represents the average of determinations done on three separate cultures. Standard errors are indicated by brackets (in one case the standard error was too small for graphic representation).
The MBH promoter mediates high-level expression of hoxA.
The increase in hyp mRNA levels in derepressed cells prompted us to search for an additional promoter. The data obtained with the translational fusion constructs suggested that the hypothetical promoter was located upstream of the hyp region. The region adjacent to hypA1 contains the genes for the MBH enzyme and its auxiliary functions. The entire 9-kb segment has been sequenced, and the genetic determinants have been mapped to the nucleotide sequence (4, 21). We inserted the Ω cassette from plasmid pGMΩ1, which carries a strong transcriptional terminator, into a plasmid-borne copy of the MBH accessory gene hoxT. The resulting mutation was substituted for the wild-type gene via an allelic exchange procedure. The site of the insertion in the resulting homogenote is located more than 8.5 kb upstream of the initiator codon of hoxA. In-frame deletions in hoxT cause only a slight reduction in MBH activity (4). The biological function of the gene product is not known. The downstream accessory gene hoxV plays a more important role in MBH biosynthesis, and polarity on hoxV should reduce MBH activity to low levels. Nonpolar hoxV mutants and even MBH null mutants are viable, and lithoautotrophic growth is only slightly slower than growth of wild-type strains, since the synthesis of the SH enzyme is not affected (4). The newly isolated hoxTΩR-B2 mutant (strain HF457) was first tested for lithoautotrophic growth on H2. After 3 days of incubation under lithoautotrophic conditions, the control strains formed zones of confluent growth, whereas no growth was apparent in the zone where the mutant had been streaked. Assays of hydrogenase activity revealed that the cells were devoid of MBH activity (Table 3). This was not unexpected, assuming a polar effect of the interposon on hoxV. Remarkably, however, SH activity was significantly lower in the mutant strain. The low levels of SH activity represented a 10-fold decrease compared to the wild type. This drastic effect on SH activity cannot be attributed to the defect in hoxT or to a polarity on hoxV, since the synthesis of SH is not dependent on MBH-related functions, and must therefore be due to polarity on one or more genes further downstream. Polarity on hypA1, hypB1, or hypF1 should have no effect on SH synthesis, since two copies of these genes are present and the second copy fully complements a knockout mutation in each case (7). A polar effect on either hypC, hypD, hypE, hypX, or hoxA should, however, have a marked effect on SH activity, since lesions in these genes either abolish or reduce both hydrogenase activities. Disrupting expression of these genes would, however, affect SH synthesis at different levels. Defects in the gene for the transcriptional activator HoxA block transcription of the SH operon, whereas lesions in any of the four hyp genes curtail maturation of the SH enzyme. To test for an effect of the polar insertion in HF457 on transcription of hoxA, which would in turn perturb transcription of the SH operon, we introduced the promoter assay plasmids pGE319 and pGE320 into the mutant and wild-type strains and monitored the β-galactosidase activity in the resulting transconjugants under hydrogenase-derepressing conditions (Table 3). The indicator plasmids produced 10-fold less β-galactosidase in the hoxTΩR-B2 background, indicating a drastic reduction in the activity of both hydrogenase promoters. The reduction in promoter activity can account for the reduction in SH activity and can in turn be explained by a curtailment of HoxA expression. The above data provide key evidence that the interposon in HF457 curtails transcription of the gene hoxA, which originates from a remote promoter. An important prediction follows from this conjecture: if transcription of hoxA in HF457 is reduced enough to cause a drastic decrease in hydrogenase activity, then the cellular levels of HoxA must be measurably lower in this strain. A Western analysis of the wild-type and mutant strains showed that this is indeed the case (Fig. 5). Not only was the HoxA content of the HF457 cells below the detection threshold of our system, but the proteins HypD and HypX were likewise undetectable. The lower levels of the Hyp proteins support our hypothesis, since a polar mutation in hoxT which exerts an effect on hoxA must necessarily also influence the expression of the genes located in between.
TABLE 3.
Lithoautotrophic growth, hydrogenase activitie, and activities of the SH and MBH promoters in wild-type and mutant strains of A. eutrophus
| Strain | Growth on H2a | Activity (U)b
|
|||
|---|---|---|---|---|---|
| Hydrogenase
|
β-Galactosidase
|
||||
| SH | MBH | PSH (pGE320) | PMBH (pGE319) | ||
| H16 | + | 2.1 ± 0.1 | 0.6 ± 0.1 | 12,961 ± 960 | 14,445 ± 773 |
| HF457 (H16 hoxTΩR-B2) | − | 0.3 ± 0 | 0.0 | 1,115 ± 32 | 942 ± 32 |
| HF491 (H16 hoxKΔ171-R4) | − | ND | ND | 4,039 ± 48 | 3,033 ± 158 |
Single colonies were picked and suspended in phosphate buffer. The optical density of the suspension was normalized, and the same volume of each was streaked on minimal salts plates. The plates were incubated for 3 days under a standard gas mixture consisting of 80% H2, 10% CO2, and 10% O2.
FGN cultures; mean values ± standard errors; n = 5. ND, not determined.
FIG. 5.
Cellular levels of HoxA, HypD, and HypX in strains of A. eutrophus under different growth conditions. Cultures of the parental strain A. eutrophus H16 (wild type [wt]) and of the isogenic derivatives HF457 (hoxTΩ) and HF491 (ΔPMBH) were grown under hydrogenase-repressing (SN) and hydrogenase-derepressing (FGN) conditions. Western immunoblots were stained with antisera directed against HoxA (A), HypD (B), and HypX (C). Arrows indicate immunoreactive species. Positions of molecular mass standards are given at the right.
The results described so far indicate that a remote promoter is responsible for elevated expression of hoxA under derepressing conditions. This promoter must be ς54 dependent and controlled by HoxA. PMBH, the promoter which directs transcription of the MBH structural genes, was an obvious candidate. However, a transcript originating at PMBH and encompassing the coding sequence of hoxA would be unusually large (16,903 nt). To resolve this question, we generated a mutant with a 171-bp deletion in the hoxK upstream region (hoxKΔ171-R4). This deletion encompasses the invariant dinucleotide at position −24 of the ς54-dependent promoter (44). The resulting mutant, designated HF491, was strongly retarded in lithoautotrophic growth. We introduced the indicator plasmids pGE319 and pGE320 into HF491 and monitored the activity of PSH and PMBH under hydrogenase-derepressing conditions. The test system showed significantly reduced activities of the two promoters (4,039 and 3,033 U versus 12,961 and 14,445 U, respectively, for the wild-type background). This shows unequivocally that PMBH contributes to the transcription of hoxA. Interestingly, activities of the two promoters were higher in HF491 than in the polar mutant HF457. This may be due to read-through transcription from upstream promoters or to the activity of a yet unidentified promoter in the MBH region. Despite the significant transcriptional activity, HF491 showed no autotrophic growth after 3 days of incubation and only slight growth after prolonged incubation. This suggests that the combined effects of lowered expression of the hyp genes, which may limit the capacity of the maturation machinery, and lowered expression of the regulator are responsible for the curtailment of autotrophic growth.
DISCUSSION
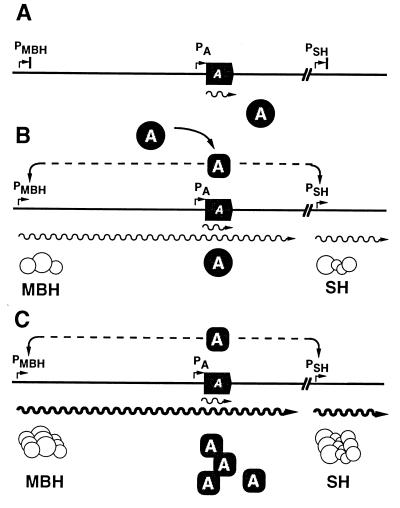
The data presented above fit into a coherent model describing expression of the regulator gene hoxA and the hydrogenase structural genes controlled by the promoters PSH and PMBH (Fig. 6). In the repressed state, low-level expression of HoxA is mediated by both a proximal (PA) and distal promoters in the hyp region. This weak expression is below the detection threshhold of the immunological assay but can be detected in vivo using the plasmid-borne translational fusions. This basal expression ensures that some regulator is present under all conditions. Thus, the system is poised to respond to the relevant physiological signal. In the absence of this cue, PSH and PMBH are inactive (Fig. 6A). Under derepressing conditions regulator molecules become competent to activate transcription at PSH and PMBH and synthesis of the hydrogenases commences (Fig. 6B). Apparently the amount of regulator present at the outset of induction is limiting and does not support levels of hydrogenase synthesis required for normal lithoautotrophic growth rates. Transcription from PMBH, however, augments expression of hoxA, leading to an increase in the pool of regulator, which in turn raises the rate of transcription from the cognate promoters (Fig. 6C). The result is a gradual amplification of the expression of both regulator and hydrogenase enzymes. This amplification is curtailed if hoxA is transcriptionally isolated from PMBH (as it is in the interposon mutant HF457) or if PMBH is eliminated altogether (as in mutant HF491).
FIG. 6.
Molecular model for the derepression of the hox regulon. The schematic shows a simplified map of the hox gene clusters, with the hoxA gene represented by a solid arrow. Gene products are represented as circles; transcripts are represented as wavy lines. The activated form of HoxA is symbolized as an oval. See text for details.
Transcriptional feedback mechanisms of this type are not uncommon in two-component regulatory systems, the classical example being the first system of this type to be described: the glnALG (ntrABC) operon of E. coli. The response regulator GlnG (NtrC; NRI) is phosphorylated in response to nitrogen limitation and activates transcription from glnAp2. This results in a 10-fold increase in the levels of GlnG over the basal level produced by transcription from glnLp (32). A similar situation is found in the bvg virulence control system of Bordetella pertussis. A weak constitutive promoter P2 provides a small cellular pool of activator protein BvgA. Phosphorylation of BvgA activates the stronger promoter P1, increasing the transcription of the bvgA gene (36). Positive autoregulation is by no means limited to two-component systems. An example from another class of transcriptional activator is the Pseudomonas aeruginosa regAB operon (42). The expression of some ς factors is controlled by a similar mechanism (12a). Positive feedback control is also found in regulatory systems which are not dependent on transcriptional activation, such as the operons encoding phosphotransferase system transporters (3, 38).
The A. eutrophus hox system differs from the examples of positive transcriptional feedback named above in an important respect: transcription of the gene hoxA by PMBH implies a transcript of at least 16,903 nt. Bacterial mRNAs in this size range are unusual but not unprecedented. A 17,000-nt transcript from the Erwinia amylovora ams locus was demonstrated by Northern mapping (5). The fla/che operon of Bacillus subtilis contains more than 30 genes and is at least 26 kb long (28a). A recent study on Bacillus brevis suggests that even larger transcriptional units exist in bacteria (31).
Membrane-bound hydrogenases are found in a wide variety of gram-negative bacteria. In several cases including A. eutrophus, the hydrogenase gene clusters have been sequenced (reviewed in reference 14). The overall arrangements of genes in these clusters are remarkably similar, pointing to conserved mechanisms of expression. In two organisms, Rhodobacter capsulatus and Bradyrhizobium japonicum, a positive regulator homologous to HoxA is encoded downstream of a promoter under its own control (35, 46). This situation is suggestive of a positive transcriptional feedback mechanism like the one reported here. In E. coli, a hyp operon consisting of the genes hypA, hypB, hypC, hypD, and hypE is transcribed from two promoters. The promoter proximal to hypA is dependent on ς54 and the positive regulator FhlA, which is encoded immediately downstream of hypE (27). FhlA is controlled by its own promoter but may also be susceptible to transcription from the FhlA-dependent hyp promoter.
Our findings also sketch a picture of the transcriptional organization of the A. eutrophus hyp region. Promoter mapping data suggest that the genes hypB1, hypF1, hypC, hypD, and hypE are expressed as a polycistronic transcript. hypA1 is controlled by a separate promoter as is hypX. We can at present only speculate on the significance of this configuration. Since almost nothing is known about the specific functions of the various proteins in the maturation of the A. eutrophus hydrogenases, we have little to go on. The former gene products may act as a set. The products of hypA1 and hypX might be required in different amounts and/or at different times and thus be subject to independent expression. However, major differences in promoter activity were not apparent under the conditions tested. The data for the hypA1 promoter test construct do not rule out the existence of two promoters: one of the ς70 type and one ς54 promoter.
hypX is also expressed from a separate promoter, suggesting that the product can act independently of the other Hyp proteins. The deduced sequence of HypX reveals motifs characteristic of N10-formyltetrahydrofolate-dependent enzymes and of enoyl-coenzyme A-hydratases/isomerases (6). It has been suggested that proteins of the HypX family are not involved in donating nickel to the nascent hydrogenase, the function postulated for the other Hyp proteins, but rather mediate the insertion of the diatomic ligands CO and CN (34). In an A. eutrophus hypX null mutant, hydrogenase activity was reduced by about 50%, whereas knockout mutants for the other hyp genes (with the exception of the two copies of hypA) blocked hydrogenase maturation (6, 7, 47). This suggests a conditional requirement for HypX in contrast to the other Hyp proteins. In B. japonicum, the arrangement of the hyp genes is similar to that in A. eutrophus, and indirect data suggest that the hypX gene is expressed from its own promoter (9).
Interestingly, activity of the hoxA promoter was higher in the HoxA− and RpoN− backgrounds (Table 2). The data for the HoxA− strain could be explained assuming that the promoter is negatively autoregulated. Since a ς54-dependent promoter (i.e., PMBH) is responsible for high-level expression of hoxA under hydrogenase-derepressing conditions, mutations in rpoN should have an effect similar to that of mutations in hoxA. The data show that this is in fact the case.
The expression of the A. eutrophus hyp genes under hydrogenase-repressing and -derepressing conditions alike seems at first paradoxical but is explained by the requirements for the synthesis of the H2-sensing apparatus. The actual H2-sensing molecule is a dimer consisting of the products of the genes hoxB and hoxC (3a, 24). HoxB and HoxC are homologous to the small and large subunits, respectively, of the dimeric hydrogenases (24). Moreover, HoxBC is, like the true hydrogenases, a nickel metalloenzyme and requires the Hyp proteins for normal maturation (6, 6a). Active HoxBC must be present in the cell at all times to enable the organism to respond to the availability of H2. This, in turn, necessitates the constitutive expression of the hyp genes.
ACKNOWLEDGMENTS
We thank P. Hübner, H. P. Schweizer, and A. Steinbüchel for the gift of plasmids, T. Eitinger for comments on the manuscript, and H. Schneeweiss for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB 344 and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Albracht S P J. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1199:167–204. doi: 10.1016/0005-2728(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- 3.Bardowski J, Ehrlich S D, Chopin A. BglR protein, which belongs to the BglG family of transcriptional antiterminators, is involved in β-glucoside utilization in Lactococcus lactis. J Bacteriol. 1994;176:5681–5685. doi: 10.1128/jb.176.18.5681-5685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bernhard, M., and B. Friedrich. Unpublished data.
- 4.Bernhard M, Schwartz E, Rietdorf J, Friedrich B. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J Bacteriol. 1996;178:4522–5429. doi: 10.1128/jb.178.15.4522-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 6.Buhrke T, Friedrich B. hoxX (hypX) is a functional member of the Alcaligenes eutrophus hyp gene cluster. Arch Microbiol. 1998;170:460–463. doi: 10.1007/s002030050667. [DOI] [PubMed] [Google Scholar]
- 6a.Buhrke, T., and B. Friedrich. Unpublished data.
- 7.Dernedde J, Eitinger T, Patenge N, Friedrich B. hyp gene products in Alcaligenes eutrophus are part of a hydrogenase-maturation system. Eur J Biochem. 1996;235:351–358. doi: 10.1111/j.1432-1033.1996.00351.x. [DOI] [PubMed] [Google Scholar]
- 8.Dernedde J, Eitinger M, Friedrich B. Analysis of a pleiotropic gene region involved in formation of catalytically active hydrogenase in Alcaligenes eutrophus H16. Arch Microbiol. 1993;159:545–553. doi: 10.1007/BF00249034. [DOI] [PubMed] [Google Scholar]
- 9.Durmowicz M C, Maier R J. Roles of HoxX and HoxA in biosynthesis of hydrogenase in Bradyrhizobium japonicum. J Bacteriol. 1997;179:3676–3682. doi: 10.1128/jb.179.11.3676-3682.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberz G, Friedrich B. Three trans-acting functions control hydrogenase expression in Alcaligenes eutrophus. J Bacteriol. 1991;173:1845–1854. doi: 10.1128/jb.173.6.1845-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberz G, Hogrefe C, Kortlüke C, Kamienski A, Friedrich B. Molecular cloning of structural and regulatory hydrogenase genes (hox) of Alcaligenes eutrophus H16. J Bacteriol. 1986;168:636–641. doi: 10.1128/jb.168.2.636-641.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eitinger T, Friedrich B. Cloning, nucleotide sequence, and heterologous expression of a high-affinity nickel transport gene from Alcaligenes eutrophus. J Biol Chem. 1991;266:3222–3227. [PubMed] [Google Scholar]
- 12a.Estacio W, Santa Anna-Arriola S, Adedipe M, Márquez-Magaña L M. Dual promoters are responsible for the transcription initiation of the fla/che operon in Bacillus subtilis. J Bacteriol. 1998;180:3548–3555. doi: 10.1128/jb.180.14.3548-3555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey M. Nickel-iron hydrogenases: structural and functional properties. Struct Bonding. 1998;90:97–126. [Google Scholar]
- 14.Friedrich B, Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich B, Bernhard M, Dernedde J, Eitinger T, Lenz O, Massanz C, Schwartz E. Hydrogen oxidation by Alcaligenes. In: Lindstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 110–117. [Google Scholar]
- 16.Friedrich C G. Derepression of hydrogenase during limitation of electron donors and derepression of ribulosebisphosphate carboxylase during carbon limitation of Alcaligenes eutrophus. J Bacteriol. 1982;149:203–210. doi: 10.1128/jb.149.1.203-210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich C G, Bowien B, Friedrich B. Formate and oxalate metabolism in Alcaligenes eutrophus. J Gen Microbiol. 1979;115:185–192. [Google Scholar]
- 18.Friedrich C G, Friedrich B, Bowien B. Formation of enzymes of autotrophic metabolism during heterotrophic growth of Alcaligenes eutrophus. J Gen Microbiol. 1981;122:69–78. [Google Scholar]
- 19.Gerischer U, Dürre P. mRNA analysis of the adc gene region of Clostridium acetobutylicum during the shift to solventogenesis. J Bacteriol. 1992;174:426–433. doi: 10.1128/jb.174.2.426-433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hübner P, Willison J C, Vignais P M, Bickel T A. Expression of regulatory nif genes in Rhodobacter capsulatus. J Bacteriol. 1991;173:2993–2999. doi: 10.1128/jb.173.9.2993-2999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortlüke C, Horstmann K, Schwartz E, Rohde M, Binsack R, Friedrich B. A gene complex coding for the membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992;174:6277–6289. doi: 10.1128/jb.174.19.6277-6289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kortlüke C, Friedrich B. Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992;174:6290–6293. doi: 10.1128/jb.174.19.6290-6293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krüger N, Steinbüchel A. Identification of acoR, a regulatory gene for the expression of genes essential for acetoin catabolism in Alcaligenes eutrophus H16. J Bacteriol. 1992;174:4391–4400. doi: 10.1128/jb.174.13.4391-4400.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenz O, Friedrich B. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1998;95:12474–12479. doi: 10.1073/pnas.95.21.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 27.Lutz S, Böhm R, Beier A, Böck A. Characterization of divergent NtrA-dependent promoters in the anaerobically expressed gene cluster coding for hydrogenase 3 components of Escherichia coli. Mol Microbiol. 1990;4:13–20. doi: 10.1111/j.1365-2958.1990.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 28.Maier T, Böck A. Nickel incorporation into hydrogenases. In: Hausinger R P, Eichhorn G L, Marzilli L G, editors. Advances in inorganic biochemistry: mechanisms of metallocenter assembly. New York, N.Y: VHC Publishers, Inc.; 1996. pp. 173–192. [Google Scholar]
- 28a.Márquez-Magaña L M, Chamberlin M J. Characterization of the sigD transcriptional unit of Bacillus subtilis. J Bacteriol. 1994;176:2427–2434. doi: 10.1128/jb.176.8.2427-2434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massanz C, Fernandez V M, Friedrich B. C-terminal extension of the H2-activating subunit, HoxH, directs maturation of the NAD-reducing hydrogenase in Alcaligenes eutrophus. Eur J Biochem. 1997;245:441–448. doi: 10.1111/j.1432-1033.1997.t01-3-00441.x. [DOI] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Mootz H D, Marahiel M A. The tyrocidin biosynthesis operon of Bacillus brevis: Complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pahel G, Rothstein D M, Magasanik B. Complex glnA-glnL-glnG operon of Escherichia coli. J Bacteriol. 1982;150:202–213. doi: 10.1128/jb.150.1.202-213.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierik A J, Schmelz M, Lenz O, Friedrich B, Albracht S P J. Characterization of the active site of a hydrogen sensor from Alcaligenes eutrophus. FEBS Lett. 1998;438:231–235. doi: 10.1016/s0014-5793(98)01306-4. [DOI] [PubMed] [Google Scholar]
- 34.Rey L, Fernández D, Brito B, Hernando Y, Palacios J-M, Imperial J, Ruiz-Argüeso T. The hydrogenase gene cluster of Rhizobium leguminosarum bv. viciae contains an additional gene (hypX), which encodes a protein with sequence similarity to the N10-formyltetrahydrofolate-dependent enzyme family and is required for nickel-dependent hydrogenase processing and activity. Mol Gen Genet. 1996;252:237–248. doi: 10.1007/BF02173769. [DOI] [PubMed] [Google Scholar]
- 35.Richaud P, Colbeau A, Toussaint B, Vignais P M. Identification and sequence analysis of the hupR1 gene, which encodes a response regulator of the NtrC family required for hydrogenase expression in Rhodobacter capsulatus. J Bacteriol. 1991;173:5928–5932. doi: 10.1128/jb.173.18.5928-5932.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarlato V, Prugnola A, Aricò B, Rappuoli R. Positive transcriptional feedback at the bvg locus controls expression of virulence factors in Bordetella pertussis. Proc Natl Acad Sci USA. 1990;87:6753–6757. doi: 10.1073/pnas.87.17.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schink B, Schlegel H G. Mutants of Alcaligenes eutrophus defective in autotrophic metabolism. Arch Microbiol. 1978;117:123–129. doi: 10.1007/BF00402299. [DOI] [PubMed] [Google Scholar]
- 38.Schnetz K, Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988;7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz E, Gerischer U, Friedrich B. Transcriptional regulation of Alcaligenes eutrophus hydrogenase genes. J Bacteriol. 1998;180:3197–3204. doi: 10.1128/jb.180.12.3197-3204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweizer H P. Small broad-host-range gentamycin resistance gene cassette for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 41.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 42.Storey D G, Raivio T L, Frank D W, Wick M J, Kaye S, Iglewski B. Effect of regB on expression from the P1 and P2 promoters of the Pseudomonas aeruginosa regAB operon. J Bacteriol. 1991;173:6088–6094. doi: 10.1128/jb.173.19.6088-6094.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiemermann S, Dernedde J, Bernhard M, Schroeder W, Massanz C, Friedrich B. Carboxyl-terminal processing of the cytoplasmic NAD-reducing hydrogenase of Alcaligenes eutrophus requires the hoxW gene product. J Bacteriol. 1996;178:2368–2374. doi: 10.1128/jb.178.8.2368-2374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thöny B, Hennecke H. The −24/−12 promoter comes of age. FEMS Microbiol Rev. 1989;63:341–358. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 45.Towbin N, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Soom C, de Wilde P, Vanderleyden J. HoxA is a transcriptional regulator for expression of the hup structural genes in free-living Bradyrhizobium japonicum. Mol Microbiol. 1997;23:967–977. doi: 10.1046/j.1365-2958.1997.2781648.x. [DOI] [PubMed] [Google Scholar]
- 47.Wolf I, Buhrke T, Dernedde J, Pohlmann A, Friedrich B. Duplication of hyp genes involved in maturation of [NiFe] hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol. 1998;170:451–459. doi: 10.1007/s002030050666. [DOI] [PubMed] [Google Scholar]
- 48.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]