Abstract
Alzheimer’s disease (AD) is an age-related neurodegenerative disease that is characterized by irreversible memory loss and cognitive decline. The deposition of amyloid-β (Aβ), especially aggregation-prone Aβ42, is considered to be an early event preceding neurodegeneration in AD. Sirtuins (SIRT1–7 in mammals) are nicotinamide adenine dinucleotide-dependent lysine deacetylases/deacylases, and several sirtuins play important roles in AD. However, the involvement of SIRT7 in AD pathogenesis is not known. Here, we demonstrate that SIRT7 mRNA expression is increased in the cortex, entorhinal cortex, and prefrontal cortex of AD patients. We also found that Aβ42 treatment rapidly increased NADPH oxidase 4 (NOX4) expression at the post-transcriptional level, and induced reactive oxygen species (ROS) production and apoptosis in neuronal SH-SY5Y cells. In contrast, SIRT7 knockdown inhibited Aβ42-induced ROS production and apoptosis by suppressing the upregulation of NOX4. Collectively, these findings suggest that the inhibition of SIRT7 may play a beneficial role in AD pathogenesis through the regulation of ROS production.
Keywords: SIRT7, Alzheimer’s disease, amyloid-β, apoptosis, reactive oxygen species, NADPH oxidase
1. Introduction
Alzheimer’s disease (AD) is an age-related neurodegenerative disease that is characterized by irreversible memory loss and cognitive decline. AD is the leading cause of dementia and is one of the great healthcare challenges of the 21st century [1]. Senile plaques, which are extracellular deposits of fibrils and amorphous aggregates of amyloid-β (Aβ), are an important pathological feature of AD. Aβ peptides are cleaved products of an amyloid precursor protein (APP) consisting of 36–43 amino acids by the sequential enzymatic action of β-secretase 1 and γ-secretase. Alternatively, APP may be cleaved by α-secretase and γ-secretase to generate non-amyloidogenic peptides. Under normal conditions, Aβ is degraded or eliminated in the cerebral spinal fluid; however, an imbalance between production and clearance causes Aβ to accumulate and aggregate. Although the mechanisms of AD are not fully understood, an excess of Aβ, especially aggregation-prone Aβ42, is generally considered to be the initiating step [1,2,3,4,5].
Sirtuins (SIRT1–7 in mammals), evolutionarily conserved nicotinamide adenine dinucleotide (NAD+)-dependent lysine deacetylases/deacylases, are involved in a wide variety of biological processes and age-related diseases [6]. Recent studies also revealed that sirtuins play pivotal roles in neurodegenerative diseases including AD [7,8,9]. For example, SIRT1 reduces the pathological accumulation of Aβ through the activation of the nonamyloidogenic APP processing pathway [10]. SIRT3 plays a neuroprotective role by deacetylating p53; however, SIRT3 expression is decreased in the brain of AD patients [11]. SIRT6 protects neuronal cells from Aβ42-induced DNA damage [12]. In contrast, SIRT2 promotes Aβ production by increasing β-secretase 1 expression, and the inhibition of SIRT2 has a beneficial effect on AD mouse models [13]. Thus, sirtuins contribute to AD in multiple ways.
SIRT7 is expressed ubiquitously, including in the brain, and regulates various cellular processes, including the metabolism, inflammation, oncogenesis, and genomic stability. SIRT1 and SIRT6 have beneficial effects against metabolic diseases, but we demonstrate that Sirt7 knockout mice are protected from high-fat-diet–induced obesity, glucose intolerance, and fatty liver [14,15], suggesting that SIRT7 deficiency plays a beneficial role in metabolic disorders. SIRT1 and SIRT6 exert anti-inflammatory roles by suppressing nuclear factor kappa B (NF-κB). In contrast, SIRT7 promotes inflammation by inhibiting the export of NF-κB from the nucleus [16,17]. In cancer, SIRT1 and SIRT6 act as tumor suppressors [18,19], whereas SIRT7 is responsible for tumor phenotype maintenance, and its expression is increased in many cancers [20,21]. Therefore, SIRT7 and SIRT1/SIRT6 play opposite roles in the metabolism, inflammation, and cancer. However, the involvement of SIRT7 in AD remains unexplored.
In this study, we found that SIRT7 mRNA expression is upregulated in the cortex of AD patients. Furthermore, we discovered that SIRT7 deficiency prevents Aβ42-induced neurotoxicity through the regulation of reactive oxygen species (ROS) production in SH-SY5Y cells. These findings suggest that the inhibition of SIRT7 may play a beneficial role in AD pathogenesis.
2. Results
2.1. SIRT7 Expression Is Increased in the Brain of AD Patients
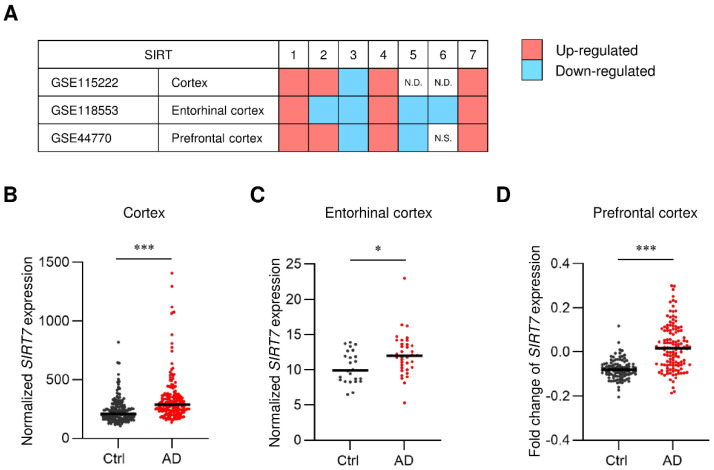
Our previous study demonstrated that SIRT7 is expressed widely in the mouse brain [22]. To evaluate the potential roles of SIRT7 in AD, we first assessed the expression profiles of sirtuin genes in the brain of AD patients using previously published datasets (GSE15222, GSE118553, and GSE44770) [23,24,25]. Amyloid deposition occurs in the cortex, entorhinal cortex, and prefrontal cortex during the progression of AD [26,27]. Consistent with a previous report [11], the decreased expression of SIRT3 mRNA was detected in these regions (Figure 1A). In contrast, SIRT7 mRNA was increased in these regions of AD patients (Figure 1A–D). These results suggest the involvement of SIRT7 in the pathogenesis of AD.
Figure 1.
SIRT7 expression is increased in the brain of AD patients. (A) Meta-analysis comparing three public microarray datasets of patients with AD (GSE15222, GSE118553, and GSE44770). The mRNA expression of SIRT1–7 was compared between AD and control patients. Red and blue boxes represent the upregulation or downregulation of the indicated gene, respectively. N.D., not detected. N.S., not significant. (B–D) Scatter plot of SIRT7 mRNA expression in (B) cortex from 176 non-AD samples and 187 AD samples (GSE15222), (C) entorhinal cortex from 27 non-AD samples and 52 AD samples (GSE118553), and (D) prefrontal cortex from 101 non-AD samples and 129 AD samples (GSE44770). Solid lines indicate the median value for each group. All data are shown as the mean ± SEM. Statistical significance was determined by Student’s t-test. * p < 0.05; *** p < 0.001.
2.2. SIRT7 Knockdown Prevents Aβ42-Induced Apoptosis in SH-SY5Y Cells
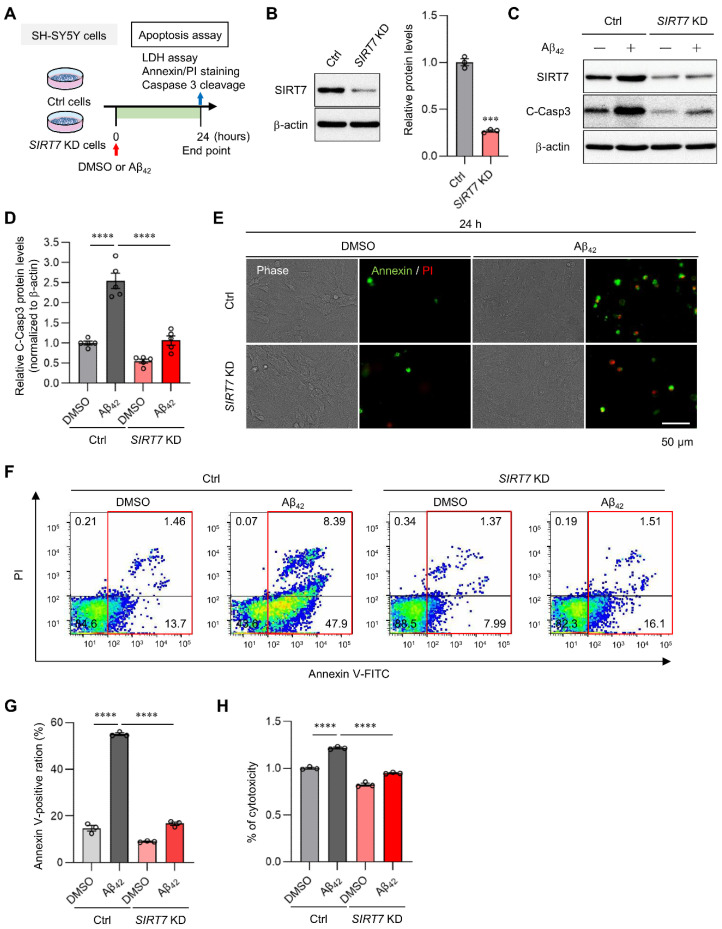
Aβ oligomerization exerts neurotoxic effects through the activation of caspase 3 [28,29]. To elucidate the role of SIRT7 in AD, we examined Aβ42-induced toxicity in SIRT7 knockdown (KD) neuronal SH-SY5Y cells, a human neuroblastoma cell line (Figure 2A,B). Aβ42 oligomer treatment induced cleaved (activated) caspase 3 expression in control SH-SY5Y cells, whereas the activation of caspase 3 was significantly suppressed in SIRT7 KD SH-SY5Y cells (Figure 2C,D). Immunocytochemical and flow cytometry analyses revealed that Aβ42 increased the number of annexin V-positive apoptotic cells in control SH-SY5Y cells (Figure 2E–G). In agreement with the decrease in caspase 3 expression, the number of annexin V-positive apoptotic cells was significantly decreased under the condition of SIRT7 KD (Figure 2F,G). A lactate dehydrogenase (LDH) assay confirmed that the cytotoxicity of Aβ42 oligomers was decreased in SIRT7 KD SH-SY5Y cells (Figure 2H). Collectively, these results indicate that SIRT7 deficiency inhibits Aβ42-induced apoptosis in SH-SY5Y cells.
Figure 2.
SIRT7 KD improves Aβ-induced apoptosis. (A) Experimental scheme for analyzing Aβ42-induced apoptosis in SIRT7 KD SH-SY5Y cells. (B) SIRT7 KD efficiency was confirmed by Western blot analysis when SH-SY5Y cells were transfected with SIRT7 siRNA for 48 h. (C) After SH-SY5Y cells had been transfected with control and SIRT7 siRNA for 48 h, cells were treated with 5 μM Aβ42 for 24 h. Western blot analysis of cleaved caspase 3 was performed. (D) The value of cleaved caspase 3 was normalized to that of β-actin. (E) Representative microscopy images of fluorescent annexin V (green)- and PI (red)-stained cells are shown for cells treated in the same condition as that in Figure 2A. Scale bar, 50 μm. (F) Flow cytometry analysis was performed on cells treated in the same condition as that in Figure 2A. Representative flow cytometry plots using annexin V-FITC/PI staining for apoptosis. (G) Percentage of total annexin V-positive cells was calculated. (H) Cell death was evaluated by an LDH activity assay on cells treated in the same condition as in Figure 2A. All data are shown as the mean ± SEM. Statistical significance was determined by either Student’s t-test or two-way ANOVA with Tukey’s post hoc test. *** p < 0.001; **** p < 0.0001.
2.3. SIRT7 Controls the Aβ42-Induced Increase in Intracellular ROS in SH-SY5Y Cells
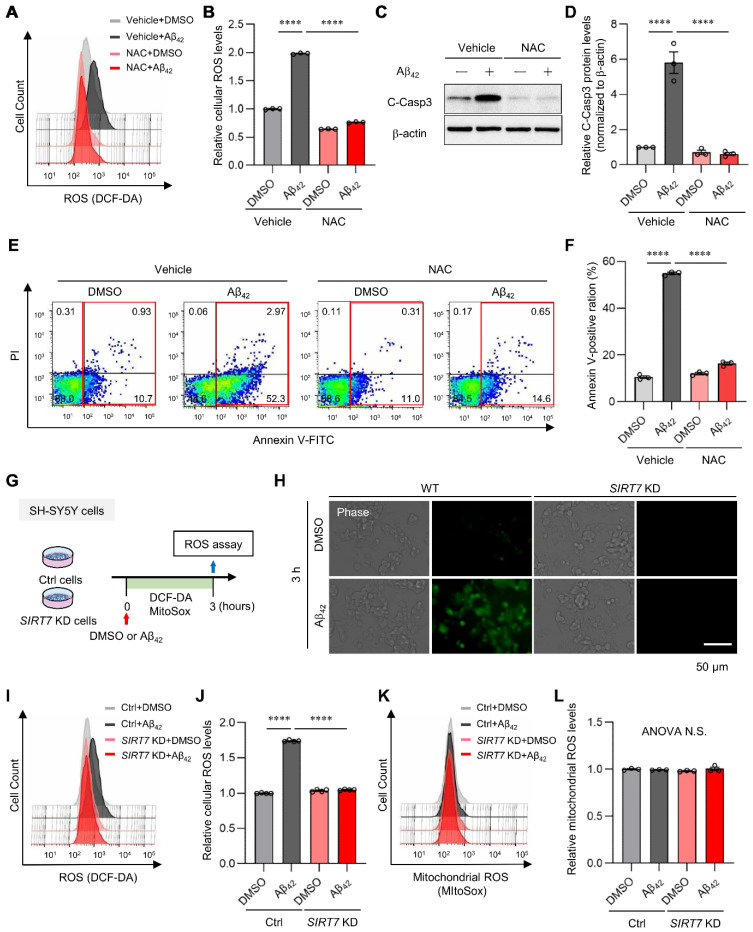
Increased ROS production plays a critical role in the onset and progression of AD [2,3,30]. Thus, we examined the effect of Aβ42 oligomer treatment on ROS production in SH-SY5Y cells. A marked increase in intracellular ROS, as measured using a DCF-DA probe, was detected following exposure to Aβ42 for 3 h, but this increase in ROS was completely repressed by the addition of general antioxidant N-acetyl L-cysteine (NAC) (Figure 3A,B). NAC treatment also inhibited Aβ42-induced activated caspase 3 expression (Figure 3C,D) and apoptosis (Figure 3E,F), indicating that the increase in ROS plays a major role in Aβ42-induced neurotoxicity in SH-SY5Y cells. We next investigated whether SIRT7 affected Aβ42-induced ROS production in SH-SY5Y cells (Figure 3G). Fluorescent staining analysis demonstrated the marked generation of intracellular ROS in control SH-SY5Y cells (Figure 3H). In sharp contrast, ROS production was not detected after Aβ42 treatment in SIRT7 KD cells. Flow cytometry analysis confirmed the inhibition of ROS production by SIRT7 KD (Figure 3I,J). These results indicate that SIRT7 controls the Aβ42-induced elevation of intracellular ROS in SH-SY5Y cells. We then investigated the cellular pathway involved in increased ROS production by Aβ42. Mitochondria are an important source of intracellular ROS production [5,31]. However, Aβ42 treatment for 3 h did not increase the fluorescence intensity of MitoSOX Red, an indicator of mitochondrial ROS, in either control or SIRT7 KD SH-SY5Y cells (Figure 3K,L), indicating that mitochondrial ROS are not involved in the increase in Aβ42-induced intracellular ROS, at least in these experimental conditions.
Figure 3.
SIRT7 deficiency inhibits Aβ-induced ROS generation. (A) Intracellular ROS levels were quantified by flow cytometry after SH-SY5Y cells were loaded with 10 μM CM-H2DCFHDA, pretreated with 1 mM NAC for 1 h, and then treated with 5 μM Aβ42 for a further 3 h in the presence of 1 mM NAC. Histogram of DCF-DA intensity of a representative experiment. (B) Vertical lines indicate the mean fluorescence values with the control cells set as 1. The geometric mean fluorescence intensity (MFI) ± SEM of three independent experiments was analyzed. (C) SH-SY5Y cells were pretreated with 1 mM NAC for 1 h, and then incubated in the presence of 5 μM Aβ42 and 1 mM NAC for a further 24 h. The intensity of cleaved caspase 3 was determined by Western blot analysis. (D) The value of cleaved caspase 3 was normalized to that of β-actin. (E) Flow cytometry analysis was performed on cells treated in the same condition as that in Figure 3C. Representative flow cytometry plots using annexin V-FITC/PI staining for apoptosis. (F) The percentage of total annexin V-positive cells was calculated. (G) Experimental scheme for analyzing Aβ42-induced ROS in SIRT7 KD SH-SY5Y cells. (H) Aβ42-induced ROS generation in control and SIRT7 KD SH-SY5Y cells was evaluated after they had been treated with Aβ42 for 3 h. Intracellular ROS levels after DCF-DA loading were visualized by fluorescence microscopy. Scale bar, 50 μm. (I) Intracellular ROS levels were quantified by flow cytometry on cells treated in the same condition as that in Figure 3G. Histogram of DCF-DA intensity in a representative experiment. (J) The geometric MFI ± SEM of three independent experiments was analyzed. (K) Mitochondrial ROS levels were assessed by flow cytometry after the cells had been treated with Aβ42 for 3 h and stained with 5 μM MitoSOX Red for a further 15 min. Histogram of MitoSOX Red intensity in a representative experiment. (L) The geometric MFI ± SEM of three independent experiments was analyzed. All data are shown as the mean ± SEM. Statistical significance was determined by two-way ANOVA with Tukey’s post hoc test. **** p < 0.0001. ANOVA N.S., p > 0.05.
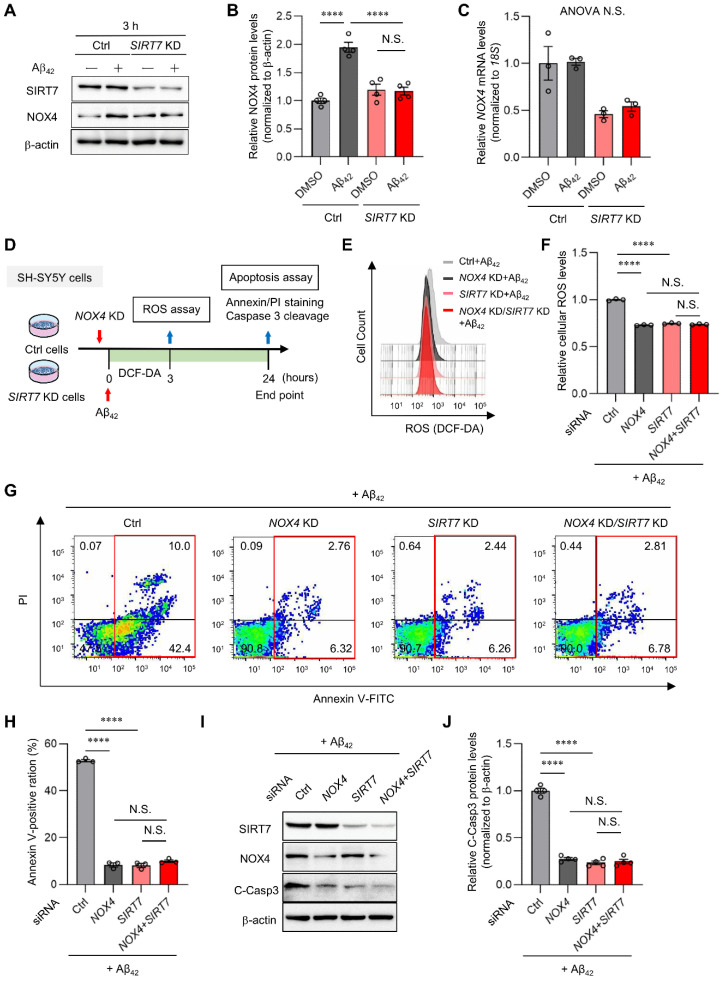
2.4. NOX4 Contributes to Aβ42-Induced ROS Production in SH-SY5Y Cells
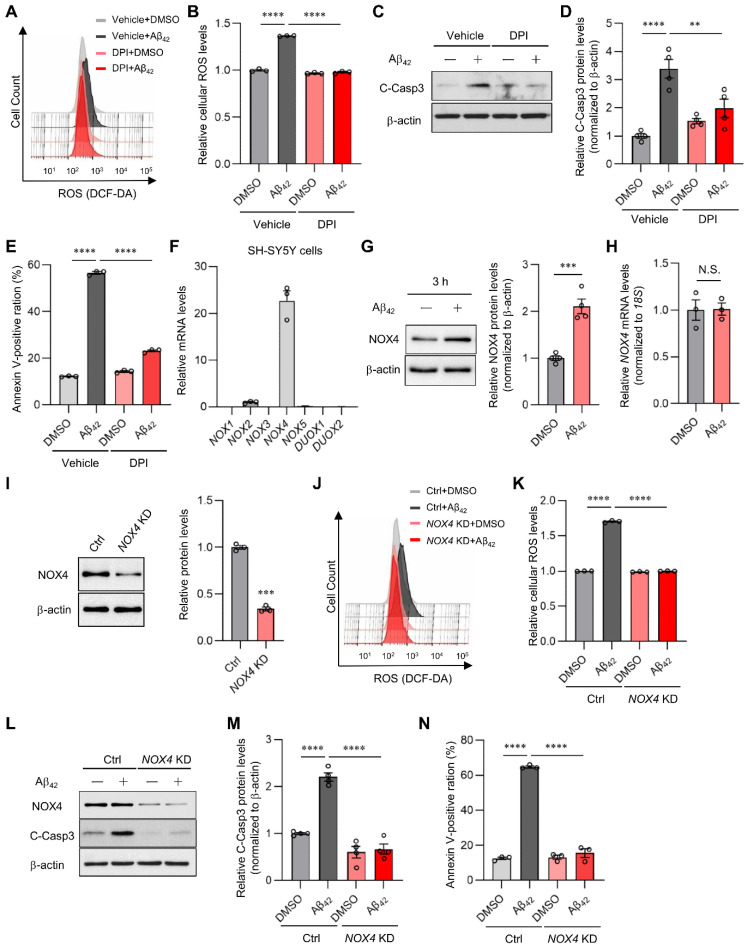
Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (NOXs) are also major sources of ROS. They are multisubunit membrane-bound enzymes that catalyze oxygen reduction into superoxide [32,33]. Intriguingly, Aβ42 oligomer-induced ROS production was markedly suppressed by diphenyleneiodonium (DPI), a pan-nhibitor of NOXs, in SH-SY5Y cells (Figure 4A,B). As in the case of NAC treatment, DPI treatment also significantly inhibited Aβ42 oligomer-induced caspase 3 activation and apoptosis (Figure 4C–E). These results strongly suggest that NOXs are a major source of Aβ42-induced ROS generation and play an important role in cell toxicity. In mammals, the NOX family comprises NOX1–5, dual oxidase 1 (DUOX1), and DUOX2. Among the seven isoforms, NOX4 is expressed in cortical neurons [34]. In line with this, NOX4 mRNA was almost exclusively expressed in SH-SY5Y cells (Figure 4F). Interestingly, Aβ42 treatment for 3 h significantly increased NOX4 protein levels by 2.1-fold without affecting NOX4 mRNA expression (Figure 4G,H), indicating that Aβ42 rapidly regulates NOX4 expression at the post-transcriptional level. Given that NOX4 generates ROS constitutively [35], these findings suggest that NOX4 may contribute to Aβ42-induced ROS production and neurotoxicity in SH-SY5Y cells. To this end, we investigated the impact of NOX4 suppression. NOX4 KD abolished ROS generation by Aβ42 in SH-SY5Y cells (Figure 4I–K). NOX4 KD also significantly suppressed caspase 3 activation (Figure 4L,M) and apoptosis (Figure 4N). These results strongly suggest that NOX4 is a major mediator of Aβ42-induced ROS production and apoptosis in SH-SY5Y cells.
Figure 4.
SIRT7 deficiency suppresses NOX-derived ROS generation. (A) After 0.1 μM DPI pretreatment for 1 h, SH-SY5Y cells were incubated with 5 μM Aβ42 and 0.1 μM DPI for a further 3 h. ROS production was assessed by flow cytometry using DCF-DA. Histogram of DCF-DA intensity in a representative experiment. (B) The geometric mean fluorescence intensity (MFI) ± SEM of three independent experiments was analyzed. (C) SH-SY5Y cells were pretreated with 0.1 μM DPI and then treated with 5 μM Aβ42 for a further 24 h in the presence of 0.1 μM DPI. Cleaved caspase 3 was examined with Western blot analysis. (D) The value of cleaved caspase 3 was normalized to that of β-actin. (E) Flow cytometry analysis was performed using annexin V-FITC/PI staining to assess apoptosis in cells treated in the same condition as in Figure 4C. The percentages of total annexin V-positive cells were calculated. (F) Quantitative RT-PCR analyses were conducted to examine the mRNA levels of the NOX family in SH-SY5Y cells. The expression level of the NOX family was normalized to that of 18S rRNA. (G) SH-SY5Y cells were incubated with 5 μM Aβ42 for 3 h. NOX4 was examined with Western blot analysis. (H) NOX4 mRNA expression was determined by the quantitative real-time RT-PCR analysis of cells treated in the same condition as that in Figure 4G. The value of NOX4 mRNA was normalized to that of 18S rRNA. (I) NOX4 KD efficiency was confirmed with Western blot analysis when SH-SY5Y cells were transfected with NOX4 siRNA for 48 h. (J) Intracellular ROS levels were evaluated by flow cytometry after control, and NOX4 KD SH-SY5Y cells were treated with Aβ42 for 3 h. Histogram of DCF-DA intensity of a representative experiment. (K) For the quantification of intracellular ROS levels, the geometric MFI ± SEM of three independent experiments was analyzed. (L) After SH-SY5Y cells had been transfected with control and NOX4 siRNA for 48 h, the cells were treated with Aβ42 for 24 h. Cleaved caspase 3 protein was evaluated with Western blot analysis. (M) The value of cleaved caspase 3 was normalized to that of β-actin. (N) Flow cytometry analysis was performed using annexin V-FITC/PI staining to assess apoptosis in cells treated in the same condition as that in Figure 4L. The percentage of total annexin V-positive cells was calculated. All data are shown as the mean ± SEM. Statistical significance was determined by either Student’s t-test or two-way ANOVA with Tukey’s post hoc test. N.S., not significant; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
2.5. SIRT7 Controls Aβ42-Induced Apoptosis through the Regulation of NOX4 in SH-SY5Y Cells
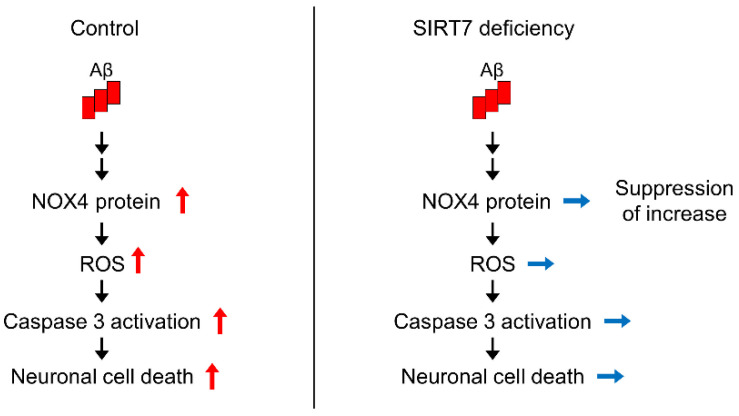
To explore the mechanism by which SIRT7 KD suppresses Aβ42-induced neurotoxicity, we investigated the role of SIRT7 in NOX4 expression. Aβ42 treatment for 3 h significantly increased NOX4 protein expression without affecting NOX4 mRNA levels in control SH-SY5Y cells, whereas the increase in protein expression was abrogated by SIRT7 KD (Figure 5A–C), indicating that SIRT7 deficiency suppresses the upregulation of NOX4 protein levels. We next investigated whether the neuroprotective effects of the loss of SIRT7 are dependent on NOX4 (Figure 5D). Aβ42 oligomer-induced ROS production was significantly reduced by NOX4 KD, as described earlier (Figure 5E,F). Notably, concomitant KD of SIRT7 and NOX4 did not result in an additional decrease in ROS production in SH-SY5Y cells compared with NOX4 KD SH-SY5Y cells (Figure 5F). Similarly, NOX4 KD resulted in a reduction in caspase 3 activation and the number of apoptotic cells, but there was no additional suppression of caspase 3 activation and apoptosis by concomitant NOX4 and SIRT7 KD (Figure 5G–J). Collectively, these results strongly suggest that the inhibition of SIRT7 alleviates Aβ42-induced neuronal damage by decreasing the expression of NOX4 (Figure 6).
Figure 5.
SIRT7 deficiency inhibits Aβ-induced NOX4 protein expression. (A) Western blot analysis of NOX4 was performed when control and SIRT7 KD SH-SY5Y cells had been treated with Aβ42 for 3 h. (B) The value of NOX4 was normalized to that of β-actin. (C) NOX4 mRNA expression was determined with the quantitative real-time RT-PCR analysis of cells treated in the same condition as that in Figure 5A. The value of NOX4 mRNA was normalized to that of 18S rRNA. (D) Experimental scheme for the effect of double KD of SIRT7 and NOX4 on Aβ42-induced ROS and apoptosis. (E) Control and NOX4 KD SH-SY5Y cells transfected with either control siRNA or SIRT7 siRNA were treated in the presence of 5 μM Aβ42 for 3 h. Intracellular ROS levels were evaluated by flow cytometry. (F) For quantification of intracellular ROS levels, the geometric MFI ± SEM of three independent experiments was analyzed. (G) Flow cytometry analysis was performed on cells treated in the same condition as that in Figure 5D. Representative flow cytometry plots using annexin V-FITC/PI staining for apoptosis. (H) The percentage of total annexin V-positive cells was calculated. (I) Western blot analysis of cleaved caspase 3 was performed on cells treated in the same condition as that in Figure 5D. (J) The value of cleaved caspase 3 was normalized to that of β-actin. All data are shown as the mean ± SEM. Statistical significance was determined by either one-way ANOVA with Tukey’s post hoc test or two-way ANOVA with Tukey’s post hoc test. N.S., not significant; **** p < 0.0001. ANOVA N.S., p > 0.05.
Figure 6.
Proposed model of how SIRT7 deficiency protects against Aβ42-induced neuronal cell death.
3. Discussion
Previous studies showed that SIRT1, SIRT3, and SIRT6 play protective roles against AD [11,12], but the involvement of SIRT7 in AD remains unknown. Here, we report for the first time that SIRT7 deficiency prevented Aβ42-induced neurotoxicity in SH-SY5Y cells. Furthermore, we found that SIRT7 mRNA expression was upregulated in the cortex, entorhinal cortex, and prefrontal cortex of AD patients. Our findings strongly suggest that SIRT7 and SIRT1/SIRT3/SIRT6 play opposite roles in the pathogenesis of AD, and the inhibition of SIRT7 plays a protective role against AD. The investigation of the roles of SIRT7 knockout in AD mouse models might provide additional information about the contribution of SIRT7 to the pathogenesis of AD.
NOXs are electron-transporting membrane proteins that are responsible for ROS production. Increased NOX activity in the brain of patients with AD and those with mild cognitive impairment was reported [36,37]. A significant correlation between NOX activity and cognitive impairment was also observed in humanized APP × PS1 mice, an animal model of AD [38]. Furthermore, recent studies have revealed that the inhibition of NOX4 by GLX351322, a selective NOX4 inhibitor, protects neuronal cells against Aβ42-induced neurotoxicity in APP × PS1 mice [39]. These results strongly support a critical role for NOX4 in AD pathogenesis. Consistent with these findings, we demonstrated that Aβ42 oligomer-induced ROS production, caspase 3 activation, and apoptosis were significantly reduced by NOX4 KD. Moreover, we demonstrated that the short (3 h) exposure of SH-SY5Y cells to the Aβ42 oligomer increased NOX4 expression at the post-transcriptional level, whereas SIRT7 deficiency abolished this upregulation. The concomitant KD of NOX4 and SIRT7 did not result in a further decrease in ROS production and apoptosis in SH-SY5Y cells compared with NOX4 KD SH-SY5Y cells. Despite not showing causality, our findings strongly support the notion that SIRT7 deficiency protects against Aβ42-induced apoptosis by regulating NOX4 protein levels. Lysine acetylation affects protein stability and protein–protein interactions [40]. Because database analysis (http://pail.biocuckoo.org/index.php, accessed on 30 May 2022) revealed that NOX4 has nine potential lysine acetylation sites, we hypothesized that SIRT7 may control NOX4 expression through its deacetylation. However, this scenario is unlikely, given that no interaction between SIRT7 and NOX4 was detected by a coimmunoprecipitation assay (data not shown). RNA-binding protein ELAV-like protein 1 (ELAVL1) increases NOX4 expression levels by binding to the 3′-untranslated region of NOX4 mRNA [41]. Therefore, SIRT7 might regulate NOX4 protein expression by deacetylating NOX4-interacting proteins, such as ELAVL1. Further studies are necessary to address how the loss of SIRT7 regulates NOX4 expression levels.
We demonstrated that SIRT7 mRNA expression was increased in the brain of AD patients. To explore the possibility that Aβ42 increases SIRT7 mRNA expression levels, we examined the effect of Aβ42 oligomer exposure on SIRT7 mRNA expression in SH-SY5Y cells. However, Aβ42 oligomer did not affect the levels of SIRT7 mRNA (data not shown), suggesting that the accumulation of Aβ42 is not a direct cause of SIRT7 upregulation in AD patients. Several studies indicated the occurrence of endoplasmic reticulum (ER) stress in the brain of AD patients [42], and ER stress enhances SIRT7 transcription [43]. Thus, increased ER stress might be involved in the altered expression of SIRT7 in AD patients.
In conclusion, SIRT7 mRNA expression is upregulated in the cortex of AD patients, and SIRT7 deficiency prevents Aβ42-induced neurotoxicity through the regulation of NOX4 expression in SH-SY5Y cells. Further studies are necessary to determine whether inhibition of SIRT7 shows beneficial roles in vivo. In addition, the identification of the exact mechanisms underlying the regulation of NOX4 expression by SIRT7 may lead to a better understanding of the pathogenesis of AD.
4. Materials and Methods
4.1. Gene Expression Analysis of Human AD Brain
Three expression datasets were used in the analysis. These microarray gene expression datasets from human brain samples (cortex, entorhinal cortex, and prefrontal cortex) were obtained from the Gene Expression Omnibus (GEO) (GEO ID: GSE15222, GSE118553, and GSE44770, respectively). The GSE15222 dataset was composed of expression data from 176 non-AD and 187 AD cases [23]. The GSE118553 dataset consisted of expression data from 27 non-AD and 52 AD cases [24]. The GSE44770 dataset comprised expression data from 101 non-AD and 129 AD cases [25]. All data were analyzed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/, accessed on 30 May 2022), and genes of the SIRT family with a p-value < 0.05 were considered to be significantly up- or downregulated.
4.2. Cell Culture
The SH-SY5Y human neuroblastoma cell line was obtained from the American Type Culture Collection (CRL-2266™). The cells were cultured in a mixed medium of Ham’s F12 and Dulbecco’s Modified Eagle’s Medium (1:1) (11320-033; Gibco™, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum and antibiotics (50 U/mL penicillin and 50 U/mL streptomycin) at 37 °C in a humidified incubator with a 5% CO2 atmosphere. The cell medium was replaced every 2 days, and the cells were reseeded after they reached 80% confluence.
4.3. Small Interfering RNA (siRNA) Transfection
For SIRT7 or NOX4 KD, SH-SY5Y cells were transfected with control scrambled siRNA (4390843; Ambion, Thermo Fisher Scientific) and either human SIRT7 Silencer Select siRNA (s28303 and s28304; Ambion, Thermo Fisher Scientific) or human NOX4 Silencer Select siRNA (s224159 and s224160; Ambion, Thermo Fisher Scientific). Transfection was performed with Opti-MEM serum-free medium (31985-070; Gibco™, Thermo Fisher Scientific) and RNAiMAX reagent (13778-150; Thermo Fisher Scientific). Cells (1.0 × 106 cells/well) in a 6-well plate were transfected with a mixture of two types of siRNA (each 5 nM) at a final concentration of 10 nM. The cells were used for experiments after 48 h of siRNA transfection.
4.4. Aβ Preparation
Synthetic Aβ42 was obtained from the Peptide Institute Inc. (4349-v; Osaka, Japan). Aβ42 oligomers were prepared as previously described [44]. In brief, Aβ42 peptides were dissolved (2 mM) in anhydrous DMSO, aliquoted, and stored at −80 °C. To yield oligomeric assemblies of Aβ42, the stock peptide solution was diluted to 100 μM in phosphate-buffered saline (PBS; 10 mM NaH2PO4, 137 mM NaCl, 2.7 mM KCl, 1.8 mM KH2PO4, pH 7.4), immediately vortexed for 30 s, and incubated at 4 °C for 24 h. SH-SY5Y cells were treated with a final concentration of 5 μM Aβ42 oligomers according to previous reports [45,46].
4.5. Cytotoxicity Assay
Cytotoxicity was determined by the release of LDH in the culture medium. Control and SIRT7 KD cells (5.0 × 104 cells/well) were seeded in 96-well plates and treated with a 5 μM Aβ42-containing medium for 24 h. LDH activity in the culture supernatant was measured using a Cytotoxicity LDH Assay Kit-WST (CK17; Dojindo Molecular Technologies, Rockville, MD, USA). The absorbance of each sample was measured at a wavelength of 490 nm by a microplate reader (Bio-Rad, Hercules, CA, USA).
4.6. Immunocytochemical Staining
An Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (K101-100; BioVision, Milpitas, CA, USA) and CM-H2DCFHDA (C6827; Invitrogen, Waltham, MA, USA) were used for a cell death assay and cellular ROS imaging, respectively. In brief, SH-SY5Y cells (5.0 × 105 cells/well) were seeded on 35-mm glass-bottom dishes (Greiner Bio-One, Kremsmünster, Austria). After being cultured in medium containing 5 μM Aβ42 for 24 h, the cells were washed twice with PBS, and then incubated with annexin V-FITC and propidium iodide (PI) for 5 min at room temperature in the dark. For the detection of ROS, the cells (5.0 × 105 cells/well) were seeded in 35 mm glass-bottom dishes and pretreated with CM-H2DCFHDA (10 μM) for 30 min at 37 °C in the dark, and then treated with 5 μM Aβ42 for 3 h. The cells were washed twice with PBS. Fluorescent images were visualized immediately using a confocal laser scanning microscope (BZ-X700; Keyence, Osaka, Japan).
4.7. Flow Cytometry Analysis
Flow cytometry was used for the quantitative determination of apoptosis and cellular ROS levels. In brief, SH-SY5Y cells (2.0 × 105 cells/well) were seeded in a 24-well plate and treated the same way as for immunocytochemistry. For mitochondrial ROS detection, the cells were incubated in the presence of 5 μM MitoSOX™ Red (M36008; Invitrogen) for 10 min after Aβ42 treatment. After trypsin/EDTA digestion, the medium containing the cells was centrifuged in 1.5 mL tubes (200× g, 3 min, 4 °C). The centrifuged cells were resuspended in 2% fetal bovine serum/PBS. Fluorescence intensity was analyzed using a flow cytometer (BD FACS Verse™ Flow Cytometer; BD Biosciences). All experiments were performed in triplicate.
4.8. Western Blot Analysis
Western blot analysis was performed as previously described [47]. Total protein lysates of cells were obtained by lysis in RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% NP-40, 5 mM EDTA, 0.5% sodium deoxycholate, 20 mg/mL Na3VO4, 10 mM NaF, and 1 mM PMSF) with a protease inhibitor cocktail (04080-11; Nacalai Tesque, Kyoto, Japan). Total protein lysates (60 µg) from SH-SY5Y cells were subjected to gel electrophoresis and protein transfer onto a PVDF membrane (IPVH00010, Immobilon-P; Millipore, Burlington, MA, USA). The following primary antibodies were used: anti-β-actin (clone AC15; Sigma-Aldrich, St. Louis, MI, USA), anti-SIRT7 (clone D3K5A, #5360; Cell Signaling Technology, Danvers, MA, USA), anti-cleaved caspase 3 (#9661; Cell Signaling Technology), and anti-NOX4 (14347-1-AP; Proteintech). After reaction with secondary antibodies, the signals were detected by using Chemi-Lumi One Super Type (Nacalai Tesque) and a ChemiDoc imaging system (BR170-8265; Bio-Rad). All experiments were performed at least three times, and band intensities were quantified using Image Lab Software version 6.0 (Bio-Rad, Hercules, CA, USA).
4.9. Treatment of NAC and DPI
To assess the effects of NAC (A9165; Sigma-Aldrich) and DPI (4673-26-1; Cayman Chemical, Ann Arbor, MN, USA), SH-SY5Y cells (1.5 × 106 cells/well) were seeded in a 6-well plate. Cells were preincubated in medium containing 1 mM NAC or 0.1 μM DPI for 1 h, followed by treatment with 5 μM Aβ42 in the presence of 1 mM NAC or 0.1 μM DPI for 3 h (ROS assay) or 24 h (apoptosis assay).
4.10. Quantitative Real-Time RT-PCR
SH-SY5Y cells were homogenized in Sepasol-RNA I Super G Solution (Nacalai Tesque), and total RNA was isolated using a conventional phenol-chloroform-based RNA extraction method. cDNA was prepared using a PrimeScript RT Reagent Kit and gDNA Eraser (RR047A; TaKaRa Bio, Inc., Shiga, Japan). Quantitative PCR was performed using SYBR Premix Ex Taq II (RR820A; TaKaRa Bio, Inc.) and an ABI 7300 thermal cycler (Applied Biosystems, Foster City, CA, USA; software version 1.4). For each gene, mRNA levels were determined by the comparative cycle threshold (Ct) method (ΔΔCt), and the levels of each mRNA were normalized to ribosomal 18S (r18S). The sequences of the primers were as follows. hr18S: forward, 5′-GGAGAACTCACTGAGGATGA-3′, reverse, 5′-CCAGTGGTCTTGGTGTGCTG-3′; hSIRT7: forward, 5′-ACTTGGTCGTCTACACAGGC-3′, reverse, 5′-CAGACGGGTGATGCTCATGT-3′; hNOX1: forward, 5′-CCTGAGTCTTGGAAGTGGATC-3′, reverse, 5′-ACGCTTGTTCATCTGCAATTC-3′; hCYBB: forward, 5′-AGCTGAACGAATTGTACGTG-3′, reverse, 5′-ACCCACTATCCATTTCCAAG-3′; hNOX3: forward, 5′-TGTGGTCTTGTATGCATGTG-3′, reverse, 5′-CACGCTTTTTCATGTGAAGT-3′; hNOX4: forward, 5′-CCAGCTGTACCTCAGTCAAA-3′, reverse, 5′-CCACAACAGAAAACACCAAC-3′; hNOX5: forward, 5′-TCTTTCGAGTGGTTTGTGAG-3′, reverse, 5′-ACTTTCTGGAACACCTTGCT-3′; hDUOX1: forward, 5′-GTCTTCATGAAAGGCTCTCC-3′, reverse, 5′-AATCTTCCCATGTCAGTTCC-3′; hDUOX2: forward, 5′-GACATGGGAGGATTTTCACT-3′, reverse, 5′-CTCGACAGCTGATGTTTTGT-3′.
4.11. Statistical Analysis
Results are presented as the mean ± standard error of the mean (SEM) of the indicated number of experiments (n). Statistical analysis was performed with an unpaired two-tailed Student’s t-test Figure 2B and Figure 4G–I), one-way ANOVA with Tukey’s post hoc test (Figure 5F,H,J), or two-way ANOVA with Tukey’s post hoc test (Figure 2D,G,H, Figure 3B,D,F,J,L, Figure 4B,D,E,K,M,N, and Figure 5B,C). A value of p < 0.05 was considered to be statistically significant. All analyses were performed using GraphPad Prism 9 software (GraphPad Software Inc., San Diego, CA, USA; software version 9.4.0).
5. Conclusions
SIRT7 deficiency protects against Aβ42-induced apoptosis through the regulation of NOX4-derived ROS production in SH-SY5Y cells.
Acknowledgments
We thank the members of Yamagata Laboratory for discussions and technical assistances.
Author Contributions
H.M., Y.S. and K.Y. conceived the project, and designed the experiments; H.M., Y.S., M.Y., T.Y., Y.A. and M.U. carried out the experiments, analyzed the data, and provided useful suggestions. H.M., Y.S. and K.Y. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no competing interest.
Funding Statement
This study was supported by a grant from the Japan Agency for Medical Research and Development under grant number JP21gm5010002: K.Y.; by Grants-in-Aid for Scientific Research (B) (19H03711 and 22H03129; K.Y.); by a Grant-in-Aid for Challenging Research (Exploratory) (19K22639; K.Y.); by a Grant-in-Aid for Scientific Research (C) (19K09008; Y.S.); and by a grant from the Naito Foundation (K.Y.) and Takeda Science Foundation (K.Y.).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Scheltens P., Blennow K., Breteler M.M.B., de Strooper B., Frisoni G.B., Salloway S., Van der Flier W.M. Alzheimer’s Disease. Lancet. 2016;388:505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 2.Wood A.J.J., Cummings J.L. Alzheimer’s Disease. N. Engl. J. Med. 2004;351:56–67. doi: 10.1056/nejmra040223. [DOI] [PubMed] [Google Scholar]
- 3.Mattson M.P. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzi L., Roriz-Cruz M. Sirtuin 1 and Alzheimer’s disease: An up-to-date review. Neuropeptides. 2018;71:54–60. doi: 10.1016/j.npep.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Querfurth H.W., LaFerla F.M. Alzheimer’s Disease. N. Engl. J. Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 6.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. doi: 10.1101/gad.14.9.1021. [DOI] [PubMed] [Google Scholar]
- 7.Gan L., Mucke L. Paths of Convergence: Sirtuins in Aging and Neurodegeneration. Neuron. 2008;58:10–14. doi: 10.1016/j.neuron.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonda D.J., Lee H.-G., Camins A., Pallàs M., Casadesus G., Smith M.A., Zhu X. The sirtuin pathway in ageing and Alzheimer disease: Mechanistic and therapeutic considerations. Lancet Neurol. 2011;10:275–279. doi: 10.1016/S1474-4422(11)70013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herskovits A.Z., Guarente L. SIRT1 in Neurodevelopment and Brain Senescence. Neuron. 2014;81:471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin W., Yang T., Ho L., Zhao Z., Wang J., Chen L., Zhao W., Thiyagarajan M., MacGrogan D., Rodgers J.T., et al. Neuronal SIRT1 Activation as a Novel Mechanism Underlying the Prevention of Alzheimer Disease Amyloid Neuropathology by Calorie Restriction. J. Biol. Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Kim Y., Liu T., Hwang Y.J., Hyeon S.J., Im H., Lee K., Alvarez V.E., McKee A.C., Um S.-J., et al. SIRT3 deregulation is linked to mitochondrial dysfunction in Alzheimer’s disease. Aging Cell. 2017;17:e12679. doi: 10.1111/acel.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung E.S., Choi H., Song H., Hwang Y.J., Kim A., Ryu H., Mook-Jung I. p53-dependent SIRT6 expression protects Aβ42-induced DNA damage. Sci. Rep. 2016;6:25628. doi: 10.1038/srep25628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Yang J., Hong T.-T., Sun Y., Huang H., Chen F., Chen X., Chen H., Dong S., Cui L., et al. RTN4B-mediated suppression of Sirtuin 2 activity ameliorates β-amyloid pathology and cognitive impairment in Alzheimer’s disease mouse model. Aging Cell. 2020;19:e13194. doi: 10.1111/acel.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshizawa T., Karim F., Sato Y., Senokuchi T., Miyata K., Fukuda T., Go C., Tasaki M., Uchimura K., Kadomatsu T., et al. SIRT7 Controls Hepatic Lipid Metabolism by Regulating the Ubiquitin-Proteasome Pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Yamagata K., Yoshizawa T. Transcriptional Regulation of Metabolism by SIRT1 and SIRT7. Int. Rev. Cell Mol. Biol. 2018;335:143–166. doi: 10.1016/bs.ircmb.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Miyasato Y., Yoshizawa T., Sato Y., Nakagawa T., Miyasato Y., Kakizoe Y., Kuwabara T., Adachi M., Ianni A., Braun T., et al. Sirtuin 7 Deficiency Ameliorates Cisplatin-induced Acute Kidney Injury Through Regulation of the Inflammatory Response. Sci. Rep. 2018;8:5927. doi: 10.1038/s41598-018-24257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobuz S.U., Sato Y., Yoshizawa T., Karim F., Ono K., Sawa T., Miyamoto Y., Oka M., Yamagata K. SIRT7 regulates the nuclear export of NF-κB p65 by deacetylating Ran. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:1355–1367. doi: 10.1016/j.bbamcr.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Chang H.-C., Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2013;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tasselli L., Zheng W., Chua K.F. SIRT6: Novel Mechanisms and Links to Aging and Disease. Trends Endocrinol. Metab. 2016;28:168–185. doi: 10.1016/j.tem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barber M.F.M., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R.I., Paredes S., Young N.L., Chen K., et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu D., Li Y., Zhu K.S., Wang H., Zhu W.-G. Advances in Cellular Characterization of the Sirtuin Isoform, SIRT7. Front. Endocrinol. 2018;9:652. doi: 10.3389/fendo.2018.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam S., Wei F.-Y., Ohta K., Shigematsu N., Fukuda T., Tomizawa K., Yoshizawa T., Yamagata K. Sirtuin 7 is involved in the consolidation of fear memory in mice. Biochem. Biophys. Res. Commun. 2018;495:261–266. doi: 10.1016/j.bbrc.2017.10.159. [DOI] [PubMed] [Google Scholar]
- 23.Webster J.A., Gibbs J.R., Clarke J., Ray M., Zhang W., Holmans P., Rohrer K., Zhao A., Marlowe L., Kaleem M., et al. Genetic Control of Human Brain Transcript Expression in Alzheimer Disease. Am. J. Hum. Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel H., Hodges A.K., Curtis C., Lee S.H., Troakes C., Dobson R.J., Newhouse S.J. Transcriptomic analysis of probable asymptomatic and symptomatic alzheimer brains. Brain Behav. Immun. 2019;80:644–656. doi: 10.1016/j.bbi.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Zhang B., Gaiteri C., Bodea L.-G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., et al. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimer’s Disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swarbrick S., Wragg N., Ghosh S., Stolzing A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019;56:6156–6167. doi: 10.1007/s12035-019-1500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goedert M. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 28.Deshpande A., Mina E., Glabe C., Busciglio J. Different Conformations of Amyloid beta Induce Neurotoxicity by Distinct Mechanisms in Human Cortical Neurons. J. Neurosci. 2006;26:6011–6018. doi: 10.1523/JNEUROSCI.1189-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pate K.M., Rogers M., Reed J.W., van der Munnik N., Vance S.Z., Moss M.A. Anthoxanthin Polyphenols Attenuate AβOligomer-induced Neuronal Responses Associated with Alzheimer’s Disease. CNS Neurosci. Ther. 2016;23:135–144. doi: 10.1111/cns.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheignon C., Tomas M., Bonnefont-Rousselot D., Faller P., Hureau C., Collin F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Biol. 2018;14:450–464. doi: 10.1016/j.redox.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng J., Kaur H., Collier T., Chang K., Brooks A.E.S., Allison J.R., Brimble M.A., Hickey A., Birch N.P. Site-specific glycation of Aβ1–42 affects fibril formation and is neurotoxic. J. Biol. Chem. 2019;294:8806–8818. doi: 10.1074/jbc.RA118.006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nayernia Z., Jaquet V., Krause K.-H. New Insights on NOX Enzymes in the Central Nervous System. Antioxid. Redox Signal. 2014;20:2815–2837. doi: 10.1089/ars.2013.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarafdar A., Pula G. The Role of NADPH Oxidases and Oxidative Stress in Neurodegenerative Disorders. Int. J. Mol. Sci. 2018;19:3824. doi: 10.3390/ijms19123824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casas A.I., Geuss E., Kleikers P.W.M., Mencl S., Herrmann A.M., Buendia I., Egea J., Meuth S.G., Lopez M.G., Kleinschnitz C., et al. NOX4-dependent neuronal autotoxicity and BBB breakdown explain the superior sensitivity of the brain to ischemic damage. Proc. Natl. Acad. Sci. USA. 2017;114:12315–12320. doi: 10.1073/pnas.1705034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Fórró L., Schlegel W., Krause K.-H. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406:105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansari M.A., Scheff S.W. NADPH-oxidase activation and cognition in Alzheimer disease progression. Free Radic. Biol. Med. 2011;51:171–178. doi: 10.1016/j.freeradbiomed.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruce-Keller A.J., Gupta S., Parrino T.E., Knight A.G., Ebenezer P.J., Weidner A.M., LeVine H., Keller J.N., Markesbery W.R. NOX Activity Is Increased in Mild Cognitive Impairment. Antioxid. Redox Signal. 2010;12:1371–1382. doi: 10.1089/ars.2009.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruce-Keller A.J., Gupta S., Knight A.G., Beckett T.L., McMullen J.M., Davis P.R., Murphy M.P., Van Eldik L.J., Clair D.S., Keller J. Cognitive impairment in humanized APP×PS1 mice is linked to Aβ1–42 and NOX activation. Neurobiol. Dis. 2011;44:317–326. doi: 10.1016/j.nbd.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao W., Yu L., Shu S., Liu Y., Zhuang Z., Xu S., Bao X., Gu Y., Cai F., Song W., et al. miR-204-3p/Nox4 Mediates Memory Deficits in a Mouse Model of Alzheimer’s Disease. Mol. Ther. 2020;29:396–408. doi: 10.1016/j.ymthe.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X.-J., Seto E. Lysine Acetylation: Codified Crosstalk with Other Posttranslational Modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Q., Lee D.-Y., Féliers D., Abboud H.E., Bhat M.A., Gorin Y. Interplay between RNA-binding protein HuR and Nox4 as a novel therapeutic target in diabetic kidney disease. Mol. Metab. 2020;36:100968. doi: 10.1016/j.molmet.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerakis Y., Hetz C. Emerging roles of ER stress in the etiology and pathogenesis of Alzheimer’s disease. FEBS J. 2018;285:995–1011. doi: 10.1111/febs.14332. [DOI] [PubMed] [Google Scholar]
- 43.Shin J., He M., Liu Y., Paredes S., Villanova L., Brown K., Qiu X., Nabavi N., Mohrin M., Wojnoonski K., et al. SIRT7 Represses Myc Activity to Suppress ER Stress and Prevent Fatty Liver Disease. Cell Rep. 2013;5:654–665. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stine W.B., Jr., Dahlgren K.N., Krafft G.A., LaDu M.J. In Vitro Characterization of Conditions for Amyloid-β Peptide Oligomerization and Fibrillogenesis. J. Biol. Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 45.Ahmed M.E., Selvakumar G.P., Kempuraj D., Thangavel R., Mentor S., Dubova I., Raikwar S.P., Zaheer S., Iyer S., Zaheer A. Synergy in Disruption of Mitochondrial Dynamics by Aβ (1–42) and Glia Maturation Factor (GMF) in SH-SY5Y Cells is Mediated through Alterations in Fission and Fusion Proteins. Mol. Neurobiol. 2019;56:6964–6975. doi: 10.1007/s12035-019-1544-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manterola L., Hernando-Rodríguez M., Ruiz A., Apraiz A., Arrizabalaga O., Vellón L., Alberdi E., Cavaliere F., Lacerda H.M., Jimenez S., et al. 1–42 β-Amyloid peptide requires PDK1/nPKC/Rac 1 pathway to induce neuronal death. Transl. Psychiatry. 2013;3:e219. doi: 10.1038/tp.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato Y., Hatta M., Karim F., Sawa T., Wei F.-Y., Sato S., Magnuson M.A., Gonzalez F.J., Tomizawa K., Akaike T., et al. Anks4b, a Novel Target of HNF4α Protein, Interacts with GRP78 Protein and Regulates Endoplasmic Reticulum Stress-induced Apoptosis in Pancreatic β-Cells. J. Biol. Chem. 2012;287:23236–23245. doi: 10.1074/jbc.M112.368779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.