Abstract
In this study we report the identification and role of the alkyl hydroperoxide reductase (ahp) gene in Bacteroides fragilis. The two components of ahp, ahpC, and ahpF, are organized in an operon, and the deduced amino acid sequences revealed that B. fragilis AhpCF shares approximately 60% identity to orthologues in other gram-positive and gram-negative bacteria. Northern blot hybridization analysis of total RNA showed that the ahpCF genes were transcribed as a polycistronic 2.4-kb mRNA and that ahpC also was present as a 0.6-kb monocistronic mRNA. ahpC and ahpCF mRNAs were induced approximately 60-fold following H2O2 treatment or oxygen exposure of the parent strain but were constitutive in a peroxide-resistant strain. Further investigation using an ahpCF′::β-xylosidase gene transcriptional fusion confirmed that ahpCF had lost normal regulation in the peroxide-resistant strain compared to the parent. The ahpCF mutant was more sensitive to growth inhibition and mutagenesis by organic peroxides than the parent strain, as determined by disk inhibition assays and the frequency of mutation to fusidic acid resistance. This finding suggests that the ahp genes play an important role in peroxide resistance in B. fragilis. Under anaerobic conditions, we observed increases in the number of spontaneous fusidic acid-resistant mutants of five- and sevenfold in ahpCF and ahpF strain backgrounds, respectively, and eightfold in the ahpCF katB double mutant strain compared to the parent and katB strains. In addition, ahpCF, ahpF, and ahpCF katB mutants were slightly more sensitive to oxygen exposure than the parent strain. Moreover, the isolation of a strain with enhanced aerotolerance and high-level resistance to alkyl hydroperoxides from an ahpCF katB parent suggests that the physiological responses to peroxide toxicity and to the toxic effects of molecular oxygen are overlapping and complex in this obligate anaerobe.
The peroxide response in facultative enteric bacteria results in synthesis of at least 30 proteins, of which 9 are under OxyR regulatory control. These include catalase (KatG), alkyl hydroperoxide reductase (Ahp), glutathione reductase (GorA), and a nonspecific DNA-binding protein (Dps) involved in protection of DNA against oxidative damage (1, 8). oxyR gene deletion mutants are hypersensitive to spontaneous mutations compared to isogenic oxyR+ control strains (27), yet single mutations in the genes encoding catalase and alkyl hydroperoxide reductase had little effect on the spontaneous-mutation frequency (27). Overexpression of oxyR-controlled genes such as KatG and Ahp in an oxyR deletion mutant significantly decrease the oxygen-dependent mutation frequency due to oxidative damage to DNA (15, 27). This apparent discrepancy may reflect the fact that there is considerable physiological redundancy built into the protective systems. In living cells, elimination of alkyl hydroperoxides is particularly important since they can initiate lipid peroxidation chain reaction and consequently propagate free radicals, leading to DNA and membrane damage (13).
In facultative and aerobic bacteria, the role in protecting cells against organic peroxides is exerted in part by the peroxide-scavenging enzyme Ahp, which consists of two components, a small 22,000-Da protein (AhpC) with peroxidase activity and a larger 57,000-Da flavoprotein (AhpF) (15, 28). These two proteins act together; AhpF uses NADH or NADPH as electron donor to AhpC, which reduces physiological lipid peroxides such as linoleic acid hydroperoxide and thymine hydroperoxide and nonphysiological alkyl hydroperoxides to their respective nontoxic alcohol forms (15). AhpC is a component of a large family of thiol-specific antioxidant (TSA) proteins whose roles generally are not well understood (5, 6). AhpC, however, has been demonstrated to act as specific alkyl hydroperoxide-scavenging enzyme for protection against oxygen radical damage (15), though elimination of reactive nitrogen intermediates also has been demonstrated to occur (7). AhpF is related to thioredoxin reductases possessing an extended additional N-terminal fragment essential to specifically reduce AhpC (4, 29).
In the obligate anaerobe Bacteroides fragilis, the defense against oxidative damage is poorly understood, although recent studies have shown that the oxidative stress response is similar in several respects to the response of aerobic and facultative bacteria. B. fragilis synthesizes two very similar and overlapping sets of approximately 28 new proteins following stress induced by either oxygen exposure or addition of hydrogen peroxide (18). Pretreatment with sublethal concentrations of hydrogen peroxide has been shown to induce catalase (katB) expression and protection against further treatment with high concentrations of hydrogen peroxide. Consistent with this observation, the katB mutant was highly sensitivity to hydrogen peroxide under anaerobic conditions (18). Evidence for the existence of independent peroxide and oxygen stress response comes from a study showing that a mutant strain (IB263, hpr [hydrogen peroxide resistance]) with constitutive expression of KatB, AhpC, and Dps homologues is highly resistant to hydrogen peroxide and cumene hydroperoxide; however, susceptibility to atmospheric oxygen was the same as in the parent strain (20). The isolation of an AhpC homologue in B. fragilis suggested that detoxification of organic peroxide in anaerobic bacteria is also an important aspect to protecting the cells against the toxic effects of reactive oxygen species (ROS) by-products. Thus, to further investigate this matter, we report the cloning, sequencing, and expression of the B. fragilis ahpCF gene as well as the role it plays in protecting this anaerobic organism against exogenous toxic organic hydroperoxides.
MATERIALS AND METHODS
Strains and growth conditions.
B. fragilis strains used in this study are listed in Table 1. All strains were grown anaerobically in brain heart infusion broth supplemented with hemin, cysteine, and NaHCO3 (BHIS broth) for routine cultures and genetic procedures (25). Cysteine was omitted where noted, and rifampicin (20 μg/ml), gentamicin (50 μg/ml), tetracycline (5 μg/ml), and erythromycin (10 μg/ml) were added to the media when required. For cell survival determination under aerobic conditions, cultures were grown in BHIS broth to mid-log phase and then split into two equal volumes. One half was placed on a rotatory incubator at 37°C and shaken at 250 rpm in air for up to 5 days, and the other half was kept in the anaerobic chamber for the same period. At specific time points, appropriate dilutions of cultures were plated on BHIS agar in duplicate and incubated anaerobically for 3 to 5 days to determine cell viability. The procedure used for the isolation of a cell population with increased oxygen tolerance was performed by selecting colonies that survived at the later time points of oxygen exposure and then repeating this enrichment procedure for several rounds of oxygen exposure.
TABLE 1.
Relevant properties of B. fragilis strains used in this study
| Strain | Relevant propertiesa | Reference |
|---|---|---|
| 638R | Clinical strain, Rifr | 17 |
| IB260 | 638R katB::tetQ Tetr | 18 |
| IB263 | 638R hpr | 20 |
| IB274 | 638R ahpCF::pFD516 Ermr | This study |
| IB276 | 638R ahpF::pFD516 Ermr | This study |
| IB277 | 638R ahpCF+ahpCF′::XA Ermr | This study |
| IB278 | IB263 ahpCF+ahpCF′::XA Ermr | This study |
| IB281 | IB260 ahpCF::pFD516 Ermr Tetr | This study |
| IB286 | IB263 ahpCF::pFD516 Ermr | This study |
| IB287 | IB263 katB::tetQ Tetr | This study |
| IB291 | IB260 ahpCF+ahpCF′::XA Ermr Tetr | This study |
| IB292A2 | IB281, highly aerotolerant isolate, Ermr Tetr | This study |
Rifr, rifamicin resistance; Tetr, tetracycline resistance; Ermr, erythromycin resistance.
Cloning and DNA sequencing of ahpCF.
All DNA modifications and manipulations were carried out according to standard protocols (2, 23). In an effort to amplify ahpC homologues from the B. fragilis chromosome, two oligonucleotide primers (sense [5′-GAY TTY ACI TTY GTY TGY CCI ACI GAR] and antisense [5′-CCA YTT IGC IGG RCA IAC YTC ICC IGG]) were designed based on conserved amino acid sequence adjacent to two cysteine residues (DFTFVCPTE and PGEVCPAKW) of bacterial AhpCs available from GenBank. A 387-bp fragment was then amplified by Taq polymerase, using a PCR amplification kit (Qiagen, Valencia, Calif.). The thermocycling conditions were set with touchdown annealing temperatures as follow: 5 cycles at 65°C, 5 cycles at 60°C, and 35 cycles at 55°C. The denaturing and extension temperatures for all reaction cycles were set at 94°C for 15 s and 72°C for 30 s, respectively. The amplified fragment was extracted from an agarose gel, ligated into the cloning vector pGEM-T (Promega, Madison, Wis.), and electrotransformed into Escherichia coli DH5α, resulting in pFD689. Southern blot hybridization analysis using the cloned fragment as a probe revealed homology to a 0.9-kb EcoRI DNA fragment in the B. fragilis chromosome. Then the 0.9-kb EcoRI fragment was amplified by inverse PCR (10) using the specific oligonucleotide primers 5′-GTG TGA GTC GGT GCT TAC CGA GTA TA and 5′-GAG ATA CAG GAT AAC AAC ATC GGA CG, based on known sequence. The amplified fragment was then cloned into pGEM-T vector for further nucleotide sequencing. Nucleotide sequence of the entire EcoRI fragment revealed 179 C-terminus codons of ahpC and the first 15 N-terminus codons of ahpF (Fig. 1). The strategy to isolate the entire ahpCF gene region was to rescue the suicide vector pFD690 from the chromosome of strain IB274 an ahpC insertional mutant (see below). Briefly, the chromosome extracted from the strain IB274 was digested with either BglII or PstI and then ligated to produce circular constructs. The ligation reaction was then electroporated into E. coli DH10B, and the transformants were selected on Luria-Bertani agar plates containing spectinomycin (50 μg/ml) to select for pFD690. The new constructs, pFD685 and pFG685A, were then used for automated nucleotide sequencing performed on double-stranded DNA templates (Molecular Biology Resource Facility, University of Tennessee, Knoxville). Additional oligonucleotide primers were designed based on available sequence information to extend and confirm existing sequence.
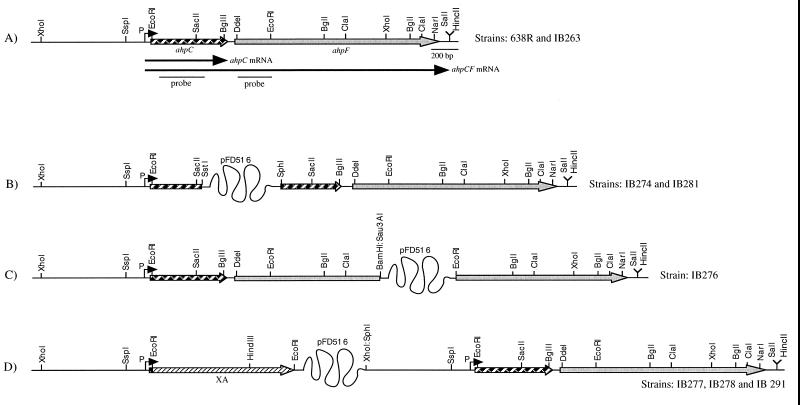
FIG. 1.
Functional and genetic maps of the ahpCF locus in B. fragilis strains. (A) Wild-type gene in strains 638R (wild type) and IB263 (hpr). The striped and grey arrows represent the ahpC and ahpF genes, respectively; The letter P and a dark arrowhead mark the promoter region and the transcription start site. Arrows under the map represent the length and orientation of the transcripts. Also shown are the ahpC and ahpF double-stranded dsDNA fragments used as probes for Northern blot hybridizations. (B to D) Schematic representations of the single-crossover insertional disruptions of the ahpC and ahpF genes and the ahpC′::XA construct in the chromosome. A partial restriction map is shown above the diagrams. The twisted line is not drawn to scale and represents the suicide vector pFD516 (7.7 kb). The bifunctional XA reporter gene is depicted as the hatched arrow.
Construction of ahpC and ahpF insertional mutants.
The SphI/SstI ahpC fragment from pFD689 was cloned into SphI/SstI-digested pFD516 (26). The new construct, pFD690, was mobilized from E. coli DH5α into B. fragilis strains by aerobic triparental filter mating protocols (24). The transconjugants were selected on BHIS agar plates containing rifampicin (20 μg/ml), gentamicin (100 μg/ml), and erythromycin (10 μg/ml). The same procedure was used to construct an ahpF::pFD516 insertional mutant except that a 903-bp EcoRI/Sau3AI internal fragment of ahpF was cloned into EcoRI/BamHI-digested pFD516. Southern blot hybridization and nucleotide sequence analysis of the chromosomal DNA flanking the suicide vector region were used to confirm the single-crossover disruption of the targeted genes. Diagrams of the constructs inserted into B. fragilis chromosome are shown in Fig. 1.
RNA extraction, Northern blot hybridization, and primer extension.
Total RNA extraction and Northern blot analysis of mRNA were carried out as previously described (19). Internal fragments of ahpC and ahpF were used as specific probes. Densitometry analysis of the autoradiograph was normalized to the relative intensity of total 23S and 16S rRNA detected on the ethidium bromide-stained agarose gel to correct for any loading differences.
Primer extension analysis was performed on total RNA obtained from mid-log-phase cells of B. fragilis 638R grown anaerobically and then subjected to oxidative stress conditions. An ahpC-specific oligonucleotide, 5′-CGT CTT CGC TGC TTA CTG, complementary to nucleotides 71 to 88 of the ahpC coding region, was labeled with [γ-32P]ATP and used as primer for the reverse transcriptase reaction as described previously (19). The extended labeled product was electrophoresed on 8% polyacrylamide gels containing urea. A nucleotide sequence ladder was prepared with Sequenase (U.S. Biochemical, Cleveland, Ohio), using a template covering the transcription start site region and the oligonucleotide used for the reverse transcription reactions. The dried gels were exposed to X-ray films.
Mutagenicity assays.
To determine the mutagenicity of peroxides, resistance to the protein synthesis inhibitor fusidic acid was measured following exposure to mutagens in a modified liquid incubation assay (14). Fusidic acid was chosen for these studies because the parent strain is rifampin resistant and it was shown previously that spontaneous fusidic acid-resistant strains could be readily isolated (25). Except for the centrifugations of the cultures in sealed tubes, all procedures were carried out in an anaerobic chamber (the O2 level was <1 ppm). B. fragilis strains were grown overnight in BHIS broth without addition of cysteine. These cultures were used to inoculate 100 ml of same medium and allowed to grow to an optical density at 550 nm of 0.5 (approximately 6 × 108 to 7 × 108 cells/ml). Ten milliliters was removed and centrifuged as described below to obtain a baseline frequency of spontaneous mutation. The remaining culture was treated with 50 μM H2O2 for 15 min twice to induce the peroxide response. Then the culture was divided into 10-ml samples, treated with appropriate concentrations of cumene hydroperoxide (Sigma) or t-butyl hydroperoxide (Sigma) for 15 min at 37°C, and centrifuged for 10 min. The pellets were washed once with same volume of fresh medium, a portion of each cell suspension was diluted, and 0.1-ml aliquots from appropriate dilutions were plated on BHIS agar plates to determine cell viability. After a second centrifugation, the pellet was suspended in about 0.15 ml of fresh medium and plated on BHIS agar plates containing 6 μg of fusidic acid (Sigma) per ml. Plates were then incubated at 37°C for 3 to 4 days. Viable counts were expressed as CFU per milliliters; the mutagenicity of peroxides was expressed as the number of fusidic acid-resistant colonies per 109 CFU. The fusidic acid MIC for the parent strain B. fragilis 638R was <2.5 μg/ml.
Disk inhibition assay.
Cells were grown overnight in BHIS broth without cysteine but containing the appropriate antibiotic; 0.1 ml of culture was spread on Wilkins-Chalgren agar (Difco Laboratories, Detroit, Mich.) plates. Then 10 μl of either 3% hydrogen peroxide aqueous solution, 3% cumene hydroperoxide in dimethyl sulfoxide (DMSO), 0.5% t-butyl hydroperoxide aqueous solution, 3% menadione in DMSO, or 3% paraquat in aqueous solution was dropped on top of a 6-mm-diameter disk paper in the center of the plates. Plates were incubated for 24 h in the dark, and the growth inhibition zones were determined. All procedures described above were performed within an anaerobic chamber except that duplicate plates containing the menadione and paraquat disks were incubated aerobically in the dark for 6 h before they were brought back into the anaerobic chamber and incubated anaerobically as described above.
Construction of ahpC′::XA transcriptional fusions.
An approximately 800-bp blunted XhoI/EcoRI fragment from pFD695 containing the promoter region and the first 12 codons of the ahpC gene was cloned into the blunted SphI/EcoRI sites of pFD516. Then a 1.2-kb EcoRI fragment from pXA1 containing the promoterless xylosidase/arabinosidase (XA) bifunctional reporter gene (30) was cloned into the unique EcoRI site of the new construct. Restriction digestion was used to confirm that the XA gene was in the same orientation as ahpC (Fig. 1D). The new construct, pFD720, was mobilized from E. coli DH5α into B. fragilis strains as described above.
Enzymes assays.
The β-xylosidase assays of crude extracts were performed with p-nitrophenyl-β-d-xylopyranoside as the substrate as specified by Whitehead (30) except that reactions were constantly monitored in an automated spectrophotometer and the molar extinction coefficient of 0.0184 μM−1 cm−1 for p-nitrophenol at 405 nm was used (12). One unit of β-xylosidase is the amount of enzyme which releases 1 μmol of p-nitrophenol per min at 37°C. Catalase was assayed exactly as described previously (20).
DNA sequence analysis and database comparison.
Computer analysis of nucleotide and amino acid sequence data was performed with the University of Wisconsin Genetics Computer Group DNA sequence analysis software (9). Phylogenetic relationships were inferred by the parsimony method with the PHYLIP phylogeny interference package (version 3.5) from a multiple amino acid sequence alignment generated by Pileup. A consensus tree was constructed from 100 bootstrap replications.
Sequences used for comparison and, in parentheses, their GenBank accession numbers are Porphyromonas gingivalis AhpCF (preliminary sequence data obtained from The Institute for Genomic Research website ;[14a;]), E. coli AhpC (P26427) and AhpF (P35340), Salmonella typhimurium AhpC (P19479) and AhpF (P19480), Enterococcus faecalis AhpC (AF016233), Pseudomonas putida AhpCF (AB010689), Treponema pallidum AhpC (AE001227), and Xanthomonas campestris pv. phaseoli AhpF (U94336).
Nucleotide sequence accession number.
The nucleotide sequence of B. fragilis ahpCF has been submitted to GenBank and assigned accession no. AF129406.
RESULTS
Analysis of the ahpCF gene nucleotide sequence.
In a previous study of an hpr mutant, IB263, we observed the constitutive overproduction of a peptide whose N-terminal amino acid sequence was highly homologous to those of known AhpC proteins (20). To clone the gene for this oxidative stress enzyme, a gene fragment was amplified from the B. fragilis 638R chromosome by PCR using primers derived from the conserved regions of bacterial AhpC homologues available in the GenBank-EMBL/Swiss-Prot databases. Following nucleotide sequence analysis of this gene fragment, the complete ahpCF gene was obtained by a combination of inverse PCR and marker rescue as described in Materials and Methods. The nucleotide sequence of the ahpCF locus in B. fragilis revealed two open reading frames (ORFs) (Fig. 1A). The first ORF encodes a protein with predicted molecular weight of 21,060. Comparison of the predicted amino acid sequence to other sequences in the GenBank-EMBL/Swiss-Prot databases revealed strong homology to bacterial alkyl hydroperoxide reductase small subunit AhpC (Fig. 2A) and other members of the TSA protein family (data not shown). The first 20 predicted N-terminal amino acids from ahpC were identical to the Edman degradation N-terminal amino acid sequence obtained from partially purified B. fragilis AhpC previously reported (20). Thus, the identity of the gene and gene product was confirmed. Sequence comparison of B. fragilis AhpC with orthologues from other species revealed highest amino acid identity to P. gingivalis (63% identity and 76% similarity), P. putida (64% identity and 74% similarity), S. typhimurium AhpC (62% identity and 70% similarity), E. coli AhpC (61% identity and 70% similarity), E. faecalis AhpC (62% identity and 68% similarity), and T. pallidum AhpC (55% identity and 65% similarity) AhpCs. The two functional cysteine residues of bacterial AhpC and TSA-like proteins (C46 and C165) as well as the consensus sequence around the N-terminal region cysteine (PXDFTFVCPTE) (6, 11) are highly conserved in B. fragilis AhpC (Fig. 2A). In addition, the AhpC C-terminal amino acid sequence (TLKPSIDLVGKI) closely matched the highly conserved bacterial AhpC C-terminal motif (TL(AK)PSLD(LI)VGKI) (6).
FIG. 2.
Alignment of the B. fragilis (Bf) deduced amino acid sequences for AhpC (A) and AhpF (B) with sequences of proteins from E. coli (Ec), S. typhimurium (St), P. putida (Pp), P. gingivalis (Pg), E. faecalis (Ef), and T. pallidum (Tp), and X. campestris (Xc). Asterisks above the sequences indicate functional redox-active cysteine residues (4, 11). Predicted adenine dinucleotide binding sites in AhpF are underline according to the putative assignment of bacterial AhpF functional domains (29). Consensus of at least 50% identical amino acid residues is denoted by black boxes; conserved amino acid substitutions are depicted by grey boxes. Only a partial alignment of the most conserved AhpF regions is shown. For comparison, amino acid residue positions are shown for E. coli AhpF and B. fragilis AhpF.
The phylogenetic relationship between B. fragilis AhpC and 34 proteins from the procaryotic and eucaryotic AhpC/TSA family was determined from a progressive multiple alignment of the amino acid sequences. Parsimony analysis of this alignment showed that AhpC from the gram-negative anaerobic bacteria B. fragilis and P. gingivalis together with T. pallidum and E. faecalis formed a unique cluster diverged from the main groups containing the gram-positive and gram-negative eubacteria (data not shown).
The first codon of ahpF starts 101 bp downstream of the ahpC. The ahpF ORF encodes 516 amino acids with a predicted molecular weight of 55,280. Pairwise alignment of the B. fragilis AhpF amino acid sequence revealed strong homology to bacterial Ahp subunit F (Fig. 2B) and to thioredoxin reductases (data not shown). When individually compared with orthologues, the B. fragilis AhpF showed highest amino acid identity to P. gingivalis AhpF (56% identity and 64% similarity), P. putida AhpF (48% identity and 59% similarity), and E. coli, S. typhimurium, and X. campestris AhpFs (46% identity and 56% similarity in each case). In addition, the B. fragilis AhpF cysteine centers (C128, C131 and C341, C344) align with the AhpF cysteine centers (C129, C132 and C345, C348) involved in the reduction of AhpC in E. coli and S. typhimurium (4). The amino acid consensus sequence around the redox-active cysteines in AhpF and TrxB involved in binding FAD cofactor and transferring of reducing equivalents to the substrate also were found in B. fragilis AhpF (Fig. 2B).
The phylogenetic relationship between B. fragilis AhpF and 25 proteins from procaryotic AhpF and thioredoxin reductases sequences obtained from the databases was also determined from a progressive multiple alignment of the amino acid sequences followed by parsimony analysis. The results showed that B. fragilis AhpF and P. gingivalis AhpF were located in a cluster diverged from other gram-positive and gram-negative facultative and aerobic bacteria (data not shown).
Regulation of ahpCF operon.
To investigate the expression of ahpCF, total RNA extracted from mid-log-phase cells of B. fragilis exposed to different oxidative stress conditions was probed with specific internal DNA fragments from either ahpC or ahpF. Northern blot hybridization analysis revealed that expression of AhpC was regulated at the transcriptional level. Transcripts of approximately 0.6 and 2.4 kb that hybridized to the ahpC probe were observed, suggesting that ahpC was transcribed both as a monocistronic mRNA and as part of a polycistronic operon (Fig. 3A). Autoradiographs of Northern blots probed with the ahpF fragment revealed an mRNA of about 2.4 kb, suggesting that ahpF was cotranscribed as part of the polycistronic ahpCF mRNA (Fig. 3B). This observation was further confirmed by Northern blot analysis of the ahpC insertional mutant IB274, in which the 2.4-kb mRNA was absent. This indicates there was a polar effect on the ahpCF mRNA due to the insertional disruption of the upstream ahpC cistron (data not shown). Moreover, the presence of an mRNA component of approximately 1.7 kb (Fig. 3B) suggests that ahpF also may be transcribed as a single cistron, but this was not further investigated. Inexplicably, a small 0.3-kb RNA which strongly hybridizes with an internal ahpC DNA probe was detected. A diagram of the major ahpC and ahpCF mRNA transcripts is shown in Fig. 1A.
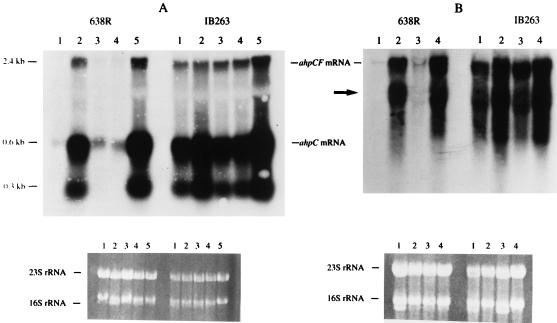
FIG. 3.
Autoradiograph of Northern hybridization filter of total RNA from mid-log-phase B. fragilis 638R and IB263 (hpr) following exposure to different oxidative stress conditions. The probe was an ahpC (A) or ahpF (B) internal gene fragment; samples consisted of anaerobic cultures (lane 1), cultures treated with 50 μM H2O2 (lane 2), cultures treated with 100 μM potassium ferricyanide (lane 3 in panel A), cultures treated with 1 mM potassium ferricyanide (lanes 4 in panel A and lane 3 in panel B), and cultures exposed to aeration for 1 h (lane 5 in panel A and lane 4 in panel B). Approximate sizes of the transcripts are indicated on the left. The arrow in panel B points to an ahpF mRNA component of approximately 1.7 kb. Bottom panels are corresponding ethidium bromide-stained agarose gels loaded with 30 μg of total RNA in each lane. Positions of 23S and 16S rRNA are also indicated.
Densitometry analysis of the Northern blots showed an approximately 60-fold increase in ahpC and ahpCF mRNA in cultures treated with H2O2 or exposed to oxygen but no increase in potassium ferricyanide-treated cultures (Fig. 3). Expression of the monocistronic ahpC mRNA was found to be approximately 3- to 3.5-fold higher than expression of the ahpCF polycistronic mRNA. Under anaerobic conditions, the levels of ahpC and ahpCF mRNA in mid-log phase and during entry into stationary phase did not change significantly, nor did limitation of glucose or addition of nonfermentable carbohydrates (acetate, fumarate, pyruvate, and succinate) have a detectable effect (data not shown).
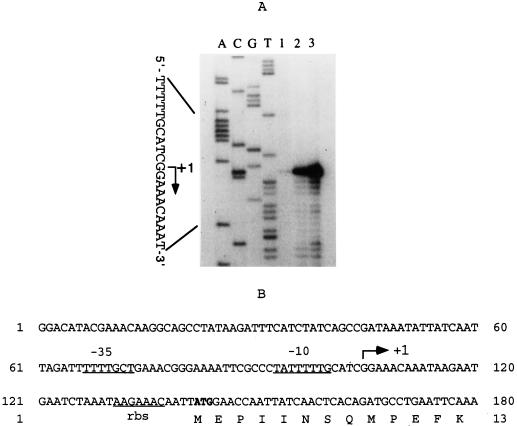
Analysis of the transcription initiation of ahpCF mRNA showed that the first base was at a guanine 37 bp upstream of the ahpC translation start codon. There was an approximately 30- or 60-fold increase in ahpCF primer extension product following treatment of the anaerobic cultures with hydrogen peroxide or oxygen exposure, respectively (Fig. 4A). A diagram of the ahpCF transcription start nucleotide as well as the predicted −10 and −35 promoter region is shown in Fig. 4B.
FIG. 4.
(A) Autoradiograph following electrophoresis of ahpCF primer extension reactions. Total RNA was obtained from mid-log-phase cells of B. fragilis 638R grown anaerobically in BHI medium and then subjected to oxidative stress conditions. Lane 1, anaerobic culture; lane 2, culture exposed to 50 μM H2O2; lane 3, culture exposed to oxygen for 1 h. The nucleotide sequence of ahpCF, using the same primer, was run in parallel. (B) Nucleotide sequence of the ahpCF regulatory region. Putative −35 and −10 regions of the promoter and the putative ribosome binding site (rbs) are underlined. The arrow indicates the initial +1 guanine nucleotide 37 bp upstream of the ahpC translation start codon. The start codon of the ahpC gene is indicated in boldface, and only the first 13 codons are shown.
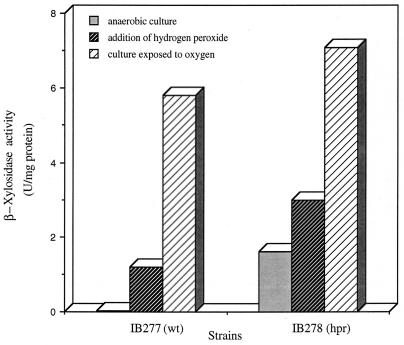
Regulation was further investigated by comparing activities of ahpC′::XA transcriptional fusions in the parent strain and a constitutive hpr mutant. It was shown previously that the hpr phenotype is constitutive expression of catalase and several other proteins including AhpC, but transcriptional control of ahpC was not known (20). Xylosidase activities in crude extracts of mid-log-phase IB277 (wild type) and IB278 (hpr) grown in BHI broth are shown in Fig. 5. There was an increase of approximately 80-fold in xylosidase activity for the peroxide mutant strain (1.6 U/mg of protein) compared to the anaerobic culture of the parent (0.021 U/mg of protein). In the parent strain IB277, treatment with H2O2 and oxygen exposure induced xylosidase activity approximately 60-fold (1.2 U/mg of protein) and 280-fold (5.8 U/mg of protein), respectively, compared to anaerobic controls. In the mutant IB278 (hpr), there was a further induction in xylosidase activity following treatment with H2O2 (twofold) and oxygen exposure (fourfold). These results are consistent with the Northern hybridization studies (Fig. 3) in which ahpC and ahpCF mRNAs were constitutively expressed in the hpr mutant strain IB263. Comparison of the parent and peroxide-resistant strains in Fig. 3 and 5 clearly shows that the peroxide-resistant mutant has lost normal regulation of both the ahpC gene and the ahpC′::XA fusion.
FIG. 5.
Analysis of ahpC′::XA transcriptional fusions in strains grown under different oxidative stress conditions. β-Xylosidase activity in crude extracts was determined for wild-type (wt) strain IB277 and constitutive hpr mutant strain IB278 grown to mid-log phase and then either shaken in air for 1 h (culture exposed to oxygen), challenged with two successive 50 μM H2O2 treatments for 15 min each (addition of hydrogen peroxide), or incubated anaerobically (anaerobic culture).
It was possible that the expression of ahpC could be modulated by the presence or absence of other detoxifying enzymes. However, we found that the ahpC′::XA fusion was not affected in the catalase mutant (katB) background under anaerobic conditions. Crude extract of anaerobic mid-log-phase cells of IB291 (katB) showed that xylosidase activity remained unaltered (0.028 U/mg of protein) compared to xylosidase activity (0.021 U/mg of protein) in the strain IB277 (wt) (data not shown). In contrast, catalase activity in the ahpC mutant IB274 (5.5 U/mg of protein) was approximately 10-fold higher than that in IB277 under anaerobic conditions (0.5 U/mg of protein) (data not shown).
Sensitivity to peroxides and superoxide-generating agents.
ahpCF mutants IB274 and IB286 were more sensitive than the parent strains to organic peroxides (Table 2). However, ahpF mutant IB276 did not show greater sensitivity to t-butyl hydroperoxide than to cumene hydroperoxide. In addition, sensitivity to hydrogen peroxide in the ahpCF and ahpF mutants was not altered in contrast to increased sensitivity to hydrogen peroxide and t-butyl hydroperoxide in the katB mutant. Surprisingly, the physiological response of the ahpCF katB double mutant to hydrogen peroxide and organic peroxides was found to be similar to that of the parent strain. Moreover, strain IB292A2, an aerotolerant strain derived from the ahpCF katB double mutant (see below), was more resistant than the parent strain to organic peroxides. The redox-cycling agent menadione had nearly the same toxic effect on all the strains tested following oxygen exposure of the plates for 6 h except for increased sensitivity to menadione in the ahpCF mutant (IB274). However, menadione also was somewhat toxic to these strains under anaerobic conditions, zones of inhibition being approximately one half of the diameters in plates exposed to oxygen. In contrast to menadione, paraquat had no effect under the same conditions. The diluent DMSO alone had no effect on growth inhibition (diameters of zones of inhibition being ≤6 mm) in the control plates (data not shown).
TABLE 2.
Sensitivity of mutant strains to inhibition by oxidants
| B. fragilis strain | Diam of disk inhibition zone (mm)a
|
||||
|---|---|---|---|---|---|
| 3% H2O2 | 3% CHP | 0.5% TBHP | 3% MD | 3% MD + oxygenb | |
| 638R | 14 | 42 | 53 | 15 | 30 |
| IB260 (katB) | 19 | 45 | 60 | 18 | 30 |
| IB274 (ahpCF) | 15 | 48 | 58 | 17 | 35 |
| IB276 (ahpF) | 14 | 47 | 51 | 17 | 30 |
| IB281 (ahpCF katB) | 16 | 42 | 50 | 14 | 29 |
| IB292A2 (ahpCF katB) | 15 | 37 | 46 | 13 | 28 |
| IB263 (hpr) | 10 | 30 | 47 | 14 | 33 |
| IB286 (ahpCF) | 10 | 39 | 55 | 16 | 31 |
| IB287 (katB) | 16 | 32 | 50 | 14 | 29 |
Average of three determinations. CHP, cumene hydroperoxide; TBHP, t-butyl hydroperoxide; MD, menadione.
Exposure of plates containing menadione to atmospheric oxygen for 6 h is described in Materials and Methods.
ahpCF protection from mutagenesis by peroxides.
We have investigated the mutagenesis of cumene hydroperoxide and t-butyl hydroperoxide on the B. fragilis genome by selection of mutants resistant to fusidic acid. Previous work has shown that at least a twofold increase in the number of mutants induced by a mutagen over the control numbers is considered to be significant (14). Table 3 shows the numbers of fusidic acid-resistant colonies per 109 CFU. In untreated cells, numbers of spontaneous mutations were approximately fivefold (ahpC), approximately sevenfold (ahpF), and approximately 8-fold (ahpC katB) higher in mutant strains than in to the parent and katB strains. Pretreatment of the cultures with a sublethal concentration of hydrogen peroxide did not significantly increase mutagenesis in most strains except for katB strains, which showed an approximately fourfold increase. t-butyl hydroperoxide at 50 μM caused a higher mutation rate in the ahpC mutant (over 10-fold increase) than observed for ahpF and ahpC katB strains (approximately 2-fold increase). After treatment with cumene hydroperoxide, the number of fusidic acid-resistant mutants increased over 20-fold in the ahpC katB (IB281) and ahpF (IB276) mutants. However, under the same conditions, 50 μM cumene hydroperoxide caused intense bacterial lysis of the ahpC mutant strain (IB274), which could account for the lack of resistant colonies after treatment with this organic peroxide. In contrast, the same effect was not observed for other strains, including the ahpC katB mutant. Bacterial cell lysis, however, occurred in all strains tested when cumene hydroperoxide was used at 100 μM (data not shown). Cumene hydroperoxide also was more mutagenic then t-butyl hydroperoxide, as it induced approximately a fourfold increase in the number fusidic acid-resistant colonies in both parent and katB mutant strains compared to controls.
TABLE 3.
Frequency of spontaneous and oxidant-induced mutations
| B. fragilis strain | Frequency of mutations (no. of fusidic acid-resistant colonies/109 cells plated)a
|
|||
|---|---|---|---|---|
| Spontaneous (control) | Induced by:
|
|||
| No addition | 50 μM TBHP | 50 μM CHP | ||
| 638R | 1.1 | 1.5 | 2.1 | 4.2 |
| IB260 (katB) | 1.0 | 4.2 | 5.2 | 15 |
| IB274 (ahpCF) | 4.8 | 5.2 | 68 | NDb |
| IB276 (ahpF) | 6.8 | 7.4 | 13.2 | 192 |
| IB281 (ahpCF katB) | 8.1 | 8.6 | 19.0 | 199 |
Average of two determinations. TBHP, t-butyl hydroperoxide; CHP, cumene hydroperoxide.
ND, not determined due to bacterial cell lysis.
Tolerance to oxygen exposure.
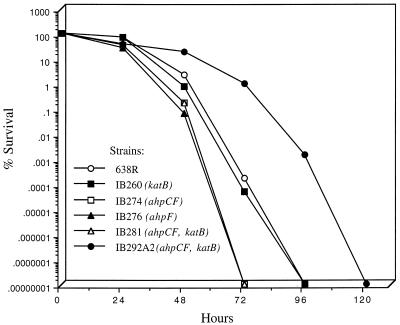
The effect of oxygen exposure on cell viability increased significantly after the first 24 h of constant culture aeration. The effect was more pronounced in the ahpCF, ahpF, and ahpC katB mutants, which declined by approximately 3 logs in cell viability during 48 h. In contrast, the parent strain 638R and the katB mutant only had a 2-log decrease in cell survival during the same period. However, the ahpCF, ahpF, and ahpCF katB strains had a complete loss of viability at 72 h, whereas 638R and IB260 remained viable until 96 h (Fig. 6). During the course of this work, we observed several colonies derived from the ahpCF katB mutant which survived more than 72 h exposure to oxygen. After two passages of these colonies in aerobic conditions (see Materials and Methods), we isolated strains that were viable for a longer period under oxygen exposure than the parent strain. One of these colonies, designated IB292A2, was highly resistant to molecular oxygen up to 48 h, with less than 1-log decrease in cell viability compared to other strains (Fig. 6). After 72 h, cell viability decreased approximately 2 logs, while its parent strain, IB281, was no longer viable in the same period of time. Moreover, total loss of viability did not occur until about 120 h of oxygen exposure. In addition, the strain IB292A2 acquired high resistance to cumene and t-butyl hydroperoxides but not hydrogen peroxide (Table 2) even though it no longer has functional catalase and alkyl hydroperoxide reductase enzymes. Using similar techniques, we were unable to isolate a cell population with altered oxygen tolerance in other mutant or wild-type strains (data not shown).
FIG. 6.
Survival of mid-log-phase anaerobic cells of B. fragilis strains shifted to aerobic conditions. Cultures of mid-log-phase cells at an optical density at 550 nm of 0.3 were divided at time zero; one half was shaken at 250 rpm in air at 37°C, and the other half was maintained anaerobically. Viable cell counts were determined at the times shown. For clarity, data for the anaerobic control cultures are not shown.
DISCUSSION
In this report we present the first study on the regulation and role of the AhpCF in the oxidative stress response of an anaerobic bacterium, B. fragilis. The findings show that ahpCF expression was up-regulated by either oxygen exposure or addition of exogenous hydrogen peroxide. Inactivation of ahpCF but not ahpF increased mutagenesis and sensitivity to alkyl hydroperoxides. Taken together, these data argue for a significant role of ahpCF in resistance to damage from peroxides. In addition, we report here the isolation of a strain derived from an ahpCF katB double-mutant background strain with altered resistance to molecular oxygen, indicating that additional mechanisms might be involved in aerotolerance in anaerobic organisms.
Transcription of ahpCF occurred at a single promoter region whose activation was greatly increased by oxidative stress, and ahpCF was constitutively expressed in the hpr mutant strain IB263. This response was similar to the expression of the katB gene, which was previously found to be under the regulation of a trans-acting regulatory mechanism (20). Evidence that ahpCF and katB are under the control of a common regulatory mechanism was obtained in transcriptional fusion experiments. The findings in Fig. 5 clearly showed that the wild-type ahp promoter was deregulated in IB263 but regulated normally in the parent strain, indicating that transcription of katB and that of ahpCF are positively regulated by a common trans-acting factor. In mid-log-phase cells of the facultative bacteria E. coli and S. typhimurium, the H2O2 redox sensor and transcriptional activator OxyR induces the synthesis of nine proteins, including KatG, AhpCF, GorA, and Dps (1, 8). A constitutive oxyR mutant confers overexpression of these proteins and resistance to hydrogen peroxide and alkyl hydroperoxides (8). In this regard, peroxide response regulation in B. fragilis may have some similarity to that in enteric gram-negative bacteria, since we have isolated an oxyR homologue in B. fragilis which is divergently transcribed from the dps gene and affects katB and ahpCF gene expression in the parent and in the constitutive hpr mutant strain IB263 (21).
In S. typhimurium and E. coli, the AhpC/AhpF system is essential for elimination of alkyl hydroperoxides (4, 15, 16). Following reduction of alkyl hydroperoxides, oxidized AhpC is reduced to the active form by an AhpF/NADH-dependent reductase system (16). In the presence of excess oxidized AhpC, AhpF becomes a limiting factor in the reduction of AhpC (16). Moreover, AhpC is specifically reduced by AhpF and cannot be reduced by other electron transfer systems such as thioredoxin reductase (4). However, inactivation of ahpF in B. fragilis did not affect sensitivity to t-butyl hydroperoxide in the disk inhibition assays (Table 2). In contrast, the ahpCF mutant was sensitive in both the disk inhibition and mutagenesis studies. This finding suggests that in B. fragilis, AhpF may not be a limiting factor for reduction of oxidized AhpC and other systems may be able to replace AhpF in electron transfer. In this regard, we observed the presence of two distinct transcripts, a monocistronic ahpC mRNA and a polycistronic ahpCF mRNA (Fig. 3). The ahpC message was clearly the more abundant of the two, indicating that they may be differentially expressed.
Under anaerobic conditions, there was an eightfold-increased frequency of spontaneous mutation to fusidic acid in the ahpCF/katB mutant. An analogous observation was made with S. typhimurium, in which spontaneous mutagenesis under anaerobic conditions in an oxyRΔ2 strain was approximately 16- and 40-fold higher than in the oxyR+ and oxyR1 (constitutively activating) strains, respectively (27). In these experiments, the spontaneous-mutation frequency was not due to low levels of hydrogen peroxide and superoxide anion present in the culture media. This indicates that even under anaerobic conditions, the presence of peroxide-scavenging enzymes may be necessary for detoxification of ROS. The source of the ROS, however, is not clear. Although no direct measurements of oxidants or oxidative damage were performed in the present study, the growth media were maintained under anaerobic conditions and the levels of molecular oxygen in the anaerobic chamber were less than 1 ppm (monitored by an oxygen detector). Thus, it is unlikely that ROS derived from molecular oxygen were formed in vivo at levels high enough to cause significant cellular damage. This suggests the possibility that ROS or other radical species may be generated internally during anaerobic growth. The role of oxygen radical-scavenging enzymes in facultative or obligate anaerobic bacteria under anaerobic conditions is not clear, but the fact that catalase activity was higher in mid-log-phase cells of an ahpCF mutant compared to the parent strain suggests that additional physiological compensatory adaptation might occur to protect B. fragilis against eventual accumulation of cellular oxidants. Similarly, disruption of the ahpCF operon in facultative bacteria causes compensatory up-regulation of genes controlled by the redox-sensitive regulator OxyR in E. coli (22) and the peroxide regulon repressor PerR in B. subtilis (3).
In previous studies we showed that neither a katB mutant nor an hpr mutant (IB263) was altered in resistance to the toxic effects of molecular oxygen exposure, which led to the suggestion that there is more than one component to the oxidative stress response in this obligate anaerobe. In contrast, the results presented here clearly indicate that while ahpCF is part of the peroxide response, mutations leading to loss of AhpC affected both oxygen sensitivity and peroxide sensitivity. This indicates that there is an important role for peroxide detoxification in aerotolerance. Further, strain IB292A2 was originally isolated for enhanced aerotolerance, yet it also displayed high-level resistance to alkyl hydroperoxides. Interestingly, the resistance of IB292A2 to a superoxide anion generator (menadione) was not significantly altered. Taken together, these observations reinforce the idea that the physiological responses to peroxide toxicity and to the toxic effects of molecular oxygen are overlapping and complex in this obligate anaerobe.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI40588 from the National Institutes of Health.
REFERENCES
- 1.Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςs in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons Inc.; 1987. [Google Scholar]
- 3.Bsat N, Chen L, Helmann J D. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol. 1996;178:6579–6586. doi: 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calzi M L, Poole L B. Requirement for the two AhpF cystine disulfide centers in catalysis of peroxide reduction by alkyl hydroperoxide reductase. Biochemistry. 1997;36:13357–13364. doi: 10.1021/bi9713660. [DOI] [PubMed] [Google Scholar]
- 5.Chae H Z, Rhee S G. A thiol-specific antioxidant and sequence homology to various proteins of unknown function. Biofactors. 1994;4:177–180. [PubMed] [Google Scholar]
- 6.Chae H Z, Robison K, Poole L B, Church G, Storz G, Rhee S G. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Xie Q W, Nathan C. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol Cell. 1998;1:795–805. doi: 10.1016/s1097-2765(00)80079-9. [DOI] [PubMed] [Google Scholar]
- 8.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieffenbach C W, Dveksler G S. PCR primer: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 11.Ellis H R, Poole L B. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry. 1997;36:13349–13356. doi: 10.1021/bi9713658. [DOI] [PubMed] [Google Scholar]
- 12.Gillard B K, Markman H C, Feig S A. Direct spectrophotometric determination of α-amylase activity in saliva, with p-nitrophenyl α-maltoside as substrate. Clin Chem. 1977;23:2279–2282. [PubMed] [Google Scholar]
- 13.Halliwell B, Gutteridge J M C. Lipid peroxidation, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan H M, Moody C S. Determination of the mutagenicity of oxygen free radicals using microbial systems. Methods Enzymol. 1984;105:254–263. doi: 10.1016/s0076-6879(84)05033-3. [DOI] [PubMed] [Google Scholar]
- 14a.The Institute for Genomic Research. 17 March 1999, revision date. [Online.] http://www.tigr.org/data/p-gingivalis. The Institute for Genomic Research, Rockville, Md. [1 April 1999, last date accessed.]
- 15.Jacobson F S, Morgan R W, Christman M F, Ames B N. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]
- 16.Poole L B, Ellis H R. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhymurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 17.Privitera G, Dublanchet A, Sebald M. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J Infect Dis. 1979;139:97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- 18.Rocha E R, Selby T, Coleman J P, Smith C J. The oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J Bacteriol. 1996;178:6895–6903. doi: 10.1128/jb.178.23.6895-6903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocha E R, Smith C J. Regulation of Bacteroides fragilis katB mRNA expression by oxidative stress and carbon limitation. J Bacteriol. 1997;179:7033–7039. doi: 10.1128/jb.179.22.7033-7039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocha E R, Smith C J. Characterization of a peroxide-resistant mutant of the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1998;180:5906–5912. doi: 10.1128/jb.180.22.5906-5912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocha E R, Smith C J. Abstracts of the 99th General Meeting of the American Society for Microbiology 1999. Washington, D.C: American Society for Microbiology; 1999. The transcriptional activator OxyR and the regulation of the oxidative stress response in the anaerobe Bacteroides fragilis, abstr. B/D-161; p. 60. [Google Scholar]
- 22.Rosner J L, Storz G. Effects of peroxides on susceptibilities of Escherichia coli and Mycobacterium smegmatis to isoniazid. Antimicrob Agents Chemother. 1994;38:1829–1833. doi: 10.1128/aac.38.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Shoemaker N B, Getty C, Gardner J F, Salyers A A. Tn4351 in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986;165:929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith C J, Spiegel H. Transposition of Tn4551 in Bacteroides fragilis: identification and properties of a new transposon from Bacteroides spp. J Bacteriol. 1987;169:3450–3457. doi: 10.1128/jb.169.8.3450-3457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith C J, Rollins L A, Parker A C. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid. 1995;34:211–222. doi: 10.1006/plas.1995.0007. [DOI] [PubMed] [Google Scholar]
- 27.Storz G, Christman M F, Sies H, Ames B N. Spontaneous mutagenesis and oxidative damage to DNA in Salmonella typhimurium. Proc Natl Acad Sci USA. 1987;84:8917–8921. doi: 10.1073/pnas.84.24.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tartaglia L A, Storz G, Brodsky M H, Lai A, Ames B N. Alkyl hydroperoxide reductase from Salmonella typhymurium. J Biol Chem. 1990;265:10535–10540. [PubMed] [Google Scholar]
- 30.Whitehead T R. Development of a bifunctional xylosidase/arabinosidase gene as a reporter gene for the Gram-negative anaerobes Bacteroides and Porphyromonas, and Escherichia coli. Curr Microbiol. 1997;35:282–286. doi: 10.1007/s002849900255. [DOI] [PubMed] [Google Scholar]