Abstract
The health-promoting Parabacteroides distasonis, which is part of the core microbiome, has recently received a lot of attention, showing beneficial properties for its host and potential as a new biotherapeutic product. However, no study has yet investigated the cell surface molecules and structures of P. distasonis that allow its maintenance within the gut microbiota. Moreover, although P. distasonis is strongly recognized as an intestinal commensal species with benefits for its host, several works displayed controversial results, showing it as an opportunistic pathogen. In this study, we reported gene clusters potentially involved in the synthesis of capsule, fimbriae-like and pili-like cell surface structures in 26 P. distasonis genomes and applied the new RfbA-typing classification in order to better understand and characterize the beneficial/pathogenic behavior related to P. distasonis strains. Two different types of fimbriae, three different types of pilus and up to fourteen capsular polysaccharide loci were identified over the 26 genomes studied. Moreover, the addition of data to the rfbA-type classification modified the outcome by rearranging rfbA genes and adding a fifth group to the classification. In conclusion, the strain variability in terms of external proteinaceous structure could explain the inter-strain differences previously observed of P. distasonis adhesion capacities and its potential pathogenicity, but no specific structure related to P. distasonis beneficial or detrimental activity was identified.
Keywords: gut microbiota, Parabacteroides distasonis, capsular polysaccharide, fimbriae, pilus, O-antigen, pathogenicity, probiotic, comparative genomics
1. Introduction
Gut microbiota (GM) is now considered as a new organ system mainly due to the microorganisms’ specific biochemical interaction with their hosts and their systemic integration into the host biology [1,2]. Bacteria that are predominant in the GM are mainly defined by anaerobic bacteria part of the Firmicutes and Bacteroidetes phyla [3]. Advances in sequencing methods have facilitated the characterization and understanding of the contribution of the GM to the host well-being, which is now indisputable. In fact, it is now well-defined that the cooperation between the GM and its host is essential to regulate the development and function of the immune, metabolic and nervous system. In turn, one of the major roles of the immune system is to control and maintain its relationships with the GM. The intestinal microbiota, in addition to contributing to the development of the immune system and to intervene into host metabolic and nervous function, also creates a protective barrier against external pathogens and participate in maintaining the structure and integrity of the gastrointestinal tract [4,5,6]. In the long run, the GM can modulate host behavior and nervous system function through dynamic and bidirectional communication along the gut–brain axis [7].
Although mechanisms underlying host–microbiota interactions are not fully described, it is now well-established that cell surface molecules and structures of the GM play a key role in such relationships via conserved microbe-associated molecular patterns (MAMPs) that will be recognized by pattern recognition receptors (PRRs) of immune system cells, including Toll-like receptors (TLRs). An interaction between MAMP and TLR will then initiate the immune response if the MAMP is identified as pathogenic [8,9]. The study of secreted and surface molecules of microbiota members is also fundamental for their involvement in the establishment of species in the versatile and competitive environment of the gut and their key role as a potential virulence factor [10]. Among cell surface markers are capsular polysaccharide (CPS), fimbriae and pili, all well-described for their crucial role in microorganism colonization of the host epithelium.
In Gram-negative anaerobic bacteria, various systems have been described for each of these cell surface markers, including the CPS of Bacteroides fragilis, the fimbriae system (Fim) of Porphyromonas gingivalis, the type V pilus system (Mfa) of P. gingivalis and Bacteroides thetaiotaomicron and the immunogenic component of lipopolysaccharide (LPS); O-antigen (Rfb) is well-described in the facultative anaerobic Escherichia coli [11,12,13,14,15].
Among the gut microbiota members is Parabacteroides distasonis, a Gram-negative bacterium strictly anaerobe belonging to the Tannerellaceae family within the Bacteroidetes phylum. This bacterial species, part of the core microbiome, has recently received a lot of attention, showing beneficial properties for its host. In fact, although strain-dependent, P. distasonis display anti-inflammatory/cancer properties and activities on decreasing weight gain, hyperglycemia and hepatic steatosis in ob/ob and high-fat diet-fed mice [16,17,18]. The importance of P. distasonis membrane in these disease treatments has been pointed out in numerous studies. Notably, it has been shown to largely suppress production of pro-inflammatory cytokines in obese animal models [19] and induce apoptosis in colon cancer cell lines, suggesting anti-inflammatory and anti-cancer effects [20]. The membrane components of P. distasonis have also been reported to decrease the severity of gut inflammation in the non-immunocompromised mouse models that had induced acute and chronic colitis [21]. Many studies have highlighted these abilities to promote P. distasonis as a new potential biotherapeutic product [22,23,24]. In our previous work, we explored P. distasonis capacities related to its maintenance within the digestive tract and the electrokinetic properties of its cell peripheral regions to provide a first qualitative picture of its surface structure [25]. This work evidenced a strain-dependent ability to adhere and to form a biofilm related to the putative presence of cell surface structures such as CPS, fimbriae, pili or capsule.
Although numerous studies described the beneficial aspects of P. distasonis or its ability to colonize the intestine, few explore mechanisms behind these aptitudes. Moreover, while P. distasonis is strongly recognized as intestinal commensal specie with benefits for its host, several studies displayed controversial results, showing P. distasonis as an opportunistic pathogen [26,27,28,29]. In this study, we investigated the cell surface structures of P. distasonis that may influence host–P. distasonis crosstalk and play an essential role in its maintenance and stability within the GM. We reported gene clusters potentially involved in the synthesis of capsule, fimbriae-like and pili-like outer membrane structure and applied the new rfbA-typing classification on 26 genomes of P. distasonis including 13 new clinical strains (CS) in order to investigate its maintenance within the digestive tract and its potential pathogenicity [30]. In this study, the designation “pilus” is used to describe the external cell surface structure originating from the “minor fimbriae” Mfa system [31,32]. “Fimbriae” refer to structures arising from the Fim system. However, the use of this designation does not mean that Mfa structures are minor and short in comparison with Fim fimbriae [33]; rather, it serves to better clarify the origin of the external appendages described.
2. Results
2.1. P. distasonis Genomes Characterization
Thirteen nonredundant P. distasonis CS were isolated (Table 1) by the Clinical Microbiology Laboratory of the University Hospital of Nancy, France, and sequenced using Illumina technology. All genomes were then integrated in the Microbial Genome Annotation and Analysis Platform (MaGe) in addition to 13 other public P. distasonis genomes (Figure 1).
Table 1.
P. distasonis strain isolation.
| Strain | Type of Sample | Host Status | Isolation Date | Isolation Country | References | |
|---|---|---|---|---|---|---|
| Parabacteroides distasonis | ATCC 8503T | Human feces | Apparently normal | 1933 | USA | [34] |
| APCS2/PD | Human feces | Unknown | 2017 | Ireland | NCBI | |
| CavFT-hAR46 | Human intramural gut wall | Severe Crohn’s disease | 2019 | USA | [35] | |
| CBBP-1 | Feces | Unknown | Unknown | Unknown | [36] | |
| CL03T12C09 | Unknown | Unknown | Unknown | Unknown | NCBI | |
| CL06T03C10 | Human feces | Unknown | 2009 | USA | [37] | |
| CL09T03C24 | Unknown | Unknown | Unknown | Unknown | NCBI | |
| CL11T00C22 | Human feces | Unknown | 2009 | USA | [37] | |
| FDAAROS_1234 | Unknown | Unknown | Unknown | Unknown | NCBI | |
| FDAARGOS_615 | Human feces | Unknown | Unknown | Unknown | Not Published | |
| FDAARGOS_759 | Human feces | Unknown | Unknown | USA | [38] | |
| NRBC 113806 | Human feces | Normal | Unknown | Unknown | NCBI | |
| 82G9 | Human feces | Unknown | Unknown | Japan | NCBI | |
| CS1 | Peritoneal fluid | Peritonitis | 2016 | France | [25] | |
| CS2 | Peritoneal fluid | Peritonitis | 2016 | France | [25] | |
| CS4 | Vulvectomy | Vulvar infection | 2016 | France | [25] | |
| CS5 | Peritoneal fluid | Peritonitis | 2016 | France | [25] | |
| CS6 | Sterility control of mesenchymal stem cells | Unknown | 2016 | France | [25] | |
| CS7 | Peritoneal fluid | Peritonitis | 2016 | France | [25] | |
| CS8 | Blood culture | Bacteremia | 2016 | France | [25] | |
| CS12 | Bone, sacrum | Osteo-articular infection | 2016 | France | [25] | |
| CS13 | Peritoneal fluid | Peritonitis | 2016 | France | [25] | |
| CS15 | Peritoneal fluid | Peritonitis | 2016 | France | [25] | |
| CS17 | Small intestine collection | Abdominal abscess | 2017 | France | [25] | |
| CS18 | Abdominal collection | Abdominal abscess | 2017 | France | [25] | |
| CS20 | Peritoneal fluid | Peritonitis | 2017 | France | [25] |
T—type strain in microbiology.
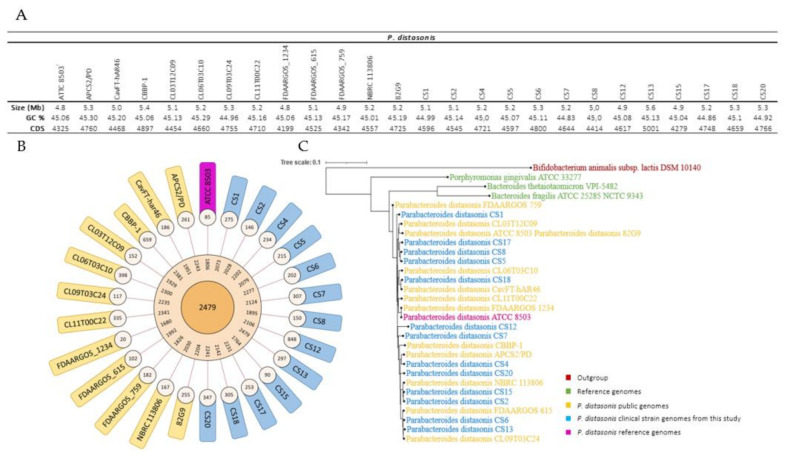
Figure 1.
P. distasonis genomes characterization. (A) General features of P. distasonis genomes used in this study. (B) Graphical representations of the pangenome characteristics. From the center outward: core, dispensable and specific genome. (C) Phylogenetic analysis of 26 strains of P. distasonis. B. thetaiotaomicron VPI-5482T [39], P. gingivalis ATCC 33277T [40] and B. fragilis ATCC 25285T [41] were added as reference genomes used in this study. Bifidobacterium animalis subsp. lactis DSM 10140T was used as outgroup genome.
The length of P. distasonis CS genomes range from ~4.8 to 5.6 Mb with an average GC content of 45.00% and a percentage of protein coding density of approximately 91.00%. The pan-genome analysis revealed 2479 functional genes presented in all strains (core-genome), between 1680 and 2479 genes presented in at least two strains function (dispensable genomes) and an average of 253 genes specific to one strain (specific genomes).
The evolutionary relationships among these strains were then investigated by constructing a phylogenetic tree based on the pairwise distances using a neighbor joining algorithm (MaGe).
The tree revealed a partial evolution of P. distasonis strains and some similarities notably with FDAARGOS_1234 and ATCC 8503T genomes that appear to be relatively closed. This genome similarity is notably highlighted by the poor specific genomes of both strains.
2.2. Identification of P. distasonis Genes Potentially Involved in Capsule, Fimbriae-like and Pilus-like Synthesis
In order to determine the potential presence of capsule, fimbriae or pili at the surface of P. distasonis, reference genes involved in their synthesis were selected from B. fragilis (gut), B. thetaiotaomicron (gut) and P. gingivalis (oral cavity), as three strictly anaerobe Gram-negative bacteria part of the Bacteroidetes phyla, and referenced as opportunistic pathogens [39,40,41]. Indeed, B. fragilis is well-known for its numerous divergent polysaccharides loci all starting by genes designated as UpxY and UpxZ families, where x goes from a to h depending on the locus. upxY genes are transcriptional antitermination factors essential to the CS synthesis, while upxZ genes inhibit their secretions [11]. P. gingivalis, for its part, is well described for its proteinaceous, filamentous appendages at its surface including fimbriae and pili, synthesized through the Fim (fimA-E) and Mfa (mfa1-5) systems, respectively [15]. A similar Mfa system including only mfa1 and mfa2 has also been described in B. thetaiotaomicron [13].
Synteny analysis of reference genes on P. distasonis genomes revealed a set of genes whose function possibly approaches that of the reference genes. To refine the search, only genes with an automatic functional assignation linked to the synthesis of the sought structures were listed (Table 2).
Table 2.
P. distasonis genes sharing synteny with reference genes and auto-assigned as part of CPS, fimbriae or pili synthesis.
| Structure | Reference Gene | Pdist Strain | Label | Length (aa) | Automatic Assignation of Biological Function | % Homology |
|---|---|---|---|---|---|---|
| Capsule | up(a-g)Y | No match | ||||
| up(a-g)Z | No match | |||||
| uphY | CS12 | PDI_v1_160022 | 185 | Transcription antitermination protein UpdY | 36.90 | |
| CL09T03C24 | AGZN01_v1_510002 | 192 | Transcription antitermination protein UpdY | 36.00 | ||
| CS4 | PDI_v1_220060 | 192 | Transcription antitermination protein UpdY | 36.00 | ||
| FDAARGOS_615 | FOB23_12755 | 179 | UpxY family transcription antiterminator | 33.72 | ||
| APCS2/PD | FQN59_13885 | 179 | UpxY family transcription antiterminator | 33.70 | ||
| CS2 | PDI_v1_140109 | 179 | Transcription antitermination protein UpdY | 33.70 | ||
| CS5 | PDI_v1_140028 | 179 | Transcription antitermination protein UpdY | 33.70 | ||
| CS6 | PDI_v1_170031 | 179 | Transcription antitermination protein UpdY | 33.70 | ||
| CS8 | PDI_v1_150106 | 179 | Transcription antitermination protein UpdY | 33.70 | ||
| CS15 | PDI_v1_340019 | 179 | Transcription antitermination protein UpdY | 33.70 | ||
| CL03T12C09 | AGZM01_v1_20031 | 179 | Transcription antitermination protein UpdY | 33.14 | ||
| FDAARGOS_759 | FIU22_01625 | 179 | UpxY family transcription antiterminator | 33.14 | ||
| 82G9 | E0E49_RS00075 | 179 | UpxY family transcription antiterminator | 33.14 | ||
| CS1 | PDI_v1_140105 | 179 | Transcription antitermination protein UpdY | 33.10 | ||
| CS7 | PDI_v1_130113 | 179 | Transcription antitermination protein UpdY | 33.10 | ||
| upgZ | No match | |||||
| Fimbriae | fimA | 82G9 | E0E49_RS19850 | 444 | fimbrial protein | 26.21 |
| ATCC 8503T | BDI_3514 | 444 | putative fimbrial protein precursor | 25.99 | ||
| CavFT-hAR46 | FE931_00755 | 444 | fimbrial protein | 25.99 | ||
| FDAARGOS_759 | FIU22_19490 | 444 | fimbrial protein | 25.99 | ||
| CS6 | PDI_v1_70115 | 432 | Fimbrial protein | 25.60 | ||
| CS13 | PDI_v1_70087 | 432 | Fimbrial protein | 25.60 | ||
| CL11T00C22 | INE94_02450 | 431 | Major fimbrium subunit FimA type-2 | 25.30 | ||
| CS12 | PDI_v1_10340 | 431 | Major fimbrial subunit protein (FimA) | 25.10 | ||
| CS1 | PDI_v1_20076 | 434 | Major fimbrial subunit protein type II | 24.90 | ||
| CS2 | PDI_v1_300040 | 419 | Fimbrial protein | 24.20 | ||
| CS15 | PDI_v1_330008 | 419 | Fimbrial protein | 24.20 | ||
| CS20 | PDI_v1_10539 | 419 | Fimbrial protein | 24.20 | ||
| APCS2/PD | FQN59_10875 | 419 | fimbrial protein | 24.10 | ||
| CS4 | PDI_v1_10167 | 419 | Fimbrial protein | 24.10 | ||
| CL06T03C10 | INE86_01122 | 420 | Major fimbrium subunit FimA type-2 | 24.00 | ||
| CS18 | PDI_v1_50210 | 420 | Fimbrial protein | 24.00 | ||
| CS8 | PDI_v1_30239 | 421 | P_gingi_FimA domain-containing protein | 23.70 | ||
| CS5 | PDI_v1_240063 | 421 | P_gingi_FimA domain-containing protein | 23.70 | ||
| CS17 | PDI_v1_20464 | 421 | P_gingi_FimA domain-containing protein | 23.70 | ||
| FDAARGOS_1234 | I6J64_10580 | 421 | fimbrial protein | 23.50 | ||
| CS7 | PDI_v1_30250 | 437 | Fimbrial protein | 23.20 | ||
| fimB | 82G9 | E0E49_RS19870 | 303 | FimB/Mfa2 family fimbrial subunit | 29.90 | |
| CBBP-1 | HHO38_19050 | 303 | FimB/Mfa2 family fimbrial subunit | 29.90 | ||
| CL06T03C10 | INE86_01123 | 303 | Fimbrillin-A associated anchor proteins Mfa1 and Mfa2 | 29.90 | ||
| FDAARGOS_1234 | I6J64_10575 | 303 | FimB/Mfa2 family fimbrial subunit | 29.90 | ||
| FDAARGOS_759 | FIU22_19510 | 303 | FimB/Mfa2 family fimbrial subunit | 29.90 | ||
| CS1 | PDI_v1_20075 | 303 | Fimbrillin-A associated anchor proteins Mfa1 and Mfa2 | 29.90 | ||
| CS6 | PDI_v1_70114 | 303 | FimB/Mfa2 family fimbrial subunit | 29.90 | ||
| CS12 | PDI_v1_10341 | 305 | Fimbrillin-A associated anchor proteins Mfa1 and Mfa2 | 29.90 | ||
| CS13 | PDI_v1_70088 | 303 | FimB/Mfa2 family fimbrial subunit | 29.90 | ||
| CL11T00C22 | INE94_02449 | 305 | Fimbrillin-A associated anchor proteins Mfa1 and Mfa2 | 29.00 | ||
| fimC | CL11T00C22 | INE94_02448 | 375 | Putative fimbrium tip subunit Fim1C | 22.50 | |
| fimD | CS2 | PDI_v1_10054 | 684 | P_gingi_FimA domain-containing protein | 26.40 | |
| CS15 | PDI_v1_140036 | 684 | P_gingi_FimA domain-containing protein | 26.40 | ||
| CS12 | PDI_v1_60229 | 685 | P_gingi_FimA domain-containing protein | 26.10 | ||
| CS17 | PDI_v1_40059 | 685 | P_gingi_FimA domain-containing protein | 26.10 | ||
| CS18 | PDI_v1_40032 | 685 | P_gingi_FimA domain-containing protein | 26.10 | ||
| CL03T12C09 | AGZM01_v1_210059 | 684 | P_gingi_FimA domain-containing protein | 26.02 | ||
| CS5 | PDI_v1_120056 | 675 | P_gingi_FimA domain-containing protein | 25.70 | ||
| CS8 | PDI_v1_160055 | 675 | P_gingi_FimA domain-containing protein | 25.70 | ||
| CS4 | PDI_v1_100056 | 677 | P_gingi_FimA domain-containing protein | 25.10 | ||
| CL09T03C24 | AGZN01_v1_280002 | 678 | P_gingi_FimA domain-containing protein | 24.76 | ||
| fimE | CL11T00C22 | INE94_03253 | 632 | Major fimbrium tip subunit FimE | 27.10 | |
| CL06T03C10 | INE86_00220 | 632 | Major fimbrium tip subunit FimE | 25.30 | ||
| CBBP-1 | HHO38_14390 | 688 | FimB/Mfa2 family fimbrial subunit | 23.01 | ||
| Pilus | Bt mfa1 | No match | ||||
| Bt mfa2 | FDAARGOS_759 | FIU22_05440 | 350 | FimB/Mfa2 family fimbrial subunit | 28.98 | |
| Pg mfa1 | CS12 | PDI_v1_130034 | 509 | Fimbrillin_C domain-containing protein | 26.50 | |
| CS18 | PDI_v1_30088 | 509 | Fimbrillin_C domain-containing protein | 26.50 | ||
| CL06T03C10 | INE86_02000 | 392 | Minor fimbrium subunit Mfa1 | 25.40 | ||
| CL11T00C22 | INE94_00002 | 509 | Major fimbrial subunit protein type IV | 25.40 | ||
| Pg mfa2 | CL06T03C10 | INE86_02001 | 329 | Minor fimbrium anchoring subunit Mfa2 | 31.20 | |
| CL11T00C22 | INE94_00003 | 329 | Minor fimbrium anchoring subunit Mfa2 | 31.20 | ||
| CS12 | PDI_v1_130033 | 329 | FimB/Mfa2 family fimbrial subunit | 30.90 | ||
| CS18 | PDI_v1_30089 | 329 | putative Minor fimbrium anchoring subunit Mfa2 | 30.40 | ||
| FDAARGOS_759 | FIU22_15640 | 300 | FimB/Mfa2 family fimbrial subunit | 24.32 | ||
| 82G9 | E0E49_RS15860 | 300 | FimB/Mfa2 family fimbrial subunit | 24.32 | ||
| Pg mfa3 | No match | |||||
| Pg mfa4 | FDAARGOS_759 | FIU22_15635 | 463 | Mfa1 fimbrilin C-terminal domain-containing protein | 20.83 | |
| ATCC 8503T | BDI_2708 | 463 | putative outer membrane protein | 20.51 | ||
| CL03T12C09 | AGZM01_v1_210028 | 463 | Fimbrillin_C domain-containing protein | 20.51 | ||
| 82G9 | E0E49_RS15855 | 463 | Mfa1 fimbrilin C-terminal domain-containing protein | 20.20 | ||
| Pg mfa5 | No match | |||||
aa: amino acid; Bt: B. thetaiotaomicron; Pg: P. gingivalis..
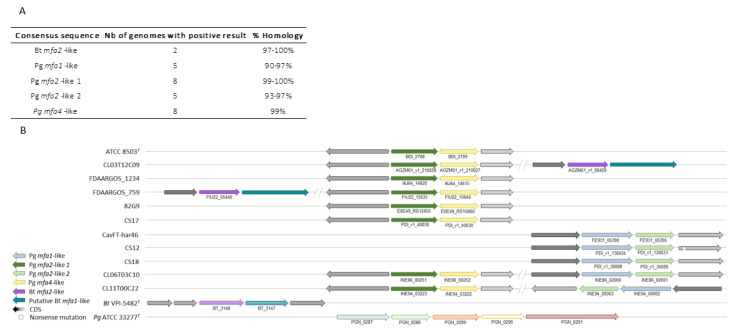
No result was found for up(a-g)Y and up(a-h)Z, while 15 genes from 15 distinct strains were referred to as potentially uphY-like with homologies ranging from 33.10% to 36.90%. Concerning the fim gene cluster, although homologies are relatively low (from 22.50% to 27.10%), all reference genes possess a synteny in at least one genome of P. distasonis with an auto-assigned function related to the fimbriae synthesis. The synteny analysis between the mfa cluster of B. thetaiotaomicron VPI-5482T and P. distasonis genomes revealed only one positive result for B. thetaiotaomicron mfa2, while the mfa gene cluster of P. gingivalis ATCC 33277T permitted the listing of multiple genes for P. gingivalis mfa1, mfa2 and mfa4. No result was found for P. gingivalis mfa3 and mfa5 genes.
Multiple sequence alignments of the listed genes (Figure S1) revealed either the conservation of one sequence (fimB-like, fimC-like, fimD-like, Bt mfa2-like, Pg mfa1-like and Pg mfa4-like) or the presence of two distinct sequences (uphY-like 1/2, fimA-like 1/2, fimE-like 1/2 and Pg mfa2-like 1/2).
2.2.1. P. distasonis Gene Cluster Potentially Involved in Capsule Synthesis
BLAST of the consensus sequences uphY-like 1 against P. distasonis genomes revealed genes with high similarity (from 99% to 100%) in 21 of the 26 studied genomes (Figure 2A). Among these genes, 15 are from the syntenic analysis while 6 are from BLAST. These last six sequences were not found during the syntenic analysis probably due to variations in their genomic organizations. On the contrary, the uphY-like 2 was identified in only three genomes with still an important sequence conservation (from 86% to 100%). Each uphY-like genomic region was then analyzed to allow the discovery of very conserved regions with a high gene homology (Figure 2B). Among them are genes linked to the CPS synthesis including glycosyltransferase, polysaccharide export, polysaccharide biosynthesis and CPS biosynthesis genes. Each CPS cluster is also composed of downstream gene encoding an integrase. The three genes similar to uphY-like 2 were analyzed and integrated at the syntenic analysis. The genomic environment of uphY-like 2 appear to be relatively close to the first loci identified with uphY-like 1, including an integrase, a glycosyltransferase, a polysaccharide export, a polysaccharide biosynthesis and a CPS biosynthesis gene, too.
Figure 2.
Identification of upxY-like gene clusters on P. distasonis genomes. (A) BLAST of upxY-like consensus sequences against P. distasonis genomes. (B) Syntenic analysis of P. distasonis capsular polysaccharide gene clusters centered on uphY-like 1 gene. Vertical and diagonal striped arrows refer to genes having a high homology and sharing synteny with reference gene. (*) indicate genes with low homology but sharing synteny over P. distasonis genomes. CDS: coding sequences are represented by gray arrows.
Specific research on P. distasonis ATCC 8503T CPS loci genomes allowed us to find a 14th CPS loci, in addition to the 13 already identified [42]. All CPS loci were then explored on other P. distasonis genomes (Table 3, gene details in Table S1). Among the 26 P. distasonis genomes, only ATCC 8503T and FDAARGOS_1234 possess the 14 CPS loci identified. Loci 3, 6, 9, 13 and 14 are shared between all P. distasonis, while only few strains possess loci 10, 11 and 12. Loci 7 and 8 are also conserved over genomes, but important intra-variations have been identified within these loci. Moreover, not all gene loci are different: 2 and 8 show high gene sequence conservation with a similar upxY-like gene. Locus 5 appear to be relatively close to 2 and 8 too, but with more variations. In the same way, the locus 4 shows some similarities with 2, 5 and 8 but has a different upxY. On the contrary, locus 13 possesses a similar upxY to 2, 5 and 8 but a different locus. Loci 3, 6, 7 and 1, 11, 12 also display similarities, especially between 6, 7 and 11, 12. Locus 1, although close to 11 and 12, presents a distinct upxY. Moreover, the conserved part of locus 9 does not always seem to be the one involved in the CPS synthesis.
Table 3.
Identification of CPS loci in 26 P. distasonis genomes and phage insertion within clusters. Color code: presence (green), partial presence (orange) or absence (red) of the CPS locus by comparison with ATCC 8503T CPS loci. Partial clusters include loci either possessing similar genes compared to ATCC 8503T loci but no upxY-like gene or an identical upxY-like gene to ATCC 8503T but a different gene locus. ● indicate loci containing phage gene insertions.
| P. distasonis | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATTC 8503T | APCS2/PD | CavFT-hAR46 | CBBP-1 | CL03T12C09 | CL06T03C10 | CL09T03C24 | CL11T00C22 | FDAARGOS_1234 | FDAARGOS_615 | FDAARGOS_759 | NBRC 113806 | 82G9 | CS1 | CS2 | CS4 | CS5 | CS6 | CS7 | CS8 | CS12 | CS13 | CS15 | CS17 | CS18 | CS20 | ||
| Capsular polysaccharide loci | 1 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |||||
| 2 | ● | ● | ● | ||||||||||||||||||||||||
| 3 | ● | ||||||||||||||||||||||||||
| 4 | |||||||||||||||||||||||||||
| 5 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |||||||||||||||
| 6 | |||||||||||||||||||||||||||
| 7 | |||||||||||||||||||||||||||
| 8 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |||||||||||||||
| 9 | ● | ● | ● | ● | |||||||||||||||||||||||
| 10 | |||||||||||||||||||||||||||
| 11 | ● | ||||||||||||||||||||||||||
| 12 | |||||||||||||||||||||||||||
| 13 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |
| 14 | |||||||||||||||||||||||||||
In addition to upxY genes, several of these CPS loci contain a phage insertion (N-acetylmuramoyl-L-alanine amidase, homolog of phage T7 lysozyme) that may modulate its expression (Table 3). Among them, CPS loci 1 and 13 of the 26 P. distasonis genomes all harbor these insertions. For CPS loci 1, this inserted segment (light blue arrows in Figure 2) is oriented in the opposite direction to upxY-like gene downstream of the CPS biosynthesis genes (red arrows in Figure 2).
2.2.2. P. distasonis Gene Cluster Potentially Involved in Fimbriae-like Synthesis
Almost all the fim-like genes investigated have been identified in the 26 P. distasonis genomes (Figure 3). The few genes not found by BLAST have been highlighted in the syntenic analysis showing fimA-E on every P. distasonis genome. Notably, BLAST of fimC-like allowed the identification of another fimC-like gene possessed by 24 of the 26 studied genomes. The identified fim-like gene cluster is composed of a various gene blocks, including one main block of four genes (fimA-like, fimB-like and fimC-like); a second block of two genes (fimD-like and fimE-like) that are always together but not located in the same region as fimA-C; and several genes showing a slight homology but a synteny with fimA-like 2, which are located sometimes in and sometimes out of the main block of genes. One nonsense mutation was found on the fimE-like gene of the CL03T12C09 that probably avoid its synthesis. Compared to the P. gingivalis fim cluster, whose genes all follow each other, P. distasonis fim-like cluster appeared to be relatively close in terms of organization, with only fimD-like and fimE-like displaying a different location.
Figure 3.
Identification of fim-like gene clusters on P. distasonis genomes compared to P. gingivalis cluster. (A) BLAST of fim-like consensus sequences against P. distasonis genomes. (B) Syntenic analysis of P. distasonis fimbriae gene clusters centered on fimA-like 1 gene. Vertical striped arrows refer to genes having a high homology and sharing synteny with reference gene. (*) indicate genes with low homology but sharing synteny over P. distasonis genomes. CDS: coding sequences are represented by gray and black arrows. Horizontal striped arrows are used for the Fim cluster of the reference genome: P. gingivalis ATCC 33277T.
2.2.3. P. distasonis Gene Cluster Potentially Involved in Pili-like Synthesis
Homologue sequences of Bt mfa2-like genes were found in only two P. distasonis genomes, while Pg mfa1-like/Pg mfa2-like 2 and Pg mfa2-like 1/Pg mfa4-like genes were found on five and eight genomes, respectively (Figure 4A). Interestingly, the five genomes containing Pg mfa1-like gene correspond to the five genomes holding Pg mfa2-like 2. In the same way, the eight genomes positive to the BLAST are the same for Pg mfa2-like 1 and Pg mfa4-like genes. The syntenic analysis of Bt mfa2-like gene (Figure 4B) revealed a conserved gene downstream of Bt mfa2-like gene showing similarities with Bt mfa1, identified as putative Bt mfa1-like gene. Some strains harbor several mfa-like clusters, such as putative Bt mfa1-like/Bt mfa2-like + Pg mfa2-like 1/Pg mfa4-like genes or Pg mfa1-like/Pg mfa2-like 2 + Pg mfa2-like 1/Pg mfa4-like genes.
Figure 4.
Identification of pili-like gene clusters on P. distasonis genomes compared to P. gingivalis and B. thetaiotaomicron cluster. (A) BLAST of mfa-like consensus sequences against P. distasonis genomes. (B) Syntenic analysis of P. distasonis pilus gene clusters centered on fimA-like 1 gene. CDS: coding sequences are represented by gray arrows. Horizontal striped arrows are used for the Mfa cluster of the reference genomes: B. thetaiotaomicron VPI-5482T and P. gingivalis ATCC 33277T.
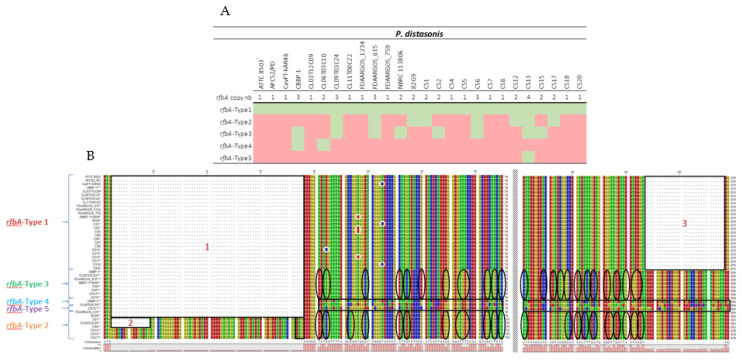
2.3. rfbA Classification and Investigation
In order to determine the potential pathogenicity of P. distasonis, the new rfbA-type classification was applied to the 26 studied genomes (Figure 5A, gene details in Table S2). The addition of new data modified the classification. A fifth group was identified and the previous gene repartition changed, notably with the presence of a rfbA-type 1 gene in all the 26 P. distasonis strains. In order to better understand the variation between each rfbA-type gene, the multiple sequence alignment of all the rfbA genes was explored (Figure 5B). The analysis revealed the presence of three gaps, two in 5′ and one in 3′. The rfbA-type 1 seems to be characterized by the presence of gaps 1 and 2, leading to a shorter rfbA sequence (876 nucleotides) with some point mutation observable. The rfbA-type 2, in addition to being characterized by the gaps 1 and 2, shows specific variations compared to the rfbA-type 1. Interestingly, a start codon ATG is observable in position 73 of every rfbA-type 2 gene that could lead to the suppression of the gap 1. The rfbA-type 3 is also identified by the gap 1 and variations from rfbA-type 1 that are relatively closed to rfbA-type 2. The gap 1 is also present for the types 4 and 5 which, however, display very unique sequences.
Figure 5.
Classification and characterization of rfbA genes of P. distasonis. (A) rfbA copy number and classification of each P. distasonis strain. Color code: presence (green) or absence (red) of the rfbA gene of the indicated type. (B) rfbA-type nucleotide sequences and gaps analysis. Gaps are framed and numerated from 1 to 3. rfbA-type 1 single-nucleotide polymorphism are surrounded in white. Variation of rfbA-type 2 and 3 from rfbA-type 1 are highlighted by black circles. rfbA-type 4 and 5 are framed in black due to the important number of variations compared to the rfbA-type 1.
2.4. Implication of P. distasonis Cell Surface Structures in Its Potential Pathogenicity
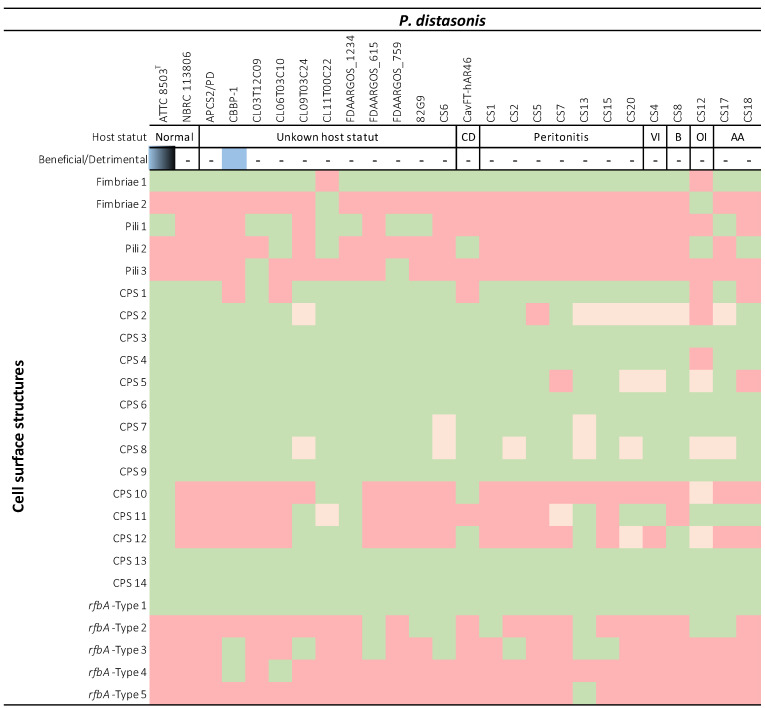
All the data generated in this study were compiled in order to determine the implication of P. distasonis cell surface markers in its potential pathogenicity (Table 4). Strains were classified as commensal (ATCC 8503T and NBRC 113806) or potential pathogens (CavFT-hAR46 and CS1-20 except CS6) on the basis of the health status of their original host (based on the isolation source of each strain, Table 1), and as beneficial or detrimental based on the literature [16,20,26,27,36]. The comparison of outer membrane structure from both categories does not bring to the fore any specific structure. Indeed, all the external structures harbored by the potential pathogen strains are identified in at least one of the commensal strains. In the same way, all structures absent from the surface of commensal strains are not systematically carried by the potential pathogens.
Table 4.
Identification of cell surface structures present on 26 P. distasonis strains based on host status. The beneficial or detrimental activity of strains (based on the literature) was added in order to compare potential pathogen from probiotic strains. Color code: beneficial properties (blue), detrimental properties (black), presence (green), partial presence (orange), absence (red). ATCC 8503T is represented as blue/black for its beneficial/detrimental activities due to various results found in the literature. Dashes (-) have been added for unknown status.
3. Discussion
The human GM and its trillion of bacteria are now well-known for their commensal and symbiotic relationships with the host. One of the GM members is P. distasonis, a Gram-negative anaerobe part of the core microbiome. While a large number of studies promotes this species as a new potential biotherapeutic product due to its multiple benefits provided to its host, controversial results have identified it as an opportunistic pathogen [22,23,24,26,27,28,29]. Although there is still a lot to understand about the mechanisms involved in the GM–host interaction, the implication of cell surface structures of GM members is now well-defined [9,43,44]. In the present study, we investigated cell surface structures of 26 P. distasonis genomes in order to better understand its maintenance within the digestive tract and its potential virulence. Among the 26 genomes, 13 new clinical strain genomes of the member of the distal gut microbiome P. distasonis were sequenced and computed on the MaGe platform. The general features of new genomes were very similar to other P. distasonis genomes already available, with an average size of 5.2 Mb and a core-genome of 2479 CDS. The phylogenetic analysis did not highlight any special difference between CS genome from this study and other P. distasonis genome with a homogenous distribution of CS genomes over the tree.
A previous investigation of CPS loci revealed the presence of the UpxY regulator on P. distasonis ATCC 8503T genome, leading to the identification of 13 putative CPS loci over its genome [42]. In our study, a 14th putative locus was identified on the ATCC 8503T genome. Although well conserved, not all 14 CPS loci are conserved over the 26 P. distasonis strains investigated in this study. Surprisingly, any of the upxY genes identified seem to be coupled with a upxZ regulator genes. However, if UpxY positively regulates B. fragilis CPS synthesis by preventing premature transcription termination in the untranslated region, UpxZ is indispensable to limit production of multiple CPSs, as described in B. fragilis and B. thetaiotaomicron [45,46]. Consequently, P. distasonis surface polysaccharide seems to result in the combination of multiple CPS loci whose expression is potentially controlled by inversions of the promoter region, leading to phase variable synthesis [47]. Th presence of phage insertions within several of these CPS loci may also modulate its expression. In addition, P. distasonis strains do not all display the same number of CPS loci and can also have sequence variations over the loci, emphasizing the strain-dependent nature of P. distasonis CPS.
In addition to external polysaccharides, another proteinaceous surface structure involved in host–microbiota interaction is the fimbriae. One of the most described fimbriae organization is the P. gingivalis Fim system strongly identified as a virulence factor [15]. A previous study identified an analogous typical pilin encoding operon on the P. distasonis ATCC 8503T genome [48]. In our work, almost all the fim-like genes investigated were identified in the 26 P. distasonis genomes, revealing the important conservation of a gene cluster involved in the fimbriae-like synthesis. Among these clusters, two distinct type of fimbriae were identified. The first one is present on 24 of the 26 studied genomes and seems to be conserved, while the second one is only harbored by two genomes. Both clusters are composed of fimA-B-C-D-E-like genes with some variations, including the putative presence of other fimA-like genes through the Fim clusters or different gene sequences such as the fimD-E of CBBP-1 and CS20 strains that display low similarity with others fimD-E. Fimbriae do not necessarily mean pathogenicity by contributing to host epithelium colonization, thus forming a protective barrier against external pathogens and stimulating the host immune system, as recently demonstrated by the recombinant pLA-K88/Lactobacillus casei strain [49].
Pili, as well as capsular polysaccharides and fimbriae, are external proteinaceous structures involved in host–GM interaction. P. gingivalis that display fimbriae also harbor pili, also called “minor fimbriae” [33]. The Mfa system of P. gingivalis involved in the pilus synthesis has also been identified in the gut commensal B. thetaiotaomicron [13]. Although partially found, no complete mfa-like gene cluster has been identified in the studied P. distasonis strains. However, 11 of the 26 genomes possess a pair of genes composed of either Bt mfa1/mfa2, Pg mfa1/mfa2 or Pg mfa4/mfa2 with mfa1/mfa4 encoding for an external polymer and mfa2 involved in the anchoring of the pilus and length regulation of Mfa1. The absence of pilus gene cluster on other genomes could be explained either by a greater diversity of pilus with pili showing important differences from the investigated ones or by the absence of pili on more than half of the studied strains. As for fimbriae, the presence of pili does not necessarily imply pathogenicity. The well-studied probiotic Lactobacillus rhamnosus GG and its spaCBA-encoded pili confirmed this by showing multiple benefits for its host despite its proteinaceous heteropolymeric extracellular appendages [50].
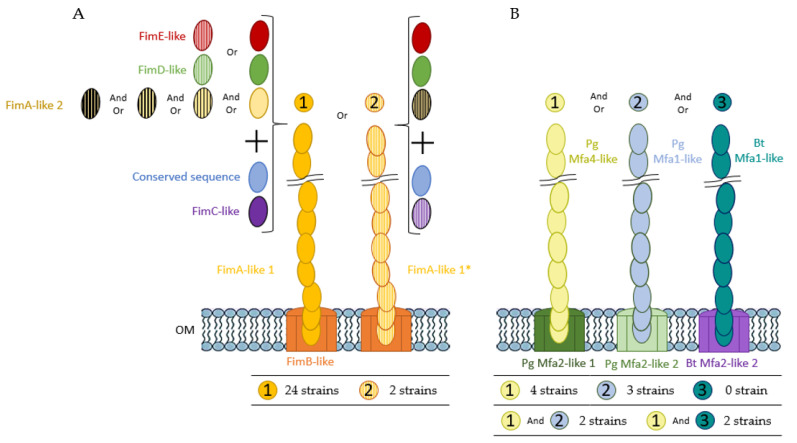
The identification of fimbriae-like and pili-like gene clusters allowed the representation of cell surface markers potentially present at the surface of P. distasonis (Figure 6). Two distinct fimbriae-like and three pili-like gene clusters have been represented depending on the gene clusters found. The first type of fimbriae (left) is harbored by 24 P. distasonis while the second one (right) is harbored by the last two studied strains (CL11T00C22 and CS12). Concerning the pili, four strains (ATCC 8503T, FDAARGOS_1234, 82G9 and CS17) harbor only the first type (left), three (CavFT-har46, CS12 and CS18) harbor only the second type (middle) and none harbor only the third type (right). Some strains also presented combination of several pilus: two (CL06T03C10 and CL11T00C22) harbor the first and second pili type and two (CL03T12C09 and FDAARGOS_759) harbor the first and third type.
Figure 6.
Hypothetical schematic representation of P. distasonis (A) fimbriae and (B) pili from the Fim and Mfa system, respectively. The different type of fimbriae and pili have been identified from 1 to 2 and from 1 to 3, respectively. The table below each structure represents the number of strains harboring the gene cluster encoding the hypothetical structure. The color code corresponds to the syntenic analysis. “Or” indicates that one P. distasonis strain can harbor only one of the proteins encoding genes concerned. For example, type 1 Fim cluster of P. distasonis contains either fimE-like or striped fimE-like genes but never both in the 24 identified clusters. “And Or” indicates that one P. distasonis strain can harbor one or several of the protein encoding genes. For example, various fimA-like 2 genes combinations can be found within type 1 Fim cluster of P. distasonis.
In order to discriminate strains regarding their LPS, all the rfbA genes of the 26 P. distasonis genomes have here been referenced, and RfbA-type classification has been applied. As described in Bank et al., 2022 [30], most of the listed rfbA genes belong to type I, highlighting the conservation of this LPS type with the ATCC 8503T rfbA belonging to the type I, and that was isolated more than 80 years ago. However, the addition of new data modified the classification with the identification of a fifth type and a new repartition of the rfbA genes within the five types. The analysis of rfbA-type variation revealed the presence of three main gaps and multiple sequence variations that shape the rfbA-type organization, including one major gap potentially non-existent due to the presence of a start codon within some rfbA gene sequences. Unlike the previous classification, the new rbfA gene repartition shows CS possessing type 1 rfbA and no specific rfbA-type allowing the distinction of CS from other P. distasonis strains. Thus, this typing does not seem to be adequate to differentiate pathogenic from non-pathogenic strains.
The comparison of outer membrane structures from commensal to potential pathogenic strains does not allow the identification of specific surface markers responsible for the putative pathogenicity of P. distasonis. The inter-strain variability observed for P. distasonis properties and potential pathogenicity could be explained by the association of all differences observed in this study, including the presence/absence of cell surface markers, loci/clusters organization and gene sequences. These variations are correlated with the phylogenetical analysis where, for example, P. distasonis ATCC 8503T and FDAARGOS_1234 strains, which are genetically close, display the exact same external structures. In the same way, FDAARGOS_759 and CL09T03C24 that are the most genetically different strains appear to differ from each other in the presence/absence of seven outer structures. Moreover, the synthesis of these external structures seems to depend on numerous factors, including genetic regulators themselves potentially contingent on environment conditions in which bacteria are evolving [47,51,52]. Thus, one plausible response to the pathogenic effects of P. distasonis is the involvement of other mechanisms than its CPS, pili, fimbriae or LPS/O-antigen membrane fractions and to the dissemination of this species from the GM to sterile sites, as an opportunistic pathogen.
Concerning P. distasonis maintenance within the GM, the presence of such external proteinaceous structures could explain its ability to adhere and persist in this complex and competitive environment. These results are consistent with our previous study that illustrates the adhesion and biofilm formation capacity of the 13 P. distasonis [25]. Although all the strains were able to adhere, an inter-strain variation was observable. These differences could be explained by a different shape of the external surface of the strains. Interestingly, the CS12 that displays the lowest adhesion capacity is also the only CS strain that does not harbor the type 1 fimbriae gene cluster identified in this study. This result could highlight the potential involvement of the type 1 fimbriae in the maintenance of P. distasonis within the GM. However, it does not seem that there is a link between the presence of a special cell surface marker for the higher adhesion or biofilm abilities. In fact, the CS1 that displays the most important adhesion capacity does not show a specific cell surface appendage that could explain this adhesion capacity. In the same way, the CS8 that has the strongest biofilm formation capacity does not show a particular cell surface structure explaining its greater capacity.
In conclusion, this work permitted the identification of several gene clusters involved in the capsule, fimbriae and pili synthesis. The presence or absence of these cell surface structures coupled with variations in gene sequences could explain P. distasonis maintenance within the GM and the inter-strain variability observed for its beneficial capacities and potential pathogenicity. However, no specific external cell surface structure that could explain P. distasonis behavior was identified. This study provides a better comprehension of the preservation of P. distasonis through the human gut and tools to better understand and characterize the beneficial/pathogenic behavior related to P. distasonis strains.
4. Materials and Methods
4.1. Whole-Genome Sequencing
The genomic DNA of the 13 CS of P. distasonis was extracted by the QiaAmpDNA MiniKit (Qiagen, Courtaboeuf, France). De Novo Microbial Genome Sequencing using Illumina technology was used to sequence the 13 CS of P. distasonis (Eurofins Genomics, Ebersberg, Germany). All genomes were integrated in the MaGe platform [53] (v3.15.3; The LABGeM, CEA/Genoscope and CNRS UMR8030).
4.2. Genome Data Used
In silico analyses of cell surface structures were performed on the 13 CS of P. distasonis and 13 public genomes available on the MaGe platform.
4.3. Pan and Core-Genome Analysis
The pan and core-genome of P. distasonis were calculated with the Pan/Core-Genome tool of MaGe, based on MicroScope gene families (MICFAM) which are computed using an algorithm implemented in the SiLiX software. The following were used as stringent parameters: 80% amino acid identity and 80% alignment coverage.
4.4. Phylogenetic Analysis
P. distasonis whole-genome sequences were used to determine the phylogenetic relationship among the isolates and public databases. Reference genomes (P. gingivalis ATCC 33277T, B. thetaiotaomicron VPI-5482T and B. fragilis ATCC 25285T) used in this study were added to the tree to demonstrate their closeness with P. distasonis. The phylogenetic tree was computed on MaGe using the Genome Clustering tool and reworked on the Interactive Tree Of Life online tool [54] (iTOL).
4.5. Comparative Genome Analysis
In order to determine the potential presence of fimbriae, pili and/or capsular polysaccharides at the surface of P. distasonis, reference genes involved in their synthesis were selected from species related to P. distasonis (Table 5).
Table 5.
Reference genes used to determine external structures of P. distasonis.
| Structure | Reference Genome | Gene | Label | Length (Aa) | Reference |
|---|---|---|---|---|---|
| Capsule | Bacteroides fragilis ATCC 25285T | upaY | BF1367 | 172 | [11,42,55] |
| upaZ | BF1368 | 157 | |||
| upbY | BF1893 | 174 | |||
| upbZ | BF1894 | 161 | |||
| upcY | BF1009 | 172 | |||
| upcZ | BF1010 | 130 | |||
| updY | BF3699 | 179 | |||
| updZ | BF3698 | 161 | |||
| upeY | BF2606 | 172 | |||
| upeZ | BF2605 | 160 | |||
| upfY | BF1549 | 199 | |||
| upfZ | BF1550 | 160 | |||
| upgY | BF0731 | 178 | |||
| upgZ | BF0732 | 162 | |||
| uphY | BF3466 | 179 | |||
| uphZ | BF3465 | 161 | |||
| Fimbriae | Porphyromonas gingivalis ATCC 33277T | fimA | PGN_0180 | 383 | [12,15] |
| fimB | PGN_0181 | 118 | |||
| fimC | PGN_0183 | 462 | |||
| fimD | PGN_0184 | 670 | |||
| fimE | PGN_0185 | 550 | |||
| Pilus | Bacteroides thetaiotaomicron VPI-5482T | mfa1 | BT_3147 | 388 | [13] |
| mfa2 | BT_3148 | 430 | |||
| Porphyromonas gingivalis ATCC 33277T | mfa1 | PGN_0287 | 563 | [14,33,48] | |
| mfa2 | PGN_0288 | 324 | |||
| mfa3 | PGN_0289 | 446 | |||
| mfa4 | PGN_0290 | 333 | |||
| mfa5 | PGN_0291 | 1228 |
Synteny enabled us to identify the conservation of homologous genes and gene order between genomes of different strains or species. Synteny blocks between references and P. distasonis genomes were investigated using the Genome Browser/Syntonome tools of MaGe and allowed the selection of a pool of genes potentially involved in the synthesis of the targeted structures. To reduce the number of P. distasonis genes and refine the search, only genes with an auto-assignation function related to the synthesis of the sought element were preserved and listed. The automatic functional assignation of MaGe follows an algorithm based on homologous relations with model organisms and completion of gene editor (gene name, product, EC numbers, roles…) using various programs or databases (RefGen, SwissProt, UniFIRE, TrEMBL…).
Multiple sequence alignments of each pool of genes related to each reference gene were then performed using CLC Viewer 8.0 to obtain one or several consensus sequences related to each reference gene.
Consensus sequences were then used for BLAST investigation against the 26 P. distasonis genomes using the Blast and Pattern Searches tool of MaGe.
Matching genes were then used to generate a syntenic block analysis between P. distasonis genome for each cell surface structure studied.
4.6. rfbA-Type Determination and Analysis
In order to determine the rfbA-type genes of the latest sequenced P. distasonis genomes, the classification method recently described by Bank et al. was used [30].
rfbA genes of P. distasonis were first referenced and aligned using CLC Viewer 8.0. The multiple sequence alignment was then used to generate a phylogenetic tree, allowing the classification of new rfbA genes.
As the rfbA-type genes obtained in this study were different from the previous classification, analyses of nucleotide sequences and gaps of distinct rfbA-type genes were performed in order to determine and better understand the variation between each type.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23169411/s1.
Author Contributions
Conceptualization, J.C., C.C.-G. and L.C.; methodology, J.C. and C.C.-G.; software, J.C. and C.C.-G.; formal analysis, J.C.; investigation, J.C. and C.C.-G.; writing—original draft preparation, J.C., C.C.-G. and L.C.; writing—review and editing, J.C., C.C.-G., L.C. and C.A.; supervision, C.C.-G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequencing data generated are indexed under the BioProject accession number PRJNA838851.
Conflicts of Interest
No potential conflict of interest was reported by the author(s).
Funding Statement
This research was funded by the French PIA project Lorraine Université d’Excellence, reference ANR-15-IDEX-04-LUE.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.O’Hara A.M., Shanahan F. The Gut Flora as a Forgotten Organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anwar H., Irfan S., Hussain G., Naeem Faisal M., Muzaffar H., Mustafa I., Mukhtar I., Malik S., Irfan Ullah M. Gut Microbiome: A New Organ System in Body. In: Antonio Bastidas Pacheco G., Ali Kamboh A., editors. Parasitology and Microbiology Research. IntechOpen; Rijeka, Croatia: 2020. [Google Scholar]
- 3.Wang H.-X., Wang Y.-P. Gut Microbiota-Brain Axis. Chin. Med. J. 2016;129:2373–2380. doi: 10.4103/0366-6999.190667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talham G.L., Jiang H.-Q., Bos N.A., Cebra J.J. Segmented Filamentous Bacteria Are Potent Stimuli of a Physiologically Normal State of the Murine Gut Mucosal Immune System. Infect. Immun. 1999;67:1992–2000. doi: 10.1128/IAI.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jandhyala S.M. Role of the Normal Gut Microbiota. WJG. 2015;21:8787. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch J.B., Hsiao E.Y. Microbiomes as Sources of Emergent Host Phenotypes. Science. 2019;365:1405–1409. doi: 10.1126/science.aay0240. [DOI] [PubMed] [Google Scholar]
- 7.Morais L.H., Schreiber H.L., Mazmanian S.K. The Gut Microbiota–Brain Axis in Behaviour and Brain Disorders. Nat. Rev. Microbiol. 2021;19:241–255. doi: 10.1038/s41579-020-00460-0. [DOI] [PubMed] [Google Scholar]
- 8.Nie L., Cai S.-Y., Shao J.-Z., Chen J. Toll-Like Receptors, Associated Biological Roles, and Signaling Networks in Non-Mammals. Front. Immunol. 2018;9:1523. doi: 10.3389/fimmu.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tytgat H.L.P., Van Teijlingen N.H., Sullan R.M.A., Douillard F.P., Rasinkangas P., Messing M., Reunanen J., Satokari R., Vanderleyden J., Dufrêne Y.F., et al. Probiotic Gut Microbiota Isolate Interacts with Dendritic Cells via Glycosylated Heterotrimeric Pili. PLoS ONE. 2016;11:e0151824. doi: 10.1371/journal.pone.0151824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligthart K., Belzer C., De Vos W.M., Tytgat H.L.P. Bridging Bacteria and the Gut: Functional Aspects of Type IV Pili. Trends Microbiol. 2020;28:340–348. doi: 10.1016/j.tim.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Patrick S., Blakely G.W., Houston S., Moore J., Abratt V.R., Bertalan M., Cerdeño-Tárraga A.M., Quail M.A., Corton N., Corton C., et al. Twenty-Eight Divergent Polysaccharide Loci Specifying within- and amongst-Strain Capsule Diversity in Three Strains of Bacteroides Fragilis. Microbiology. 2010;156:3255–3269. doi: 10.1099/mic.0.042978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano K., Hasegawa Y., Abiko Y., Yoshida Y., Murakami Y., Yoshimura F. Porphyromonas Gingivalis FimA Fimbriae: Fimbrial Assembly by FimA Alone in the Fim Gene Cluster and Differential Antigenicity among FimA Genotypes. PLoS ONE. 2012;7:e43722. doi: 10.1371/journal.pone.0043722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mihajlovic J., Bechon N., Ivanova C., Chain F., Almeida A., Langella P., Beloin C., Ghigo J.-M. A Putative Type V Pilus Contributes to Bacteroides Thetaiotaomicron Biofilm Formation Capacity. J. Bacteriol. 2019;201:e00650-18. doi: 10.1128/JB.00650-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa Y., Iwami J., Sato K., Park Y., Nishikawa K., Atsumi T., Moriguchi K., Murakami Y., Lamont R.J., Nakamura H., et al. Anchoring and Length Regulation of Porphyromonas Gingivalis Mfa1 Fimbriae by the Downstream Gene Product Mfa2. Microbiology. 2009;155:3333–3347. doi: 10.1099/mic.0.028928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa Y., Nagano K. Porphyromonas Gingivalis FimA and Mfa1 Fimbriae: Current Insights on Localization, Function, Biogenesis, and Genotype. Jpn. Dent. Sci. Rev. 2021;57:190–200. doi: 10.1016/j.jdsr.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh G.Y., Kane A.V., Wu X., Crott J.W. Parabacteroides Distasonis Attenuates Tumorigenesis, Modulates Inflammatory Markers and Promotes Intestinal Barrier Integrity in Azoxymethane-Treated A/J Mice. Carcinogenesis. 2020;41:909–917. doi: 10.1093/carcin/bgaa018. [DOI] [PubMed] [Google Scholar]
- 17.Wang K., Liao M., Zhou N., Bao L., Ma K., Zheng Z., Wang Y., Liu C., Wang W., Wang J., et al. Parabacteroides Distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019;26:222–235.e5. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 18.Li M., Wang S., Li Y., Zhao M., Kuang J., Liang D., Wang J., Wei M., Rajani C., Ma X., et al. Gut Microbiota-Bile Acid Crosstalk Contributes to the Rebound Weight Gain after Calorie Restriction in Mice. Nat. Commun. 2022;13:2060. doi: 10.1038/s41467-022-29589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfalzer A.C., Nesbeth P.-D.C., Parnell L.D., Iyer L.K., Liu Z., Kane A.V., Chen C.-Y.O., Tai A.K., Bowman T.A., Obin M.S., et al. Diet- and Genetically-Induced Obesity Differentially Affect the Fecal Microbiome and Metabolome in Apc1638N Mice. PLoS ONE. 2015;10:e0135758. doi: 10.1371/journal.pone.0135758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh G.Y., Kane A., Lee K., Xu Q., Wu X., Roper J., Mason J.B., Crott J.W. Parabacteroides Distasonis Attenuates Toll-like Receptor 4 Signaling and Akt Activation and Blocks Colon Tumor Formation in High-fat Diet-fed Azoxymethane-treated Mice. Int. J. Cancer. 2018;143:1797–1805. doi: 10.1002/ijc.31559. [DOI] [PubMed] [Google Scholar]
- 21.Kverka M., Zakostelska Z., Klimesova K., Sokol D., Hudcovic T., Hrncir T., Rossmann P., Mrazek J., Kopecny J., Verdu E.F., et al. Oral Administration of Parabacteroides Distasonis Antigens Attenuates Experimental Murine Colitis through Modulation of Immunity and Microbiota Composition. Clin. Exp. Immunol. 2011;163:250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuffaro B., Assohoun A.L.W., Boutillier D., Súkeníková L., Desramaut J., Boudebbouze S., Salomé-Desnoulez S., Hrdý J., Waligora-Dupriet A.-J., Maguin E., et al. In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides Distasonis Strains Alleviating TNBS-Induced Colitis in Mice. Cells. 2020;9:2104. doi: 10.3390/cells9092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuffaro B., Assohoun A.L.W., Boutillier D., Peucelle V., Desramaut J., Boudebbouze S., Croyal M., Waligora-Dupriet A.-J., Rhimi M., Grangette C., et al. Identification of New Potential Biotherapeutics from Human Gut Microbiota-Derived Bacteria. Microorganisms. 2021;9:565. doi: 10.3390/microorganisms9030565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiippala K., Kainulainen V., Suutarinen M., Heini T., Bowers J.R., Jasso-Selles D., Lemmer D., Valentine M., Barnes R., Engelthaler D.M., et al. Isolation of Anti-Inflammatory and Epithelium Reinforcing Bacteroides and Parabacteroides Spp. from A Healthy Fecal Donor. Nutrients. 2020;12:935. doi: 10.3390/nu12040935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamarande J., Cunat L., Caillet C., Mathieu L., Duval J.F.L., Lozniewski A., Frippiat J.-P., Alauzet C., Cailliez-Grimal C. Surface Properties of Parabacteroides Distasonis and Impacts of Stress-Induced Molecules on Its Surface Adhesion and Biofilm Formation Capacities. Microorganisms. 2021;9:1602. doi: 10.3390/microorganisms9081602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dziarski R., Park S.Y., Kashyap D.R., Dowd S.E., Gupta D. Pglyrp-Regulated Gut Microflora Prevotella Falsenii, Parabacteroides Distasonis and Bacteroides Eggerthii Enhance and Alistipes Finegoldii Attenuates Colitis in Mice. PLoS ONE. 2016;11:e0146162. doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blacher E., Bashiardes S., Shapiro H., Rothschild D., Mor U., Dori-Bachash M., Kleimeyer C., Moresi C., Harnik Y., Zur M., et al. Potential Roles of Gut Microbiome and Metabolites in Modulating ALS in Mice. Nature. 2019;572:474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 28.Moreno-Arrones O.M., Serrano-Villar S., Perez-Brocal V., Saceda-Corralo D., Morales-Raya C., Rodrigues-Barata R., Moya A., Jaen-Olasolo P., Vano-Galvan S. Analysis of the Gut Microbiota in Alopecia Areata: Identification of Bacterial Biomarkers. J. Eur. Acad. Derm. Venereol. 2020;34:400–405. doi: 10.1111/jdv.15885. [DOI] [PubMed] [Google Scholar]
- 29.Zhou C., Zhao H., Xiao X., Chen B., Guo R., Wang Q., Chen H., Zhao L., Zhang C., Jiao Y., et al. Metagenomic Profiling of the Pro-Inflammatory Gut Microbiota in Ankylosing Spondylitis. J. Autoimmun. 2020;107:102360. doi: 10.1016/j.jaut.2019.102360. [DOI] [PubMed] [Google Scholar]
- 30.Bank N.C., Singh V., Rodriguez-Palacios A. Classification of Parabacteroides Distasonis and Other Bacteroidetes Using O- Antigen Virulence Gene: RfbA -Typing and Hypothesis for Pathogenic vs. Probiotic Strain Differentiation. Gut Microbes. 2022;14:1997293. doi: 10.1080/19490976.2021.1997293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamada N., Sojar H.T., Cho M.I., Genco R.J. Isolation and Characterization of a Minor Fimbria from Porphyromonas Gingivalis. Infect. Immun. 1996;64:4788–4794. doi: 10.1128/iai.64.11.4788-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamont R.J., El-Sabaeny A., Park Y., Cook G.S., Costerton J.W., Demuth D.R. Role of the Streptococcus Gordonii SspB Protein in the Development of Porphyromonas Gingivalis Biofilms on Streptococcal Substrates. Microbiology. 2002;148:1627–1636. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 33.Nagano K., Hasegawa Y., Yoshida Y., Yoshimura F. A Major Fimbrilin Variant of Mfa1 Fimbriae in Porphyromonas Gingivalis. J. Dent. Res. 2015;94:1143–1148. doi: 10.1177/0022034515588275. [DOI] [PubMed] [Google Scholar]
- 34.Eggerth A.H., Gagnon B.H. The Bacteroides of Human Feces. J. Bacteriol. 1933;25:389–413. doi: 10.1128/jb.25.4.389-413.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F., Kumar A., Davenport K.W., Kelliher J.M., Ezeji J.C., Good C.E., Jacobs M.R., Conger M., West G., Fiocchi C., et al. Complete Genome Sequence of a Parabacteroides Distasonis Strain (CavFT HAR46) Isolated from a Gut Wall-Cavitating Microlesion in a Patient with Severe Crohn’s Disease. Microbiol. Resour. Announc. 2019;8:e00585-19. doi: 10.1128/MRA.00585-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caballero S., Kim S., Carter R.A., Leiner I.M., Sušac B., Miller L., Kim G.J., Ling L., Pamer E.G. Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus Faecium. Cell Host Microbe. 2017;21:592–602.e4. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Bayona L., Coyne M.J., Comstock L.E. Mobile Type VI Secretion System Loci of the Gut Bacteroidales Display Extensive Intra-Ecosystem Transfer, Multi-Species Spread and Geographical Clustering. PLoS Genet. 2021;17:e1009541. doi: 10.1371/journal.pgen.1009541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sichtig H., Minogue T., Yan Y., Stefan C., Hall A., Tallon L., Sadzewicz L., Nadendla S., Klimke W., Hatcher E., et al. FDA-ARGOS Is a Database with Public Quality-Controlled Reference Genomes for Diagnostic Use and Regulatory Science. Nat. Commun. 2019;10:3313. doi: 10.1038/s41467-019-11306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornton R.F., Murphy E.C., Kagawa T.F., O’Toole P.W., Cooney J.C. The Effect of Environmental Conditions on Expression of Bacteroides Fragilis and Bacteroides Thetaiotaomicron C10 Protease Genes. BMC Microbiol. 2012;12:190. doi: 10.1186/1471-2180-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bostanci N., Belibasakis G.N. Porphyromonas Gingivalis: An Invasive and Evasive Opportunistic Oral Pathogen. FEMS Microbiol. Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 41.Wexler H.M. Bacteroides: The Good, the Bad, and the Nitty-Gritty. Clin. Microbiol. Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J., Mahowald M.A., Ley R.E., Lozupone C.A., Hamady M., Martens E.C., Henrissat B., Coutinho P.M., Minx P., Latreille P., et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmela C., Chevarin C., Xu Z., Torres J., Sevrin G., Hirten R., Barnich N., Ng S.C., Colombel J.-F. Adherent-Invasive Escherichia Coli in Inflammatory Bowel Disease. Gut. 2018;67:574–587. doi: 10.1136/gutjnl-2017-314903. [DOI] [PubMed] [Google Scholar]
- 44.Garber J.M., Hennet T., Szymanski C.M. Significance of Fucose in Intestinal Health and Disease. Mol. Microbiol. 2021;115:1086–1093. doi: 10.1111/mmi.14681. [DOI] [PubMed] [Google Scholar]
- 45.Chatzidaki-Livanis M., Weinacht K.G., Comstock L.E. Trans Locus Inhibitors Limit Concomitant Polysaccharide Synthesis in the Human Gut Symbiont Bacteroides Fragilis. Proc. Natl. Acad. Sci. USA. 2010;107:11976–11980. doi: 10.1073/pnas.1005039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bechon N., Mihajlovic J., Vendrell-Fernández S., Chain F., Langella P., Beloin C., Ghigo J.-M. Capsular Polysaccharides Cross-Regulation Modulates Bacteroides Thetaiotaomicron biofilm formation. Microbiology. 2020;11:e00729-20. doi: 10.1128/mBio.00729-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patrick S., Parkhill J., McCoy L.J., Lennard N., Larkin M.J., Collins M., Sczaniecka M., Blakely G. Multiple Inverted DNA Repeats of Bacteroides Fragilis That Control Polysaccharide Antigenic Variation Are Similar to the Hin Region Inverted Repeats of Salmonella Typhimurium. Microbiology. 2003;149:915–924. doi: 10.1099/mic.0.26166-0. [DOI] [PubMed] [Google Scholar]
- 48.Xu Q., Shoji M., Shibata S., Naito M., Sato K., Elsliger M.-A., Grant J.C., Axelrod H.L., Chiu H.-J., Farr C.L., et al. A Distinct Type of Pilus from the Human Microbiome. Cell. 2016;165:690–703. doi: 10.1016/j.cell.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin D., Bai Y., Li Y., Huang Y., Li L., Wang G., Qu Y., Wang J., Yu L.-Y., Hou X. Changes in Gut Microbiota by the Lactobacillus Casei Anchoring the K88 Fimbrial Protein Prevented Newborn Piglets From Clinical Diarrhea. Front. Cell. Infect. Microbiol. 2022;12:842007. doi: 10.3389/fcimb.2022.842007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vargas García C.E., Petrova M., Claes I.J.J., De Boeck I., Verhoeven T.L.A., Dilissen E., Von Ossowski I., Palva A., Bullens D.M., Vanderleyden J., et al. Piliation of Lactobacillus Rhamnosus GG Promotes Adhesion, Phagocytosis, and Cytokine Modulation in Macrophages. Appl. Environ. Microbiol. 2015;81:2050–2062. doi: 10.1128/AEM.03949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruzzo C., Dainelli B., Ricchetti M. Piliated Bacteroides Fragilis Strains Adhere to Epithelial Cells and Are More Sensitive to Phagocytosis by Human Neutrophils Than Nonpilated Strains. Infect. Immun. 1984;43:6. doi: 10.1128/iai.43.1.189-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fletcher C.M., Coyne M.J., Bentley D.L., Villa O.F., Comstock L.E. Phase-Variable Expression of a Family of Glycoproteins Imparts a Dynamic Surface to a Symbiont in Its Human Intestinal Ecosystem. Proc. Natl. Acad. Sci. USA. 2007;104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vallenet D., Belda E., Calteau A., Cruveiller S., Engelen S., Lajus A., Le Fèvre F., Longin C., Mornico D., Roche D., et al. MicroScope—An Integrated Microbial Resource for the Curation and Comparative Analysis of Genomic and Metabolic Data. Nucleic Acids Res. 2013;41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letunic I., Bork P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coyne M.J., Kalka-Moll W., Tzianabos A.O., Kasper D.L., Comstock L.E. Bacteroides Fragilis NCTC9343 Produces at Least Three Distinct Capsular Polysaccharides: Cloning, Characterization, and Reassignment of Polysaccharide B and C Biosynthesis Loci. Infect. Immun. 2000;68:6. doi: 10.1128/IAI.68.11.6176-6181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequencing data generated are indexed under the BioProject accession number PRJNA838851.