Abstract
Prostate cancer (PCa) is the second most common cause of mortality among men. Tumor secretome is a promising strategy for understanding the biology of tumor cells and providing markers for disease progression and patient outcomes. Here, transcriptomic-based secretome analysis was performed on the PCa tumor transcriptome of Genetically Engineered Mouse Model (GEMM) Pb-Cre4/Ptenf/f mice to identify potentially secreted and membrane proteins—PSPs and PMPs. We combined a selection of transcripts from the GSE 94574 dataset and a list of protein-coding genes of the secretome and membrane proteome datasets using the Human Protein Atlas Secretome. Notably, nine deregulated PMPs and PSPs were identified in PCa (DMPK, PLN, KCNQ5, KCNQ4, MYOC, WIF1, BMP7, F3, and MUC1). We verified the gene expression patterns of Differentially Expressed Genes (DEGs) in normal and tumoral human samples using the GEPIA tool. DMPK, KCNQ4, and WIF1 targets were downregulated in PCa samples and in the GSE dataset. A significant association between shorter survival and KCNQ4, PLN, WIF1, and F3 expression was detected in the MSKCC dataset. We further identified six validated miRNAs (mmu-miR-6962-3p, mmu-miR- 6989-3p, mmu-miR-6998-3p, mmu-miR-5627-5p, mmu-miR-15a-3p, and mmu-miR-6922-3p) interactions that target MYOC, KCNQ5, MUC1, and F3. We have characterized the PCa secretome and membrane proteome and have spotted new dysregulated target candidates in PCa.
Keywords: prognostic biomarkers, prostate cancer, transcriptomic-based secretome
1. Introduction
Prostate cancer (PCa) is the most frequent cancer and has the second-highest morbidity and mortality rate among men, with 1,276,106 (7.1%) new cases globally and 358,989 (3.8%) deaths by cancer [1,2]. In the United States, the estimated number of new cases of PCa diagnosed in 2021 was 248,530, with 34,130 deaths. PCa in the United States accounts for 26% of all new cancer cases [3]. Current statistics show that one in seven men will be diagnosed with prostate cancer during their life and that one in 39 men will die of the disease [4].
The introduction of novel androgen receptor (AR) antagonists for clinical treatment has improved outcomes; however, most metastatic castration-resistant prostate cancer (mCRPC) patients ultimately develop resistance to these therapies. Patients with localized and advanced prostate tumors are sensitive to androgen deprivation therapy (ADT) and are highly curable; patients with metastatic prostate cancer acquire resistance to ADT and succumb to this disease [5]. While a large number of prostate cancer cases are diagnosed at a localized stage and are curable, metastatic prostate cancer remains fatal. In the last decade, large-scale omics analysis has revealed well-established and new master regulators and pathways involved in the metastatic and lethal behavior of PCa [6]. mCRPC is incurable, with a median survival rate f two years from diagnosis, and available treatments extend life for few months [7].
mCRPC commonly exhibits genetic alterations involving the AR, cell cycle and cell survival pathways such as the phosphatidylinositol-3-kinase (PI3K) and protein kinase B (PKB/AKT) [8,9]. One of the most frequently deleted genes in PCa, which negatively regulates PI3K-AKT signaling, is the tumor suppressor phosphatase and tensin homolog (PTEN) that is consistently associated with more aggressive forms and worse prognosis of PCa [10,11]. Loss of PTEN function has been well documented in PCa and PTEN mutations have been found in 40% metastatic PCa tumors [12,13]. Genetically Engineered Mouse Model (GEMM) Pb-Cre4/Ptenf/f mice have been used since 2003, exhibiting pathological features similar with human prostate cancers, which includes the progression from intraepithelial neoplasia to invasive well- and poor-differentiated adenocarcinoma [14]. Moreover, this model has been used to produce new GEMM by combining mutations and to explore diet manipulation effects on prostate cancer progression [15,16].
Gene expression analysis is an important tool for understanding the behavior of tumors. Gene expression signatures have been successfully applied to define subclasses of different types of cancers with different biological behaviors and responses to therapies [17,18,19,20,21]. Several studies have revealed gene expression signatures of PCa tumors that correlate with poor prognosis in retrospective analyses [22,23,24]. Some of these molecular signatures help stratify patients with a Gleason score of 7, improve prognostic prediction, and provide appropriate management plans for patients after radical prostatectomy [23,24,25].
Comprehensive studies of histological, genomic, and transcriptome analyses and their relationship with PCa are necessary. Abida et. al. (2019) [26] presented an integrative analysis of genomic alterations with expression and histological evaluation of tumors from patients with mCRPC, representing the clinical spectrum of advanced disease, and with tissues collected before and after treatment with androgen signaling inhibitors [26]. However, most molecular signatures do not require validation before clinical use. In addition, some signatures include too many genes, which are expensive and hard to use in the clinic [23]. In addition, the list of genes generated in these signatures generally does not overlap between studies, and no gene sets have been validated for clinical use [27,28,29]. The search for molecular gene signatures is based on the assumption that a clear distinction between tumors that will relapse and those that will not is possible using gene expression profiles [29]. Therefore, more studies are needed to identify and validate prognostic markers.
Therapeutic and diagnostic options for PCa are limited, and progress in drug development is delayed because most cancers are highly complex at different levels, including cellular, genomic, and metabolic. The current challenge in PCa diagnosis is the lack of alternative screening to replace the existing PCa biomarker, prostate-specific antigen (PSA). Although PSA is widely used, it cannot distinguish between indolent and aggressive PCa [30,31,32]. Therefore, exploring new types of biomarkers beyond the conventional AR and PI3K pathways and/or altered genes, such as PTEN, P53, and RB1, are highly important in prostate cancer research.
The tumor microenvironment plays an important role in the initiation and progression of tumors [33,34,35]. Transcriptome analysis revealed that stromal regions adjacent to the tumor express genes that allow for re-stratification of the tumor microenvironment [36]. Secreted and membrane proteins play an important role in cancer metastasis by stimulating cancer cell migration and invasion, consequently increasing cancer metastasis [35,36,37,38]. Therefore, investigating potential targets for diagnosis and prognosis that are available in PCa tumor stroma provides an opportunity to reframe and help treat this disease.
In this study, we used available data to perform an integrative analysis of the PCa secretome and tumor membrane proteome. Our design consisted of identifying potential biomarker targets at different stages of PCa progression, demonstrated here by the early stages of mouse Prostatic Intraepithelial Neoplasia (mPIN), Middle-stage tumor (MT), and Advanced-stage tumor (AT), focusing on the tumor microenvironment of PCa. From the list of targets (genes and proteins) available in the Human Protein Atlas (HPA) secretome, we investigated a commonly deregulated gene network in the transcriptome of Pb-Cre4/Ptenf/f mice. Enrichment analysis, protein–protein interaction (PPI) network, and in silico tools allowed us to identify nine membranes and secreted proteins that were either downregulated or upregulated in PCa. We also compared the transcriptomic profiles of prostate adenocarcinoma (PRAD) and normal tissue samples using The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) data, which revealed that four genes that encode secreted proteins were downregulated in PRAD. Finally, the gene expression patterns and prognosis of patients with PCa were analyzed by comparing four published datasets with disease outcomes (decreased relapse-free survival, overall survival, and probability of freedom from biochemical recurrence), with subsequent validation of HPA protein expression in human PCa and normal prostate samples.
2. Results
2.1. Identification of Gene Expression Profile in Prostate Cancer of Pb-Cre4/Ptenf/f Mice
We performed an integrative analysis of the prostate cancer secretome and membrane proteome data to identify clinically relevant diagnostic and prognostic biomarkers. According to the criteria used and from the list of genes included, our analysis identified upregulated and downregulated genes in the mPIN, MT, and AT distributed in all Anterior Prostate (AP), Dorsal Prostate (DP), Lateral Prostate (LP), and Ventral Prostate (VP) lobes.
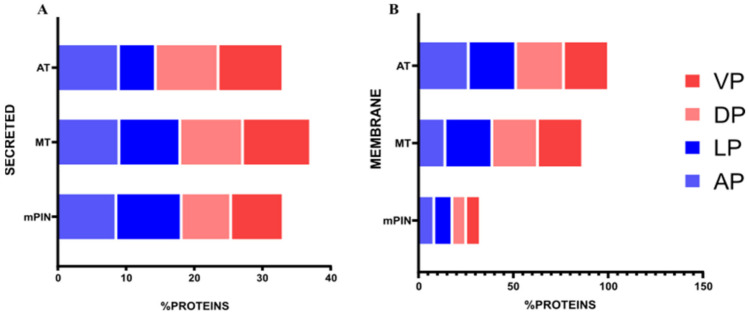
Analysis of membrane protein targets showed upregulated genes: in the AP lobe (mPIN = 403 genes; MT = 510 genes; AT = 536 genes), DP lobe (mPIN = 263 genes; MT = 319 genes; AT = 377 genes), LP lobe (mPIN = 261 genes; MT = 369 genes; AT = 421 genes), and VP lobe (mPIN = 127 genes; MT = 316 genes; AT = 303 genes). The downregulated genes for membrane proteins were as follows: in the AP lobe (mPIN = 184 genes; MT = 237 genes; AT = 192 genes), DP lobe (mPIN = 146 genes; MT = 204 genes; AT = 347 genes), LP lobe (mPIN = 245 genes; MT = 264 genes; AT = 308 genes), and VP lobe (mPIN = 88 genes; MT = 207 genes; AT = 181 genes) (Figure 1A).
Figure 1.
The proportion of membrane and secreted proteins downregulated and upregulated found in transcriptomics data of GEMM Pb-Cre4/Ptenf/f PCa. (A) Bar plot showing the proportion of membrane protein in GEMM Pb-Cre4/Ptenf/f PCa in VP, DP, LP, and AP; (B) Bar plot showing the proportion of secreted protein in GEMM Pb-Cre4/Ptenf/f PCa in VP, DP, LP, and AP. VP = Ventral prostate; DP = Dorsal prostate; LP = Lateral prostate; AP = Anterior prostate. mPIN = mouse Prostatic Intraepithelial Neoplasia; MT = Middle-stage tumor; AT = Advanced-stage tumor.
Analyses of the secreted protein targets were performed for each prostate lobe, and we found upregulated and downregulated genes in the three stages, mPIN, MT, and AT, of PCa progression. The number of upregulated genes was as follows: in the AP lobe (mPIN = 120 genes; MT = 155 genes; AT = 196 genes), DP lobe (mPIN = 49 genes; MT = 103 genes; AT = 163 genes), LP lobe (mPIN = 88 genes; MT = 141 genes; AT = 158 genes), and VP lobe (mPIN = 55 genes; MT = 128 genes; AT = 122 genes). The downregulated genes for secreted proteins were as follows: AP lobe (mPIN = 86 genes; MT = 100 genes; AT = 66 genes), DP lobe (mPIN = 69 genes; MT = 97 genes; AT = 101 genes), LP lobe (mPIN = 112 genes; MT = 87 genes; AT = 99 genes), and VP lobe (mPIN = 49 genes; MT = 69 genes; AT = 72 genes) (Figure 1B).
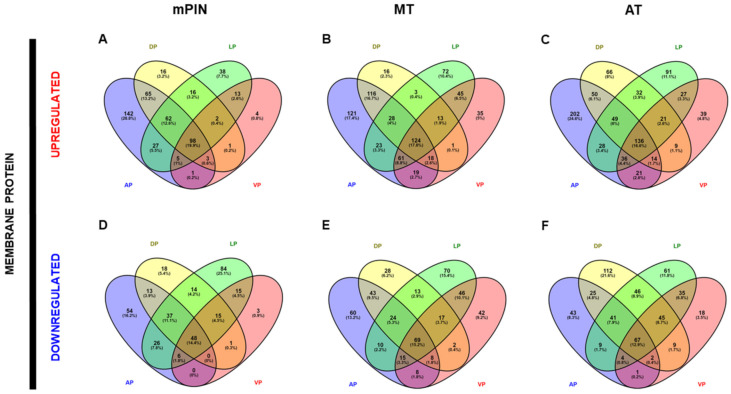
After identifying the list of upregulated and downregulated genes in each prostate lobe, we checked for shared genes present in the AP, DP, LP, and VP lobes. We identified a list of genes common to the four prostate lobes. The number of upregulated common genes for membrane proteins was as follows: in mPIN = 98 genes, MT = 124 genes, and AT = 136 genes (Figure 2A–C). The number of downregulated common genes for membrane proteins present in the four lobes was as follows: in mPIN = 48 genes, MT = 69 genes, and AT = 67 genes (Figure 2D–F).
Figure 2.
Venn’s diagrams depicting common genes of membrane proteins from Anterior Prostate (AP), Dorsal Prostate (DP), Lateral Prostate (LP), and Ventral Prostate (VP) PCa in Pten knockout mice. Venn’s diagrams of exclusive upregulated genes in mPIN (A), MT (B), and AT (C), and downregulated genes in mPIN (D), MT (E), and HT (F). mPIN = mouse Prostatic Intraepithelial Neoplasia. MT = Middle-stage tumor. AT = Advanced-stage tumor.
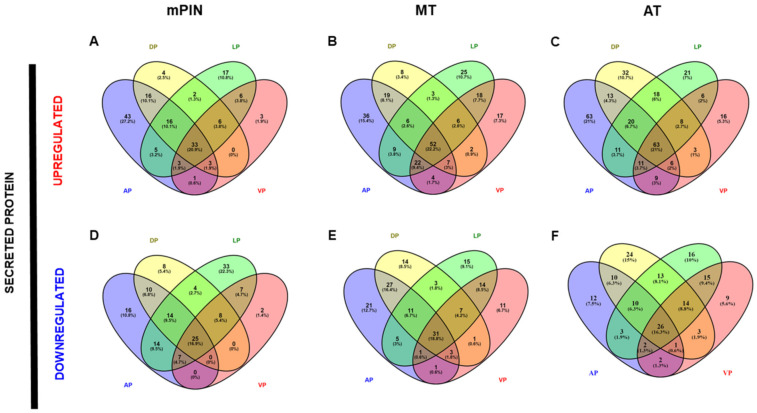
A list of secreted protein genes common to all four prostate lobes was also performed. The number of genes commonly upregulated for secreted proteins was as follows: in mPIN = 33 genes, MT = 52 genes, and AT = 63 genes (Figure 3A–C). The number of downregulated common genes for secreted proteins present in the four lobes was as follows: in mPIN = 25 genes, MT = 31 genes, and AT = 26 genes (Figure 3D–F).
Figure 3.
Venn’s diagrams depicting common genes of secreted proteins from Anterior Prostate (AP), Dorsal Prostate (DP), Lateral Prostate (LP), and Ventral Prostate (VP) PCa in Pten knockout mice. Venn’s diagrams of exclusive Upregulated genes in mPIN (A), MT (B), and AT (C), and downregulated genes in mPIN (D), MT (E), and HT (F). mPIN = mouse Prostatic Intraepithelial Neoplasia. MT = Middle-stage tumor. AT = Advanced-stage tumor.
Our list of downregulated and upregulated PCa genes in mPIN, MT, and AT was analyzed using the EnrichR platform to identify enriched ontological terms. The most significant categories were tyrosine kinase activity, an integral component of the plasma membrane for upregulated genes (Table 1 and Table 2). The most enriched terms for downregulated genes were inorganic cation transmembrane transport, potassium channel activity, and ion transmembrane transport (Table 3 and Table 4). This analysis also showed their involvement in biological processes, such as glycolysis, carbohydrate biosynthetic processes, glycosaminoglycan metabolic processes, and extracellular organization.
Table 1.
A gene ontology (GO) enrichment analysis of membrane proteins upregulated in mPIN, MT, and AT of prostate cancer.
| mPIN Upregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0005887 | Integral component of plasma membrane | 32 | 1.73 × 10−11 | 10.76299509 |
| GO:0004713 | Protein tyrosine kinase activity | 5 | 0.012138352 | 1.915840284 |
| GO:0016758 | Hexosyltransferase activity | 5 | 0.014297294 | 1.844746144 |
| GO:0043190 | ATP-binding cassette (ABC) transporter complex | 2 | 0.019180885 | 1.717131357 |
| GO:0008194 | UDP-glycosyltransferase activity | 4 | 0.024662147 | 1.607969117 |
| GO:0009312 | Oligosaccharide biosynthetic process | 3 | 0.034162979 | 1.466444268 |
| GO:0016051 | Carbohydrate biosynthetic process | 3 | 0.034162979 | 1.466444268 |
| GO:0019375 | Galactolipid biosynthetic process | 2 | 0.034162979 | 1.466444268 |
| GO:0006682 | Galactosylceramide biosynthetic process | 2 | 0.034162979 | 1.466444268 |
| GO:0046476 | Glycosylceramide biosynthetic process | 2 | 0.0382352 | 1.417536631 |
| GO:0006681 | Galactosylceramide metabolic process | 2 | 0.041917255 | 1.377607169 |
| GO:0030165 | PDZ domain binding | 3 | 0.048747995 | 1.31204324 |
| MT Upregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0005887 | Integral component of plasma membrane | 43 | 9.77 × 10−17 | 16.01024386 |
| GO:0016323 | Basolateral plasma membrane | 10 | 0.00000211 | 5.675736848 |
| GO:0004713 | Protein tyrosine kinase activity | 7 | 0.001099549 | 2.9587853 |
| GO:0019199 | Transmembrane receptor protein kinase activity | 5 | 0.002610742 | 2.5832361 |
| GO:0004714 | Transmembrane receptor protein tyrosine kinase activity | 5 | 0.002610742 | 2.5832361 |
| GO:0005254 | Chloride channel activity | 5 | 0.00268003 | 2.571860416 |
| GO:0045121 | Membrane raft | 7 | 0.002867281 | 2.542529695 |
| GO:0043005 | Neuron projection | 11 | 0.01394385 | 1.855617311 |
| GO:0007167 | Enzyme linked receptor protein signaling pathway | 7 | 0.024380271 | 1.612961464 |
| AT Upregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0005887 | Integral component of plasma membrane | 46 | 1.74 × 10−17 | 16.75829697 |
| GO:0030667 | Secretory granule membrane | 13 | 0.00000273 | 5.564593362 |
| GO:0006954 | Inflammatory response | 11 | 0.000308 | 3.511189697 |
| GO:0071345 | Cellular response to cytokine stimulus | 15 | 0.000421 | 3.375939369 |
| GO:0002283 | Neutrophil activation involved in immune response | 13 | 0.00509802 | 2.292598474 |
| GO:0004713 | Protein tyrosine kinase activity | 6 | 0.00518138 | 2.285554596 |
| GO:0070372 | Regulation of ERK1 and ERK2 cascade | 9 | 0.005657676 | 2.247361946 |
| GO:0032103 | Positive regulation of response to external stimulus | 7 | 0.006311353 | 2.199877562 |
| GO:0043410 | Positive regulation of MAPK cascade | 9 | 0.008909311 | 2.050155885 |
| GO:0030198 | Extracellular matrix organization | 9 | 0.012342256 | 1.908605459 |
| GO:0005925 | Focal adhesion | 9 | 0.013649783 | 1.864874257 |
| GO:0005789 | Endoplasmic reticulum membrane | 12 | 0.028337729 | 1.547634959 |
Table 2.
A gene ontology (GO) enrichment analysis of secreted proteins upregulated in mPIN, MT, and AT of prostate cancer.
| mPIN Upregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0019221 | Cytokine-mediated signaling pathway | 10 | 0.0000154 | 4.813747101 |
| GO:0062023 | Collagen-containing extracellular matrix | 7 | 0.0000354 | 4.451038902 |
| GO:0016019 | Peptidoglycan immune receptor activity | 2 | 0.000428 | 3.368891105 |
| GO:1902533 | Positive regulation of intracellular signal transduction | 7 | 0.001214858 | 2.915474599 |
| GO:0042834 | Peptidoglycan binding | 2 | 0.002962979 | 2.528271463 |
| GO:0008284 | Positive regulation of cell population proliferation | 6 | 0.003344125 | 2.475717449 |
| GO:0001774 | Microglial cell activation | 2 | 0.008863214 | 2.052408777 |
| GO:0030855 | Epithelial cell differentiation | 3 | 0.008882339 | 2.051472641 |
| GO:0033628 | Regulation of cell adhesion mediated by integrin | 2 | 0.01496553 | 1.824907884 |
| GO:0030198 | Extracellular matrix organization | 4 | 0.01496553 | 1.824907884 |
| GO:0010811 | Positive regulation of cell-substrate adhesion | 2 | 0.037254018 | 1.428826881 |
| GO:0060252 | Positive regulation of glial cell proliferation | 1 | 0.04201877 | 1.376556662 |
| GO:0033631 | Cell-cell adhesion mediated by integrin | 1 | 0.04201877 | 1.376556662 |
| GO:0071902 | positive regulation of protein serine/threonine kinase activity | 2 | 0.046363298 | 1.333825674 |
| MT Upregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0034774 | Secretory granule lumen | 14 | 4.44 × 10−12 | 11.35278878 |
| GO:0006954 | Inflammatory response | 13 | 2.06 × 10−11 | 10.6853178 |
| GO:0062023 | Collagen-containing extracellular matrix | 12 | 9.21 × 10−9 | 8.035890283 |
| GO:1905517 | Macrophage migration | 3 | 0.000185 | 3.732564754 |
| GO:0007160 | Cell-matrix adhesion | 3 | 0.011210456 | 1.950376724 |
| GO:0030855 | Epithelial cell differentiation | 3 | 0.011480631 | 1.940034241 |
| GO:0098632 | Cell-cell adhesion mediator activity | 2 | 0.025492739 | 1.5935835 |
| GO:0030203 | Glycosaminoglycan metabolic process | 2 | 0.031920127 | 1.495935395 |
| GO:0045545 | Syndecan binding | 1 | 0.045268101 | 1.344207727 |
| GO:0048708 | Astrocyte differentiation | 1 | 0.048279814 | 1.316234416 |
| GO:0008347 | Glial cell migration | 1 | 0.048279814 | 1.316234416 |
| AT Upregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0048018 | Receptor ligand activity | 19 | 3.70 × 10−18 | 17.43149267 |
| GO:0019221 | Cytokine-mediated signaling pathway | 23 | 5.23 × 10−16 | 15.28185137 |
| GO:0006954 | Inflammatory response | 15 | 1.64 × 10−13 | 12.78451214 |
| GO:0062023 | Collagen-containing extracellular matrix | 13 | 1.23 × 10−8 | 7.90940544 |
| GO:0008284 | Positive regulation of cell population proliferation | 13 | 2.40 × 10−7 | 6.620158329 |
| GO:0070374 | Positive regulation of ERK1 and ERK2 cascade | 9 | 3.14 × 10−7 | 6.503756602 |
| GO:0030198 | Extracellular matrix organization | 7 | 0.0000857 | 4.066941739 |
| GO:0000165 | MAPK cascade | 7 | 0.0000906 | 4.043108741 |
| GO:1905517 | Macrophage migration | 2 | 0.000194 | 3.712020847 |
| GO:0043062 | Extracellular structure organization | 5 | 0.000676 | 3.170015558 |
| GO:0042834 | Peptidoglycan binding | 1 | 0.006637813 | 2.177975013 |
Table 3.
A gene ontology (GO) enrichment analysis of membrane proteins downregulated in mPIN, MT, and AT of prostate cancer.
| mPIN Downregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0098662 | Inorganic cation transmembrane transport | 11 | 2.10 × 10−8 | 7.677498224 |
| GO:0005887 | Integral component of plasma membrane | 19 | 2.38 × 10−8 | 7.623546608 |
| GO:0016529 | Sarcoplasmic reticulum | 6 | 3.99 × 10−8 | 7.399562462 |
| GO:0042383 | Sarcolemma | 6 | 6.56 × 10−8 | 7.18327432 |
| GO:0051480 | Regulation of cytosolic calcium ion concentration | 8 | 3.05 × 10−7 | 6.516291269 |
| GO:0006874 | Cellular calcium ion homeostasis | 7 | 0.00000312 | 5.505384291 |
| GO:0070588 | Calcium ion transmembrane transport | 5 | 0.0000635 | 4.197087214 |
| GO:0005267 | Potassium channel activity | 4 | 0.002683985 | 2.571219909 |
| GO:0005217 | Intracellular ligand-gated ion channel activity | 2 | 0.004871735 | 2.312316306 |
| GO:0006939 | Smooth muscle contraction | 2 | 0.005857764 | 2.232268112 |
| GO:0005790 | Smooth endoplasmic reticulum | 2 | 0.007141408 | 2.146216169 |
| GO:0015081 | Sodium ion transmembrane transporter activity | 2 | 0.017072923 | 1.76769213 |
| MT Downregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0005267 | Potassium channel activity | 9 | 3.84 × 10−9 | 8.415844939 |
| GO:0006813 | Potassium ion transport | 10 | 8.24 × 10−9 | 8.084282366 |
| GO:0016529 | Sarcoplasmic reticulum | 7 | 9.40 × 10−9 | 8.027036253 |
| GO:0005887 | Integral component of plasma membrane | 20 | 0.00000197 | 5.704434553 |
| GO:0005789 | Endoplasmic reticulum membrane | 11 | 0.000418 | 3.37836809 |
| GO:0015085 | Calcium ion transmembrane transporter activity | 3 | 0.010602036 | 1.974610737 |
| GO:0005790 | Smooth endoplasmic reticulum | 2 | 0.018509381 | 1.732608106 |
| GO:0005355 | Glucose transmembrane transporter activity | 2 | 0.019938461 | 1.700308368 |
| AT Downregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0005887 | Integral component of plasma membrane | 25 | 2.28 × 10−10 | 9.642075486 |
| GO:0098662 | Inorganic cation transmembrane transport | 13 | 1.98 × 10−9 | 8.703210974 |
| GO:0006813 | Potassium ion transport | 10 | 2.03 × 10−9 | 8.693400143 |
| GO:0010232 | Vascular transport | 6 | 0.0000439 | 4.357272375 |
| GO:0035725 | Sodium ion transmembrane transport | 6 | 0.0000439 | 4.357272375 |
| GO:0005267 | Potassium channel activity | 6 | 0.0001 | 3.999508817 |
| GO:0016529 | Sarcoplasmic reticulum | 4 | 0.000734 | 3.134317147 |
| GO:0015079 | Potassium ion transmembrane transporter activity | 3 | 0.003470428 | 2.459616983 |
Table 4.
A gene ontology (GO) enrichment analysis of secreted proteins downregulated in mPIN, MT and AT of prostate cancer.
| mPIN Downregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0062023 | Collagen-containing extracellular matrix | 6 | 0.000153 | 3.814225913 |
| GO:0008237 | Metallopeptidase activity | 3 | 0.008113586 | 2.090787142 |
| GO:0032027 | Myosin light chain binding | 1 | 0.049310437 | 1.307061149 |
| GO:0008270 | Zinc ion binding | 3 | 0.049310437 | 1.307061149 |
| MT Downregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0062023 | Collagen-containing extracellular matrix | 7 | 0.0000450 | 4.34654643 |
| GO:0005604 | Basement membrane | 2 | 0.037653103 | 1.424199233 |
| GO:1903561 | extracellular vesicle | 2 | 0.037653103 | 1.424199233 |
| AT Downregulated Gene Ontology (GO) | ||||
| Pathway ID | Pathway Description | Gene Count | Adjusted p-Value | Log10 |
| GO:0062023 | Collagen-containing extracellular matrix | 6 | 0.000226 | 3.645111972 |
| GO:0001823 | Mesonephros development | 2 | 0.028927408 | 1.538690472 |
| GO:0004518 | Nuclease activity | 2 | 0.048024251 | 1.318539402 |
| GO:0004540 | Ribonuclease activity | 2 | 0.048024251 | 1.318539402 |
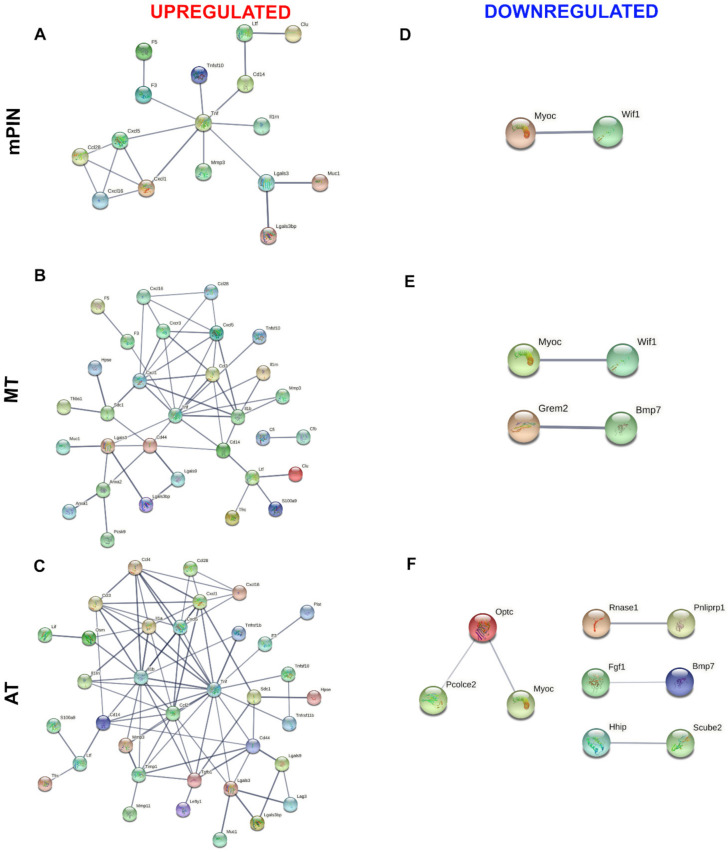
2.2. Protein–Protein Interaction (PPI) Network of Membrane and Secreted Proteins Enriched in Prostate Cancer
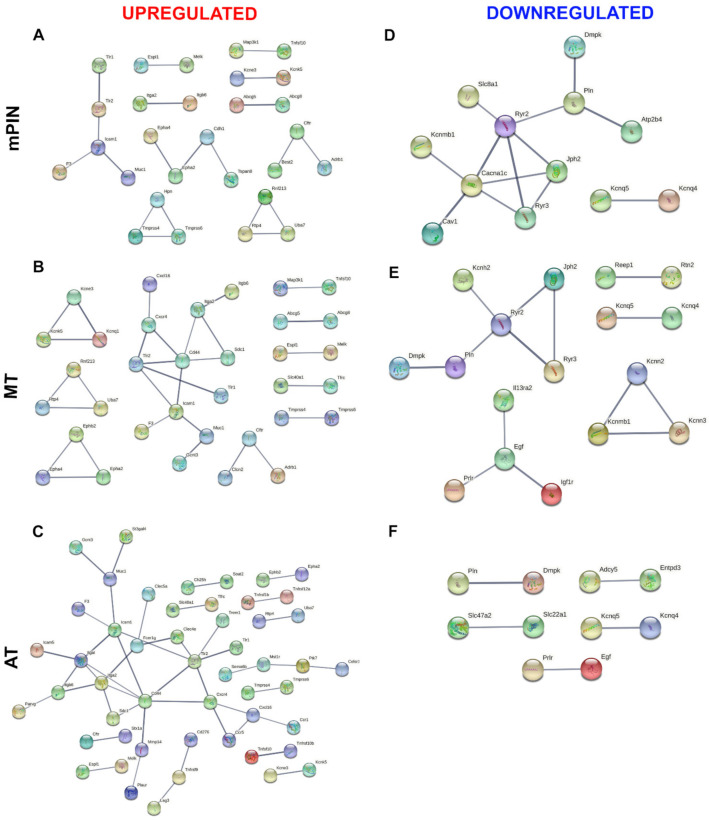
Venn diagrams showing the list of common genes identified in the four prostatic lobes of predicted membrane proteins are represented in Figure 2; upregulated and downregulated genes are shown in Figure 2A–C, and Figure 2D–F, respectively. The common genes identified in the predicted secreted proteins are represented in Figure 3 for upregulated (Figure 3A–C), and downregulated (Figure 3D–F) genes. Next, the common genes were used to generate a PPI network and GO using the STRING database (Supplementary Data). Data from this database revealed a complex interaction network among membrane proteins (Figure 4) and secreted proteins (Figure 5) with strong associations represented by thick lines of commonly upregulated genes in mPIN (Figure 4A and Figure 5A), MT (Figure 4B and Figure 5B), and AT (Figure 4C and Figure 5C); a complex interaction network with strong associations of commonly downregulated genes in mPIN (Figure 4D and Figure 5D), MT (Figure 4E and Figure 5E), and AT (Figure 4F and Figure 5F) is noted. The disconnected nodes in the network were hidden.
Figure 4.
The PPI network involves common genes analysis of a cluster of membrane proteins of GEMM Pb-Cre4/Ptenf/f PCa. PPI of commonly upregulated genes in mPIN (A), MT (B), and AT (C), and downregulated genes in mPIN (D), MT (E), and AT (F). Analysis using STRING [39] illustrates potential interactions. Proteins in the interaction network are represented as nodes connected by lines whose thickness reflects a confidence index higher than 0.7. mPIN = mouse Prostatic Intraepithelial Neoplasia; MT = Middle-stage tumor; AT = Advanced-stage tumor.
Figure 5.
The PPI network involves common genes analysis of a cluster of secreted proteins of PCa in Pten knockout mice. PPI of commonly upregulated genes in mPIN (A), MT (B), and AT (C), and downregulated genes in mPIN (D), MT (E), and AT (F). Analysis using STRING [39] illustrates potential interactions. Proteins in the interaction network are represented as nodes connected by lines whose thickness reflects a confidence index higher than 0.7. mPIN = mouse Prostatic Intraepithelial Neoplasia; MT = Middle-stage tumor; AT = Advanced-stage tumor.
GO analysis of upregulated membrane proteins revealed significant protein enrichment in the integral components of the membrane, cell surface, immune system processes, and cell surface receptor signaling pathways (Supplementary Materials Data; Figure S1). The GO terms of downregulated membrane proteins revealed significant protein enrichment in the sarcoplasmic reticulum membrane, transmembrane transporter activity, transmembrane transporter activity, and ion transport (Supplementary Materials; Figure S1).
GO analysis of upregulated secreted proteins revealed significant protein enrichment in the categories of immune system process, glycosaminoglycan metabolic process, and cell migration (Supplementary Materials; Figure S2). The GO terms of downregulated secreted proteins revealed significant protein enrichment in the extracellular region, glycosaminoglycan binding, and extracellular space (Supplementary Materials; Figure S2).
The PPI network for membrane proteins showed 28 (mPIN), 34 (MT), and 51 (AT) upregulated proteins and 12 (mPIN), 17 (MT), and 10 (AT) downregulated proteins (Figure 4). Whilst in the secreted proteins the PPI network showed 16 (mPIN), 31 (MT), and 34 (AT) upregulated proteins, while the PPI network showed 2 (mPIN), 4 (MT), and 9 (AT) downregulated proteins (Figure 5).
2.3. Differential Gene Expression of Transcripts Translated into The Membrane and Secreted Proteins in Prostate Cancer
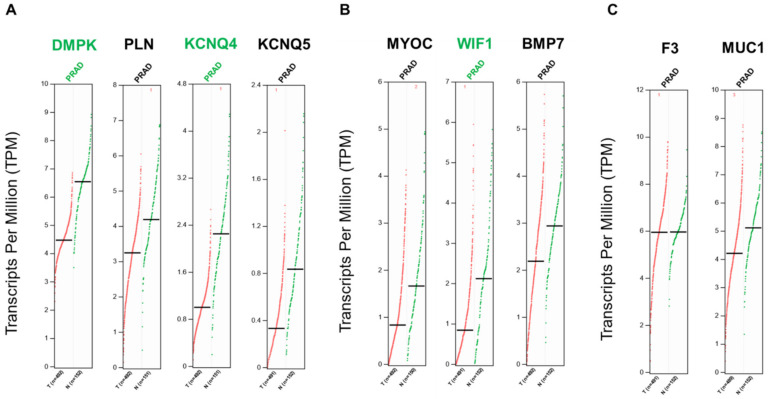
After identifying all proteins present in the prostatic PPI network in mPIN, MT, and AT and among lobes, we selected the common upregulated and downregulated protein clusters in the three stages of PCa progression. The results identified 4 downregulated membrane proteins: Myotonin-protein kinase or myotonic dystrophy protein kinase (DMPK); Phospholamban (PLN); Potassium voltage-gated channel subfamily KQT member 5 (KCNQ5) and Potassium voltage-gated channel subfamily KQT member 4 (KCNQ4). We have also found 3 downregulated secreted proteins: Myocilin (MYOC); Wnt inhibitory factor 1 (WIF1); and Bone morphogenetic protein 7 (BMP7). The results identified Tissue Factor (F3) and Mucin-1 (MUC1) common upregulated proteins were present in secreted and membrane proteins.
The gene expression levels of the targets identified in our final list were analyzed using the online Gene Expression Profiling Interactive Analysis (GEPIA) tool (Tang et al., 2017). This tool allows the comparison of transcriptome profiles from TCGA and GTEx using uniformly processed and unified RNA sequencing data from the Toil Pipeline. The expression profiles of genes encoding secreted and membrane proteins were analyzed using the GEPIA tool to identify prognostic biomarkers of PRAD. The analysis showed that three genes (DMPK, KCNQ4, and WIF1) were significantly downregulated in PRAD (Log2 fold change cutoff = 1 and q-value cutoff = 0.01) when compared to normal tissues (Figure 6).
Figure 6.
Differential gene expressions of transcripts translated into the membrane and secreted proteins in prostate cancer. (A)—Genes from downregulated membrane proteins. (B)—Genes from downregulated secreted proteins. (C)—Genes from upregulated proteins are commonly present in secreted and membrane proteins. Differential expression levels were calculated using the web based GEPIA tool [40]. GEPIA analysis revealed that genes of the proteins identified were negatively regulated in PRAD. Genes were considered positively or negatively regulated written in red and green, respectively, in PRAD (n = 489–492) relative to normal tissue (n = 150–152) when absolute values of fold-change were >1.0 and the q-value < 0.01 (ANOVA). Red dots: indicate prostate cancer tumor samples; green dots: indicate normal prostate samples.
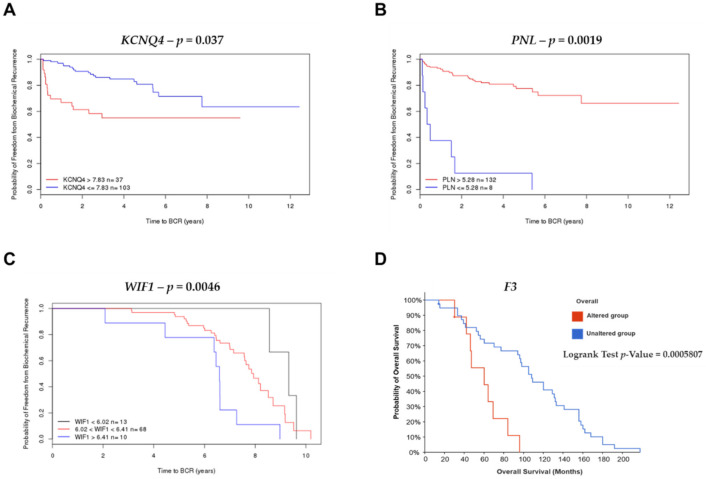
2.4. Survival Analysis and Risk Assessment
The gene (KCNQ4, PNL, F3, and WIF1) expression patterns and prognosis of patients with PCa were analyzed by comparing four published datasets (MSKCC, Cambridge, Stockholm, and MCTP). In these analyses, overexpression of the KCNQ4 gene expression with cut-off >7.83 (red line) and £7.83 (blue line) was associated with a reduced time of biochemical recurrence (p = 0.037) in the MSKCC dataset (Figure 7A). In the MSKCC and Stockholm datasets PLN (p = 0.019) expression with cut-off >5.28 (red line) and £5.28 (blue line), and WIF1 (p = 0.0046) overexpression with cut-off <6.41 (red line), and patients with WIF1 gene expression with cut-off >6.41 (blue line) were associated with a good prognosis for biochemical recurrence (Figure 7B,C). In metastatic PCa, the expression of F3 showed the worst probability of overall survival (p = 0.0005807) in the metastatic prostate adenocarcinoma (MCTP, Nature 2012) study (Figure 7D). The other genes, DMPK, KCNQ5, MYOC, BPM7, and MUC1 did not show statistical differences in the analysis of Kaplan–Meier curves for the probability of freedom from biochemical recurrence (BRC) and overall survival to PCa. A histogram of expression level of KCNQ4, PLN and WIF1 were generated with a line to indicate the recursive partitioning (RP) cut-off (Supplementary Materials).
Figure 7.
Kaplan–Meier curves displaying the probability of freedom from BRC and overall survival of PCa with (red) or without (blue) Differentially Expressed Genes (DEGs) overexpression. Analyzed by the Cambridge Carcinoma of the Prostate App (CamcAPP) (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/) [41] (accessed on 21 December 2021) and the cBioPortal for Cancer Genomics database (https://www.cbioportal.org/) (accessed on 21 December 2021) [42,43] from an integrative study. (A)—Kaplan–Meier curve with the probability of freedom from biochemical recurrence of PCa with (red) or without (blue) KCNQ4 overexpression with cut-off = 7.83 from the MSKCC study [8]; the difference was statistically significant, p = 0.037. (B)—Kaplan–Meier curve with the probability of freedom from biochemical recurrence of PCa with (red) or without (blue) PLN overexpression with cut-off = 5.28 from the MSKCC study [8]; the difference was statistically significant, p = 0.0019. (C)—Kaplan–Meier curve with the probability of freedom from biochemical recurrence of PCa with WIF1 expression with cut-off <6.02 (black line), WIF1 gene expression with cut-off <6.41 (red line), and patients with WIF1 gene expression with cut-off >6.41 (blue line) (p = 0.0046) from the Stockholm study [44]; the difference was statistically significant, p = 0.0046. (D)—Kaplan–Meier curve with the probability of overall survival of PCa patients with (red) or without (blue) F3 alteration from the metastatic prostate adenocarcinoma (MCTP, Nature 2012) study [45]; the difference was statistically significant, p = 0.0005807.
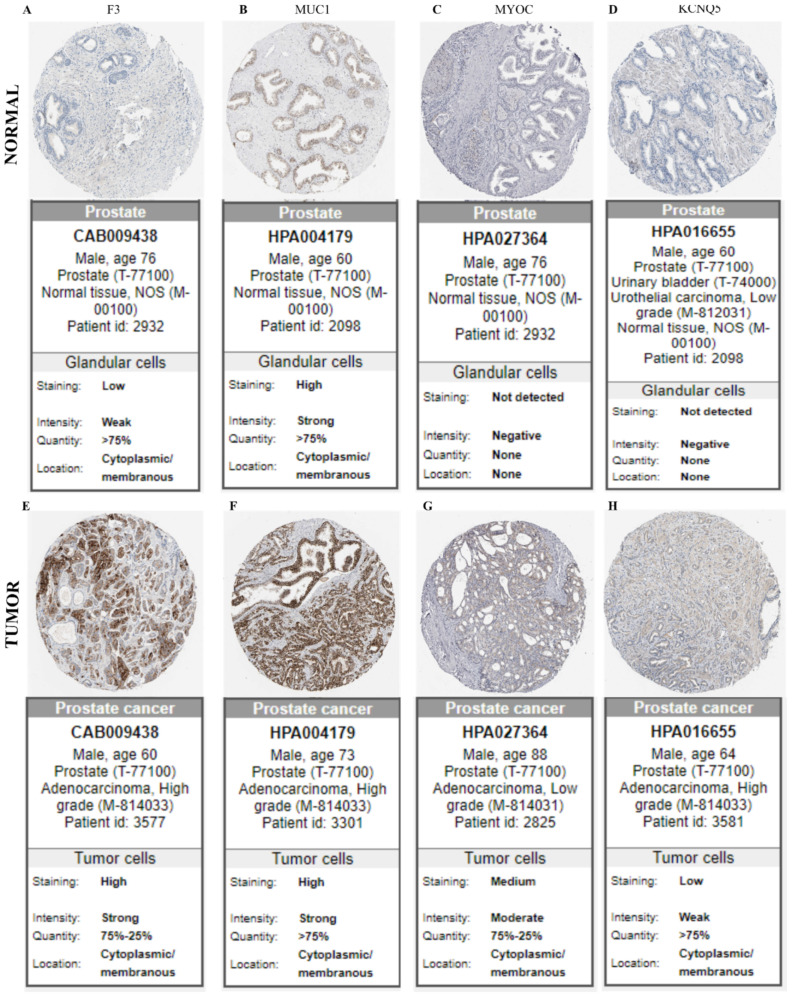
2.5. In Silico Validation of Protein Expression in Membrane and Secreted Proteins in Human Prostate Cancer
We analyzed the protein expression in human PCa and normal prostate samples from two upregulated (F3 and MUC1) and downregulated (MYOC and KCNQ5) targets found in the membrane and secreted protein list. The expression of F3 and MUC1 proteins using the HPA database showed increased (high or medium) immunostaining intensity in PCa tumor tissues; however, the expression remained low or undetectable in normal prostate tissues (Figure 8). Other dysregulated proteins identified in the HPA analyses were KCNQ4, DMPK, and PLN. At the time of this study, there was no immunohistochemistry tissue data available in the HPA database on PCa samples for BMP7 and WIF1 proteins.
Figure 8.
The validation of target genes in normal and tumor tissues. Immunostaining of normal and prostate tumor samples, respectively, for targets in our samples are: F3 (A,E); MUC1 (B,F); MYOC (C,G); and KCNQ5 (D,H), using immunohistochemical data available in the Human Protein Atlas database (https://proteinatlas.org/ accessed on 21 December 2021).
2.6. In Silico Prediction of miRNAs-mRNA Regulatory Modules in Prostate Cancer
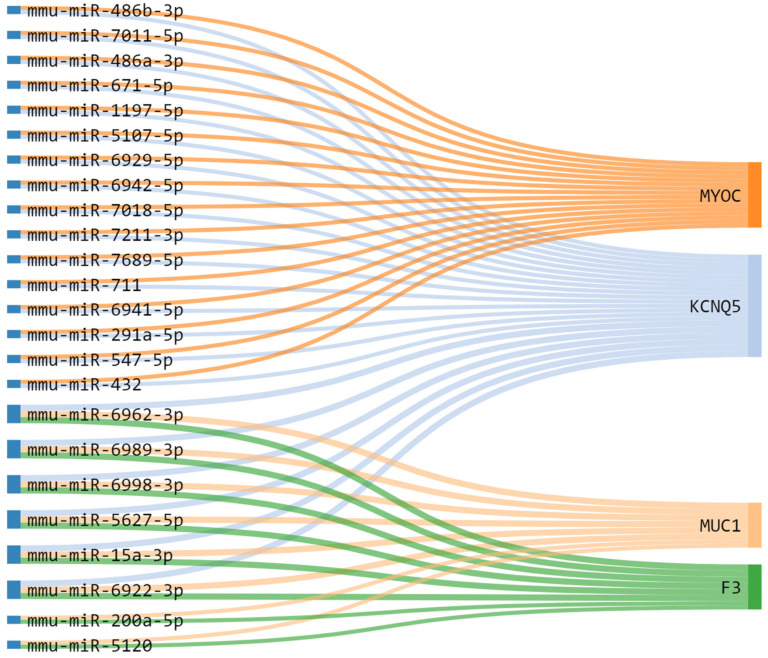
In order to obtain additional mechanistic information about the regulation of MYOC, KCNQ5, MUC1, and F3, we evaluated micro-RNAs (miRNAs) that can regulate the expression of these genes. After identifying the regulated miRNAs, using miRWalk 3.0, we selected only validated miRNAs interactions in at least three of the four DEGs. After constructing miRNA-mRNA interaction networks (Figure 9), we identified six miRNAs: mmu-miR-6962-3p, mmu-miR-6989-3p, mmu-miR-6998-3p, mmu-miR-5627-5p, mmu-miR-15a-3p, and mmu-miR-6922-3p that have a potential interaction that regulate three DEGs KCNQ5, MUC1, and F3 in PCa (Figure 9).
Figure 9.
Dysregulated targets genes and potential association with micro-RNA (miRNA) in prostate tumor tissues. Alluvial diagram connecting the representative miRNA-mRNA targets, associated with genes clusters upregulated (MUC1 and F3) and downregulated (MYOC and KCNQ5) in PRAD. Note the Highlight for mmu-miR-6962-3p, mmu-miR-6989-3p, mmu-miR-6998-3p, mmu-miR-5627-5p, mmu-miR-15a-3p, and mmu-miR-6922-3p regulating MUC1, F3 and KCNQ5.
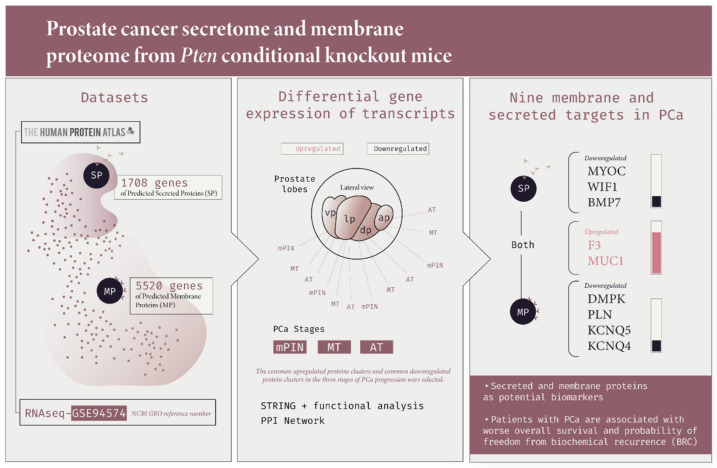
Figure 10 summarizes the main findings of the functional analysis and main results of our study.
Figure 10.
An overview of the main methodological strategies and results of prostate cancer secretome and membrane proteome from Pten conditional knockout mice and human prostate cancer. Here, we combined the selection of transcripts from the list of protein-coding genes of the secretome and membrane proteome dataset using The Human Protein Atlas Secretome and the Pten conditional knockout mice dataset (GSE 94574). After finding the upregulated and downregulated genes in the mPIN, MT and AT stages of PCa, we performed the GO enrichment analysis, PPI network using the STRING database, and the gene expression levels of the targets using the online GEPIA tool. Finally, we obtained nine (DMPK, PLN, KCNQ5, KCNQ4, MYOC, WIF1, BMP7, F3, and MUC1) deregulated PMPs and PSPs potential biomarker targets in PCa.
3. Discussion
In this study, we first analyzed the PCa transcriptome from a Pten knockout mouse model for genes encoding membrane proteins and secreted proteins in PCa. We identified important DEGs in the PCa extracellular matrix (ECM) pertaining to hitherto unexplored pathways. From an integrative analysis of the secretome and membrane proteome, we identified 9 altered targets in PCa progression stages. The gene expression profile of these markers was altered in human PCa patients with worse overall survival and a worse probability of biochemical recurrence. Our strategy was to identify potential prognostic biomarker targets at different stages of PCa progression, focusing on the tumor microenvironment of PCa.
Tumors in patients with PCa present large histological, genetic, and molecular heterogeneity. A patient may harbor more than one genomic and phenotypically distinct prostate cancer; that is, these tumors appear independently and follow separate evolutionary trajectories. These clonally independent tumors exhibit biological differences and contribute differently to disease progression and clinical outcomes [46,47,48]. Currently, data on PCa proteomics and transcriptomes, using different GEMM and human patient samples, have been explored and integrated to identify potential targets against this disease.
The tumor microenvironment is a dynamic network of cells and structures, including tumor cells. The surrounding stroma is comprised of cancer-associated fibroblasts (CAFs), immune cells, mesenchymal stem cells (MSCs), ECM, cytokines, chemokines, and growth factors secreted by these cells [33,49]. It is already known that the tumor microenvironment plays an important role in the formation and progression of metastasis. CAFs deposit and degrade ECM components and thus remodel it during cancer progression, promoting immune cell infiltration and cancer cell proliferation, migration, and invasion [33,34]. CAFs can significantly promote proliferation and migration of prostate cancer cell lines [50,51]. Studies seeking to understand and identify precise biomarker signatures are necessary to identify effective targeted therapeutics to reduce the clinical and lethal diseases related to the role of inflammation in PCa progression [52,53]. It is important to note that studies that identified biomarkers for PCa, derived from markers of stromal infiltration or stromal transcriptomic and proteomic profiles, have not pointed to any of the markers that we found in our study [54,55,56,57]. Our targets are not directly related to the inflammatory profile but rather to another class of proteins related to the tumor microenvironment and ECM.
Of note, the findings of these targets were from the PCa transcriptome of knockout animals for Pten (Pb-Cre4/Ptenf/f GEMM), in which they present important histopathological characteristics [58,59]. In addition to prostatic intraepithelial neoplastic (PIN) lesions, larger heterogeneous areas of fully invasive, both well- and poorly differentiated adenocarcinomas associated with reactive stroma are present in Pb-Cre4/Ptenf/f GEMM. This model also presents a loss of the basal membrane structure and disorganization of the smooth muscle cells but shows rare metastasis. Additionally, infiltrated inflammatory cells are commonly identified in these tumors [58,59,60].
We believe that we found a set of proteins downregulated in PCa that are biologically important in (sub)types of human cancers. Myotonin-protein kinase or myotonic dystrophy protein kinase (DMPK) is a serine/threonine-protein kinase necessary for the maintenance of muscle structure and function [61]. DMPK is mainly expressed in smooth, skeletal, and cardiac muscles, and overexpression of DMPK mediated by p53 promotes contraction of the actomyosin cortex, which leads to the activation of caspases and concomitant cell death by apoptosis [61,62]. DMPK also phosphorylated phospholamban (PLN), another downregulated protein determined in our analyses. PLN is a small, and reversibly phosphorylated transmembrane protein found in the sarcoplasmic reticulum. Depending on its phosphorylation state, PLN binds to and regulates the activity of Ca2+ pumps [63]. These two proteins are downregulated during PCa progression. We believe that this was due to the loss of smooth muscle cells [58]. Our enrichment analysis also showed changes in the sarcoplasmic reticulum membrane and ion transport, which may be related to autophagy processes, calcium homeostasis, and endoplasmic reticulum stress, as previously reported [64,65].
We also identified potassium voltage-gated channel subfamily KQT member 5 (KCNQ5) and member 4 (KCNQ4), both of which are important in regulating neuronal excitability. Voltage-gated potassium channels are responsible for the repolarization phase of the membrane action potential and play crucial roles in the excitability of neurons and other cells (Li et al., 2021). Several studies have proposed the use of KCNQ5 gene for the early clinical detection of colorectal precancerous lesions and cancer [66,67,68]. Downregulated expression of KCNQ5 has also been observed in other diseases [69,70]. These proteins play an important role in potassium homeostasis and are related to the enriched terms of potassium channel activity and potassium ion transport at different levels of PCa progression presented in our results. In our analysis, patients with altered KCNQ4 and PLN genes showed the shortest time for biochemical recurrence. The downregulation of these genes may be related to PCa progression.
In addition to membrane proteins, Myocilin (MYOC) was identified in our study. MYOC is a secreted glycoprotein that regulates the activation of different signaling pathways in adjacent cells to control different processes, including cell adhesion, cell-matrix adhesion, cytoskeleton organization, and cell migration [71]. Mutations in the MYOC gene are an important cause of glaucoma with dominant inheritance (Liuska [72,73]. However, other types of cancer, such as thymoma, exhibit MYOC downregulation, thereby corroborating our results [74].
Wnt inhibitory factor 1 (WIF1) is a secreted protein that binds to WNT proteins and inhibits their activities. WNT signaling mainly controls cell proliferation, differentiation, and maintenance of stem cells (β-catenin-dependent pathway), cell polarity, and migration (β-catenin-independent signaling). The WNT/Ca2+ signalling pathway is also associated with the release of Ca2+ from intracellular stores [75,76]. A large body of evidence has shown that activation of the WNT signaling pathway contributes to the proliferation and transformation of malignant cells with metastatic activity [77,78]. The WNT protein is regulated by a variety of secreted extracellular proteins that interfere with the formation of the WNT-receptor complexes. Extracellular inhibition of the WNT signaling pathway, WIF1, plays an important role in controlling cell proliferation and acts as a tumor suppressor [79]. Owing to its biological function, interest in using WIF1 as a biomarker for the early detection, diagnosis, and prognosis of cancer has increased in recent years [80,81,82,83]. As shown here, WIF1 was associated with favorable overall survival in PCa, corroborating other studies [84].
Bone morphogenetic protein 7 (BMP7) (https://www.uniprot.org/uniprot/P18075 (accessed on 21 December 2021)) is a growth factor of the TGF-β superfamily that plays important role in various biological processes, including proliferation, differentiation, and apoptosis in many different cell types [85,86]. Bone morphogenetic proteins can act as either tumor suppressors or oncogenes, depending on the cellular context and tumor type [87,88]. Studies have suggested that BPM7 inhibition may represent a target for overcoming resistance to cancer immunotherapies [85], and the use of BPM7 overexpression is a strong predictor of the risk of tumor recurrence in gastric cancer [87].
The upregulated gene in the common membrane and secreted proteins, tissue factor (F3) (https://www.uniprot.org/uniprot/P13726 (accessed on 21 December 2021)), is a transmembrane glycoprotein and primary initiator of the extrinsic blood coagulation cascade and ensures rapid hemostasis in case of organ damage [89]. F3 has been associated with strong tumor growth enhancement and poor prognosis in cancer [90]. In our analyses, we found that PCa patients with altered F3 gene expression had reduced survival rate. F3 expression is increased in tumors and is associated with tumor progression, particularly in pancreatic [91,92], cervical [93], breast [94], and prostate cancers [95].
The transmembrane glycoprotein Mucin-1 (MUC1) is highly glycosylated and is normally expressed in glandular and luminal epithelial cells. MUC1 provides protection and creates a physical barrier to negatively charged sugars, limiting accessibility, and preventing pathogenic colonization [96,97]. MUC1 is overexpressed and has been identified as a potential target for diagnosis, prognosis, and therapy in most human cancers and plays an important role in tumor progression [96,97,98,99,100]. Recently, we reported a family of deregulated mucins, including MUC1, in PCa progression, where it was shown that mucin cells (mucinous metaplasia) are in AR-negative areas of proliferation, and that mucin-associated genes have a worse prognosis in PCa and have significant prognostic value for PCa patients [58].
miRNA controls gene expression by targeting mRNA based on sequence complementarity and can serve as oncomiR or tumor suppressor miRs by targeting mRNA that encode oncoproteins or tumor suppressor proteins [101,102]. Using miRNA-mRNA tumor expression data, we identified deregulated miRNA that was validated in the regulatory networks of the four target genes. Some miRNAs found in our analysis that regulate MYOC, KCNQ5, MUC1, and F3 mRNAs has been also described in other cancers, such as colorectal cancer cells [103] and identified as biomarkers in lung cancer [104], osteosarcoma [105], ovarian cancer [106] and penile cancer [107].
The miR-15a-3p miRNA has been associated with the three DEGs (KCNQ5, MUC1, and F3) in PCa in our analysis. The miR-15a-3p has been shown to suppress proliferation and migration inhibiting the expression of BCL2 and MCL1 in epithelial cells [108] and restrains the growth and metastasis of ovarian cancer cells by regulating Twist1 [106]. Evidence has shown that miR-15a-3p overexpression also suppressed cell proliferation via down-regulating Wnt/β-catenin signaling in PCa cells [109]. Moreover, the mir-671-5p was previously described to function as a tumor suppressor, inhibiting tumor proliferation by blocking cell cycle in osteosarcoma and negatively regulates SMAD3 to inhibit migration and invasion of osteosarcoma cells [110,111]. The mir-671-5p interacts with MYOC and KCNQ5 and may be down-regulating the expression of these genes in PCa.
Here, our strategy consisted of selecting upregulated and downregulated membrane and secreted proteins identified in PCa transcriptome analysis, which stratifies a group of unexplored proteins with high prognostic value and a potential target for therapy in a subgroup of patients. Although we tried to avoid bias in our study, certain limitations still need to be considered. Experimental in vivo and in vitro analysis should be performed to confirm our findings. Investigate the role and function of these miRNAs in PCa and their regulation of these genes are required. The experimental validation of the membrane and secreted proteins identified in this study could help correlate the results obtained herein with another group of patients’ prognoses, diagnosis, and/or overall survival. Despite the above limitations, we have demonstrated a well-characterization of secretome and membrane proteome and have spotted new dysregulated target candidates in PCa.
4. Materials and Methods
4.1. Analysis of RNA-Seq Data of the Genetically Engineered Mouse Model (GEMM) for PCa: The Pten Conditional Knockout
We used RNA-seq data derived from the analysis of samples from the four prostatic lobes obtained from the GEMM Ptenf/f, control, and Pb-Cre4/Ptenf/f mice. We accessed RNA sequencing data derived from all prostate lobes using the NCBI Gene Expression Omnibus platform (GEO, https://www.ncbi.nlm.nih.gov/geo/ (accessed on 21 December 2021)), reference number GSE94574. Briefly, 72 samples were submitted for RNA-seq analysis, including 20 prostate samples from wild-type (WT), 16 mouse prostatic intraepithelial neoplasia (mPIN), 20 well-differentiated tumors (middle-stage tumor, MT), and 16 poorly differentiated tumors (advanced-stage tumor, AT). A minimum of four samples for each prostatic lobe and pathological condition for RNA-seq analysis were used. A detailed description of the histopathological aspects of each prostatic lobe and tumor stage of the mouse model has been previously described [59,112]. First, we explored genes that were differentially expressed in each lobe and at different stages of tumor progression (mPIN, MT and AT); we used Log2FC ≥ |+1| ≤ |−1| and adjusted the p-value < 0.05. The transcriptome used in this study was generated from animals provided by Dr. David Neal with experiments approved by the CRUK Institute Ethics Committee of the Cambridge University, UK, under design license 80/2435, and by the Ethics Committee on Animal Experimentation of the Institute of Biosciences of Botucatu (IBB)—UNESP, Brazil (Protocol CEUA 613/2014, 1145/2019 and 4921200721).
4.2. Integration of Secretome and Membrane Proteome Analyses to Identify Prostate Cancer Biomarkers
The DEGs from RNA-seq were used to predict membrane and secretome proteins using a known list of 5520 genes of Predicted Membrane Proteins and 1708 genes of Predicted Secreted Proteins available in The Human Protein Atlas Secretome (https://www.proteinatlas.org/humanproteome/tissue/secretome) [113,114] (accessed on 21 December 2021). The Predicted Membrane Proteins are a selection of seven prediction algorithms used to create a majority decision-based method (MDM) using the combined results from the chosen tools to estimate the human membrane proteome [115]. The human secretome was predicted by a whole-proteome scan using three methods for signal peptide prediction: SignalP4.0, Phobius, and SPOCTOPUS, which have all been shown to produce reliable prediction results in comparative analysis and selected the genes that were altered in at least three prostatic lobes in mPIN, MT, and/or AT.
4.3. Protein–Protein Interaction Network and Functional Enrichment Analysis
We used EnrichR software from Ma’ayan Lab (https://maayanlab.cloud/Enrichr/) (accessed on 21 December 2021) to determine the enrichment of ontological terms and molecular pathways related to DEGs [116,117]. The cutoff criteria used for both analyses were adjusted to a p-value of ≤ 0.05. Gene ontology (GO) enrichment analysis of secreted and membrane proteins was grouped into a single list. The ontological terms downregulated and upregulated in the biological process, molecular function, and cellular component categories with the lowest adjusted p-values were selected.
We used the STRING database (https://string-db.org/) [39] to identify the protein–protein interaction (PPI) network by individually analyzing the upregulated and downregulated genes. The minimum interaction score required was 0.700 (high confidence), and the nodes disconnected from the network were hidden to simplify the display. The PPI enrichment p-value indicated the statistical significance provided by STRING (accessed on 21 December 2021).
The ShinyGO application (version 0.741) (http://bioinformatics.sdstate.edu/go/) [118] (accessed on 21 December 2021) was used to explore the enrichment of ontological terms in GO (http://geneontology.org/) (accessed on 21 December 2021) categories for the biological process of proteins from PPI. The cut-off criterion used for both analyses was a false discovery rate (FDR) p-value < 0.05.
4.4. Gene Expression Profile in Prostate Cancer
Differential expression levels were calculated using a web-based gene expression profiling analysis (GEPIA) tool [40]. GEPIA analysis revealed that genes encoding secreted proteins are regulated in PRAD (http://gepia.cancer-pku.cn/detail.php) (accessed on 21 December 2021). DEGs between tumor and normal samples were determined by one-way analysis of variance (ANOVA), applying the log2 fold-change > 1 and q-value < 0.01. Genes were considered positively or negatively regulated and indicated in red and green, respectively, in PRAD (n = 489–492) relative to normal tissue (n = 150–152).
4.5. Survival Analysis and Risk Assessment
After identifying the secretome and membrane proteome targets of knockout mice, we performed analyses using data from publicly available databases. We investigated gene expression using the Cambridge Carcinoma of the Prostate App (CamcAPP) database and developed the CamcAPP (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/) [41] (accessed on 21 December 2021) and the cBioPortal for Cancer Genomics database (https://www.cbioportal.org/) [42,43] (accessed on 21 December 2021), to determine the association of gene alterations with patient clinical data, such as tumor risk development, prognosis, and survival rates. Survival curves were constructed using the Kaplan–Meier method. The expression of genes was associated with disease outcomes (decreased relapse-free survival and an increased expression level of genes in advanced prostate cancer) in several published PCa datasets, namely Memorial Sloan-Kettering Cancer Center (MSKCC) [8] and Cambridge and Stockholm integrative studies [44] performed using CamcAPP [41]. The published PCa datasets used was Metastatic Prostate Adenocarcinoma (MCTP, Nature 2012) [45] performed using the cBioPortal for Cancer Genomics database [42,43]. The grouping of samples found by recursive partitioning (RP) was used to construct a Kaplan-Meier plot by CamcAPP.
4.6. In Silico Validation of Differentially Expressed Genes (DEGs)
After gene expression analysis, the deregulated genes identified in our analysis of GEMM Pb-Cre4/Ptenf/f PCa were assessed using the HPA (https://www.proteinatlas.org/) database (accessed on 21 December 2021) [113,114] to identify the distribution and localization of proteins in normal and tumor prostate samples via immunohistochemistry.
4.7. Prediction of Commonly Dysregulated miRNAs-mRNA Targets
We used the miRWalk 3.0 tool (http://mirwalk.umm.uni-heidelberg.de/) [119] to perform the regulatory interaction between miRNA and mRNA (MYOC, KCNQ5, MUC1, and F3); the algorithm for target validation was used in other available databases of Homo sapiens. The miRNA was considered significant when involved with at least three of the four genes selected. An alluvial plot diagram was generated using the online tool SankeyMATIC (http://sankeymatic.com/) to demonstrate the interaction networks between the miRNA and mRNA.
4.8. Data Representation and Analysis
Venn diagrams were plotted using Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/) [120]. The STRING database was used (https://string-db.org/) [39] to construct a protein-protein interaction network. Differential expression levels were calculated using the web based GEPIA [40] (http://gepia.cancer-pku.cn/detail.php). The CamcAPP database (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/) [41] and cBioPortal for Cancer Genomics database (https://www.cbioportal.org/) [42,43] were used for survival analysis.
Acknowledgments
We thank David Neal’s Uro-Oncology group (CRUK Cambridge Institute, University of Cambridge, UK) for providing genetically engineered mouse model FFPE samples. We thank Carolina Frandsen Pereira da Costa (@Clorofreela_Sci—https://clorofreela.com/) for the great work on the illustrations. This article represents part of NJS’s Ph.D. thesis (Protocol 4921200721 and CNPq grant number: 142485/2019-5) conducted at the Institute of Biosciences of Botucatu, São Paulo State University (UNESP), Botucatu, São Paulo, Brazil, and Institute of Biology (IB), University of Campinas (UNICAMP), Campinas, São Paulo, Brazil.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23169224/s1. Reference [121] are cited in the supplementary materials.
Author Contributions
Conception and design: N.J.S. and A.C.L.C.; development of methodology: N.J.S. and A.C.L.C.; data acquisition: N.J.S. and A.C.L.C.; analysis and interpretation of data: N.J.S., H.F.C., L.A.J. and S.L.F.; manuscript writing and editing: N.J.S., L.A.J., H.F.C. and S.L.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The animal study protocol was approved by the CRUK Institute Ethics Committee of the Cambridge University, UK, under design license 80/2435, and by the Ethics Committee on Animal Experimentation of the Institute of Biosciences of Botucatu (IBB)—UNESP, Brazil (Protocol CEUA 613/2014, 1145/2019 and 4921200721).
Informed Consent Statement
Not applicable.
Data Availability Statement
The results presented in this article are partly based on data generated by the Cambridge Carcinoma of the Prostate App (CamcAPP) (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/) from the Cancer Research UK Cambridge Institute, cBioPortal for Cancer Genomics database (https://www.cbioportal.org/), and Human Protein Atlas (HPA) (https://www.proteinatlas.org/) database. The RNA sequencing data derived from all prostatic lobes of Pten knockout are available in the NCBI Gene Expression Omnibus platform (GEO, https://www.ncbi.nlm.nih.gov/geo/, accessed on 21 December 2021), reference number GSE94574. The datasets presented in this study can be found in online repositories on gene expression profiling interactive analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html, accessed on 21 December 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The Brazilian National Council for Scientific and Technological Development (CNPq) [grant numbers: 310805/2018-0; 142485/2019-5; 465699/2014-6] and São Paulo State Research Foundation (FAPESP) [grant numbers: 2019/19644-1; 2019/05952-6] financially supported this work. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Schatten H. Cell & Molecular Biology of Prostate Cancer. Volume 1095. Springer; Cham, Switzerland: 2018. Brief Overview of Prostate Cancer Statistics, Grading, Diagnosis and Treatment Strategies; pp. 1–14. [DOI] [PubMed] [Google Scholar]
- 5.Litwin M.S., Tan H.J. The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA. 2017;317:2532–2542. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- 6.Quigley D.A., Dang H.X., Zhao S.G., Lloyd P., Aggarwal R., Alumkal J.J., Foye A., Kothari V., Perry M.D., Bailey A.M., et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell. 2018;174:758–769.e9. doi: 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain M., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K.N., Sartor O., Agarwal N., Olmos D., et al. Survival with Olaparib in Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020;383:2345–2357. doi: 10.1056/NEJMoa2022485. [DOI] [PubMed] [Google Scholar]
- 8.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhury A.D. PTEN-PI3K pathway alterations in advanced prostate cancer and clinical implications. Prostate. 2022;82((Suppl. 1)):S60–S72. doi: 10.1002/pros.24372. [DOI] [PubMed] [Google Scholar]
- 10.Pungsrinont T., Kallenbach J., Baniahmad A. Role of PI3K-AKT-mTOR Pathway as a Pro-Survival Signaling and Resistance-Mediating Mechanism to Therapy of Prostate Cancer. Int. J. Mol. Sci. 2021;22:1088. doi: 10.3390/ijms222011088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi Z., Xu Z., Zhang L., Zou Y., Li J., Yan W., Li C., Liu N., Wu H. Overcoming resistance to immune checkpoint therapy in PTEN-null prostate cancer by intermittent anti-PI3Kα/β/δ treatment. Nat. Commun. 2022;13:182. doi: 10.1038/s41467-021-27833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamaspishvili T., Berman D.M., Ross A.E., Scher H.I., De Marzo A.M., Squire J.A., Lotan T.L. Clinical implications of PTEN loss in prostate cancer. Nat. Rev. Urol. 2018;15:222–234. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnham D.J., Bullock N., Dass M.S., Staffurth J.N., Pearson H.B. The PTEN Conundrum: How to Target PTEN-Deficient Prostate Cancer. Cells. 2020;9:2342. doi: 10.3390/cells9112342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S., Gao J., Lei Q., Rozengurt N., Pritchard C., Jiao J., Thomas G.V., Li G., Roy-Burman P., Nelson P.S., et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/S1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 15.Irshad S., Abate-Shen C. Modeling prostate cancer in mice: Something old, something new, something premalignant, something metastatic. Cancer Metastasis Rev. 2013;32:109–122. doi: 10.1007/s10555-012-9409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M., Zhang J., Sampieri K., Clohessy J.G., Mendez L., Gonzalez-Billalabeitia E., Liu X.S., Lee Y.R., Fung J., Katon J.M., et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 2018;50:206–218. doi: 10.1038/s41588-017-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Oliveira G., Freire P.P., Cury S.S., de Moraes D., Oliveira J.S., Dal-Pai-Silva M., Reis P.P., Carvalho R.F. An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma. Cancers. 2020;12:716. doi: 10.3390/cancers12030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latha N.R., Rajan A., Nadhan R., Achyutuni S., Sengodan S.K., Hemalatha S.K., Varghese G.R., Thankappan R., Krishnan N., Patra D., et al. Gene expression signatures: A tool for analysis of breast cancer prognosis and therapy. Crit. Rev. Oncol. Hematol. 2020;151:102964. doi: 10.1016/j.critrevonc.2020.102964. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Wu L., Ao H., Zhao M., Leng X., Liu M., Ma J., Zhu J. Prognostic implications of autophagy-associated gene signatures in non-small cell lung cancer. Aging. 2019;11:11440–11462. doi: 10.18632/aging.102544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tonella L., Giannoccaro M., Alfieri S., Canevari S., De Cecco L. Gene Expression Signatures for Head and Neck Cancer Patient Stratification: Are Results Ready for Clinical Application? Curr. Treat. Options Oncol. 2017;18:32. doi: 10.1007/s11864-017-0472-2. [DOI] [PubMed] [Google Scholar]
- 21.Venet D., Dumont J.E., Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput. Biol. 2011;7:e1002240. doi: 10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu D., Jiang L., Luo S., Zhao X., Hu H., Zhao G., Tang W. Development of an autophagy-related gene expression signature for prognosis prediction in prostate cancer patients. J. Transl. Med. 2020;18:160. doi: 10.1186/s12967-020-02323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penney K.L., Sinnott J.A., Fall K., Pawitan Y., Hoshida Y., Kraft P., Stark J.R., Fiorentino M., Perner S., Finn S., et al. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J. Clin. Oncol. 2011;29:2391–2396. doi: 10.1200/JCO.2010.32.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao N., Tang H., Mi Y., Zhu Y., Wan F., Ye D. A novel gene signature to predict immune infiltration and outcome in patients with prostate cancer. Oncoimmunology. 2020;9:1762473. doi: 10.1080/2162402X.2020.1762473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong C.W., Maxwell P., Alvi M.A., McQuaid S., Waugh D., Mills I., Salto-Tellez M. A gene signature associated with PTEN activation defines good prognosis intermediate risk prostate cancer cases. J. Pathol. Clin. Res. 2018;4:103–113. doi: 10.1002/cjp2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abida W., Cyrta J., Heller G., Prandi D., Armenia J., Coleman I., Cieslik M., Benelli M., Robinson D., Van Allen E.M., et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA. 2019;116:11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chibon F. Cancer gene expression signatures—The rise and fall? Eur. J. Cancer. 2013;49:2000–2009. doi: 10.1016/j.ejca.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 28.Cooper C.S., Campbell C., Jhavar S. Mechanisms of Disease: Biomarkers and molecular targets from microarray gene expression studies in prostate cancer. Nat. Clin. Pract. Urol. 2007;4:677–687. doi: 10.1038/ncpuro0946. [DOI] [PubMed] [Google Scholar]
- 29.Michiels S., Koscielny S., Hill C. Interpretation of microarray data in cancer. Br. J. Cancer. 2007;96:1155–1158. doi: 10.1038/sj.bjc.6603673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cary K.C., Cooperberg M.R. Biomarkers in prostate cancer surveillance and screening: Past, present, and future. Ther. Adv. Urol. 2013;5:318–329. doi: 10.1177/1756287213495915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saini S. PSA and beyond: Alternative prostate cancer biomarkers. Cell Oncol. 2016;39:97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szeliski K., Adamowicz J., Gastecka A., Drewa T., Pokrywczyńska M. Modern urology perspectives on prostate cancer biomarkers. Cent. Eur. J. Urol. 2018;71:420–426. doi: 10.5173/ceju.2018.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denton A.E., Roberts E.W., Fearon D.T. Stromal Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2018;1060:99–114. doi: 10.1007/978-3-319-78127-3_6. [DOI] [PubMed] [Google Scholar]
- 34.Lalonde E., Ishkanian A.S., Sykes J., Fraser M., Ross-Adams H., Erho N., Dunning M.J., Halim S., Lamb A.D., Moon N.C., et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: A retrospective cohort study. Lancet Oncol. 2014;15:1521–1532. doi: 10.1016/S1470-2045(14)71021-6. [DOI] [PubMed] [Google Scholar]
- 35.Lunt S.J., Chaudary N., Hill R.P. The tumor microenvironment and metastatic disease. Clin. Exp. Metastasis. 2009;26:19–34. doi: 10.1007/s10585-008-9182-2. [DOI] [PubMed] [Google Scholar]
- 36.Berglund E., Maaskola J., Schultz N., Friedrich S., Marklund M., Bergenstråhle J., Tarish F., Tanoglidi A., Vickovic S., Larsson L., et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018;9:2419. doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bacac M., Provero P., Mayran N., Stehle J.C., Fusco C., Stamenkovic I. A mouse stromal response to tumor invasion predicts prostate and breast cancer patient survival. PLoS ONE. 2006;1:e32. doi: 10.1371/journal.pone.0000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Planche A., Bacac M., Provero P., Fusco C., Delorenzi M., Stehle J.C., Stamenkovic I. Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS ONE. 2011;6:e18640. doi: 10.1371/journal.pone.0018640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Z., Li C., Kang B., Gao G., Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunning M.J., Vowler S.L., Lalonde E., Ross-Adams H., Boutros P., Mills I.G., Lynch A.G., Lamb A.D. Mining Human Prostate Cancer Datasets: The “camcAPP” Shiny App. EBioMedicine. 2017;17:5–6. doi: 10.1016/j.ebiom.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross-Adams H., Lamb A.D., Dunning M.J., Halim S., Lindberg J., Massie C.M., Egevad L.A., Russell R., Ramos-Montoya A., Vowler S.L., et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine. 2015;2:1133–1144. doi: 10.1016/j.ebiom.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haffner M.C., Zwart W., Roudier M.P., True L.D., Nelson W.G., Epstein J.I., De Marzo A.M., Nelson P.S., Yegnasubramanian S. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 2021;18:79–92. doi: 10.1038/s41585-020-00400-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labrecque M.P., Coleman I.M., Brown L.G., True L.D., Kollath L., Lakely B., Nguyen H.M., Yang Y.C., da Costa R.M.G., Kaipainen A., et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Investig. 2019;129:4492–4505. doi: 10.1172/JCI128212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X., Guo J., Liu K., Chen L., Liu D., Dong S., Xia J., Long Q., Yue Y., Zhao P., et al. Identification of a distinct luminal subgroup diagnosing and stratifying early stage prostate cancer by tissue-based single-cell RNA sequencing. Mol. Cancer. 2020;19:147. doi: 10.1186/s12943-020-01264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu T., Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 50.Ippolito L., Morandi A., Taddei M.L., Parri M., Comito G., Iscaro A., Raspollini M.R., Magherini F., Rapizzi E., Masquelier J., et al. Cancer-associated fibroblasts promote prostate cancer malignancy via metabolic rewiring and mitochondrial transfer. Oncogene. 2019;38:5339–5355. doi: 10.1038/s41388-019-0805-7. [DOI] [PubMed] [Google Scholar]
- 51.Sun D.Y., Wu J.Q., He Z.H., He M.F., Sun H.B. Cancer-associated fibroblast regulate proliferation and migration of prostate cancer cells through TGF-β signaling pathway. Life Sci. 2019;235:116791. doi: 10.1016/j.lfs.2019.116791. [DOI] [PubMed] [Google Scholar]
- 52.Archer M., Dogra N., Kyprianou N. Inflammation as a Driver of Prostate Cancer Metastasis and Therapeutic Resistance. Cancers. 2020;12:2984. doi: 10.3390/cancers12102984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larionova I., Tuguzbaeva G., Ponomaryova A., Stakheyeva M., Cherdyntseva N., Pavlov V., Choinzonov E., Kzhyshkowska J. Tumor-Associated Macrophages in Human Breast, Colorectal, Lung, Ovarian and Prostate Cancers. Front. Oncol. 2020;10:566511. doi: 10.3389/fonc.2020.566511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen M.K., Rise K., Giskeødegård G.F., Richardsen E., Bertilsson H., Størkersen Ø., Bathen T.F., Rye M., Tessem M.B. Integrative metabolic and transcriptomic profiling of prostate cancer tissue containing reactive stroma. Sci. Rep. 2018;8:14269. doi: 10.1038/s41598-018-32549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia Z., Rahmatpanah F.B., Chen X., Lernhardt W., Wang Y., Xia X.Q., Sawyers A., Sutton M., McClelland M., Mercola D. Expression changes in the stroma of prostate cancer predict subsequent relapse. PLoS ONE. 2012;7:e41371. doi: 10.1371/annotation/7de63575-e5c9-4f1d-bb45-fc6420e92c71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karkampouna S., De Filippo M.R., Ng C.K.Y., Klima I., Zoni E., Spahn M., Stein F., Haberkant P., Thalmann G.N., Kruithof-de Julio M. Stroma Transcriptomic and Proteomic Profile of Prostate Cancer Metastasis Xenograft Models Reveals Prognostic Value of Stroma Signatures. Cancers. 2020;12:3786. doi: 10.3390/cancers12123786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mo F., Lin D., Takhar M., Ramnarine V.R., Dong X., Bell R.H., Volik S.V., Wang K., Xue H., Wang Y., et al. Stromal Gene Expression is Predictive for Metastatic Primary Prostate Cancer. Eur. Urol. 2018;73:524–532. doi: 10.1016/j.eururo.2017.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos N.J., Ximenes P.P., Constantino F.B., Carvalho H.F., Felisbino S.L. Mucinous metaplasia in Pten conditional knockout mice and mucin family genes as prognostic markers for prostate cancer. Life Sci. 2022;293:120264. doi: 10.1016/j.lfs.2021.120264. [DOI] [PubMed] [Google Scholar]
- 59.Santos N.J., Barquilha C.N., Barbosa I.C., Macedo R.T., Lima F.O., Justulin L.A., Barbosa G.O., Carvalho H.F., Felisbino S.L. Syndecan Family Gene and Protein Expression and Their Prognostic Values for Prostate Cancer. Int. J. Mol. Sci. 2021;22:8669. doi: 10.3390/ijms22168669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurmeister S., Ramos-Montoya A., Sandi C., Pertega-Gomes N., Wadhwa K., Lamb A.D., Dunning M.J., Attig J., Carroll J.S., Fryer L.G., et al. Identification of potential therapeutic targets in prostate cancer through a cross-species approach. EMBO Mol. Med. 2018;10:e8274. doi: 10.15252/emmm.201708274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaliman P., Llagostera E. Myotonic dystrophy protein kinase (DMPK) and its role in the pathogenesis of myotonic dystrophy 1. Cell. Signal. 2008;20:1935–1941. doi: 10.1016/j.cellsig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 62.Itoh K., Ebata T., Hirata H., Torii T., Sugimoto W., Onodera K., Nakajima W., Uehara I., Okuzaki D., Yamauchi S., et al. DMPK is a New Candidate Mediator of Tumor Suppressor p53-Dependent Cell Death. Molecules. 2019;24:3175. doi: 10.3390/molecules24173175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacLennan D.H., Kranias E.G. Phospholamban: A crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 64.Gautier M., Trebak M., Fleig A., Vandier C., Ouadid-Ahidouch H. Ca2+ homeostasis and cancer. Cell Calcium. 2019;84:102084. doi: 10.1016/j.ceca.2019.102084. [DOI] [PubMed] [Google Scholar]
- 65.Kania E., Pająk B., Orzechowski A. Calcium homeostasis and ER stress in control of autophagy in cancer cells. Biomed. Res. Int. 2015;2015:352794. doi: 10.1155/2015/352794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao Y., Zhao G., Yuan M., Liu X., Ma Y., Miao B., Zhao S., Li D., Xiong S., Zheng M., et al. KCNQ5 and C9orf50 Methylation in Stool DNA for Early Detection of Colorectal Cancer. Front. Oncol. 2020;10:621295. doi: 10.3389/fonc.2020.621295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jensen S., Øgaard N., Ørntoft M.W., Rasmussen M.H., Bramsen J.B., Kristensen H., Mouritzen P., Madsen M.R., Madsen A.H., Sunesen K.G., et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer—A clinical biomarker discovery and validation study. Clin. Epigenetics. 2019;11:158. doi: 10.1186/s13148-019-0757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y., Wu Q., Xu L., Wang H., Liu X., Li S., Hu T., Liu Y., Peng Q., Chen Z., et al. Sensitive detection of colorectal cancer in peripheral blood by a novel methylation assay. Clin. Epigenetics. 2021;13:90. doi: 10.1186/s13148-021-01076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouzid A., Smeti I., Dhouib L., Roche M., Achour I., Khalfallah A., Gibriel A.A., Charfeddine I., Ayadi H., Lachuer J., et al. Down-expression of P2RX2, KCNQ5, ERBB3 and SOCS3 through DNA hypermethylation in elderly women with presbycusis. Biomarkers. 2018;23:347–356. doi: 10.1080/1354750X.2018.1427795. [DOI] [PubMed] [Google Scholar]
- 70.Zimmer J., Takahashi T., Hofmann A.D., Puri P. Downregulation of KCNQ5 expression in the rat pulmonary vasculature of nitrofen-induced congenital diaphragmatic hernia. J. Pediatr. Surg. 2017;52:702–705. doi: 10.1016/j.jpedsurg.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 71.Goldwich A., Scholz M., Tamm E.R. Myocilin promotes substrate adhesion, spreading and formation of focal contacts in podocytes and mesangial cells. Histochem. Cell Biol. 2009;131:167–180. doi: 10.1007/s00418-008-0518-4. [DOI] [PubMed] [Google Scholar]
- 72.Liuska P.J., Harju M., Kivelä T.T., Turunen J.A. Prevalence of MYOC risk variants for glaucoma in different populations. Acta Ophthalmol. 2021;99:e1090–e1097. doi: 10.1111/aos.14738. [DOI] [PubMed] [Google Scholar]
- 73.Wiggs J.L., Pasquale L.R. Genetics of glaucoma. Hum. Mol. Genet. 2017;26:R21–R27. doi: 10.1093/hmg/ddx184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu L., Ke J., Du X., Yu Z., Gao D. Genetic characterization of thymoma. Sci. Rep. 2019;9:2369. doi: 10.1038/s41598-019-38878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gajos-Michniewicz A., Czyz M. WNT Signaling in Melanoma. Int. J. Mol. Sci. 2020;21:4852. doi: 10.3390/ijms21144852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nusse R., Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 77.Galluzzi L., Spranger S., Fuchs E., López-Soto A. WNT Signaling in Cancer Immunosurveillance. Trends Cell Biol. 2019;29:44–65. doi: 10.1016/j.tcb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ring A., Kim Y.M., Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem. Cell Rev. Rep. 2014;10:512–525. doi: 10.1007/s12015-014-9515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kerekes K., Bányai L., Trexler M., Patthy L. Structure, function and disease relevance of Wnt inhibitory factor 1, a secreted protein controlling the Wnt and hedgehog pathways. Growth Factors. 2019;37:29–52. doi: 10.1080/08977194.2019.1626380. [DOI] [PubMed] [Google Scholar]
- 80.Beggs A.D., Mehta S., Deeks J.J., James J.D., Caldwell G.M., Dilworth M.P., Stockton J.D., Blakeway D., Pestinger V., Vince A., et al. Validation of epigenetic markers to identify colitis associated cancer: Results of module 1 of the ENDCAP-C study. EBioMedicine. 2019;39:265–271. doi: 10.1016/j.ebiom.2018.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu H., Li B., Zhou C., Ying X., Chen M., Huang T., Chen Y., Ji H., Pan R., Wang T., et al. Diagnostic value of WIF1 methylation for colorectal cancer: A meta-analysis. Oncotarget. 2018;9:5378–5386. doi: 10.18632/oncotarget.23870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu S., Chen X., Chen R., Wang J., Zhu G., Jiang J., Wang H., Duan S., Huang J. Diagnostic role of Wnt pathway gene promoter methylation in non small cell lung cancer. Oncotarget. 2017;8:36354–36367. doi: 10.18632/oncotarget.16754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu X., Fu J., Bi H., Ge A., Xia T., Liu Y., Sun H., Li D., Zhao Y. DNA methylation of SFRP1, SFRP2, and WIF1 and prognosis of postoperative colorectal cancer patients. BMC Cancer. 2019;19:1212. doi: 10.1186/s12885-019-6436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang S., Zhong X., Gao J., Song R., Wu H., Zi S., Yang S., Du P., Cui L., Yang C., et al. Coexpression of SFRP1 and WIF1 as a prognostic predictor of favorable outcomes in patients with colorectal carcinoma. Biomed. Res. Int. 2014;2014:256723. doi: 10.1155/2014/256723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cortez M.A., Masrorpour F., Ivan C., Zhang J., Younes A.I., Lu Y., Estecio M.R., Barsoumian H.B., Menon H., Caetano M.D.S., et al. Bone morphogenetic protein 7 promotes resistance to immunotherapy. Nat. Commun. 2020;11:4840. doi: 10.1038/s41467-020-18617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perron J.C., Rodrigues A.A., Surubholta N., Dodd J. Chemotropic signaling by BMP7 requires selective interaction at a key residue in ActRIIA. Biol. Open. 2019;8:bio042283. doi: 10.1242/bio.042283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aoki M., Ishigami S., Uenosono Y., Arigami T., Uchikado Y., Kita Y., Kurahara H., Matsumoto M., Ueno S., Natsugoe S. Expression of BMP-7 in human gastric cancer and its clinical significance. Br. J. Cancer. 2011;104:714–718. doi: 10.1038/sj.bjc.6606075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen J., Ye L., Xie F., Yang Y., Zhang L., Jiang W.G. Expression of bone morphogenetic protein 7 in lung cancer and its biological impact on lung cancer cells. Anticancer Res. 2010;30:1113–1120. [PubMed] [Google Scholar]
- 89.Witkowski M., Landmesser U., Rauch U. Tissue factor as a link between inflammation and coagulation. Trends Cardiovasc. Med. 2016;26:297–303. doi: 10.1016/j.tcm.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 90.Hisada Y., Mackman N. Tissue Factor and Cancer: Regulation, Tumor Growth, and Metastasis. Semin. Thromb. Hemost. 2019;45:385–395. doi: 10.1055/s-0039-1687894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khorana A.A., Ahrendt S.A., Ryan C.K., Francis C.W., Hruban R.H., Hu Y.C., Hostetter G., Harvey J., Taubman M.B. Tissue factor expression, angiogenesis, and thrombosis in pancreatic cancer. Clin. Cancer Res. 2007;13:2870–2875. doi: 10.1158/1078-0432.CCR-06-2351. [DOI] [PubMed] [Google Scholar]
- 92.Kobayashi S., Koizume S., Takahashi T., Ueno M., Oishi R., Nagashima S., Sano Y., Fukushima T., Tezuka S., Morimoto M., et al. Tissue factor and its procoagulant activity on cancer-associated thromboembolism in pancreatic cancer. Cancer Sci. 2021;112:4679–4691. doi: 10.1111/cas.15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao X., Cheng C., Gou J., Yi T., Qian Y., Du X. Expression of tissue factor in human cervical carcinoma tissue. Exp. Ther. Med. 2018;16:4075–4081. doi: 10.3892/etm.2018.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shaker H., Bundred N.J., Landberg G., Pritchard S.A., Albadry H., Nicholson S.L., Harries L.J., Heah J.Y.E., Castle J., Kirwan C.C. Breast cancer stromal clotting activation (Tissue Factor and thrombin): A pre-invasive phenomena that is prognostic in invasion. Cancer Med. 2020;9:1768–1778. doi: 10.1002/cam4.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hell L., Däullary T., Burghart V., Mauracher L.M., Grilz E., Moser B., Kramer G., Schmid J.A., Ay C., Pabinger I., et al. Extracellular Vesicle-Associated Tissue Factor Activity in Prostate Cancer Patients with Disseminated Intravascular Coagulation. Cancers. 2021;13:1487. doi: 10.3390/cancers13071487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen W., Zhang Z., Zhang S., Zhu P., Ko J.K., Yung K.K. MUC1: Structure, Function, and Clinic Application in Epithelial Cancers. Int. J. Mol. Sci. 2021;22:6567. doi: 10.3390/ijms22126567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nath S., Mukherjee P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bouillez A., Rajabi H., Pitroda S., Jin C., Alam M., Kharbanda A., Tagde A., Wong K.K., Kufe D. Inhibition of MUC1-C Suppresses MYC Expression and Attenuates Malignant Growth in KRAS Mutant Lung Adenocarcinomas. Cancer Res. 2016;76:1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hagiwara M., Yasumizu Y., Yamashita N., Rajabi H., Fushimi A., Long M.D., Li W., Bhattacharya A., Ahmad R., Oya M., et al. MUC1-C Activates the BAF (mSWI/SNF) Complex in Prostate Cancer Stem Cells. Cancer Res. 2021;81:1111–1122. doi: 10.1158/0008-5472.CAN-20-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yasumizu Y., Rajabi H., Jin C., Hata T., Pitroda S., Long M.D., Hagiwara M., Li W., Hu Q., Liu S., et al. MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer. Nat. Commun. 2020;11:338. doi: 10.1038/s41467-019-14219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Acunzo M., Romano G., Wernicke D., Croce C.M. MicroRNA and cancer—A brief overview. Adv. Biol. Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 102.Rupaimoole R., Slack F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 103.Feng L., Jing L., Han J., Wang G., Liu Y., Zhang X., Wang Y., Wang F., Ma H. MicroRNA 486-3p directly targets BIK and regulates apoptosis and invasion in colorectal cancer cells. Onco Targets Ther. 2018;11:8791–8801. doi: 10.2147/OTT.S180354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hu L., Ai J., Long H., Liu W., Wang X., Zuo Y., Li Y., Wu Q., Deng Y. Integrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget. 2016;7:8441–8454. doi: 10.18632/oncotarget.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo H., Ye Z. Identification of Serum miR-337-3p, miR-484, miR-582, and miR-3677 as Promising Biomarkers for Osteosarcoma. Clin. Lab. 2021;67 doi: 10.7754/Clin.Lab.2020.200455. [DOI] [PubMed] [Google Scholar]
- 106.Fan B., Chen L.P., Yuan Y.H., Xiao H.N., Lv X.S., Xia Z.Y. MiR-15a-3p suppresses the growth and metastasis of ovarian cancer cell by targeting Twist1. Eur. Rev. Med. Pharmacol. Sci. 2019;23:1934–1946. doi: 10.26355/eurrev_201903_17232. [DOI] [PubMed] [Google Scholar]
- 107.Furuya T.K., Murta C.B., Murillo Carrasco A.G., Uno M., Sichero L., Villa L.L., Cardilli L., Coelho R.F., Guglielmetti G.B., Cordeiro M.D., et al. Disruption of miRNA-mRNA Networks Defines Novel Molecular Signatures for Penile Carcinogenesis. Cancers. 2021;13:4745. doi: 10.3390/cancers13194745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu S.J., Wang W.T., Zhang F.L., Yu Y.H., Yu H.J., Liang Y., Li N., Li Y.B. miR-15a-3p affects the proliferation, migration and apoptosis of lens epithelial cells. Mol. Med. Rep. 2019;19:1110–1116. doi: 10.3892/mmr.2018.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cui Y., Yang Y., Ren L., Yang J., Wang B., Xing T., Chen H., Chen M. miR-15a-3p Suppresses Prostate Cancer Cell Proliferation and Invasion by Targeting SLC39A7 via Downregulating Wnt/β-Catenin Signaling Pathway. Cancer Biother. Radiopharm. 2019;34:472–479. doi: 10.1089/cbr.2018.2722. [DOI] [PubMed] [Google Scholar]
- 110.Xin C., Lu S., Li Y., Zhang Y., Tian J., Zhang S., Yang S., Gao T., Xu J. miR-671-5p Inhibits Tumor Proliferation by Blocking Cell Cycle in Osteosarcoma. DNA Cell Biol. 2019;38:996–1004. doi: 10.1089/dna.2019.4870. [DOI] [PubMed] [Google Scholar]
- 111.Hu Y., Liang D., Chen X., Chen L., Bai J., Li H., Yin C., Zhong W. MiR-671-5p negatively regulates SMAD3 to inhibit migration and invasion of osteosarcoma cells. Nan Fang Yi Ke Da Xue Xue Bao. 2021;41:1562–1568. doi: 10.12122/j.issn.1673-4254.2021.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barquilha C.N., Santos N.J., Monção C.C.D., Barbosa I.C., Lima F.O., Justulin L.A., Pértega-Gomes N., Felisbino S.L. Sulfiredoxin as a Potential Therapeutic Target for Advanced and Metastatic Prostate Cancer. Oxidative Med. Cell. Longev. 2020;2020:2148562. doi: 10.1155/2020/2148562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M., et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 114.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 115.Fagerberg L., Jonasson K., von Heijne G., Uhlén M., Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 116.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ge S.X., Jung D., Yao R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sticht C., De La Torre C., Parveen A., Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13:e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oliveros J.C. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. [(accessed on 21 December 2021)]. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 121.Hothorn T., Hornik K., Zeileis A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J. Comput. Graph. Stat. 2006;15:651–674. doi: 10.1198/106186006X133933. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results presented in this article are partly based on data generated by the Cambridge Carcinoma of the Prostate App (CamcAPP) (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/) from the Cancer Research UK Cambridge Institute, cBioPortal for Cancer Genomics database (https://www.cbioportal.org/), and Human Protein Atlas (HPA) (https://www.proteinatlas.org/) database. The RNA sequencing data derived from all prostatic lobes of Pten knockout are available in the NCBI Gene Expression Omnibus platform (GEO, https://www.ncbi.nlm.nih.gov/geo/, accessed on 21 December 2021), reference number GSE94574. The datasets presented in this study can be found in online repositories on gene expression profiling interactive analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html, accessed on 21 December 2021).