Abstract
Major lipoprotein antigens, known as variable membrane surface lipoproteins (Vsps), on the surface of the bovine pathogen Mycoplasma bovis were shown to spontaneously undergo noncoordinate phase variation between ON and OFF expression states. The high rate of Vsp phenotypic switching was also shown to be linked with DNA rearrangements that occur at high frequency in the M. bovis chromosome (I. Lysnyansky, R. Rosengarten, and D. Yogev, J. Bacteriol. 178:5395–5401, 1996). In the present study, 13 single-copy vsp genes organized in a chromosomal cluster were identified and characterized. All vsp genes encode highly conserved N-terminal domains for membrane insertion and lipoprotein processing but divergent mature Vsp proteins. About 80% of each vsp coding region is composed of reiterated coding sequences that create a periodic polypeptide structure. Eighteen distinct repetitive domains of different lengths and amino acid sequences are distributed within the products of the various vsp genes that are subject to size variation due to spontaneous insertions or deletions of these periodic units. Some of these repeats were found to be present in only one Vsp family member, whereas other repeats recurred at variable locations in several Vsps. Each vsp gene is also 5′ linked to a highly homologous upstream region composed of two internal cassettes. The findings that rearrangement events are associated with Vsp phenotypic switching and that multiple regions of high sequence similarity are present upstream of the vsp genes and within the vsp coding regions suggest that modulation of the Vsp antigenic repertoire is determined by recombination processes that occur at a high frequency within the vsp locus of M. bovis.
Mycoplasmas belong to the class Mollicutes, which includes more than 170 distinct species that are phylogenetically related to gram-positive eubacteria and are the smallest microorganisms capable of self-replication and autonomous life (18, 19). Despite the fact that these organisms lack a cell wall and contain a remarkably small genome (5), the mycoplasmas are widespread in nature and many species are recognized as pathogens of humans, animals, and plants (25, 29). The persistence of these wall-less agents in different environments as well as in various hosts indicates that mycoplasmas possess a capability to successfully adapt and respond to environmental fluctuations and to the defense mechanisms of the animal hosts. Studies in recent years have shown that populations of several pathogenic mycoplasmal species spontaneously and randomly generate distinct progenies with varied antigenic phenotypes (3, 6, 14, 16, 19, 21, 28, 30–32, 34, 37, 38). These antigenic variants may efficiently escape the host immune response and subsequently may play an important role in the chronic nature of mycoplasmal infections (19, 20, 32). The importance of diversifying the antigenic repertoire of the cell surface in these minute microorganisms is reflected by the fact that, despite their limited genetic material, in mycoplasmas the number of genes that are exclusively dedicated to this purpose is unexpectedly large (19).
Mycoplasma bovis is widely known to be the most important etiological agent of various bovine diseases, such as mastitis in cows and pneumonia and arthritis in calves as well as genital disorders (10, 17). M. bovis infections, which tend to be chronic, are responsible for considerable economic losses in cattle and milk production.
The antigenic repertoire of the M. bovis cell surface was found to be subject to rapid changes due to the presence of a set of antigenically and structurally related variable membrane surface lipoproteins designated Vsps. Three members (VspA, VspB, and VspC) have so far been characterized (1, 22). Each Vsp was shown to possess the following features: (i) independent high-frequency phase variation between ON and OFF expression states, (ii) independent high-frequency size variation, (iii) membrane anchorage via the N-terminal domain and a C-terminal region which is surface exposed, (iv) extensive repetitive domains over the full length of the Vsp molecule, and (v) regions of shared epitopes.
The extensive Vsp phenotypic switching in M. bovis was recently shown to be linked with high-frequency chromosomal rearrangements that occur within the vsp genomic locus (12). However, the genomic organization and the structural features of the vsp genes have not yet been described, nor has the precise nature of the vsp ON-OFF switching mechanism.
In the present study we identified and characterized a chromosomal region containing a large family of multiple related but divergent lipoprotein-encoding genes comprising the vsp locus of M. bovis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The sequence analysis of the vsp locus was done on clonal isolate 6 of the M. bovis type strain PG45, which has the phenotype VspA+ VspB+. Its origin and growth conditions have been described elsewhere (22). The Escherichia coli strains used were DH5αMCR (Gibco BRL Life Technologies, Inc., Gaithersburg, Md.), KW251, and LE392 (Promega, Madison, Wis.). Recombinant clones were constructed in the plasmid vector pBluescript II KS(+) (Stratagene, La Jolla, Calif.) or in pGEM-7Z (Promega).
Chemicals, media, and growth conditions.
E. coli cultures for plasmid and bacteriophage isolation were grown with shaking at 37°C in Luria-Bertani broth (LB) (23). E. coli cultures for expression of proteins under T7 promoter control (27) were grown at 30°C with shaking in M9 media (23) supplemented with an amino acid mixture. Restriction enzymes, T4 ligase, and T4 polynucleotide kinase were purchased from Promega and used according to the recommendations of the manufacturer. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), ampicillin, kanamycin, and rifampin were purchased from Sigma Chemicals, St. Louis, Mo. [γ-32P]ATP and [α-32P]CTP were purchased from Amersham, Little Chalfont, United Kingdom.
DNA preparation and manipulation.
Genomic DNAs from M. bovis PG45 clonal isolates were extracted and purified as previously described (15). The DNAs were digested by restriction enzymes and electrophoresed as previously described (34).
Labelling of oligonucleotide or DNA probes and hybridization conditions.
Vsp sequence-specific oligonucleotides were synthesized at the interdepartmental facility of the Hebrew University—Hadassah Medical School on a model 380B DNA synthesizer (Applied Biosystems, Inc., Foster City, Calif.). A sequence 18 nucleotides (nt) long, 5′-TTAGCTTCAATTCCCTTT-3′, was designated sig-oligonucleotide. A sequence 19 nt long, 5′-GGAGAGGATAAATTTATGA-3′, was designated pro-oligonucleotide. About 100 ng of each oligonucleotide was 32P labeled by using 25 U of T4 polynucleotide kinase at 37°C for 1 h in 25 μl of a reaction mixture containing 40 mM Tris (pH 7.5), 10 mM MgCl2, 5 mM dithiothreitol, and 2.5 μl of [γ-32P]ATP (3,000 Ci/nmole). The conditions for oligonucleotide hybridization as well as for DNA hybridization have been described elsewhere (12, 34).
DNA sequence analysis.
DNA sequence analysis of both strands was performed by the dideoxy chain termination method (24). Overlapping sets of deletion mutants were generated from the recombinant plasmids carrying the vsp genes by graded directional exonuclease III digestion by using the Erase-A-Base deletion kit (Promega). The T7 promoter sequence or the SP6 or T3 sequence located on the various plasmid vectors as well as vsp-related sequences were used as primers for sequencing. Sequencing was done by using the automatic sequencer dye terminator cycle sequencing model IBA PRISM 377 (Perkin Elmer, Foster City, Calif.). Sequence data were analyzed by using the computer software Assembly LIGN and MacVector 6.0.
Nucleotide sequence accession numbers.
The nucleotide sequences of the vsp genes reported in this study have been assigned the following numbers: vspB, AF162138; vspE, AF162139; vspF, AF162140; vspG, AF162141; vspH, AF162142; vspI, AF162143; vspJ, AF162144; vspK, AF162145; vspL, AF162146; vspM, AF162147; vspN, AF162148; and vspO, AF162149.
RESULTS
Cloning, sequence analysis, and gene organization of the vsp locus in a clonal isolate of the M. bovis type strain PG45.
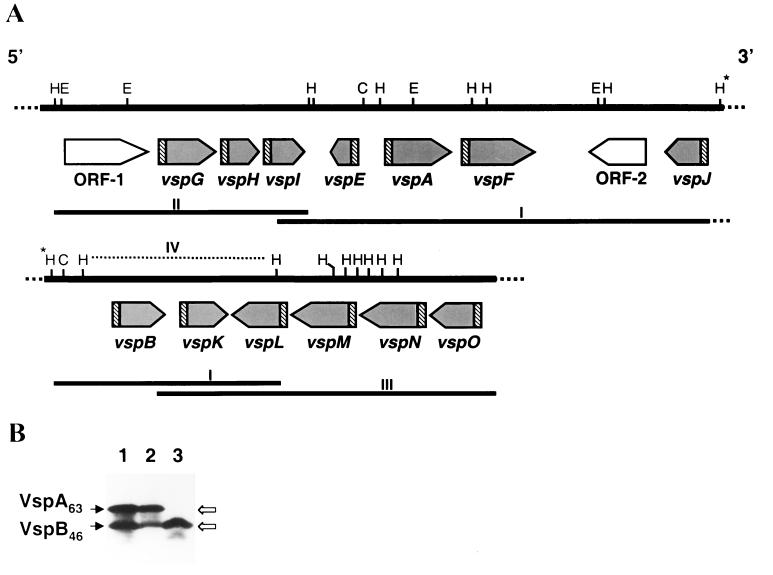
A recombinant phage, designated λMbA1, which showed strong immunostaining with monoclonal antibody (MAb) 1E5, was isolated from a previously described M. bovis genomic library constructed from a clonal isolate with the expression phenotype VspA+ VspB+ (63-kDa VspA protein and 46-kDa VspB protein) (12). MAb 1E5 was previously shown to recognize a common epitope present on three distinct Vsps: VspA, VspB, and VspC (1). An 11.8-kb DNA insert fragment from this phage was digested with the restriction enzyme HindIII to give seven identifiable authentic fragments of about 0.3, 0.6, 1.2, 1.5, 2.0, 2.1, and 3.2 kb (Fig. 1A, fragment I). An adjoining overlapping HindIII genomic fragment of about 4.5 kb located upstream of the 5′ end of the 11.8-kb phage insert (Fig. 1A, fragment II) was identified by Southern blot hybridization by using the vspA gene as a probe and excised from the gel. Another adjoining overlapping genomic fragment of about 6-kb length located downstream of the 3′ end of the 11.8-kb phage insert (Fig. 1A, fragment III) was identified by screening the genomic library by using the vspK gene as a probe. As a result, a region of about 23 kb of the M. bovis chromosome containing putative genes encoding Vsp proteins was identified and sequenced.
FIG. 1.
(A) Schematic representation, restriction map, and genomic organization of the M. bovis vsp locus. The solid line labeled I represents an 11.8-kb M. bovis DNA insert obtained from the recombinant bacteriophage λMbA1, which showed strong immunostaining with MAb 1E5. Two additional solid lines (II and III) represent adjoining and overlapping cloned genomic fragments spanning the vsp region. The positions of HindIII (H), EcoRI (E), and ClaI (C) restriction sites are marked. The locations and the directions of 13 Vsp ORFs are indicated by shaded arrows. Two additional non-Vsp-related ORFs (ORF-1 and ORF-2) are indicated by open arrows. Highly homologous regions 5′ of each vsp gene are indicated by hatched boxes. The broken line (IV) above the vspB, vspK, and vspL gene region represents a 3.2-kb HindIII cloned fragment with which expression of the 46-kDa VspB product was obtained. The asterisks above the two HindIII sites indicate that these sites are the same site and the continuation of the genomic locus. (B) Expression of mycoplasma-encoded VspB protein in E. coli. Proteins from cells expressing, under the selective induction of the T7 promoter, the recombinant plasmid pKB46 carrying the vspB gene (lane 3) or proteins from the recombinant, λMbA phage lysate (lane 2) were separated by SDS-PAGE and immunoblotted with MAb 1E5. Total proteins of a clonal isolate of M. bovis PG45 exhibiting the phenotype VspA+ VspB+ (63-kDa VspA protein and 46-kDa VspB protein) and used for cloning of the vsp locus served as a positive control (lane 1). The authentic products VspA and VspB expressed in the mycoplasma are indicated on the left. The recombinant polypeptides of 63-kDa VspA and 46-kDa VspB expressed in E. coli are indicated on the right.
Within that genomic region, a cluster of 15 open reading frames (ORFs) that were not all similarly oriented was deduced to exist from the nucleotide sequences analyzed. The genomic organization and orientation of these ORFs are shown in Fig. 1A. Thirteen ORFs possessing features characteristic of known Vsp products (1, 12, 22) were designated as putative vsp genes. Two additional ORFs, designated ORF-1 and ORF-2, exhibited high homology to the prokaryotic mobile genetic elements IS-4 (homologous to ORF-1) and IS-30 (homologous to ORF-2). These IS-related ORFs were localized upstream of the vspG gene and between the vspF and vspJ genes, respectively (Fig. 1A).
To identify ORFs encoding the known VspA, VspB, and VspC proteins recognized by MAb 1E5, recombinant vsp genes were placed downstream of the selective T7 promoter and expressed in E. coli by using the T7 RNA polymerase expression system (27). Expressed mycoplasmal proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with MAb 1E5. Two genomic fragments carrying distinct vsp genes were found to encode products recognized by MAb 1E5. A polypeptide band of 46 kDa was synthesized in E. coli from the recombinant plasmid pKB46, which contained a 3.2-kb HindIII fragment (Fig. 1A, fragment IV) bearing three vsp genes. This product was similar to the authentic 46-kDa VspB product expressed in the mycoplasmal clonal isolate as revealed both by epitope specificity and by size (Fig. 1B, lanes 1 and 3). Expression experiments with deletion mutants generated from this fragment have localized the gene encoding the 46-kDa product at the 5′ end of that fragment. We therefore designated this gene vspB (Fig. 1A). In a previous study (12), a recombinant plasmid, pKA63, carrying a 1.5-kb HindIII genomic fragment was shown to express in E. coli a polypeptide of 63 kDa similar to the authentic 63-kDa VspA product expressed in the mycoplasma. This fragment was shown to carry the entire vspA gene and to be localized between the vspE and the vspF genes (Fig. 1A). Notably, Northern blot analysis of total RNA using DNA and oligonucleotide probes representing vsp conserved regions, as well as analysis of freshly broth-grown organisms metabolically labeled with [3H]palmitate, clearly identified the VspA and VspB products but failed to detect expression of additional Vsps (data not shown).
Structural features and comparison of the vsp genes and their deduced proteins.
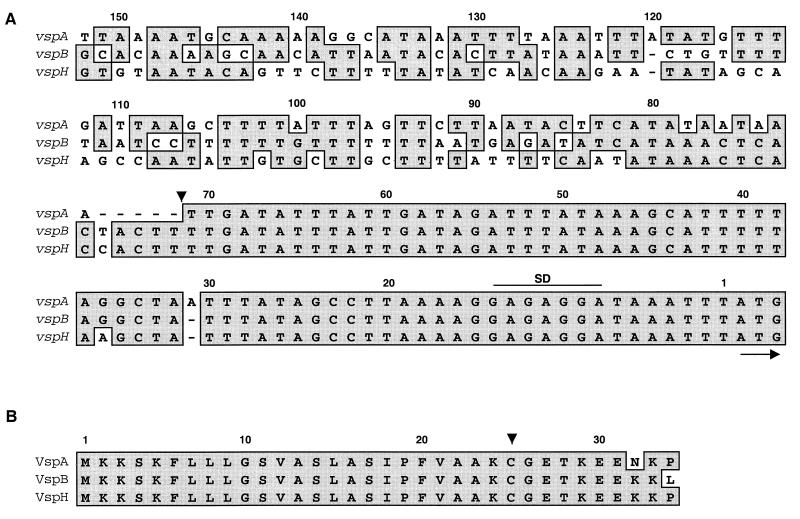
The nucleotide sequences and the deduced proteins of the 13 vsp genes were compared. A schematic representation of Vsp structure is shown in Fig. 3. Several striking aspects of Vsp structural similarity, sequence divergence, and variability were revealed. Each vsp gene is preceded by a highly conserved 5′ noncoding sequence that can be divided into two internal cassettes (Fig. 2A). The first, a 71-bp region upstream of the ATG initiation codon, contains a putative ribosome binding site and exhibits 99% homology among all vsp genes. The second region, of about 80 bp, is more divergent. The comparison of the 5′ upstream regions of three vsp genes (vspA, vspB, and vspH) as representatives of the vsp gene family is shown in Fig. 2A.
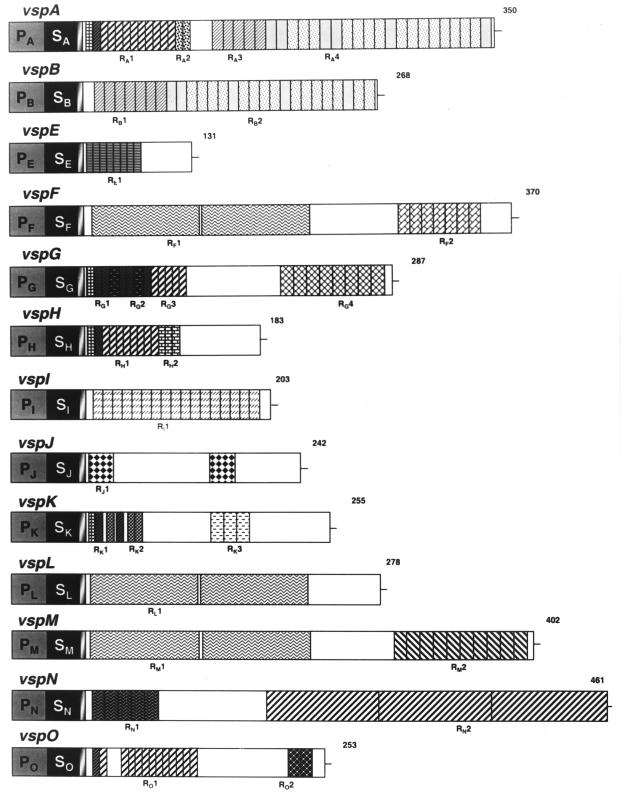
FIG. 3.
Structural features and comparison of vsp genes and Vsp protein products. The structures of each vsp gene and predicted Vsp protein are schematically presented by aligned rectangles. A highly conserved 5′ upstream region extending about 150 bp 5′ of each vsp gene is represented by the first block (gray and labeled P). The second block (solid and labeled S) represents a highly homologous 75-bp DNA sequence encoding a conserved prolipoprotein signal peptide. A sequence of six aa common to all Vsps is shown by the third block (shaded). In-frame reiterated coding sequences extending from the N termini to the C termini of the Vsp proteins and encoding periodic amino acid sequences are shown by different hatched blocks. Distinctive repetitive domains within each Vsp are labeled with R and the letter of the corresponding vsp gene. Repetitive units present in more than one Vsp protein are similarly hatched. The number on the right end of each Vsp indicates the length of the Vsp polypeptide chain.
FIG. 2.
Sequence alignments of vsp 5′ upstream regions (A) and of Vsp N-terminal regions (B). Three representatives of the vsp gene family (vspA, vspB, and vspH) are shown. The alignment was done with the MacVector 6.0 software program. Position numbers are given above the sequences. The name of each vsp gene or Vsp protein is indicated on the left of each row. Nucleotides representing a putative ribosome-binding site (SD) are spanned by a line. The initiation codon (ATG) is marked by an arrow. The division of the vsp 5′ upstream region into two distinct cassettes is shown by an arrowhead at nucleotide position 71. Identical nucleotides or amino acid residues are shown by dark shaded boxes. The single Cys residue within the lipoprotein box is indicated by an arrow.
The Vsp N-terminal region of 25 amino acids (aa) is also a highly homologous domain showing 99% amino acid identity among Vsps and representing a typical prokaryotic lipoprotein signal peptide. It begins with a sequence containing three positively charged Lys residues followed by a core of 20 hydrophobic aa and terminated with the tetrapeptide Ala-Ala-Lys-Cys. The comparison of the N-terminal regions of three Vsp proteins (VspA, VspB, and VspH) studied as representatives of the Vsp protein family is shown in Fig. 2B. The presence of the Ala and Cys residues is consistent with a prokaryotic prolipoprotein signal peptidase recognition sequence (4, 8). However, the presence of the Lys residue preceding the Cys residue within the lipoprotein box of each of the 13 Vsp proteins is surprising and rarely encountered in bacterial lipoproteins. The Cys residue in the lipoprotein box is the only one that occurs in each Vsp sequence and is consistent with the predicted acylation site and point of anchorage of a mature processed prokaryotic lipoprotein (4, 8). The presence of only a single Cys residue on one hand and the intense autoradiographic signals of Vsps in Triton X-114 phase proteins from [35S]cysteine-labeled mycoplasma proteins (1) on the other hand indicate that Vsps are the most abundant Cys-containing amphiphilic proteins in this organism. A block of 6 aa following the Cys residue is also highly conserved among all Vsps (Fig. 2B).
In contrast to the highly conserved nature of the region 5′ upstream of vsp and of the N-terminal domain, there is a considerable sequence divergence among mature Vsp molecules. Examination of the deduced Vsp amino acid sequences revealed an unusual structural motif. Most of the Vsp molecules are composed of reiterated coding sequences extending from the N terminus to the C terminus of the protein chain, thus creating a periodic polypeptide structure (Fig. 3). The substantial sequence divergence of the mature protein in the Vsp family is generated by 18 distinct reiterated units, of different amino acid sequences and lengths, which are distributed within the Vsp molecules. The majority of these repeated sequences are arranged in the form of tandem domains comprising up to 80% of the entire Vsp molecules and carrying repetitive units of 6, 8, 10, 11, 12, 26, 84, or 87 aa (Fig. 3 and Table 1). Some of these repeats were found to be present only in one Vsp member (e.g., RA2, RE1, RF2, RG2, RH2, RI1, RJ1, RK2, RK3, RM2, RN1, RN2, and RO2), whereas other repeats recurred at variable locations in several Vsps (Fig. 3 and Table 1). For example, the repeat RA1 was present in tandem 10 times in VspA, 5 times in VspG, and 8 times in VspH. In the protein VspO the RA1 repeat appeared 12 times, but the repetitions were distributed in two locations.
TABLE 1.
Repetitive sequences in M. bovis Vsp proteins
| Protein designation | Length of ORF (no. of amino acids) | Repeat designation, length (no. of amino acids, no. of Vsp repeats) | Repeat amino acid sequencec | Other Vsps in which the repeat is present |
|---|---|---|---|---|
| VspAa | 350 | RA1, 6, 10 | PGENKT | VspG, VspH, VspO |
| RA2, 6, 2 | PEENKK | |||
| RA3, 8, 5 | GTPANPDQ | VspB | ||
| RA4.1, 8, 6 | GAGTKPGQ | VspB | ||
| RA4.2, 8, 15 | GAGTNS(T)QQ | VspB | ||
| VspBb | 268 | RB1, 8, 7 | GTPA(T)NPDQ | VspA |
| RB2.1, 8, 5 | GAGTKPGQ | VspA | ||
| RB2.2, 8, 15 | GAGTNS(T)QQ | VspA | ||
| VspEb | 131 | RE1, 6, 8 | PETPKG | |
| VspFb | 370 | RF1, 84, 2 | KPKLSETLKSITGNDLGKVQVAEQDKSNKEKIET AIKEAIVTKVPA(T)LKDKELNLNADLSKKSVT VSAKGFEGEVTLKFEIETQS | VspL, VspM |
| RF2, 10, 7 | GTGAPKA(S)PQQ | |||
| VspGb | 287 | RG1, 6, 5 | PGGDKN | VspK |
| RG2, 10, 2 | PGENTEPDKN | |||
| RG3, 6, 5 | PG(E)E(G)NKT | VspA, VspH, VspO | ||
| RG4, 11, 8 | SGTMSKGPGAQ | |||
| VspHb | 183 | RH1, 6, 8 | PGENKT | VspA, VspG, VspO |
| RH2, 6, 3 | PEGEKK | |||
| VspIb | 203 | RI1, 8, 17 | PD(G)QGTPA(T)N | |
| VspJb | 242 | RJ1, 26, 2 | KEEKKPEADKPADKQPGDDM(I)KKDNDK | |
| VspKb | 255 | RK1, 6, 2 | PGGDKN | VspG |
| RK2, 6, 3 | KKPEGD(E) | |||
| RK3, 11, 3 | KGSDTESSKKD | |||
| VspLb | 278 | RL1, 84, 2 | KPKLSETLKSITGNDLGKVQVAEQDKSNKEKIEA AIKEAIVT(A)KVPTLKDKELKLNADLSKKSVT VSAKDFEGEVTLKFEIEAKS | VspF, VspM |
| VspMb | 402 | RM1, 84, 2 | KPKLSETLKSITGNDLGKVQVAEQDKSNKEKIET AIKEAIVTKVPTLKDKELNLNADLSKKSVTVSA KGFEGEVTLKFEIETQS | VspF, VspL |
| RM2, 11, 10 | QGTE(G)ANSGT(K)KD(V) | |||
| VspNb | 461 | RN1, 10, 5 | NTEPGKNPGG(E) | |
| RN2, 87, 3 | KQKLEDALKSISGKNLGKVQVSKETEKKDKAKIEAS IKETIILKVPALAGKDLQFKTDLTKNEVNVSSNDFQ GEVVLKFEVEVKSSE | |||
| VspOb | 253 | RO1, 6, 12 | PG(E)ENKT | VspA, VspG, VspH |
| RO2, 12, 2 | GQGTSAKSGANS |
Data are from reference 12.
Data are from this study.
Parentheses indicate amino acid substitution within the repeat.
An unusual feature displayed by the known Vsps, i.e., their abnormal migration in SDS-PAGE gels, generated significant discrepancies between the molecular weights calculated from the deduced sequences and the molecular masses estimated for the proteins in gels. For example, the mature VspB protein reported here (Fig. 3) contains 268 aa, predicting a mass of 25 kDa, whereas the electrophoretic migration of the product expressed in E. coli and in the mycoplasma suggests that the mass is 46 kDa (Fig. 1B).
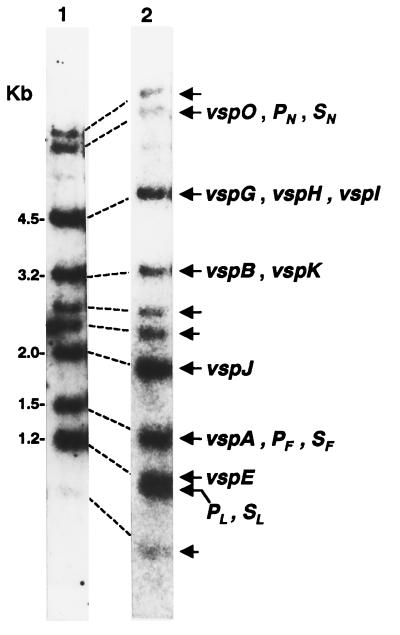
Since all 13 vsp genes clustered within the cloned vsp locus were shown to possess highly conserved sequences, we examined the possibility that additional vsp genes may be present in the M. bovis chromosome. Synthetic oligonucleotides representing the highly conserved region 5′ upstream of vsp or sequences complementary to the highly conserved signal peptide-encoding region were used as probes in Southern blot hybridization against HindIII-restricted genomic DNA of the clonal isolate. The HindIII restriction enzyme was chosen since it is capable of segregating the vsp locus into several known genomic fragments (Fig. 1). vsp-related genomic fragments carrying vsp genes corresponding to those localized within the 23-kb cloned locus were detected by both oligonucleotide probes (Fig. 4, lanes 1 and 2). The vspM gene was not detected due to the presence of several HindIII sites within the highly conserved region generating small and unidentifiable fragments (Fig. 1). Interestingly, however, four additional HindIII genomic fragments that hybridized with both oligonucleotide probes were observed (Fig. 4, lanes 1 and 2, indicated by unlabeled arrows). These results indicate that additional nucleotide sequences with homology to the highly conserved vsp domains are present and may also represent additional vsp genes comprising a larger gene family.
FIG. 4.
Identification of vsp-related genomic fragments in the M. bovis PG45 clonal isolate. HindIII-digested chromosomal DNA (about 4 μg) from the M. bovis clonal isolate (PG45) with the expression phenotype VspA+ VspB+ (63-kDa VspA and 46-kDa VspB) (11) was subjected to Southern blot hybridization with an oligonucleotide probe representing the highly conserved region 5′ upstream of vsp (lane 1) or with an oligonucleotide representing a sequence complementary to the highly homologous Vsp signal peptide-encoding sequence (lane 2). Genomic fragments bearing known vsp genes identified by the oligonucleotide probes are indicated by labeled arrows. Fragments carrying only the highly conserved 5′ upstream region or the signal-encoding region are labeled P and S, respectively, together with the letter of the corresponding gene. Unlabeled arrows indicate additional vsp-related genomic fragments. Molecular size markers are shown on the left.
DISCUSSION
This study identified in the bovine pathogen M. bovis a genomic locus bearing a large family of multiple related vsp genes utilized for generating and maintaining surface antigenic variation. All vsp genes encode a highly conserved N-terminal region mediating membrane translocation and anchorage of surface-exposed lipoproteins (Fig. 2B). Since these are wall-less bacteria and a periplasmic space is absent, the use of acylated proteins with long-chain fatty acids is an effective way to anchor and expose surface antigens (19, 32). Lipoproteins are therefore remarkably abundant in mycoplasmal membranes (19) and are increasingly identified as key antigens in systems capable of diversifying the antigenic character of the mycoplasmal cell surface (1, 6, 14, 19, 21, 26, 35). Interestingly, the presence of a hydrophilic residue, Lys, preceding the Cys residue (−1 position) within the lipoprotein box of every Vsp is very uncommon. Analysis of the signal sequences of 75 distinct lipid-modified precursors has revealed that glycine (55%), alanine (38%), and serine (7%) residues are strongly biased at the −1 position with respect to the Cys residue (4, 8). None of these lipoproteins was found to contain a Lys residue at the −1 position. Notably, this peculiar Lys residue was found in the 13 vsp genes in the M. bovis strain PG45 reported in this study, in 10 vsp genes of the M. bovis strain 422 (11), and in 3 distinct vsp-related lipoprotein-encoding genes of the Mycoplasma agalactiae strain PG2, which is known to be closely related to M. bovis (7). Despite the presence of such a peculiar amino acid residue within the Vsp lipoprotein box, the migration of the recombinant products VspA, VspB, and VspC expressed in E. coli on SDS-PAGE gels was similar to the migration of authentic products expressed in the mycoplasma (reference 12, Fig. 1B, and data not shown, respectively). Further experiments are needed to determine the ability of Vsp lipoprotein-like signal sequences to be processed by the classic prolipoprotein pathway (4, 8).
A common feature of antigens involved in surface antigenic variation is the presence of a highly mutable module composed of coding sequences reiterated in tandem. This domain is subject to frequent contraction or expansion of these intragenic repetitive units, resulting in the expression of size variants of the corresponding protein. In this respect the Vsps are no exception. However, unlike other variable lipoproteins which have reiterated coding sequences only at their carboxy-terminal ends (19), approximately 80% of all Vsp molecules are made up of periodic structures extending from the N-terminal to the C-terminal end (Fig. 3), a motif that is also present in the M protein surface antigen of group A streptococci (9). This Vsp structural motif confirmed our earlier observations that Vsp size variation is generated not only by changes within the C-terminal domain analogous to those reported for the Vlp (33), Vaa (36), and Vsa (26) systems but also by similar changes within other regions of the Vsp molecules (1).
In previous studies, three Vsp products (VspA, VspB, and VspC) were identified and shown to possess a common epitope recognized by MAb 1E5 (1, 22). Each of these three products was shown to undergo independent high-frequency changes in size as well as noncoordinate phase variation in expression. The distinction among these three translational products was based on their epitope profiles and on patterns of degradation at carboxypeptidase Y pause sites. These structural fingerprints could easily distinguish between VspA and VspB products (1). The genes encoding these products were identified based on the expression of recombinant products in E. coli. Recombinant VspA or VspB products were demonstrated to be similar to the authentic VspA products or to the VspB products expressed in the mycoplasma both by epitope specificity and by size (reference 12 and Fig. 1). As to the vspC gene, it was previously shown that the carboxypeptidase Y digestion fingerprints of the VspA and VspC proteins were remarkably identical (1). Despite this marked structural similarity between the VspA protein and the VspC protein, we could not detect the vspC gene. Southern blot hybridization using several oligonucleotide probes representing distinct regions of the vspA gene failed to detect the third, related gene, vspC, in the chromosome of the clonal isolate analyzed (data not shown). Interestingly, however, preliminary genetic analysis of another M. bovis clonal isolate expressing VspC as a single product has shown that a recombination event occurring between the vspA gene and the vspO gene results in the generation of the vspC gene (13). These intriguing findings are consistent with (i) the observed profound structural similarity between VspA and VspC proteins (ii), the fact that coexpression of VspA and VspC proteins in a single M. bovis clonal isolate was not observed, and (iii) our inability to detect the vspC gene in the genome of the M. bovis PG45 clonal isolate expressing the products VspA and VspB. Therefore, among the vsp genes, the vspC gene was not assigned a designation (Fig. 1).
Behrens et al. (2) have used electron microscopy to localize variable proteins on the surface of M. bovis PG45. In addition to VspA, VspB, and VspC, they designated a fourth protein, VspD. No evidence indicating that VspD is indeed a member of the Vsp family was provided. The fact that this protein was shown to be recognized by MAb 1E5 and by MAb 87-2 can be simply explained by the presence of a common epitope. Since we could not correlate this protein to any of the vsp genes reported in our study, the designation vspD gene within the vsp locus was not used.
As to the molecular mechanism mediating Vsp phase variation, Vsp phenotypic switching was shown to be associated with genomic rearrangement events occurring at a high frequency (12). The presence of regions of high sequence homology upstream of each vsp gene and within the vsp structural coding regions (Fig. 2 and 3 and Table 1) provides potential sites for recombination initiation within the vsp locus, which is a possible mechanism for Vsp ON-OFF switching. Data obtained in our laboratory recently have shown that apparently only two vsp genes are expressed at a given time in a single isolate. The rest of the vsp genes are transcriptionally silent (13). Oscillating phase transition of the VspA product between ON and OFF expression states was shown to be a result of a recombination event that occurs between the highly conserved sequences upstream of vsp and to involve DNA inversion of three distinct vsp genes. The consequence of this event is the replacement of one of the upstream cassettes of the vspA gene with the corresponding cassette of another, silent vsp gene (13).
Elucidation of the molecular genetic basis of Vsp phenotypic switching is currently under way. This analysis will provide an insight into genomic rearrangement processes mediating surface antigenic variation and will contribute to a better understanding of the successful persistence of pathogenic mycoplasmas in their natural hosts.
ACKNOWLEDGMENTS
This study was supported in part by the German-Israel Foundation for Scientific Research and Development (GIF), by the United States-Israel Binational Agricultural Research and Development Fund (BARD), and by the Israel Academy of Sciences and Humanities Foundation.
REFERENCES
- 1.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun. 1994;62:5075–5084. doi: 10.1128/iai.62.11.5075-5084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens A, Heller M, Rosenbusch R, Kirchhoff H. Immunoelectron microscopic localization of variable protein on the surface of Mycoplasma bovis. Microbiology. 1996;142:1863–1871. doi: 10.1099/13500872-142-7-1863. [DOI] [PubMed] [Google Scholar]
- 3.Bergonier D, de Simon F, Russo P, Solsona M, Lambert M, Poumarat F. Variable expression and geographic distribution of Mycoplasma agalactiae surface epitopes demonstrated with monoclonal antibodies. FEMS Microbiol Lett. 1996;143:159–165. doi: 10.1111/j.1574-6968.1996.tb08475.x. [DOI] [PubMed] [Google Scholar]
- 4.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J-M, Hakenbeck R, editors. New comprehensive biochemistry, vol. 27: bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 319–341. [Google Scholar]
- 5.Colman S D, Hu P C, Litaker W, Bott K F. A physical map of the Mycoplasma genitalium genome. Mol Microbiol. 1990;4:683–687. doi: 10.1111/j.1365-2958.1990.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 6.Droesse M, Tangen G, Gummelt I, Kirchhoff H, Washburn L R, Rosengarten R. Major membrane proteins and lipoproteins as highly variable immunogenic surface components and strain-specific antigenic markers of Mycoplasma arthritidis. Microbiology. 1995;141:3207–3219. doi: 10.1099/13500872-141-12-3207. [DOI] [PubMed] [Google Scholar]
- 7.Flitman-Tene, R., S. Levisohn, I. Lysnyansky, E. Rapoport, and D. Yogev. Unpublished data. [DOI] [PubMed]
- 8.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–470. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 9.Hollingshead S K, Fischetti V A, Scott J R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987;207:196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- 10.Jasper D E. The role of Mycoplasma in bovine mastitis. J Am Vet Med Assoc. 1982;181:158–162. [PubMed] [Google Scholar]
- 11.Kotzer, S., I. Lysnyansky, and D. Yogev. Unpublished data.
- 12.Lysnyansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lysnyansky, I., K. Sachse, and D. Yogev. Unpublished data.
- 14.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 16.Noormohammadi A H, Markham P F, Whithear K G, Walker I D, Ley D H, Browning G F. Mycoplasma synoviae has two distinct phase-variable major membrane antigens, one of which is a putative hemagglutinin. Infect Immun. 1997;65:2542–2547. doi: 10.1128/iai.65.7.2542-2547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfützner H, Sachse K. Mycoplasma bovis as an agent of mastitis pneumonia, arthritis and genital disorders in cattle. Rev Sci Tech Off Int Epizoot. 1996;15:1477–1494. doi: 10.20506/rst.15.4.987. [DOI] [PubMed] [Google Scholar]
- 18.Razin S. Peculiar properties of mycoplasmas: the smallest self-replicating prokaryotes. FEMS Microbiol Lett. 1992;100:423–432. doi: 10.1111/j.1574-6968.1992.tb14072.x. [DOI] [PubMed] [Google Scholar]
- 19.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson B D, Meyer T F. Antigenic variation in bacterial pathogens. In: Hormaeche C W, Penn C W, Smyth C J, editors. Molecular biology of bacterial infection. Vol. 49. Cambridge, England: Cambridge University Press; 1992. pp. 61–73. [Google Scholar]
- 21.Rosengarten R, Wise K S. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science. 1990;247:315–318. doi: 10.1126/science.1688663. [DOI] [PubMed] [Google Scholar]
- 22.Rosengarten R, Behrens A, Stetefeld A, Heller M, Ahrens M, Sachse K, Yogev D, Kirchhoff H. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect Immun. 1994;62:5066–5074. doi: 10.1128/iai.62.11.5066-5074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simecka J W, Davis J K, Davidson M K, Ross S E, Stadtländer C T K-H, Cassell G H. Mycoplasma diseases of animals. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 391–416. [Google Scholar]
- 26.Simmons W L, Zuhua C, Glass J I, Simecka J W, Cassell G H, Watson H L. Sequence analysis of the chromosomal region around and within the V-1-encoding gene of Mycoplasma pulmonis: evidence for DNA inversion as a mechanism for V-1 variation. Infect Immun. 1996;64:472–479. doi: 10.1128/iai.64.2.472-479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theiss P M, Kim M F, Wise K S. Differential protein expression and surface presentation generate high-frequency antigenic variation in Mycoplasma fermentans. Infect Immun. 1993;61:5123–5128. doi: 10.1128/iai.61.12.5123-5128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tully J G, Whitcomb R F. The mycoplasmas. 2. Human and animal mycoplasmas. New York, N.Y: Academic Press; 1979. [Google Scholar]
- 30.Watson H L, McDaniel L S, Blalock D K, Fallon M T, Cassell G H. Heterogeneity among strains and a high rate of variation within strains of a major surface antigen of Mycoplasma pulmonis. Infect Immun. 1988;56:1358–1363. doi: 10.1128/iai.56.5.1358-1363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise K S, Yogev D, Rosengarten R. Antigenic variation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 473–489. [Google Scholar]
- 32.Wise K S. Adaptive surface variation in mycoplasmas. Trends Microbiol. 1993;1:59–63. doi: 10.1016/0966-842X(93)90034-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yogev D, Rosengarten R, Watson-McKown R, Wise K S. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 1991;10:4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yogev D, Menaker D, Strutzberg K, Levisohn S, Kirchhoff H, Hinz K-H, Rosengarten R. A surface epitope undergoing high-frequency phase variation is shared by Mycoplasma gallisepticum and Mycoplasma bovis. Infect Immun. 1994;62:4962–4968. doi: 10.1128/iai.62.11.4962-4968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yogev D, Watson-McKown R, Rosengarten R, Im J, Wise K S. Increased structural and combinatorial diversity in an extended family of genes encoding Vlp surface proteins of Mycoplasma hyorhinis. J Bacteriol. 1995;177:5636–5643. doi: 10.1128/jb.177.19.5636-5643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Wise K S. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect Immun. 1996;64:2737–2744. doi: 10.1128/iai.64.7.2737-2744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Wise K S. Localized reversible frameshift mutation in an adhesin gene confers a phase-variable adherence phenotype in mycoplasma. Mol Microbiol. 1997;25:859–869. doi: 10.1111/j.1365-2958.1997.mmi509.x. [DOI] [PubMed] [Google Scholar]
- 38.Zheng X, Watson H L, Waites K B, Cassell G H. Serotype diversity and antigenic variation among invasive isolates of Ureaplasma urealyticum from neonates. Infect Immun. 1992;60:3472–3474. doi: 10.1128/iai.60.8.3472-3474.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]