Abstract
Neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and Parkinson’s disease (PD) are both characterized by pathogenic protein aggregates that correlate with the progressive degeneration of neurons and the loss of behavioral functions. Both diseases lack biomarkers for diagnosis and treatment efficacy. Proteomics is an unbiased quantitative tool capable of the high throughput quantitation of thousands of proteins from minimal sample volumes. We review recent proteomic studies in human tissues, plasma, cerebrospinal fluid (CSF), and exosomes in ALS and PD that identify proteins with potential utility as biomarkers. Further, we review disease-related post-translational modifications in key proteins TDP43 in ALS and α-synuclein in PD studies, which may serve as biomarkers. We compare relative and absolute quantitative proteomic approaches in key biomarker studies in ALS and PD and discuss recent technological advancements which may identify suitable biomarkers for the early-diagnosis treatment efficacy of these diseases.
Keywords: biomarkers, neurodegeneration, ALS, PD, TDP43, proteomics, LC-MS/MS
1. Introduction
Amyotrophic lateral sclerosis (ALS) and Parkinson’s disease (PD) are neurodegenerative diseases characterized by the progressive degeneration of neurons.
In ALS, the progressive degeneration of motor neurons occurs with a debilitating loss of movement control. The majority (90–95%) of ALS cases are sporadic in origin and 5–10% of cases are of known genetic origin [1]. Of the known genetic cases, mutations in the C9ORF72, SOD1, TARDBP [2,3,4], NEK1 [5,6], UBQLN2 [7,8], KIF5A [9,10], and FUS [11,12] genes contribute to familial ALS cases [13]. Other genes that contribute to familial ALS includes VCP, ALS2, SETX, ANG, PFN1, MATR3, CHCHD10, TUBA4A, TBK1, GRN, C21orf2, and OPTN [14].
In PD, the progressive degeneration of dopaminergic neurons occurs with a loss of movement control. The majority (90%) of PD cases are sporadic in origin and 10% of cases are of known genetic origin [15]. Risk for familial PD has been associated with 28 distinct chromosomal regions called PARK genes [16,17,18,19]. Six genes are linked with monogenic familial PD. They include SNCA and LRRK2 mutations that have been linked with an autosomal dominant form and Parkin, PINK1, DJ-1, and ATP13A2, which have been linked with an autosomal recessive mode of inheritance in familial PD. Variants in certain other genes such as UCHL1, GAK, MAPT, GBA, NAT2, INOS2A, GAK, HLA-DRA, and APOE are associated with increased risk of PD, where additional factors, along with mutations, might play a role in disease causation [20].
Biomarkers are essential for the early diagnosis, prognosis, and assessment of treatment for both ALS and PD, but biomarker discovery has been challenging for these diseases.
The increased use of unbiased tools including proteomics, however, is advancing biomarker discovery. Proteomics enables biomarker discovery using traditional mass spectrometry-based platforms as well as newer platforms such as multiplexed immunoassays and aptamer techniques by identifying disease-associated protein alterations. Here, we provide a detailed review of several key proteomics studies showing promising techniques and describe the technological advancements for ALS and PD biomarker discovery.
2. Overview of Proteomics Technological Advancement for Biomarker Discovery
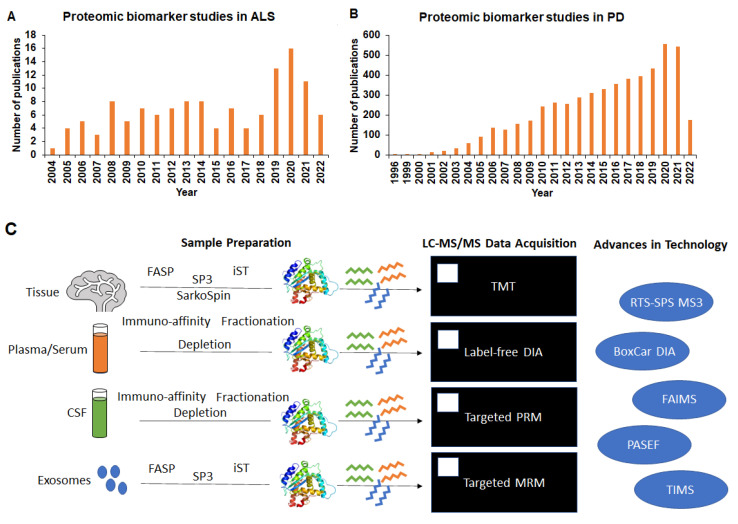
Proteomics tools enable the deep protein profiling of tissue, plasma, serum, and CSF for biomarker discovery [21]. Using proteomics, several studies have attempted to identify pathological disease mechanisms by identifying differentially expressed proteins and proteoforms (proteins and their modifications) as potential biomarkers in disease models, biofluids from patients, as well as post-mortem brain tissue. Discovery proteomics technology using mass spectrometry, immunoassays, and aptamers can identify several thousands of differentially expressed proteins and their altered forms with disease. However, only a fraction of them have translated to clinical utility due to the lack of standardized validated assays and the lack of access to instrumentation. Several studies that assessed specific brain regions for biomarker discovery fail to recognize the systemic mechanisms in neurodegenerative disease progression. Many neurodegenerative disease mechanisms converge and present differently. However, some have similar affected underlying pathways which makes the identification of disease-specific biomarkers a challenge. The gap needs to be bridged between the increasing number of publications studying biomarkers for ALS and PD (Figure 1A,B) and relatively few biomarkers in the clinic.
Figure 1.
Summary of the number of proteomic biomarker studies in (A) ALS and (B) PD. (C) Schematic of proteomics workflow and advances in mass spectrometry instrumentation for biomarker discovery.
Some of the technological advances in mass-spectrometry-based proteomics for biomarker discovery are described in Figure 1C and Table 1.
Table 1.
Summary of methods used in proteomics.
| Method | Advantages | Disadvantages |
|---|---|---|
| Sample Preparation Methods | ||
| Filter-aided sample preparation (FASP) | Unbiased filter-based approach which removes detergents |
Molecular weight cutoffs and can be challenging for aggregated proteins |
| Single-pot solid-phase-enhanced sample preparation (SP3) | Bead-based approach with low sample loss Unbiased robust recovery and can be automated using magnetic beads |
Beads have a limited capacity and should not be overloaded to cause inconsistencies |
| In-StageTip (iST) | Peptide can be fractionated on the tip to gain depth | Tips must be compatible with solubilization reagents used |
| SarkoSpin | Can isolate insoluble pathological protein aggregated | Utilize detergents that require sample clean up prior to mass spectrometry |
| Depletion | Gain depth | Low throughput, induce variability |
| Immunoenrichment | Gain depth | Low throughput, induce variability |
| Offline fractionation | Gain depth | Low throughput |
| Mass Spectrometry Quantitation Strategies | ||
| Isobaric TMT tags | Relative quantitation in MS2/MS3 dimension with multiplexing to save time | Require (SPS) MS3 for accurate quantitation to overcome challenges with ratio compression |
| Label-free DIA | Relative quantitation by area under the peak, enabling the acquisition of complete data in large cohorts | Requires expertise in MS method design for window strategy and data interpretation |
| MRM/PRM-targeted quantitation | Absolute quantitation with standard curves and the ability to monitor disease progression with time | Limited in the number of targets that can be analyzed |
| Advances in Instrumentation and Technology | ||
| BOXCAR data-independent acquisition | Improves data completeness | Requires good method design |
| Real-time search RTS-SPS-MS3 | Improves quantitation | Special feature in an instrument |
| High-field asymmetric ion mobility spectrometry FAIMS | Improves sensitivity and selectivity | In front end and not true ion mobility to separate isomers |
| Modified LC- Evosep | Improves throughput and robustness | Has defined methods and is not customizable |
| Automation | Improves reproducibility | Tedious to implement changes in workflows |
| Trapped ion mobility TIMS | PTM and isoform identification | Needs expertise to achieve good separation |
| Parallel acquisition serial fragmentation PASEF | Improves scan speed and sensitivity | Needs optimization based on gradient |
| Multiplexed immunoassay O-link | Improves dynamic range based on antibody specificity | Limited in the panel of targets based on the availability of antibodies |
| Aptamer-based assay | Improves dynamic range based on aptamer specificity | Limited in the panel of targets based on the availability of aptamers |
Improved instrumentation, including faster scan speeds, parallel accumulation serial fragmentation (PASEF) [22,23], and trapped ion mobility [24,25], enables the identification of disease-associated proteins and their modifications from patient samples using small sample volumes. While these technological advances drive the limits of quantitation and enable sensitive analysis to gain the greatest depth in proteome being measured, the need to analyze thousands of samples robustly still remains a challenge in biomarker discovery.
Improvements in data acquisition strategies include using label-free data-independent acquisition (DIA), making the relative quantitation of proteins possible from many samples. In the DIA approach, complete data are acquired using a label-free strategy matched to a large spectral library built from representative samples of the cohort. While previously DIA required building and using a spectral library for the accurate identification of the acquired MS2 spectra [26], recent advances in direct DIA and machine learning algorithms allow library-free accurate identification [27] and quantification [28]. Today, >10,000 proteins from tissue [29] or >1000 proteins from plasma can be identified by DIA [30]. To overcome the dynamic range challenge and gain depth in the number of proteins identified from complex matrices such as plasma/serum, using sample preparation strategies including depletion, immuno-enrichment, and data acquisition strategies (such as the BoxCar-segmented MS1 approach and BoxCarmax) can achieve maximized depth in the proteome [31,32,33].
An alternative relative quantitative strategy uses isobaric TMT tags. Complete deep data profiles for proteins are acquired in the MS2 dimension and enhanced quantitation using synchronous precursor selection (SPS) in the MS3 dimension overcomes inherent challenges with TMT such as ratio compression. Currently, the use of real-time search SPS MS3 gives the accurate quantitation and in-depth profiling of pooled fractionated samples without any sacrifice in the number of proteins identified, overcoming previous limitations in TMT-based approaches. Using automated sample preparation, ion mobility with high-field asymmetric waveform ion mobility spectrometry (FAIMS), and real-time search SPS MS3, deep protein profiles from challenging matrices such as plasma can be obtained [34,35].
Further, absolute quantitation strategies using targeted proteomics approaches to monitor selected peptides or proteins by parallel reaction monitoring (PRM) or multiple reaction monitoring (MRM) across large study cohorts identified biomarkers are validated for clinical use [36]. An advancement in this field includes the improved sensitivity in targeted analysis platforms.
Traditional nano-LC approaches allow a small number of samples to be analyzed per day, making rapid and robust analysis of large cohorts for biomarker discovery challenging. An advancement in throughput involves the use of novel LC platforms such as Evosep which can analyze up to 300 samples per day in a rapid and ultra-robust analysis compared to traditional nano-LC approaches [37]. Using this platform makes clinical proteomic analysis for biomarker discovery possible on a large scale.
Other non-mass-spectrometry-based proteomics techniques for biomarker discovery include O-link and aptamer-based technology. O-link technology uses a multiplexed immunoassay and has the advantage of overcoming the dynamic range challenge via the enrichment of proteins with antibodies, but is limited in the panel of proteins that can be detected because it relies on the availability of specific antibodies [38]. Aptamer technology involves small nucleic acids binding to proteins and subsequent detection by means of complementary nucleic acids and fluorescence [39,40]. While these approaches have high throughput, they rely on the availability of well-characterized reagents, unlike mass spectrometry which is an unbiased proteomic approach. Combining these complementary approaches of mass-spectrometry-based proteomics, immunoassays, and aptamers can help to gain depth in the proteins identified [41].
3. Proteomics for Biomarker Discovery in ALS
3.1. Differential Expression of Proteins and Interactome from Post-Mortem Human Tissue as Biomarkers in ALS
Post-mortem tissue proteomics is a key method used for identifying disease biomarkers in bulk tissues, tissue sections on a slide, and single cells. Several workflows have been suggested for bulk tissue analysis such as suspension trapping filter-aided sample preparation (FASP), single-pot solid-phase-enhanced sample preparation (SP3), and in-StageTip (iST) [42] for disease biomarker analysis. On a FFPE/frozen slide, proteins associated with pathological regions in the tissue can be identified by liquid chromatography–tandem mass spectrometry (LC-MS/MS) or matrix-assisted laser desorption–ionization (MALDI) approaches [43,44,45,46]. Using a combination of imaging and high-resolution mass spectrometry, the characterization of proteins at a single cell resolution and the intracellular sub-proteome has been possible [47,48]. This has been utilized to study the sub-proteome of endosomal vesicle trafficking [49]. Since these endo-lysosomal networks are known to be dysregulated in neurodegenerative diseases, the signatures of potential biomarkers can be detected [50]. These approaches enable biomarker discovery from tissue.
Studies that provide insight into potential tissue markers in ALS are summarized in Table 2. In ALS, proteomic profiling via LC-MS/MS in post-mortem spinal cord tissue compared sporadic ALS and controls, revealed ATP5D, and calmodulin was downregulated in sporadic ALS [51]. Here, 2D-gel electrophoresis combined with LC-MS/MS was used in identification of biomarkers. While 2D gel electrophoresis enables the isoelectric separation of proteins, the method is relatively low throughput and requires expertise to isolate spots consistently. By assessing ALS mutation carriers (C9orf72, SOD1, and TARDBP), sporadic ALS and controls identified several proteins that were upregulated, including UCHL1, MAP2, CAPG, GPNMB, HIST1H4A, HIST1H2B, NEFL, NEFH, NEFM, CHIT1, and CHI3L1 in both spinal cord and CSF in ALS [52]. The upregulation of four of these proteins (UCHL1, MAP2, CAPG, and GPNMB) in both CSF and spinal cord was confirmed in an independent cohort by MRM [52]. CSF from symptomatic and asymptomatic ALS mutation carriers was assessed, and NEFL, NEFM, NEFH, CHIT1, and CHI3L1 were upregulated in symptomatic individuals [52]. The strength of this analysis lies in the use of both post-mortem spinal cord and CSF in discovery proteomics. The CSF proteomics method was modified to enhance depth. Further, candidate markers were validated by absolute quantitation using a targeted proteomic approach in an independent cohort. Using both discovery and targeted proteomics approaches to validate markers in an independent cohort, this study provides a comprehensive and validated approach in identifying biomarkers. Proteomics is also valuable in assessing the interactome. A proximity-based ligation BioID assay of di-peptide repeats in ALS patients with C9ORF72 mutations identified chaperone proteins to be associated with poly-GA, while ribosomal and nucleolar proteins were associated with poly-GR and poly-GP di-peptide repeats in ALS patients [53,54]. Toxic repeats can be identified using BioID to analyze proteins which provide insight into disease mechanisms. A disadvantage of the method is the need to express a vector and identify interacting proteins. This approach is possible in model systems and primary cells, but not in patient samples. In these references, primary proteins interacting with poly-GA, poly-GR, and poly-GP repeats were identified in cortical neurons and cells, and these were validated in patient samples by mass spectrometry. This is a balanced approach in addressing disease mechanisms in model systems and confirms the findings in patient samples.
Table 2.
Proteomic studies in ALS focused on biomarker discovery.
| Disease | Marker | Quantitation | Tissue | Summary | Reference |
|---|---|---|---|---|---|
| Tissue-based proteomic markers in ALS | |||||
| ALS | TDP43 | PRM absolute quantitation |
Prefrontal/motor cortex and spinal cord | An increase in C: N-terminal TDP43 peptide ratio > 1.5, new truncation site-specific trend observed in ALS-TDP | [51,55] |
| ALS (sporadic) |
Calmodulin | Label-free | Spinal cord | Downregulated in ALS | [51] |
| ALS (sporadic) |
ATP5D | Label-free | Spinal cord | Downregulated in ALS | [51] |
| ALS | UCHL1 | Label-free and MRM | Spinal cord | Upregulated in ALS and correlated with CSF | [52] |
| ALS | MAP2 | Label-free and MRM | Spinal cord | Upregulated in ALS and correlated with CSF | [52] |
| ALS | GPNMB | Label-free and MRM | Spinal cord | Upregulated in ALS and correlated with CSF | [52] |
| Plasma/Serum proteomics biomarkers in ALS | |||||
| ALS | Gelsolin | LFQ and MRM | Plasma | Differentially expressed in ALS | [56,57,58] |
| ALS | Clusterin | MRM | Plasma | Downregulated in ALS | [57,58] |
| ALS | CD5L | MRM | Plasma | Differentially expressed in ALS | [57,58] |
| ALS | Ficolin 3 | MRM | Plasma | Upregulated in ALS | [57,58] |
| CSF proteomic biomarkers in ALS | |||||
| ALS | α-1-antichymotrypsin | LFQ | CSF | In CSF, 118 proteins were significantly altered in ALS compared to controls | [56] |
| ALS | Amyloid beta A4 protein | LFQ | CSF | In CSF, 118 proteins were significantly altered in ALS compared to controls | [56] |
| ALS | Gelsolin | LFQ | CSF | In CSF, 118 proteins were significantly altered in ALS compared to controls | [56] |
| ALS | Chitinase-3-like protein 1 (CHI3L1) |
LFQ | CSF | Upregulated | [59] |
| ALS | Chitinase-3-like protein 2 (CHI3L2) |
LFQ, TMT | CSF | Upregulated in mutated C9orf72 symptomatic ALS compared to asymptomatic controls with C9orf72 mutations | [59,60] |
| ALS | Chitotriosidase-1 (CHIT-1) | LFQ, TMT | CSF | Upregulated in mutated C9orf72 symptomatic ALS compared to asymptomatic controls with C9orf72 mutations | [59,60] |
| ALS | Ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) | LFQ, TMT, MRM | CSF | Upregulated in mutated C9orf72 symptomatic ALS compared to asymptomatic controls with C9orf72 mutations | [52,59,60] |
| ALS | MAP2 | MRM | CSF | Upregulated | [52] |
| ALS | CAPG | MRM | CSF | Upregulated | [52] |
| ALS | GPNMB | MRM | CSF | Upregulated | [52] |
| ALS | CRYAB | TMT | CSF | Upregulated in mutated C9orf72 symptomatic ALS compared to asymptomatic controls with C9orf72 mutations | [60] |
| ALS | PFN1 | TMT | CSF | Upregulated in mutated C9orf72 symptomatic ALS compared to asymptomatic controls with C9orf72 mutations | [60] |
| ALS | TFRC | TMT | CSF | Upregulated in mutated C9orf72 symptomatic ALS compared to asymptomatic controls with C9orf72 mutations | [60] |
| ALS | TREM2 | TMT | CSF | Upregulated in C9orf72 variant-associated symptomatic ALS compared to asymptomatic controls with C9orf72 variants | [60] |
| ALS | TXNDC17 | TMT | CSF | Upregulated in mutated C9orf72 variant-associated symptomatic ALS compared to asymptomatic controls with C9orf72 variants | [60] |
| ALS | NEFM | TMT | CSF | Upregulated in mutated C9orf72 symptomatic ALS compared to asymptomatic controls with C9orf72 mutations | [60] |
| Exosomal biomarkers in ALS | |||||
| ALS | Gelsolin | LFQ | CSF exosomes | Upregulated in C9orf mutated ALS cases | [61] |
| ALS | Clusterin | LFQ | CSF exosomes | Upregulated | [61] |
| ALS | UBA1 | LFQ | CSF exosomes | Upregulated in C9orf mutated ALS cases | [61] |
| ALS | NIR | LFQ | CSF exosomes | Upregulated in sporadic ALS | [62] |
| ALS | TDP43 | LFQ | Plasma exosomes | Levels correlated with longitudinal progression | [63] |
3.2. Post-Translational Modifications in TDP43 from Tissue Proteomics Studies in ALS
Post-translational modifications (PTMs) include changes in the amino acid side chains in proteins due to the covalent addition of functional groups or proteolytic cleavage [64]. PTMs are readily detected by mass spectrometry due to characteristic mass shifts caused by the biochemical modification of proteins [65]. Abnormal PTMs can disrupt normal biological processes and serve as biomarkers for disease diagnosis and progression [66,67].
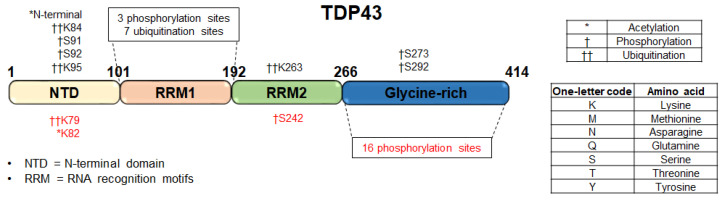
A pathological hallmark of ALS is the presence of cytoplasmic inclusions of TAR DNA-binding protein 43 (TDP43) and mislocalization of the protein from the nucleus to the cytoplasm [68]. Major modifications in TDP43 associated with protein aggregates include truncations, ubiquitination, and hyperphosphorlation in the C-terminal and N-terminal domains [69,70] (Figure 2).
Figure 2.
Schematic representation of post-translational modifications (PTMs) in TDP43 in ALS. The red font indicates putative disease-specific modifications [70] and the black font indicates all known modifications from proteomics databases (ProteomicsDB and PhosPhosite) and literature.
PTMs in pathological TDP43 were differentiated from endogenous TDP43 using a method called SarkoSpin (a technique for the isolation of pathological TDP43 aggregates), and characterized by mass spectrometry from ALS brains [71]. Using SarkoSpin on ALS and FTD brains, normal proteins including physiological TDP43 were exclusively found in the supernatants, while protein aggregates such as pathological TDP43 that underwent polyubiquitation and hyperphosphorylation were detected in the pellets. SarkoSpin is an excellent method used to separate insoluble disease-specific aggregates from the soluble form and could potentially be applied to other aggregated proteins common in neurodegenrative diseases. Here, after separating the pathological TDP43 aggregates, LC-MS/MS with spectral counting was used for the relative quantitation of proteins in different groups. A label-free approach allows intact PTMs to be evaluated, in turn idenitfying and quantifying the relative abundance of proteins in different disease groups. Phosphorylated TDP43 leads to an increase in cytoplasmic and mitochondrial mislocalization and aggregation in neurons. The ubiquitination of TDP43 has been associated with TDP43 aggregation [72]. These modifications that are unique to disease pathology could serve as biomarkers for ALS. Most of the modifications and cleavages occurred in the glycine-rich C-terminal and N-terminal domains of TDP43 [70]. In ALS, a measured ratio of C:N terminal TDP43 fragments >1.5 could differentiate ALS from control subjects in a study using post-mortem brain and spinal cord tissue [55].
3.3. Plasma and Serum as Sources for Proteomic Biomarkers in ALS
There are several strategies for plasma protein profiling in disease biomarker identification. The first technique presented here uses mass-spectrometry-based proteomics in plasma. To successfully identify disease biomarkers from plasma, it is important to find ways to measure the low-abundance disease-relevant proteins, while overcoming the large dynamic range of proteins in plasma. This is challenging, since most instruments can measure a range of up to 5 orders of magnitude, with bias towards the most abundant proteins, while proteins in plasma span 10 orders in magnitude, with disease-specific proteins being lower in abundance. The depletion of the most abundant proteins [73,74], the enrichment of proteins of interest with antibodies [75], and deep offline fractionation are commonly used methods [75] to identify low-abundance disease-relevant proteins. While these methods increase depth, they result in lower throughput [76]. These strategies have identified clusterin and ficolin-3 as differentially expressed proteins in ALS plasma [57,58]. Here, SWATH was applied for the identification of proteins, and levels were confirmed by Western blotting. SWATH has the advantage of being label-free, and can preserve modifications in proteins for later query if needed and provide relative quantitation. The disadvantage of SWATH is the requirement of careful design in windows to gain the maximum number of proteins identified, thus involving the need for complex software to interpret the spectra. The advantages of being unbiased and label-free, as well as providing complete data with minimal missing values, outweigh these disadvantages. The deep plasma profiling of matched plasma and CSF samples showed that the upregulation of gelsolin and several proteins such as chitinase-3-like 1 and alpha-1 antichymotrypsin was validated in ALS plasma and CSF [56]. Here, matched plasma and CSF samples were analyzed by discovery proteomics. The advantage of using matched samples enables the identification of neurological and systemic changes in proteins due to the disease. Label-free quantitation provided a relative abundance of differentially expressed proteins. Using machine learning algorithms, two candidate proteins were validated for absolute levels by a targeted proteomics approach. This strategy uses both discovery and targeted proteomics with machine learning to select candidate markers which provides a well-defined strategy for biomarker discovery.
Another strategy to gain depth without necessarily compromising throughput involves using the TMT calibrator method, where spiking the disease peptides from peripheral blood mononuclear cells (PBMCs) or brain tissue in multiplex channels of TMT boosts the associated plasma proteome being measured in the other channels. ALS studies have used this technique to study the rate of disease progression [57,58] and phenotypic variability in sporadic ALS patients [77,78], and have identified proteins in biological processes of senescence, RNA processing, cell stress, and metabolism; moreover, major histocompatibility complex-II-linked immune reactivity and apoptosis were enriched in fast-progressing ALS. While using a booster channel from tissue or PBMC is advantageous in boosting low-abundance peptide signals seen in biofluids that are otherwise masked, the validation of positive identification is required. Quantitation using isobaric tags enables multiplexing and depth gains. However, this method suffers from ratio compression which leads to inaccurate quantitation. The development of novel strategies such as real-time search MS3 overcomes some of the previous limitations in TMT-based quantitation. The known plasma biomarkers in ALS from discovery proteomics approaches are summarized in Table 2.
3.4. Cerebrospinal Fluid (CSF) Proteomic Biomarker Identification
CSF analysis is widely used in biomarker studies of neurodegenerative disease, since CSF is believed to reflect brain processes within the blood–brain barrier. Important considerations in CSF proteomics include the proper collection and storage of samples to minimize blood contamination and maximize protein stability [79]. In CSF, as with plasma, depletion [80], immuno-enrichment, and fractionation have been applied in several studies to acquire deep CSF profiles in ALS.
Using these approaches, differentially expressed proteins in CSF from ALS patients have been identified as potential biomarkers (Table 2). Using isobaric tags for relative quantitation, nine proteins were upregulated in CSF in C9orf72 variant-associated ALS compared to controls. These include chitinase-3-like protein 2 (CHI3L2), alpha-crystallin B chain (CRYAB), profilin-1 (PFN1), transferrin receptor protein 1 (TFRC), triggering receptor expressed on myeloid cells 2 (TREM2), thioredoxin domain-containing protein 17 (TXNDC17), ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1), CHIT1, and NEFM [60]. Here, isobaric tags were used for the relative quantitation of differentially expressed proteins and eight candidate proteins were validated using a targeted proteomics approach for absolute levels. Multiplexing can be used to assess large study samples, but requires a good study design to bridge the different TMT batches. Following up the discovery proteomics with validation in an independent cohort can confidently and effectively identify biomarkers. Other studies have also demonstrated the upregulation of chitotriosidase, chitinase-3 like protein 1, chitotriosidase-3 like protein 2, chitotriosidase-1 (CHIT1), alpha-1-antichymotrypsin, and amyloid beta A4 protein [56,59,81] in ALS. In these studies, a label-free strategy was applied for the discovery of biomarkers, and a small number of candidates were validated in a separate cohort using targeted proteomics or ELISA. Label-free quantitation allows post-translational modifications to be preserved and is a great method to overcome study design biases that may occur in TMT-based workflows. Validating proteins observed in discovery proteomics using a targeted or orthogonal approach provides greater confidence in the identified biomarkers.
3.5. Exosomes Proteomics in Biomarker Identification
Exosomes are extracellular vesicles released from various types of cells, including CNS cells, and are enriched in a variety of bioactive molecules such as RNAs, proteins, and lipids [82]. Exosomes carrying cell-type-specific molecules reach the periphery by crossing the blood–brain barrier (BBB), making them ideal for biomarker studies [82]. Measuring biomarkers in neuron-derived exosomes in plasma could serve to monitor neuronal health and neuroinflammation [83,84]. Indeed, profiling CSF exosomes from sporadic ALS patients identified three downregulated proteins and eleven upregulated proteins [61]. A label-free proteomics study in CSF exosomes demonstrated the upregulation of ubiquitin-like modifying-activating protein 1 (UBA1) in C9orf72-ALS patients [61]. Exosomes have been studied in neuroinflammation and disease progression biomarkers using proteomics in ALS. Here, a label-free strategy was applied for differential proteomic analysis. This strategy has the advantage of greater sensitivity in small samples, such as exosomes, and can preserve modifications seen in proteins. Here, only relative quantitation with proteomics was applied. If these results were validated in an independent cohort using a targeted or orthogonal approach, greater confidence in the markers could be established.
4. Proteomics for Biomarker Discovery in PD
4.1. Differential Expression of Proteins from Post-Mortem Human Tissue as Biomarkers in PD
Several studies investigated differentially expressed proteins that change post-mortem tissue with PD. In a proteomic study profiling the post-mortem substantia nigra of PD patients, pathway analysis found alterations in proteins associated with mitochondrial dysfunction, oxidative stress, or cytoskeleton impairment [85]. Here, the samples were analyzed using isobaric tags and some markers were validated by immunohistochemistry and Western blot. To gain depth, offline isoelectric focusing was used. While it is great that the markers were validated using an orthogonal method, all the quantitation was carried out using relative quantitative approaches. The implementation of an absolute quantitation method might be helpful to gain accuracy in the quantified biomarkers. In another proteomic study used to identify pathways that contribute to Lewy body (protein inclusions containing aggregated proteins) pathology, pathways such as Arp2/3, synaptic function, and hydrogen peroxide metabolism were found to be directly correlated with Lewy body pathology, while poly(A) RNA binding protein pathways such as TDP43 and FUS were inversely correlated with Lewy body pathology. In a comparison of Lewy body pathology with and without neuronal loss, it was found that CD59 was upregulated and RGS6 and GANAB were downregulated [86] (Table 3). Here, label-free quantitation was performed to assess pathways that contribute to Lewy body pathology. Further, pathways were identified using the quantitative proteomics results and rigorous statistics to define pathways and proteins associated with Lewy body pathology.
Table 3.
Proteomic studies in PD focused on biomarker discovery.
| Disease | Marker | Quantitation | Tissue | Summary | Reference |
|---|---|---|---|---|---|
| Tissue-based proteomic markers in PD | |||||
| PD | Mitochondrial dysfunction, oxidative stress, cytoskeleton impairment-related proteins | TMT | Substantia nigra | Significant changes in expression levels of 204 nigral proteins in human PD samples | [85] |
| PD | RGS6 | LFQ | Substantia nigra (Lewy body pathology) |
Changes in proteins related to (1) Arp2/3 complex-mediated actin nucleation; (2) synaptic function; (3) poly(A) RNA binding; (4) basement membrane and endothelium; and (5) hydrogen peroxide metabolic processes | [86] |
| PD | GANAB | LFQ | Substantia nigra (Lewy body pathology) |
Changes in proteins related to (1) Arp2/3 complex-mediated actin nucleation; (2) synaptic function; (3) poly(A) RNA binding; (4) basement membrane and endothelium; and (5) hydrogen peroxide metabolic processes | [86] |
| PD | CD59 | LFQ | Substantia nigra (Lewy body pathology) |
Changes in proteins related to (1) Arp2/3 complex-mediated actin nucleation; (2) synaptic function; (3) poly(A) RNA binding; (4) basement membrane and endothelium; and (5) hydrogen peroxide metabolic processes | [86] |
| Plasma/serum proteomic biomarkers in PD | |||||
| PD | Apolipoprotein A1 | iTRAQ | Plasma/serum | Downregulated in PD | [87,88] |
| PD | Apolipoprotein A-IV | LFQ | Plasma/serum | Downregulated in PD | [87,88] |
| PD | Apolipoprotein B | LFQ | Plasma | Downregulated in PD | [89] |
| PD | Apolipoprotein CI | LFQ | Plasma | Downregulated in PD | [89] |
| PD | Apolipoprotein CIII | LFQ | Plasma | Downregulated in PD | [89] |
| PD | Apolipoprotein C4 | LFQ | Plasma | Downregulated in PD | [89] |
| PD | Apolipoprotein C4 | LFQ | Plasma | Downregulated in PD | [89] |
| PD | Apolipoprotein M | LFQ | Plasma | Downregulated in PD | [89] |
| PD | Inter-alpha-trypsin inhibitor heavy | LFQ | Plasma/serum | Downregulated in PD | [87] |
| PD | Complement C4A | LFQ | Plasma/serum | Downregulated in PD | [87] |
| PD | Complement C4B | iTRAQ | Plasma/serum | Downregulated in PD | [87,88] |
| PD | Complement C3 | LFQ | Plasma/serum | Downregulated in PD | [87] |
| PD | Haptoglobin | LFQ | Plasma | Downregulated in PD | [89] |
| PD | Clusterin | LFQ | Plasma/serum | Upregulated in PD | [87] |
| PD | Transthyretin | LFQ | Plasma/serum | Upregulated in PD | [87] |
| PD | Zinc α-2 glycoprotein | LFQ | Plasma/serum | Upregulated in PD | [87] |
| PD | Vitamin D binding protein | LFQ | Plasma/serum | Upregulated in PD | [87] |
| PD | Afamin | LFQ | Plasma/serum | Upregulated in PD | [87] |
| CSF proteomic biomarkers in PD | |||||
| PD | α-synuclein peptide (81–96) | MRM | CSF | α-Synuclein peptide altered in PD | [90] |
| PD | α-synuclein pS129 | MRM | CSF | α-Synuclein pS129 correlates with disease severity | [91] |
| PD | Granins | DIA | CSF | Granins are downregulated in PD | [92] |
| Exosomal biomarkers in PD | |||||
| PD | α-synuclein | LFQ | Serum neuronal(L1CAM+) exosomes | Upregulated in prodromal and clinical PD compared to controls and other neurodegenerative diseases | [93] |
| PD | Clusterin | LFQ | Serum neuronal (L1CAM+) exosomes, plasma exosomes | Upregulated in FTD but not PD, served as a combined marker with α-synuclein and is downregulated in PD in plasma exosomes | [93,94] |
| PD | α-synuclein | SRM | Plasma neuronal (L1CAM+) exosomes | α-synuclein is upregulated in PD | [95,96] |
| PD | Complement C1r | LFQ | Plasma exosomes | Downregulated in PD | [94] |
| PD | Apolipoprotein A1 | LFQ | Plasma exosomes | Downregulated in PD | [94] |
4.2. Post-Translational Modifications in Key Proteins from Tissue Proteomics Studies in PD
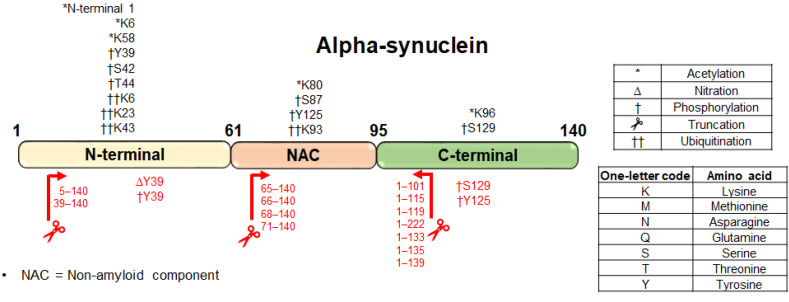
Aggregated forms of α-synuclein found in Lewy bodies are a primary hallmark of PD. α-Synuclein has been associated with many PTMs, including acetylation, phosphorylation, nitration, O-GlcNAcylation, SUMOylation, and truncations. These modifications are linked to the aggregation and toxicity of α-synuclein [97] (Figure 3).
Figure 3.
Schematic representation of post-translational modifications (PTMs) identified in α-synuclein. The red text indicates putative disease-specific modifications reported in PD [98,99,100,101,102,103,104,105,106,107,108,109,110], while black test indicates all known modifications that are reported in the literature and proteomic databases (ProteomicsDB and PhosPhosite).
Mass-spectrometry-based proteomic studies in human brains with synucleopathies reveal that the phosphorylation of α-synuclein at Serine 129 (Ser129-p) is the predominant modification method of α-synuclein in Lewy bodies, followed by ubiquitination and truncations at the C-terminus [98,101,110]. Here, by using a combination of detergents to extract the insoluble aggregates, Ser129-p was identified with MALDI. MALDI undergoes a relatively gentle ionization method, enabling the preservation of the phopho-group, and is more robust to the presence of detergents in the samples. The disadvantage of MALDI is that it can only identify few of the most abundant peptides or proteins. A study characterizing the interplay between tyrosine phosphorylation and nitration in the C-terminus found that phosphorylation at the proximal tyrosine 125 (pY125) altered metal binding and induced pathogenic aggregation [107]. Protein tyrosine nitration (PTN) at Y39 is also believed to be critical in oligomer formation [98,99,100,101,102,103,104,105,106,107,108,109,110].
PD, the process by which oligomeric proteins are seeded and spread, involves intricate interactions with the cell membrane and extra-cellular matrix (ECM), and these processes can be characterized in tissue proteomic studies. Glycosylation is the predominant post-translational modification in matrisome molecules (proteins constituting the cell membrane and ECM). Proteomics methods to study the glycosylation of matrisome molecules have been summarized by Raghunathan et al. [111]. A study in human prefrontal cortex identified that ECM molecules exhibit the highest degree of upregulation in PD. These molecules include proteoglycans associated with perineuronal nets and various collagen types [112]. They contribute to the blood–brain barrier and have important neuronal signaling implications. In addition, collagen type I has a differential hydroxyl proline state in PD compared to controls [113].
4.3. Plasma and Serum as Sources for Proteomic Biomarkers in PD
In PD, plasma levels of α-synuclein pS129 in α-synuclein are associated with motor symptom severity and disease progression [90,91]. Here, targeted proteomics is used to gain the absolute quantitation of pS129 in α-synuclein in plasma. One advantage is that it selectively monitors target peptides, overcoming other dynamic range challenges and providing absolute quantitation. However, it can be used on a limited number of targets.
A metabolomic and proteomic study in plasma using mass spectrometry revealed that all apolipoprotein isoforms were downregulated in PD [89]. Assessing both metabolomic and proteomic analysis in plasma provides valuable information about the disease. Label-free proteomics analysis was used, and all quantitation was carried out with relative abundance. The validation of these markers in an independent cohort could strengthen these findings. In a systematic blood-based biomarker review in PD, seven proteins (apolipoprotein A1, apolipoprotein-A IV, inter-alpha-trypsin inhibitor heavy, complement C4A, complement C4B, complement C3, and haptoglobulin) were consistently downregulated in PD compared to the control [87]. Five proteins (clusterin, transthyretin, zinc α-2-glycoprotein, vitamin D binding protein, and afamin) were consistently upregulated in plasma and serum in PD compared to controls [87].
Aptamer-based approaches can overcome the dynamic range challenge associated with protein measurements in plasma by specifically enriching proteins that bind aptamers. Using aptamer technology, a multicohort study of plasma blood-based biomarkers was analyzed in 96 PD patients and 45 neurological controls. Four proteins (bone sialoprotein, osteomodulin, aminoacylase-1, and growth hormone receptor) were differentially expressed in ALS [114]. Aptamer-based technology overcomes the dynamic range challenge associated with plasma proteomics. Yet, it is limited by the panel of observable proteins that are predetermined, as well as by the specificity of the aptamers. The use of well-characterized reagents is necessary to rule out false positives with the aptamers.
4.4. Cerebrospinal Fluid (CSF) as a Source for Proteomic Biomarkers in PD
Targeted proteomic identification and quantification in CSF was used as a diagnostic marker and marker of disease progression [90]. Using a targeted proteomics approach to monitor α-synuclein peptide (81–96) revealed that monitoring this peptide level in CSF can act as both a diagnostic marker and a marker of disease progression [90]. In a separate study, it was found that the level of pS129 in α-synuclein correlated with disease severity in PD [91]. Using a targeted proteomics approach enables absolute quantitation, and thus longitudinal comparisons in different cohorts. A global proteomics study using label-free quantitation in CSF identified multiple proteins downregulated in PD, including seven of eight members of the granulin family [92] (Table 3).
4.5. Exosomes Proteomics in Biomarker Identification
In PD, exosomes have been investigated as a source of biomarkers (Table 2). Neuronal exosomes isolated from serum were investigated for signatures in clinical PD, finding that α-synuclein and clusterin levels together can serve as a marker for the differential diagnosis of PD [93]. While total plasma levels of α-synuclein show no change, neuronal exosomal α-synuclein correlate with disease progression in PD, indicating that an assessment of neuronal exosomes from plasma may be a superior strategy for biomarker discovery in other neurodegenerative diseases [115]. In addition, oligomeric α-synuclein resistant to proteinase K and pSer129 in α-synuclein was identified in plasma exosomes from PD patients by proteomics. The ratio of α-synuclein oligomer–total α-synuclein and the ratio of p-α-synuclein oligomer–total p-α-synuclein in plasma exosomes served as a diagnostic biomarker in PD compared to controls in a proteomic study [116]. In a PD study, α-synuclein levels were identified in L1CAM+ exosomes and correlated with GCase activity in PBMC [96]. Here, L1CAM+ exosomes were used as a marker of neuronal exosomes. There is still uncertainty around the specificity of L1CAM as a marker of neuronal origin. The measurement of oligomeric α-synuclein in plasma exosomes is novel and can be insightful if validated as a biomarker. Total plasma exosomes from PD patients in stage II and III were profiled by proteomics, and three proteins (clusterin, complement C1r, and apolipoprotein A1) were found to be downregulated in PD compared to control [94]. Here, by using a combination of 2D gel electrophoresis and MALDI-TOF, the relative quantitation of proteins was analyzed. MALDI provides few identified proteins and a greater depth, but lower throughput could be achieved using LC-MS/MS. Further, these biomarkers only have relative quantitation. A follow-up study with targeted proteomic analysis for the absolute quantitation of the three proteins can provide greater confidence in the biomarkers. Exosomes have been studied in diagnosis and disease progression using proteomics in PD.
5. Clinical Trials Using Proteomics for Biomarker Discovery in ALS and PD
The application of proteomics to patient stratification, biomarker measurements for clinical end points, and integration with genomics for the identification of novel drug targets has immense potential to advance precision medicine. Proteomic biomarkers have been implemented as a noninvasive diagnostic tool in many studies [117] and used patient stratification [117].
Developing a validated proteomic assay is critical prior to its utilization for decision making in the clinic. Discovery proteomics can identify thousands of proteins. However, to be able to validate these identified analytes, a small number is chosen, and a targeted proteomics strategy by MRM is often applied for absolute quantitation. Sometimes, protein signature classifiers identify groups of proteins, which can be used as biomarkers for patient stratification [118]. The Clinical Proteomics Tumor Analysis Consortium (CPTAC) has developed a fit-for-purpose best-practices guideline in targeted proteomic analysis for the validation of proteomic clinical assays [119]. These guidelines can be used to standardize targeted measurements for clinical use across diseases. The authors identified three tiers of analysis and described the validation steps for each tier with respect to the analytical goal of the assay. In Tier 1, the goal of the assay is to provide decision-making information in drug development or for medical practitioners on a small number of analytes. Since the intent is to use the assay for clinical purposes, a high degree of analytical validation is warranted, including measurements of assay precision, accuracy, specificity, analytical sensitivity (including limit of detection (LOD), limit of the blank (LOB), and lower limit of quantification (LLOQ)), linearity, and parallelism. The use of stable isotope internal standards for each analyte and/or protein heavy-labeled standards is recommended to achieve accurate quantitation. The other two tiers are for non-clinical purposes. Tier 2 uses stable isotopes to validate hundreds of analytes for research purposes. Tier 3, which is semi-quantitative, is used for exploratory studies [119]. Some examples of clinical studies that have used proteomics in ALS and PD are shown in Table 4.
Table 4.
Examples of clinical trials using proteomics in ALS and PD.
| Disease | Clinical Trial | Summary | Reference |
|---|---|---|---|
| ALS | NCT01948102 | An observational study for the identification of prognostic and diagnostic markers in skin and adipose samples using proteomics to measure changes in abundance and/or post-translational modifications of proteins in the trial | [6] |
| PD | NCT00315250 | An interventional study with the aim of developing imaging, clinical, and biochemical biomarkers for PD uses proteomics in combination with metabolomics and gene expression to categorize Parkinson’s syndrome vs. non-Parkinson’s syndrome | [7] |
| PD | NCT02263235 | A study in Alzheimer’s, PD, and other neurological disorders without cognitive decline uses targeted quantitative proteomics by MRM in CSF, blood, urine, and saliva for diagnostic purposes after administering stable isotope-labelled leucine for the diagnosis of neurological disorders | [4] |
| PD | NCT02524405 | An investigational study in Alzheimer’s and PD (called the brain–eye amyloid memory study (BEAM)), MRI, and amyloid PET were used for primary and secondary outcomes, genetic analysis for ApoE4 status, and proteomics and lipidomics analyses | [2] |
| PD | NCT02387281 | An observational study in PD studying freezing of gait (FOG) proteomics on CSF is used in combination with analysis of catecholamines along with MRI and other cognitive tests to assess types of FOG and if there is a connection with cognitive differences and gait patterns presented in PD | [3] |
6. Discussion
In ALS and PD, molecular biomarkers for diagnosis and prognosis are limited, and many of the available ones seem to be useful for advanced stages of the diseases, when therapeutic intervention is likely no longer effective. Both diseases have a critical need for biomarkers, and proteomics may be able to address this need.
Current technological advancements in proteomics have made biomarker discovery for ALS and PD more efficient. Short gradients with novel LC systems [120] provide accurate and rapid quantitation without a loss of identifications with the new RTS-MS3 quantitation [121] or BoxCar DIA [31] to gain depth, range, and completeness, while addressing some of the dynamic range challenges in biofluids makes mass spectrometry a powerful and indispensable tool in biomarker discovery.
Mass spectrometry has advantages over immunoassays and aptamers because of its unbiased nature and non-reliance on antibody/aptamer specificity. This is particularly helpful in PTM analysis, in the identification of novel biomarkers, and in cases with point mutations or proteins, for which antibodies/aptamers are not available. In ALS, several PTMs have been defined in TDP43. These include hyperphosphorylation, polyubiquitination, and C-terminal truncations which are believed to play a role in aggregation and disease pathology [70]. In PD, α-synuclein is one of the best-studied proteins with several PTMs, including phosphorylation, nitration, ubiquitination, O-GlcNAcylation, and N- and C-terminal truncations. The use of mass shifts to characterize these PTMs and quantify their levels with mass spectrometry allows multiple PTMs to be analyzed in parallel in an unbiased manner without the need to synthesize and characterize antibodies/reagents. The characterization of disease-specific modifications in tissue and biofluids may serve as diagnostic markers or novel drug targets.
After comparing proteomic biomarker studies in tissue, biofluids, and exosomes in ALS, it was observed that proteins belonging to transcriptional pathways were altered in both spinal cord tissue and the CSF proteome. UCHL1, MAP2, and GPNMB were upregulated in spinal cord tissue and CSF in ALS [52]. Gelsolin was altered in ALS in plasma, and CSF was upregulated in CSF exosomes in patients with C9orf72 mutations. Clusterin was upregulated in CSF exosomes similar to CSF biofluids [56,61]. TDP43 modifications have been observed in human prefrontal, motor cortex brain tissue, and spinal cord, as well as in plasma-derived exosomes [51,55,63]. In PD, the presence of pS129 in α-synuclein is associated with the oligomeric form in brain tissues and has been observed in plasma, CSF and serum-derived neuronal exosomes (L1CAM+) in independent studies [90,101,103,116]. The total α-synuclein levels were upregulated in tissue and plasma-derived neuronal exosomes, but were unchanged in plasma measurements. The enrichment of exosomes helps to alleviate the dynamic range issue in proteomics of plasma/serum and could explain some findings where total α-synuclein levels in plasma were not significantly altered in PD, while neuronal-derived exosomes in plasma show upregulation in PD. Some studies comparing CSF and plasma in ALS demonstrated that gelsolin increased in both CSF and plasma. In PD, total and pS129 α-synuclein levels were measured in plasma and CSF [91]. An alternative is to use neuronal-derived exosomes from plasma for biomarker discovery. Further exploration and validation of neuronal specific markers in exosomes is warranted.
While several of these biomarkers have been identified in certain patient populations such as ALS with C9orf72 mutations, the specificity of these biomarkers needs to be investigated to see if these markers will be useful in sporadic ALS or ALS with other mutations. To assess if C9orf72 biomarkers will be useful in a larger population, biomarker discovery and validation will need to be performed on non-C9orf72 patients. In a similar vein, the use of matched samples from siblings with mutations who do not exhibit symptoms can provide insight into prognostic markers.
When evaluating the value of biomarkers studies, animal models that are assessed longitudinally as a measure of disease progression might be useful. Similarly, identifying markers in patient cohorts could be validated in animal models or reprogrammed iPSC cells followed longitudinally. However, the caveat with using animal models particularly in neurodegenerative diseases is the low translatability. Several models do not accurately represent the disease pathology and systemic effects observed in patients.
The other approach used to identify biomarkers that predict the onset of neurodegenerative diseases in the pre-symptomatic phase involves profiling healthy samples from people at risk of neurodegenerative diseases using mass spectrometry and by comparing signatures that change with the onset of symptoms and if these markers correlate with disease progression. The challenge lies in validating these early-onset biomarkers. Correlating molecular signatures to imaging markers is another avenue to identify early-onset markers. With growing technology and machine learning, biomarker signatures in the pre-symptomatic phase could potentially predict disease onset in the future. Efforts in the identification and validation of these markers in disease models with high translatability, as well as the assessment of early-onset samples, will remain a challenge in biomarker discovery for neurodegenerative diseases.
Proteomics has immense clinical potential for biomarker discovery, but there is only a limited number of validated proteomic biomarkers. This is due to the complexity and lack of standardized validation protocols for assays. In the cancer field, the use of validated targeted proteomic assays has been described by the CPTAC consortium. Applying these guidelines to biomarker validation in neurodegenerative diseases may be helpful in bridging the gap between discovery proteomics and the translation of biomarkers to the clinic. There are a few clinical trials that incorporate proteomics in their outcome measures in ALS and PD. Multi-omics studies integrating genomics, proteomics, transcriptomics, and metabolomics to identify markers for patient stratification may be able to address current challenges with biomarkers in diseases such as ALS and PD.
Acknowledgments
The authors thank David Litwack and Benjamin Shykind for their valuable feedback on the manuscript.
Author Contributions
R.R. conceptualized the study. R.R., K.T. and L.C.W. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
All authors are employees of Prevail Therapeutics, a wholly owned subsidiary of Eli Lilly and Company.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Talbott E.O., Malek A.M., Lacomis D. The epidemiology of amyotrophic lateral sclerosis. Handb. Clin. Neurol. 2016;138:225–238. doi: 10.1016/B978-0-12-802973-2.00013-6. [DOI] [PubMed] [Google Scholar]
- 2.Edgar S., Ellis M., Abdul-Aziz N.A., Goh K.J., Shahrizaila N., Kennerson M.L., Ahmad-Annuar A. Mutation analysis of SOD1, C9orf72, TARDBP and FUS genes in ethnically-diverse Malaysian patients with amyotrophic lateral sclerosis (ALS) Neurobiol. Aging. 2021;108:200–206. doi: 10.1016/j.neurobiolaging.2021.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Smith B.N., Newhouse S., Shatunov A., Vance C., Topp S., Johnson L., Miller J., Lee Y., Troakes C., Scott K.M., et al. The C9ORF72 expansion mutation is a common cause of ALS+/-FTD in Europe and has a single founder. Eur. J. Hum. Genet. 2013;21:102–108. doi: 10.1038/ejhg.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen P.M. Mutation in C9orf72 changes the boundaries of ALS and FTD. Lancet Neurol. 2012;11:205–207. doi: 10.1016/S1474-4422(12)70020-0. [DOI] [PubMed] [Google Scholar]
- 5.Yao L., He X., Cui B., Zhao F., Zhou C. NEK1 mutations and the risk of amyotrophic lateral sclerosis (ALS): A meta-analysis. Neurol. Sci. 2021;42:1277–1285. doi: 10.1007/s10072-020-05037-6. [DOI] [PubMed] [Google Scholar]
- 6.Naruse H., Ishiura H., Mitsui J., Takahashi Y., Matsukawa T., Yoshimura J., Doi K., Morishita S., Goto J., Toda T., et al. Loss-of-function variants in NEK1 are associated with an increased risk of sporadic ALS in the Japanese population. J. Hum. Genet. 2021;66:237–241. doi: 10.1038/s10038-020-00830-9. [DOI] [PubMed] [Google Scholar]
- 7.Riley J.F., Fioramonti P.J., Rusnock A.K., Hehnly H., Castaneda C.A. ALS-linked mutations impair UBQLN2 stress-induced biomolecular condensate assembly in cells. J. Neurochem. 2021;159:145–155. doi: 10.1111/jnc.15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin B.C., Phung T.H., Higgins N.R., Greenslade J.E., Prado M.A., Finley D., Karbowski M., Polster B.M., Monteiro M.J. ALS/FTD mutations in UBQLN2 are linked to mitochondrial dysfunction through loss-of-function in mitochondrial protein import. Hum. Mol. Genet. 2021;30:1230–1246. doi: 10.1093/hmg/ddab116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baron D.M., Fenton A.R., Saez-Atienzar S., Giampetruzzi A., Sreeram A., Shankaracharya, Keagle P.J., Doocy V.R., Smith N.J., Danielson E.W., et al. ALS-associated KIF5A mutations abolish autoinhibition resulting in a toxic gain of function. Cell Rep. 2022;39:110598. doi: 10.1016/j.celrep.2022.110598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolas A., Kenna K.P., Renton A.E., Ticozzi N., Faghri F., Chia R., Dominov J.A., Kenna B.J., Nalls M.A., Keagle P., et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron. 2018;97:1268–1283.e6. doi: 10.1016/j.neuron.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu S., Rajendra K.C., Alagar S., Bahadur R.P. Impaired nuclear transport induced by juvenile ALS causing P525L mutation in NLS domain of FUS: A molecular mechanistic study. Biochim. Biophys. Acta Proteins Proteom. 2022;1870:140766. doi: 10.1016/j.bbapap.2022.140766. [DOI] [PubMed] [Google Scholar]
- 12.Robertson J., Bilbao J., Zinman L., Hazrati L.N., Tokuhiro S., Sato C., Moreno D., Strome R., Mackenzie I.R., Rogaeva E. A novel double mutation in FUS gene causing sporadic ALS. Neurobiol. Aging. 2011;32:553.e27–553.e30. doi: 10.1016/j.neurobiolaging.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Chio A., Mazzini L., D’Alfonso S., Corrado L., Canosa A., Moglia C., Manera U., Bersano E., Brunetti M., Barberis M., et al. The multistep hypothesis of ALS revisited: The role of genetic mutations. Neurology. 2018;91:e635–e642. doi: 10.1212/WNL.0000000000005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim G., Gautier O., Tassoni-Tsuchida E., Ma X.R., Gitler A.D. ALS Genetics: Gains, Losses, and Implications for Future Therapies. Neuron. 2020;108:822–842. doi: 10.1016/j.neuron.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ascherio A., Schwarzschild M.A. The epidemiology of Parkinson’s disease: Risk factors and prevention. Lancet Neurol. 2016;15:1257–1272. doi: 10.1016/S1474-4422(16)30230-7. [DOI] [PubMed] [Google Scholar]
- 16.Li W., Fu Y., Halliday G.M., Sue C.M. PARK Genes Link Mitochondrial Dysfunction and Alpha-Synuclein Pathology in Sporadic Parkinson’s Disease. Front. Cell Dev. Biol. 2021;9:612476. doi: 10.3389/fcell.2021.612476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., Luo X., Li F., Tian X., Zhu L., Yang Y., Ren Y., Pang H. Association of Parkinson’s disease with six single nucleotide polymorphisms located in four PARK genes in the northern Han Chinese population. J. Clin. Neurosci. 2012;19:1011–1015. doi: 10.1016/j.jocn.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 18.Chung S.J., Armasu S.M., Biernacka J.M., Lesnick T.G., Rider D.N., Lincoln S.J., Ortolaza A.I., Farrer M.J., Cunningham J.M., Rocca W.A., et al. Common variants in PARK loci and related genes and Parkinson’s disease. Mov. Disord. 2011;26:280–288. doi: 10.1002/mds.23376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein C., Schneider S.A., Lang A.E. Hereditary parkinsonism: Parkinson disease look-alikes—An algorithm for clinicians to “PARK” genes and beyond. Mov. Disord. 2009;24:2042–2058. doi: 10.1002/mds.22675. [DOI] [PubMed] [Google Scholar]
- 20.Domingo A., Klein C. Genetics of Parkinson disease. Handb. Clin. Neurol. 2018;147:211–227. doi: 10.1016/B978-0-444-63233-3.00014-2. [DOI] [PubMed] [Google Scholar]
- 21.Geyer P.E., Holdt L.M., Teupser D., Mann M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017;13:942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brzhozovskiy A., Kononikhin A., Bugrova A.E., Kovalev G.I., Schmit P.O., Kruppa G., Nikolaev E.N., Borchers C.H. The Parallel Reaction Monitoring-Parallel Accumulation-Serial Fragmentation (prm-PASEF) Approach for Multiplexed Absolute Quantitation of Proteins in Human Plasma. Anal. Chem. 2022;94:2016–2022. doi: 10.1021/acs.analchem.1c03782. [DOI] [PubMed] [Google Scholar]
- 23.Lesur A., Dittmar G. The clinical potential of prm-PASEF mass spectrometry. Expert Rev. Proteom. 2021;18:75–82. doi: 10.1080/14789450.2021.1908895. [DOI] [PubMed] [Google Scholar]
- 24.Meier F., Beck S., Grassl N., Lubeck M., Park M.A., Raether O., Mann M. Parallel Accumulation-Serial Fragmentation (PASEF): Multiplying Sequencing Speed and Sensitivity by Synchronized Scans in a Trapped Ion Mobility Device. J. Proteome Res. 2015;14:5378–5387. doi: 10.1021/acs.jproteome.5b00932. [DOI] [PubMed] [Google Scholar]
- 25.Garabedian A., Benigni P., Ramirez C.E., Baker E.S., Liu T., Smith R.D., Fernandez-Lima F. Towards Discovery and Targeted Peptide Biomarker Detection Using nanoESI-TIMS-TOF MS. J. Am. Soc. Mass Spectrom. 2018;29:817–826. doi: 10.1007/s13361-017-1787-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsou C.C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A.C., Nesvizhskii A.I. DIA-Umpire: Comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinitcyn P., Hamzeiy H., Salinas Soto F., Itzhak D., McCarthy F., Wichmann C., Steger M., Ohmayer U., Distler U., Kaspar-Schoenefeld S., et al. MaxDIA enables library-based and library-free data-independent acquisition proteomics. Nat. Biotechnol. 2021;39:1563–1573. doi: 10.1038/s41587-021-00968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muntel J., Gandhi T., Verbeke L., Bernhardt O.M., Treiber T., Bruderer R., Reiter L. Surpassing 10000 identified and quantified proteins in a single run by optimizing current LC-MS instrumentation and data analysis strategy. Mol. Omics. 2019;15:348–360. doi: 10.1039/c9mo00082h. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y., Tan Z., Xue P., Wang Y., Li X., Guan F. High-throughput, in-depth and estimated absolute quantification of plasma proteome using data-independent acquisition/mass spectrometry (“HIAP-DIA”) Proteomics. 2021;21:e2000264. doi: 10.1002/pmic.202000264. [DOI] [PubMed] [Google Scholar]
- 31.Mehta D., Scandola S., Uhrig R.G. BoxCar and Library-Free Data-Independent Acquisition Substantially Improve the Depth, Range, and Completeness of Label-Free Quantitative Proteomics. Anal. Chem. 2022;94:793–802. doi: 10.1021/acs.analchem.1c03338. [DOI] [PubMed] [Google Scholar]
- 32.Meier F., Geyer P.E., Virreira Winter S., Cox J., Mann M. BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat. Methods. 2018;15:440–448. doi: 10.1038/s41592-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 33.Salovska B., Li W., Di Y., Liu Y. BoxCarmax: A High-Selectivity Data-Independent Acquisition Mass Spectrometry Method for the Analysis of Protein Turnover and Complex Samples. Anal. Chem. 2021;93:3103–3111. doi: 10.1021/acs.analchem.0c04293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaun A., Lewis Hardell K.N., Olsson N., O’Brien J.J., Gollapudi S., Smith M., McAlister G., Huguet R., Keyser R., Buffenstein R., et al. Automated 16-Plex Plasma Proteomics with Real-Time Search and Ion Mobility Mass Spectrometry Enables Large-Scale Profiling in Naked Mole-Rats and Mice. J. Proteome Res. 2021;20:1280–1295. doi: 10.1021/acs.jproteome.0c00681. [DOI] [PubMed] [Google Scholar]
- 35.Hebert A.S., Prasad S., Belford M.W., Bailey D.J., McAlister G.C., Abbatiello S.E., Huguet R., Wouters E.R., Dunyach J.J., Brademan D.R., et al. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Anal. Chem. 2018;90:9529–9537. doi: 10.1021/acs.analchem.8b02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He B., Huang Z., Huang C., Nice E.C. Clinical applications of plasma proteomics and peptidomics: Towards precision medicine. Proteom. Clin. Appl. 2022:e2100097. doi: 10.1002/prca.202100097. [DOI] [PubMed] [Google Scholar]
- 37.Bache N., Geyer P.E., Bekker-Jensen D.B., Hoerning O., Falkenby L., Treit P.V., Doll S., Paron I., Muller J.B., Meier F., et al. A Novel LC System Embeds Analytes in Pre-formed Gradients for Rapid, Ultra-robust Proteomics. Mol. Cell Proteom. 2018;17:2284–2296. doi: 10.1074/mcp.TIR118.000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar D., Hassan M.I. Ultra-sensitive techniques for detecting neurological biomarkers: Prospects for early diagnosis. Biochem Biophys. Res. Commun. 2021;584:15–18. doi: 10.1016/j.bbrc.2021.10.073. [DOI] [PubMed] [Google Scholar]
- 39.Cole K.H., Luptak A. High-throughput methods in aptamer discovery and analysis. Methods Enzymol. 2019;621:329–346. doi: 10.1016/bs.mie.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Chen L.C., Tzeng S.C., Peck K. Aptamer microarray as a novel bioassay for protein-protein interaction discovery and analysis. Biosens. Bioelectron. 2013;42:248–255. doi: 10.1016/j.bios.2012.10.082. [DOI] [PubMed] [Google Scholar]
- 41.Huang J., Chen X., Fu X., Li Z., Huang Y., Liang C. Advances in Aptamer-Based Biomarker Discovery. Front. Cell Dev. Biol. 2021;9:659760. doi: 10.3389/fcell.2021.659760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sielaff M., Kuharev J., Bohn T., Hahlbrock J., Bopp T., Tenzer S., Distler U. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. 2017;16:4060–4072. doi: 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- 43.Raghunathan R., Sethi M.K., Zaia J. On-slide tissue digestion for mass spectrometry based glycomic and proteomic profiling. MethodsX. 2019;6:2329–2347. doi: 10.1016/j.mex.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scicchitano M.S., Dalmas D.A., Boyce R.W., Thomas H.C., Frazier K.S. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J. Histochem. Cytochem. 2009;57:849–860. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsa G., Guo Q., Goncalves C., Preston S.E.J., Lacasse V., Aguilar-Mahecha A., Benlimame N., Basik M., Spatz A., Batist G., et al. A Non-Hazardous Deparaffinization Protocol Enables Quantitative Proteomics of Core Needle Biopsy-Sized Formalin-Fixed and Paraffin-Embedded (FFPE) Tissue Specimens. Int. J. Mol. Sci. 2022;23:4443. doi: 10.3390/ijms23084443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reimel B.A., Pan S., May D.H., Shaffer S.A., Goodlett D.R., McIntosh M.W., Yerian L.M., Bronner M.P., Chen R., Brentnall T.A. Proteomics on Fixed Tissue Specimens-A Review. Curr. Proteom. 2009;6:63–69. doi: 10.2174/157016409787847420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundberg E., Borner G.H.H. Spatial proteomics: A powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 2019;20:285–302. doi: 10.1038/s41580-018-0094-y. [DOI] [PubMed] [Google Scholar]
- 48.Mao Y., Wang X., Huang P., Tian R. Spatial proteomics for understanding the tissue microenvironment. Analyst. 2021;146:3777–3798. doi: 10.1039/d1an00472g. [DOI] [PubMed] [Google Scholar]
- 49.Shin J.J.H., Crook O.M., Borgeaud A.C., Cattin-Ortola J., Peak-Chew S.Y., Breckels L.M., Gillingham A.K., Chadwick J., Lilley K.S., Munro S. Spatial proteomics defines the content of trafficking vesicles captured by golgin tethers. Nat. Commun. 2020;11:5987. doi: 10.1038/s41467-020-19840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Navarro-Romero A., Montpeyo M., Martinez-Vicente M. The Emerging Role of the Lysosome in Parkinson’s Disease. Cells. 2020;9:2399. doi: 10.3390/cells9112399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engelen-Lee J., Blokhuis A.M., Spliet W.G.M., Pasterkamp R.J., Aronica E., Demmers J.A.A., Broekhuizen R., Nardo G., Bovenschen N., Van Den Berg L.H. Proteomic profiling of the spinal cord in ALS: Decreased ATP5D levels suggest synaptic dysfunction in ALS pathogenesis. Amyotroph. Lateral Scler. Front. Degener. 2017;18:210–220. doi: 10.1080/21678421.2016.1245757. [DOI] [PubMed] [Google Scholar]
- 52.Oeckl P., Weydt P., Thal D.R., Weishaupt J.H., Ludolph A.C., Otto M. Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis. Acta Neuropathol. 2020;139:119–134. doi: 10.1007/s00401-019-02093-x. [DOI] [PubMed] [Google Scholar]
- 53.Liu F., Morderer D., Wren M.C., Vettleson-Trutza S.A., Wang Y., Rabichow B.E., Salemi M.R., Phinney B.S., Oskarsson B., Dickson D.W., et al. Proximity proteomics of C9orf72 dipeptide repeat proteins identifies molecular chaperones as modifiers of poly-GA aggregation. Acta Neuropathol. Commun. 2022;10:22. doi: 10.1186/s40478-022-01322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartmann H., Hornburg D., Czuppa M., Bader J., Michaelsen M., Farny D., Arzberger T., Mann M., Meissner F., Edbauer D. Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci. Alliance. 2018;1:e201800070. doi: 10.26508/lsa.201800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feneberg E., Charles P.D., Finelli M.J., Scott C., Kessler B.M., Fischer R., Ansorge O., Gray E., Talbot K., Turner M.R. Detection and quantification of novel C-terminal TDP-43 fragments in ALS-TDP. Brain Pathol. 2021;31:e12923. doi: 10.1111/bpa.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bereman M.S., Beri J., Enders J.R., Nash T. Machine Learning Reveals Protein Signatures in CSF and Plasma Fluids of Clinical Value for ALS. Sci. Rep. 2018;8:16334. doi: 10.1038/s41598-018-34642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Z., Lee A., Nouwens A., Henderson R.D., McCombe P.A. Mass spectrometry analysis of plasma from amyotrophic lateral sclerosis and control subjects. Amyotroph. Lateral Scler. Front. Degener. 2018;19:362–376. doi: 10.1080/21678421.2018.1433689. [DOI] [PubMed] [Google Scholar]
- 58.Mohanty L., Henderson R.D., McCombe P.A., Lee A. Levels of clusterin, CD5L, ficolin-3, and gelsolin in ALS patients and controls. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2020;21:631–634. doi: 10.1080/21678421.2020.1779303. [DOI] [PubMed] [Google Scholar]
- 59.Thompson A.G., Gray E., Thezenas M.L., Charles P.D., Evetts S., Hu M.T., Talbot K., Fischer R., Kessler B.M., Turner M.R. Cerebrospinal fluid macrophage biomarkers in amyotrophic lateral sclerosis. Ann. Neurol. 2018;83:258–268. doi: 10.1002/ana.25143. [DOI] [PubMed] [Google Scholar]
- 60.Barschke P., Oeckl P., Steinacker P., Al Shweiki M.R., Weishaupt J.H., Landwehrmeyer G.B., Anderl-Straub S., Weydt P., Diehl-Schmid J., Danek A., et al. Different CSF protein profiles in amyotrophic lateral sclerosis and frontotemporal dementia with C9orf72 hexanucleotide repeat expansion. J. Neurol. Neurosurg. Psychiatry. 2020;91:503–511. doi: 10.1136/jnnp-2019-322476. [DOI] [PubMed] [Google Scholar]
- 61.Thompson A.G., Gray E., Mager I., Thezenas M.L., Charles P.D., Talbot K., Fischer R., Kessler B.M., Wood M., Turner M.R. CSF extracellular vesicle proteomics demonstrates altered protein homeostasis in amyotrophic lateral sclerosis. Clin. Proteom. 2020;17:31. doi: 10.1186/s12014-020-09294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayashi N., Doi H., Kurata Y., Kagawa H., Atobe Y., Funakoshi K., Tada M., Katsumoto A., Tanaka K., Kunii M., et al. Proteomic analysis of exosome-enriched fractions derived from cerebrospinal fluid of amyotrophic lateral sclerosis patients. Neurosci. Res. 2020;160:43–49. doi: 10.1016/j.neures.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Chen P.C., Wu D., Hu C.J., Chen H.Y., Hsieh Y.C., Huang C.C. Exosomal TAR DNA-binding protein-43 and neurofilaments in plasma of amyotrophic lateral sclerosis patients: A longitudinal follow-up study. J. Neurol. Sci. 2020;418:117070. doi: 10.1016/j.jns.2020.117070. [DOI] [PubMed] [Google Scholar]
- 64.Ramazi S., Zahiri J. Posttranslational modifications in proteins: Resources, tools and prediction methods. Database. 2021;2021:baab012. doi: 10.1093/database/baab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 66.Carbonara K., Andonovski M., Coorssen J.R. Proteomes Are of Proteoforms: Embracing the Complexity. Proteomes. 2021;9:38. doi: 10.3390/proteomes9030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Didonna A., Benetti F. Post-translational modifications in neurodegeneration. AIMS Biophys. 2016;3:27–49. [Google Scholar]
- 68.Suk T.R., Rousseaux M.W.C. The role of TDP-43 mislocalization in amyotrophic lateral sclerosis. Mol. Neurodegener. 2020;15:45. doi: 10.1186/s13024-020-00397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 70.Kametani F., Obi T., Shishido T., Akatsu H., Murayama S., Saito Y., Yoshida M., Hasegawa M. Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 2016;6:23281. doi: 10.1038/srep23281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laferrière F., Maniecka Z., Pérez-Berlanga M., Hruska-Plochan M., Gilhespy L., Hock E.M., Wagner U., Afroz T., Boersema P.J., Barmettler G., et al. TDP-43 extracted from frontotemporal lobar degeneration subject brains displays distinct aggregate assemblies and neurotoxic effects reflecting disease progression rates. Nat. Neurosci. 2019;22:65–77. doi: 10.1038/s41593-018-0294-y. [DOI] [PubMed] [Google Scholar]
- 72.Prasad A., Bharathi V., Sivalingam V., Girdhar A., Patel B.K. Molecular Mechanisms of TDP-43 Misfolding and Pathology in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2019;12:25. doi: 10.3389/fnmol.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao X., Sandberg A., Araujo J.E., Cvetkovski F., Berglund E., Eriksson L.E., Pernemalm M. Evaluation of Spin Columns for Human Plasma Depletion to Facilitate MS-Based Proteomics Analysis of Plasma. J. Proteome Res. 2021;20:4610–4620. doi: 10.1021/acs.jproteome.1c00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee P.Y., Osman J., Low T.Y., Jamal R. Plasma/serum proteomics: Depletion strategies for reducing high-abundance proteins for biomarker discovery. Bioanalysis. 2019;11:1799–1812. doi: 10.4155/bio-2019-0145. [DOI] [PubMed] [Google Scholar]
- 75.El Rassi Z., Puangpila C. Liquid-phase based separation systems for depletion, prefractionation, and enrichment of proteins in biological fluids and matrices for in-depth proteomics analysis-An update covering the period 2014–2016. Electrophoresis. 2017;38:150–161. doi: 10.1002/elps.201600413. [DOI] [PubMed] [Google Scholar]
- 76.Keshishian H., Burgess M.W., Specht H., Wallace L., Clauser K.R., Gillette M.A., Carr S.A. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc. 2017;12:1683–1701. doi: 10.1038/nprot.2017.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zubiri I., Lombardi V., Bremang M., Mitra V., Nardo G., Adiutori R., Lu C.H., Leoni E., Yip P., Yildiz O., et al. Tissue-enhanced plasma proteomic analysis for disease stratification in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018;13:60. doi: 10.1186/s13024-018-0292-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leoni E., Bremang M., Mitra V., Zubiri I., Jung S., Lu C.H., Adiutori R., Lombardi V., Russell C., Koncarevic S., et al. Author Correction: Combined Tissue-Fluid Proteomics to Unravel Phenotypic Variability in Amyotrophic Lateral Sclerosis. Sci. Rep. 2020;10:18603. doi: 10.1038/s41598-020-74974-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang J., Khademi M., Lindhe O., Jonsson G., Piehl F., Olsson T., Kockum I. Assessing the Preanalytical Variability of Plasma and Cerebrospinal Fluid Processing and Its Effects on Inflammation-Related Protein Biomarkers. Mol. Cell Proteom. 2021;20:100157. doi: 10.1016/j.mcpro.2021.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macron C., Nunez Galindo A., Cominetti O., Dayon L. A Versatile Workflow for Cerebrospinal Fluid Proteomic Analysis with Mass Spectrometry: A Matter of Choice between Deep Coverage and Sample Throughput. Methods Mol. Biol. 2019;2044:129–154. doi: 10.1007/978-1-4939-9706-0_9. [DOI] [PubMed] [Google Scholar]
- 81.Varghese A.M., Sharma A., Mishra P., Vijayalakshmi K., Harsha H.C., Sathyaprabha T.N., Bharath S.M., Nalini A., Alladi P.A., Raju T.R. Chitotriosidase-a putative biomarker for sporadic amyotrophic lateral sclerosis. Clin. Proteom. 2013;10:19. doi: 10.1186/1559-0275-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pinnell J.R., Cui M., Tieu K. Exosomes in Parkinson disease. J. Neurochem. 2021;157:413–428. doi: 10.1111/jnc.15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang N., Gu D., Meng M., Gordon M.L. TDP-43 Is Elevated in Plasma Neuronal-Derived Exosomes of Patients With Alzheimer’s Disease. Front. Aging Neurosci. 2020;12:166. doi: 10.3389/fnagi.2020.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goetzl E.J., Abner E.L., Jicha G.A., Kapogiannis D., Schwartz J.B. Declining levels of functionally specialized synaptic proteins in plasma neuronal exosomes with progression of Alzheimer’s disease. FASEB J. 2018;32:888–893. doi: 10.1096/fj.201700731R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Licker V., Turck N., Kövari E., Burkhardt K., Côte M., Surini-Demiri M., Lobrinus J.A., Sanchez J.C., Burkhard P.R. Proteomic analysis of human substantia nigra identifies novel candidates involved in Parkinson’s disease pathogenesis. Proteomics. 2014;14:784–794. doi: 10.1002/pmic.201300342. [DOI] [PubMed] [Google Scholar]
- 86.Petyuk V.A., Yu L., Olson H.M., Yu F., Clair G., Qian W.J., Shulman J.M., Bennett D.A. Proteomic Profiling of the Substantia Nigra to Identify Determinants of Lewy Body Pathology and Dopaminergic Neuronal Loss. J. Proteome Res. 2021;20:2266–2282. doi: 10.1021/acs.jproteome.0c00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chelliah S.S., Bhuvanendran S., Magalingam K.B., Kamarudin M.N.A., Radhakrishnan A.K. Identification of blood-based biomarkers for diagnosis and prognosis of Parkinson’s disease: A systematic review of proteomics studies. Ageing Res. Rev. 2022;73:101514. doi: 10.1016/j.arr.2021.101514. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X., Yin X., Yu H., Liu X., Yang F., Yao J., Jin H., Yang P. Quantitative proteomic analysis of serum proteins in patients with Parkinson’s disease using an isobaric tag for relative and absolute quantification labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. Analyst. 2012;137:490–495. doi: 10.1039/c1an15551b. [DOI] [PubMed] [Google Scholar]
- 89.Hu L., Dong M.X., Huang Y.L., Lu C.Q., Qian Q., Zhang C.C., Xu X.M., Liu Y., Chen G.H., Wei Y.D. Integrated Metabolomics and Proteomics Analysis Reveals Plasma Lipid Metabolic Disturbance in Patients With Parkinson’s Disease. Front. Mol. Neurosci. 2020;13:80. doi: 10.3389/fnmol.2020.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang L., Stewart T., Shi M., Pottiez G., Dator R., Wu R., Aro P., Schuster R.J., Ginghina C., Pan C., et al. An alpha-synuclein MRM assay with diagnostic potential for Parkinson’s disease and monitoring disease progression. Proteom. Clin. Appl. 2017;11:1700045. doi: 10.1002/prca.201700045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y., Shi M., Chung K.A., Zabetian C.P., Leverenz J.B., Berg D., Srulijes K., Trojanowski J.Q., Lee V.M., Siderowf A.D., et al. Phosphorylated alpha-synuclein in Parkinson’s disease. Sci. Transl. Med. 2012;4:121ra120. doi: 10.1126/scitranslmed.3002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rotunno M.S., Lane M., Zhang W., Wolf P., Oliva P., Viel C., Wills A.M., Alcalay R.N., Scherzer C.R., Shihabuddin L.S., et al. Cerebrospinal fluid proteomics implicates the granin family in Parkinson’s disease. Sci. Rep. 2020;10:2479. doi: 10.1038/s41598-020-59414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang C., Hopfner F., Katsikoudi A., Hein R., Catli C., Evetts S., Huang Y., Wang H., Ryder J.W., Kuhlenbaeumer G., et al. Serum neuronal exosomes predict and differentiate Parkinson’s disease from atypical parkinsonism. J. Neurol. Neurosurg. Psychiatry. 2020;91:720–729. doi: 10.1136/jnnp-2019-322588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kitamura Y., Kojima M., Kurosawa T., Sasaki R., Ichihara S., Hiraku Y., Tomimoto H., Murata M., Oikawa S. Proteomic Profiling of Exosomal Proteins for Blood-based Biomarkers in Parkinson’s Disease. Neuroscience. 2018;392:121–128. doi: 10.1016/j.neuroscience.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 95.Shi M., Liu C., Cook T.J., Bullock K.M., Zhao Y., Ginghina C., Li Y., Aro P., Dator R., He C., et al. Plasma exosomal alpha-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014;128:639–650. doi: 10.1007/s00401-014-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zou J., Guo Y., Wei L., Yu F., Yu B., Xu A. Long Noncoding RNA POU3F3 and alpha-Synuclein in Plasma L1CAM Exosomes Combined with beta-Glucocerebrosidase Activity: Potential Predictors of Parkinson’s Disease. Neurotherapeutics. 2020;17:1104–1119. doi: 10.1007/s13311-020-00842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang J., Li X., Li J.-D. The Roles of Post-translational Modifications on α-Synuclein in the Pathogenesis of Parkinson’s Diseases. Front. Neurosci. 2019;13:381. doi: 10.3389/fnins.2019.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anderson J.P., Walker D.E., Goldstein J.M., de Laat R., Banducci K., Caccavello R.J., Barbour R., Huang J., Kling K., Lee M., et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 99.Burai R., Ait-Bouziad N., Chiki A., Lashuel H.A. Elucidating the Role of Site-Specific Nitration of alpha-Synuclein in the Pathogenesis of Parkinson’s Disease via Protein Semisynthesis and Mutagenesis. J. Am. Chem. Soc. 2015;137:5041–5052. doi: 10.1021/ja5131726. [DOI] [PubMed] [Google Scholar]
- 100.Foulds P., Mann D.M., Allsop D. Phosphorylated alpha-synuclein as a potential biomarker for Parkinson’s disease and related disorders. Expert Rev. Mol. Diagn. 2012;12:115–117. doi: 10.1586/erm.12.5. [DOI] [PubMed] [Google Scholar]
- 101.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M.S., Shen J., Takio K., Iwatsubo T. alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 102.Gorbatyuk O.S., Li S., Sullivan L.F., Chen W., Kondrikova G., Manfredsson F.P., Mandel R.J., Muzyczka N. The phosphorylation state of Ser-129 in human alpha-synuclein determines neurodegeneration in a rat model of Parkinson disease. Proc. Natl. Acad. Sci. USA. 2008;105:763–768. doi: 10.1073/pnas.0711053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hasegawa M., Fujiwara H., Nonaka T., Wakabayashi K., Takahashi H., Lee V.M., Trojanowski J.Q., Mann D., Iwatsubo T. Phosphorylated alpha-synuclein is ubiquitinated i.in alpha-synucleinopathy lesions. J. Biol. Chem. 2002;277:49071–49076. doi: 10.1074/jbc.M208046200. [DOI] [PubMed] [Google Scholar]
- 104.Kellie J.F., Higgs R.E., Ryder J.W., Major A., Beach T.G., Adler C.H., Merchant K., Knierman M.D. Quantitative measurement of intact alpha-synuclein proteoforms from post-mortem control and Parkinson’s disease brain tissue by intact protein mass spectrometry. Sci. Rep. 2014;4:5797. doi: 10.1038/srep05797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kragh C.L., Lund L.B., Febbraro F., Hansen H.D., Gai W.P., El-Agnaf O., Richter-Landsberg C., Jensen P.H. Alpha-synuclein aggregation and Ser-129 phosphorylation-dependent cell death in oligodendroglial cells. J. Biol. Chem. 2009;284:10211–10222. doi: 10.1074/jbc.M809671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuwahara T., Tonegawa R., Ito G., Mitani S., Iwatsubo T. Phosphorylation of alpha-synuclein protein at Ser-129 reduces neuronal dysfunction by lowering its membrane binding property in Caenorhabditis elegans. J. Biol. Chem. 2012;287:7098–7109. doi: 10.1074/jbc.M111.237131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu Y., Prudent M., Fauvet B., Lashuel H.A., Girault H.H. Phosphorylation of alpha-Synuclein at Y125 and S129 alters its metal binding properties: Implications for understanding the role of alpha-Synuclein in the pathogenesis of Parkinson’s Disease and related disorders. ACS Chem. Neurosci. 2011;2:667–675. doi: 10.1021/cn200074d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Na C.H., Sathe G., Rosenthal L.S., Moghekar A.R., Dawson V.L., Dawson T.M., Pandey A. Development of a novel method for the quantification of tyrosine 39 phosphorylated alpha- and beta-synuclein in human cerebrospinal fluid. Clin. Proteom. 2020;17:13. doi: 10.1186/s12014-020-09277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ohrfelt A., Zetterberg H., Andersson K., Persson R., Secic D., Brinkmalm G., Wallin A., Mulugeta E., Francis P.T., Vanmechelen E., et al. Identification of novel alpha-synuclein isoforms in human brain tissue by using an online nanoLC-ESI-FTICR-MS method. Neurochem. Res. 2011;36:2029–2042. doi: 10.1007/s11064-011-0527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Walker D.G., Lue L.F., Adler C.H., Shill H.A., Caviness J.N., Sabbagh M.N., Akiyama H., Serrano G.E., Sue L.I., Beach T.G., et al. Changes in properties of serine 129 phosphorylated alpha-synuclein with progression of Lewy-type histopathology in human brains. Exp. Neurol. 2013;240:190–204. doi: 10.1016/j.expneurol.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raghunathan R., Sethi M.K., Klein J.A., Zaia J. Proteomics, Glycomics, and Glycoproteomics of Matrisome Molecules. Mol. Cell Proteom. 2019;18:2138–2148. doi: 10.1074/mcp.R119.001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Raghunathan R., Hogan J.D., Labadorf A., Myers R.H., Zaia J. A glycomics and proteomics study of aging and Parkinson’s disease in human brain. Sci. Rep. 2020;10:12804. doi: 10.1038/s41598-020-69480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Downs M., Sethi M.K., Raghunathan R., Layne M.D., Zaia J. Matrisome changes in Parkinson’s disease. Anal. Bioanal. Chem. 2022;414:3005–3015. doi: 10.1007/s00216-022-03929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Posavi M., Diaz-Ortiz M., Liu B., Swanson C.R., Skrinak R.T., Hernandez-Con P., Amado D.A., Fullard M., Rick J., Siderowf A., et al. Characterization of Parkinson’s disease using blood-based biomarkers: A multicohort proteomic analysis. PLoS Med. 2019;16:e1002931. doi: 10.1371/journal.pmed.1002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cerri S., Ghezzi C., Sampieri M., Siani F., Avenali M., Dornini G., Zangaglia R., Minafra B., Blandini F. The Exosomal/Total alpha-Synuclein Ratio in Plasma Is Associated With Glucocerebrosidase Activity and Correlates With Measures of Disease Severity in PD Patients. Front. Cell Neurosci. 2018;12:125. doi: 10.3389/fncel.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng H., Xie Z., Zhang X., Mao J., Wang M., Wei S., Fu Y., Zheng H., He Y., Chen H., et al. Investigation of alpha-Synuclein Species in Plasma Exosomes and the Oligomeric and Phosphorylated alpha-Synuclein as Potential Peripheral Biomarker of Parkinson’s Disease. Neuroscience. 2021;469:79–90. doi: 10.1016/j.neuroscience.2021.06.033. [DOI] [PubMed] [Google Scholar]
- 117.He T. Implementation of Proteomics in Clinical Trials. Proteom. Clin. Appl. 2019;13:e1800198. doi: 10.1002/prca.201800198. [DOI] [PubMed] [Google Scholar]
- 118.Gomez Ravetti M., Moscato P. Identification of a 5-protein biomarker molecular signature for predicting Alzheimer’s disease. PLoS ONE. 2008;3:e3111. doi: 10.1371/journal.pone.0003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carr S.A., Abbatiello S.E., Ackermann B.L., Borchers C., Domon B., Deutsch E.W., Grant R.P., Hoofnagle A.N., Huttenhain R., Koomen J.M., et al. Targeted peptide measurements in biology and medicine: Best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell Proteom. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Krieger J.R., Wybenga-Groot L.E., Tong J., Bache N., Tsao M.S., Moran M.F. Evosep One Enables Robust Deep Proteome Coverage Using Tandem Mass Tags while Significantly Reducing Instrument Time. J. Proteome Res. 2019;18:2346–2353. doi: 10.1021/acs.jproteome.9b00082. [DOI] [PubMed] [Google Scholar]
- 121.Fu Q., Liu Z., Bhawal R., Anderson E.T., Sherwood R.W., Yang Y., Thannhauser T., Schroyen M., Tang X., Zhang H., et al. Comparison of MS(2), synchronous precursor selection MS(3), and real-time search MS(3) methodologies for lung proteomes of hydrogen sulfide treated swine. Anal. Bioanal. Chem. 2021;413:419–429. doi: 10.1007/s00216-020-03009-5. [DOI] [PubMed] [Google Scholar]