Abstract
The Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene encodes for a chloride channel defective in Cystic Fibrosis (CF). Accordingly, upregulation of its expression might be relevant for the development of therapeutic protocols for CF. MicroRNAs are deeply involved in the CFTR regulation and their targeting with miRNA inhibitors (including those based on Peptide Nucleic Acids, PNAs)is associated with CFTR upregulation. Targeting of miR-145-5p, miR-101-3p, and miR-335-5p with antisense PNAs was found to be associated with CFTR upregulation. The main objective of this study was to verify whether combined treatments with the most active PNAs are associated with increased CFTR gene expression. The data obtained demonstrate that synergism of upregulation of CFTR production can be obtained by combined treatments of Calu-3 cells with antisense PNAs targeting CFTR-regulating microRNAs. In particular, highly effective combinations were found with PNAs targeting miR-145-5p and miR-101-3p. Content of mRNAs was analyzed by RT-qPCR, the CFTR production by Western blotting. Combined treatment with antagomiRNAs might lead to maximized upregulation of CFTR and should be considered in the development of protocols for CFTR activation in pathological conditions in which CFTR gene expression is lacking, such as Cystic Fibrosis.
Keywords: peptide nucleic acids, cystic fibrosis, microRNAs, miRNA targeting, miR-101-3p, miR-145-5p, CFTR
1. Introduction
MicroRNAs (miRNAs) are a class of short (19–25 nucleotides) noncoding RNAs that exhibit a very important role in post-transcriptional regulation of gene expression [1,2,3,4]. The control of gene expression by miRNAs is achieved by sequence-specific targeting of regulated mRNAs (most frequently within the 3′UTR), leading to a translational repression or mRNA degradation [1,2,3]. The miRNA/mRNA interaction occurs at the level of the RNA-induced Silencing Complex (RISC) [1,2,3]. The complex networks constituted by miRNAs and mRNA targets are the basis for the control of several biological functions, including cell growth, apoptosis, and differentiation [4,5,6]. A single microRNA might interact with several target mRNAs; conversely, a single 3′UTR mRNA sequence contains several functional miRNA binding sites [6]. Alterations of the miRNA/mRNA networks are associated with the onset and/or progression of several human pathologies [7,8,9,10]. In agreement, alteration of miRNA biological functions (either by miRNA inhibition or miRNA mimicking) is considered an interesting and innovative strategy with potential therapeutic implications [11,12,13,14,15]. This is not merely a theoretical possibility, since efforts aimed at translating laboratory investigations to clinical practice are in progress as demonstrated by the several ongoing clinical trials focusing on miRNA targeting in chronic hepatitis C, type 2 diabetes, cutaneous T-cell lymphoma (see, for instance, NCT01646489, NCT01200420, NCT02826525, and NCT02580552), and by an increasing number of reports and reviews related to this issue [16,17,18,19].
Several studies have been reported demonstrating that microRNAs are involved in Cystic Fibrosis (CF) [20,21,22,23,24,25,26,27,28], a genetic disease caused by alteration of production and/or biological activity of the gene coding the chloride channel CFTR (Cystic Fibrosis Transmembrane Conductance Regulator) [29,30]. Therefore, miRNA therapeutic approaches for CF are expected to have a great impact in the near future [27,28].
The role of microRNAs in CFTR regulation is well-established and has been recently explored by several research groups in different experimental model systems, including CF primary bronchial epithelial cells in vitro or bronchial brushings ex vivo [31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50]. For instance, Gillen et al. [31] identified at least 12 miRNAs (including miR-145 and miR-494) capable of repressing the expression of CFTR. In general, miRNA profiling strongly supported the concept that high expression of a set of miRNAs (directly interacting with the CFTR transcript) is associated with low expression of CFTR, as reported by Ramachandran et al. [35] in CF cells for miR-494 and miR-509. Oglesby et al. [36] confirmed and extended these observations in ex vivo analyses, demonstrating increased expression of miR-494, miR-223, and miR-145 in CF brushings of airway cells [36].
Our group has reported that miR-145-5p [38,39,45,47,49], miR-101-3p [46,49], and miR-335-5p [50] play an important role in regulating the expression of the CFTR gene. In particular, miR-145-5p has been demonstrated to regulate CFTR by several research groups, as demonstrated by Oglesby et al. [36], Fabbri et al. [38], Lutful Kabir et al. [41], and Dutta et al. [44]. A possible therapeutic application of these studies is that targeting these miRNAs (in our case miR-145-5p, miR-101-3p and miR-335-5p) with antagomiRNAs might be considered an experimental strategy to upregulate CFTR expression. Interestingly, the activity of different miRNAs on post-transcriptional CFTR gene regulation might be synergistic, as found by Megiorni et al. [32] for miR-101 and miR-494. In our recently published studies, we employed, as antagomiRNAs, Peptide Nucleic Acids (PNAs), DNA analogues of outstanding properties [51,52,53]. These molecules, despite a radical structural change with respect to DNA and RNA, are capable of sequence-specific and efficient hybridization with complementary DNA and RNA, forming Watson–Crick double helices [53,54]. Accordingly, PNAs and PNA-based analogues have been proposed as antisense molecules targeting mRNAs [55,56].
The main objective of the present study was to verify whether combined treatments with PNAs targeting miRNAs involved in CFTR regulation are associated with increased (possibly synergistic) CFTR gene expression. For this purpose, the effect of PNA mixtures on the content of miRNAs and mRNAs was analyzed by RT-qPCR, while the CFTR production by Western blotting.
The possibility of a synergistic action of antagomiRNAs targeting different microRNAs might retain important impact in the development of protocols of interest in translating laboratory findings into therapeutic applications.
2. Results
2.1. The MicroRNAs miR-145-5p, miR-101-3p, and miR-335-5p Regulate Different Target mRNAs
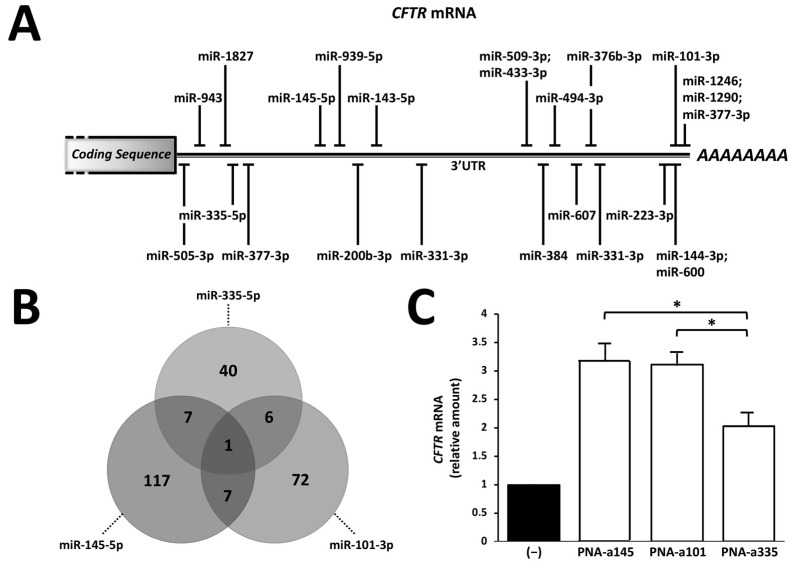
Figure 1A summarizes the binding sites for microRNAs present within the 3′UTR of the CFTR mRNA. This scheme is based on several recently published reports [31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,57]. The list of these miRNA binding sites, the location of their most probable binding site(s) within the 3′UTR CFTR mRNA sequence, and the studies originating this information are presented in Table S1. As clearly evident, several functional sites are present that can be the object of novel approaches in miRNA-therapeutics, including combined treatments.
Figure 1.
Characterization of the biological activity of miR-145-5p, miR-101-3p, and miR-335-5p on CFTR gene expression. (A) microRNA binding sites present within the 3′UTR of CFTR mRNA; (B) Venn-diagram indicating the number of mRNA targets of miR-145-5p, miR-101-3p, and miR-335-5p: only CFTR mRNA is a molecular target of all the three microRNAs. Analysis of mRNAs targeted by miRNAs was conducted using TargetScan (release 8.0) and data already in the literature. Afterwards, a Venn-diagram was performed using a Draw Venn Diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/, accessed on 9 May 2022); (C) upregulation of CFTR mRNA following treatment of Calu-3 cells with PNAs against miR-145-5p (PNA-a145), miR-101-3p (PNA-a101) and miR-335-5p (PNA-a335). n = 3 independent RT-qPCR experiments (* indicates differences with p < 0.05).
Most of these sites have been shown to regulate CFTR mRNA stability and production of CFTR protein.
Our group has demonstrated that, among these microRNAs, miR-145-5p, miR-101-3p, and miR-335-5p efficiently and selectively bind the 3′UTR sequence of CFTR mRNA. Inhibition of their interaction with CFTR mRNA leads to upregulation of CFTR mRNA content and CFTR protein accumulation. On the contrary, PNA-based miRNA inhibitors targeting other microRNAs (such as miR-433-3p and miR-509-3p) were found to be less efficient in our experimental Calu-3 cell system. For this reason, in this Short Report, we focused our attention on PNAs inhibiting miR-145-5p, miR-101-3p, and miR-335-5p. Figure 1B shows a computer-aided analysis demonstrating that miR-145-5p, miR-101-3p, and miR-335-5p are able to target different sets of mRNAs. Interestingly, CFTR mRNA was found to be targeted, unlike the other mRNAs, by these three microRNAs. This analysis was performed using the list of the mRNAs targeted by miR-145-5p, miR-101-3p, and miR-335-5p, presented in Table S2, which includes the relative publications.
2.2. CFTR Expression Depends on MicroRNAs miR-145-5p, miR-101-3p, and miR-335-5p
When bronchial epithelial Calu-3 cells were cultured in the presence of PNAs targeting miR-145-5p, miR-101-3p, and miR-335-5p (PNA-a145, PNA-a101, and PNA-a335, respectively), an upregulation of CFTR mRNA was observed when RT-qPCR was performed, as outlined in the representative example shown in Figure 1C. In order to obtain efficient transfection levels, in our studies, PNAs were functionalized with a R8-polyarginine peptide. This strategy allows the uptake of the majority of the PNA molecules within few hours, reaching a maximum level (near 100%) after 72 h, as firstly published by Brognara et al. [58] and then confirmed by several studies of our group [38,39,46,50]. The issue of PNA delivery has been fully addressed in a recent review paper by Volpi et al. [59].
These data obtained are in good agreement with published reports by Oglesby et al. [36], Fabbri et al. [38], Gambari et al. [39], Lutful Kabir et al. [41], and Dutta et al. [44] on antagomiRNAs against miR-145-5p, by Megiorni et al. [32], Hassan et al. [33], and Fabbri et al. [46] on antagomiRNAs against miR-101-3p, and by Tamanini et al. on antagomiRNAs against miR-335-5p [50]. The selectivity of the effects of PNA-a145, PNA-a101, and PNA-a335 on the expression of miR-145-5p, miR-101-3p, and miR-335-5p has been already discussed elsewhere by Fabbri et al. [38], Fabbri et al. [46], and Tamanini et al. [50]. On the basis of the data discussed in Figure 1, combined treatments based on PNA-a145 and PNA-a101 were performed on Calu-3 cells.
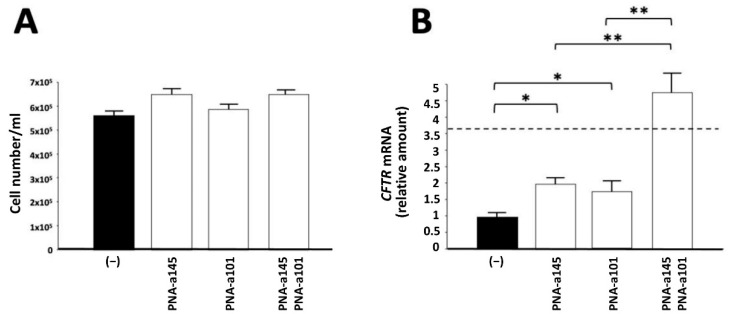
2.3. Combined Treatment Based on PNAs against miR-145-5p and miR-101-3p: Effects on Cell Growth of Calu-3 Cells and on CFTR mRNA Accumulation
In the experiment shown in Figure 2, the effects of combined treatment with PNA-a145 and PNA-a101 are reported, which are the PNAs that, when administered singularly, have been shown to increase the CFTR levels more efficiently.
Figure 2.
Effects on cell proliferation and CFTR mRNA accumulation of combined treatment of Calu-3 cells with PNAs against miR-145-5p and miR-101-3p. Calu-3 cells were treated for three days with PNA-a145 and PNA-a101, administered either singularly, or in combination. After these treatments, cell number/mL was determined, and CFTR mRNA content was analyzed by RT-qPCR. (A) effects on cell growth; (B) effects on CFTR mRNA. The results of (B) represent the fold change of the CFTR with respect to control untreated Calu-3 cells (n = 3 independent RT-qPCR experiments; * p < 0.05, ** p < 0.01). The dotted line represents the sum of the values obtained with single administrations of PNA-a145 and PNA-a101.
No major inhibitory effects on cell proliferation of Calu-3 cells were observed. The cell number/mL was determined after three days of cell culture. Each cell culture was started using a Calu-3 cell concentration of 200,000 cells/mL (day 0) (Figure 2A). On the contrary, when CFTR mRNA was quantified by RT-qPCR, an increase was detectable in the combined treatments. The fact that treatment with PNA-based antagomiRNAs leads to alteration of CFTR mRNA accumulation is expected, as elsewhere discussed [38,39,46,50]. Therefore, a more extensive analysis was performed by Western blotting, as shown in Figure 3.
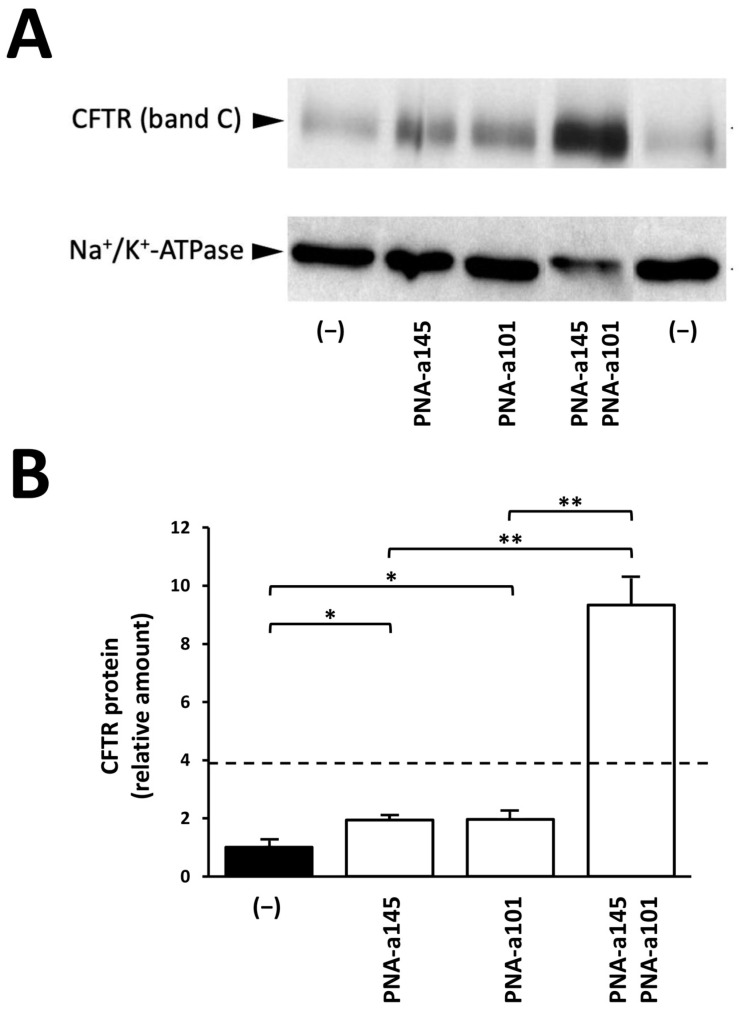
Figure 3.
Effects of combined treatment of Calu-3 cells with PNAs against miR-145-5p and miR-101-3p on CFTR protein accumulation. Calu-3 cells were treated for three days with PNA-a145 and PNA-a101, administered either singularly, or in combination. After these treatments, proteins were isolated and Western blotting was performed. (A) representative results of Western blotting performed using monoclonal antibodies recognizing CFTR or Na+/K+-ATPase, as indicated; (B) summary of the experiments performed. Results represent the mean fold changes with respect to the control untreated cells of the CFTR/Na+/K+-ATPase ratios (n = 3 independent quantifications of the same blot; * p < 0.05, ** p < 0.01). The uncut version of the Western blotting gels of (A) and the relative Ponceau staining are included in Supplementary materials (Figure S1). The dotted line represents the sum of the values obtained with single administrations of PNA-a145 and PNA-a101.
2.4. Combined Treatments Based on PNAs against miR-145-5p and miR-101-3p: Synergistic Effects on CFTR Protein Accumulation
Calu-3 cells were cultured in the presence of PNAs against miR-145-5p and miR-101-3p, administered singularly or in combination. After 72 h of treatment, proteins were isolated from the cells, and Western blotting was performed using two antibodies, one against CFTR (the mouse monoclonal antibody, clone 596, against NBD2 domain of CFTR) and the other against the house-keeping internal control Na+/K+-ATPase. A representative Western blotting is shown in Figure 3A, while the summary of multiple determinations is shown in Figure 3B. The uncut version of the Western blotting autoradiogram generating Figure 3A has been included in Supplementary materials (Figure S1).
The data obtained show that the combined treatment based on co-administration of PNA-a145 and PNA-a101 exhibits high efficiency in inducing the increased expression of CFTR. The obtained levels of CFTR content in combined PNA-a145/PNA-a101 treatments were higher than those predicted considering the sum of the respective values of CFTR content obtained using singularly administered PNA-a145 and PNA-a101. For example, the fold increase of CFTR was 1.94 and 1.96 in Calu-3 cells treated with PNA-a145 and PNA-a101, respectively. The increase of CFTR in PNA-a145 plus PNA-a101 combined treatment was 9.33, a value that is 2.4-fold higher than the addition of the values obtained using single treatments based on PNA-a145 and PNA-a101 (i.e., 3.9). According to the approach described by Chou and Talalay [60,61], these results indicate synergistic (rather than additional) effects.
3. Discussion
Since the demonstration that microRNAs are deeply involved in the regulation of the CFTR gene [31,32,33,34,35,36,37,38], a great attention has been dedicated to possible alteration of CFTR gene expression by targeting those miRNAs causing downregulation of this gene. For instance, PNA-mediated inhibition of the CFTR-regulators miR-145-5p, miR-101-3p and miR-335-5p leads to CFTR increase in the Calu-3 model system [28,39,46,50]. The upregulation of CFTR using antagomiRNA strategies has been also reported in a limited number of studies, such as those published by Oglesby et al. [36] and by Amato et al. [37]. Oglesby et al. found that transfection to CFBE41o− cells of molecules designed to knock down miR-145, miR-223, and miR-494 alone and in combination, leads to a reciprocal increase in CFTR mRNA expression. When all three miRNAs are knocked down in combination, CFTR expression is increased, but not in a synergistic fashion [36]. Amato et al. exploited a PNA targeting miR-509-3p and demonstrated that it causes an increase of luciferase activity in A549 cells co-transfected with a pLUC-CFTR-3′UTR-wt vector and miR-509-3p [37].
The possibility to upregulate CFTR expression is of great relevance in the possible personalized treatment of Cystic Fibrosis, a genetic disease that is characterized by a deep alteration of CFTR. In this context, the possibility of PNA-dependent modulation of miRNAs involved in the regulation of CFTR expression is of great impact in the case this approach will be demonstrated to be efficient in further increasing the activity of drugs already employed for personalized therapy of CF, such as, for instance, read-through molecules [62,63,64] and CFTR correctors [65,66,67]. This combined treatment might lead to unmet levels of functional CFTR production in CF cells, through the combination of two complementary therapeutic relevant activities, i.e., the miRNA-dependent upregulation of CFTR production and the use of personalized drugs.
The main result of our study is the demonstration that combined treatments of Calu-3 cells with molecules targeting different CFTR-regulating miRNAs might lead to a synergistic action on CFTR upregulation. This was found with combined treatments based on PNA-a145 and PNA-a101. Possible synergistic effects of miRNAs on CFTR have also been proposed in the past. For instance, synergistic post-transcriptional regulation of the CFTR by miR-101 and miR-494 specific binding was firstly reported by Megiorni et al. [32]. In their study, HEK293 cells were co-transfected either with a reporter construct containing 741 base pairs of the human CFTR 3′UTR in the presence of a synthetic microRNA mimic (miR-101 or miR-494). While both miR-101 and miR-494 significantly suppressed luciferase expression, when they were co-overexpressed, a synergistic effect between miRNAs was observed.
The knowledge that miRNAs might inhibit CFTR is the basis of a rational approach to upregulate CFTR by means of using anti-miRNA molecules (such as those based on PNAs used by our research group) or interfering with the miRNA/CFTR interactions using CFTR-specific target site blockers (TSBs) able to “mask” the miRNA binding sites present within the 3′UTR of the CFTR mRNA (see Figure 1A) [47,48,68]. In particular, the first report on the use of PNAs as target site blockers in cystic fibrosis was published by Zarrilli et al., who tested the ability of PNAs fully complementary to the CFTR 3′UTR sequence recognized by miR-509-3p, in order to rescue the CFTR expression in the A549 cellular model co-transfected with a pLuc-CFTR-3′UTR vector [68]. This “masking” approach was then fully validated by Sultan et al. [47] and by De Santi et al. [48].
Our study is the proof-of-principle that combined treatments (using different miRNA inhibitors or TBS) might be necessary for obtaining very high CFTR upregulation. This is a pre-requisite to develop efficient biomedicines for CFTR therapy. With respect to clinical relevance of the approach presented in this study, it is expected that decreased availability of miRNAs (as in the anti-miR approach) is associated with an accumulation and translation of target mRNAs (in our case CFTR mRNA).
Experimental trials based on miRNA targeting are ongoing. A first example is constituted by the Phase 2 trial, using miravirsen (Santaris Pharma, Copenhagen, Denmark, NCT01200420) and the Phase I trial, using RG-101 (Regulus Therapeutics, San Diego, CA, USA, NCT00980161), both based on targeting miR-122 for therapy of Hepatitis C Virus infection. Another example is a study of RGLS4326 in patients with autosomal dominant Polycystic Kidney Disease (NCT04536688, by Regulus Therapeutics, San Diego, CA, USA), targeting miR-17. In this context, PNAs represent molecules with strong anti-miRNA activity, although there are few clinical trials, in this case. Reviews on clinical trials based on miRNA therapeutics have been recently published [15,69,70].
While the results presented in this Brief Report are in our opinion promising, for our study, further improvements can be envisaged, namely: (a) since many other miRNA binding sites are present within the 3′UTR CFTR mRNA sequence (see Figure 1A), combinatorial studies can be performed in order to identify the best mixture for CFTR upregulation; (b) triple- or multi-combination might be also considered; (c) PNAs targeting multiple miRNAs might also display improved effects on CFTR; and (d) co-treatments with CFTR correctors/potentiators and/or with read-through molecules can be explored.
Regarding point (a) and in consideration of our recent interest in regulation of CFTR gene expression by miR-335-5p [50], the possible synergism between PNA-a145 and PNA-a335 was also considered (Table S3). We found that the combined treatment with PNA-a145 and PNA-a335 leads to a more efficient CFTR upregulation (4.95 fold) than the singular treatments (1.94 fold and fold 1.57, respectively). The effect on CFTR increase is less efficient than that obtained with the combination between PNA-a145 and PNA-a101. No increase was found with combination PNA-a101 and PNA-a335, compared to that derived from the single treatments.
In conclusion, despite the fact that the combinations used were limited, the present study represents a proof of principle that co-targeting of multiple miRNAs might lead to high induction of CFTR.
This might open a novel avenue in the personalized treatments of CF. In fact, in addition to personalized treatments considering the CFTR mutation of the CF patients under treatment, different dosage of the anti-miRNA components can be tailored depending on the pattern of CFTR-regulating miRNAs upregulated in the CF patients subject to further optimization of the intervention based on these precision medicines.
Several issues remain to be studied in depth, i.e., how much synergism can be obtained using different doses of PNA-a145 and PNA-a101. Moreover, a key issue is how many folds of dose-reduction for each drug (PNA-a145 and PNA-a101) can be proposed as a result of synergism. This is a key factor predicting for toxicity reduction to be considered in the design of protocols for clinical trials. The studied PNAs have been demonstrated, under the experimental conditions used in this Brief Report, to be unable to induce increase of the proportion of the dead cell population in treated Calu-3 cells. With respect to this point, these PNAs have been previously demonstrated by FACS analyses to be unable to induce apoptosis and increase of the proportion of the dead cell population in treated Calu-3 cells [38,46]. Apoptosis assays have been performed using the Muse Cell Analyzer instrument (Millipore Corporation, Billerica, MA, USA). The Muse Annexin V and Dead Cell Kit and the Muse Caspase-3/7 Kit were employed [15,17]. These assays differentiate viable non-apoptotic cells from early apoptotic, late apoptotic, and dead cells. Even more importantly, a recently published NGS study demonstrated that PNA-a145 and PNA-a101 do not alter the miRNome of treated Calu-3 cells [49], supporting the concept that they have no (or limited) effect on the overall transcriptional machinery of treated cells. Altogether, these studies suggest that the PNAs used here are not toxic to Calu-3 cells, fully in agreement with the data presented in Figure 2A, demonstrating that PNA-a145 and PNA-a101 do not alter the proliferation rate of Calu-3 cells when used either singularly or in combination. A final unanswered question is what are the best combinations considering PNAs targeting all the miRNAs regulating CFTR expression (see Table S2) through a binding to the 3′UTR (see Figure 1A). Despite the fact that this study is still ongoing, the present Brief Report sustains the concept that combined treatments targeting multiple miRNAs might be considered to maximize CFTR pharmacological upregulation.
4. Materials and Methods
4.1. Synthesis and Characterization of PNAs
Synthesis and characterizations of anti-miRNA PNAs were previously reported [38,39,46,47,50]. All PNA sequences have an octaarginine tail (R8), for the delivery into the cells without other carriers [12,59]. After synthesis and cleavage from the solid support, PNA purification was performed by HPLC using a Phenomenex Jupiter RPC18, (250 × 4.6 mm, 1.7 μm) column as described elsewhere [38]. After purification, the PNAs’ identity and purity were confirmed by UPLC/ESI-MS (Waters Acquity ultra performance LC HO6UPS-823M, with Waters SQ detector equipped with Waters UPLC BEH C18, 50 × 2.1 mm, 1.7 μm) at 35 °C. The concentration of the PNA was determined using UV-absorbance at 260 nm assuming an additive contribution of nucleobases. The sequence of the PNAs employed in this study is reported in Table 1. The PNAs were designed according to our standardized protocols, using the following criteria: (a) length of 18 bp, suitable for efficient synthesis also on large scale; (b) lack of self-complementarity both in antiparallel and parallel orientation; (c) minimal length of complementary sequences in mRNA, as evaluated by BLAST search; (d) when possible, targeting of the “seed region” which is an essential element for miRNA function [12,39]. A carrier octaargine (R8) peptide was conjugated at N-terminus of the PNA chain since it induces an efficiency in the delivery which approaches 100% (i.e., uptake in 100% of the target cell population), as elsewhere published [12,38,47,50,58]; this conjugation is easily realized during PNA solid-phase synthesis using the same reagents and solvents. We have previously reported the higher efficiency of R8-PNA conjugates (R8-PNAs) in inhibiting target miRNAs when this activity is compared to that of conventional commercially available antagomiRNAs [38,39]. The possible formation of heterodimers between the different PNA probes has been further evaluated using mfold and Multiple Primer Analyzer webtools. In all cases, no significant interactions were observed.
Table 1.
PNA sequence employed in Calu-3 cells treatment.
| PNA | Sequence | MicroRNA Target |
|---|---|---|
| PNA-a101 | H-R8-AGTTATCACAGTACTGTA-Gly-NH2 | miR-101-3p |
| PNA-a335 | H-R8-TTTCGTTATTGCTCTTGA-Gly-NH2 | miR-335-5p |
| PNA-a145 | H-R8-AGGGATTCCTGGGAAAAC-Gly-NH2 | miR-145-5p |
4.2. Cell Lines and Culture Conditions
The bronchial epithelial Calu-3 cells [38,71] were cultured in a humidified atmosphere of 5% CO2/air in DMEM/F12 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Biowest, Nauillè, France), 100 units/mL penicillin, 100 μg/mL streptomycin (Lonza, Verviers, Belgium), and 1% NEEA (100X) (Non-Essential Amino Acids Solution; Gibco, Grand Island, NY, USA). To determine the effect on proliferation, cell growth was monitored by determining the cell number/mL using a Z2 Coulter Counter (Coulter Electronics, Hialeah, FL, USA).
4.3. Cell Treatments with PNAs
Calu-3 cell lines were seeded in a 12-well plate with a concentration of 200,000 cells/mL. The day after the seeding, cells were treated with PNA-a145 2 μM, PNA-a101, and PNA-a335 4 μM, singularly or in combination. Cellular uptake with these R8-PNAs did not require additional transfection agents. After 72 h cells were collected, washed with cold sterile PBS, and total cell extracts and RNA were prepared.
4.4. RNA Extraction
Cultured cells were trypsinized and collected by centrifugation at 1500 rpm for 10 min at 4 °C, washed with PBS, lysed with Tri-Reagent (Sigma Aldrich, St. Louis, MI, USA), according to manufacturer’s instructions. The isolated RNA was washed once with cold 75% ethanol, dried, and dissolved in nuclease free pure water before use [38,39,46,50].
4.5. Analysis of CFTR Expression: RT-qPCR
Gene expression analysis was performed by RT-qPCR using 300 ng of total RNA, extracted, and reverse transcribed using the Taq-Man Reverse Transcription PCR Kit and random hexamers (Applied Biosystems, Waltham, MA, USA) as RT reaction primers. Quantitative real-time PCR (RT-qPCR) assays were carried out using gene-specific double fluorescently labeled probes. Primers and probes used to assay CFTR (Assay ID: Hs00357011_m1) gene expression were purchased from Applied Biosystems. The relative expression was calculated using the comparative cycle threshold method and, as reference genes, the human RPL13A (Assay ID: Hs03043885_g1).
4.6. Analysis of CFTR Expression: Western Blotting
CFTR expression was measured using Western blotting analyses. Cell pellets were lysed in RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA) and sonicated for 30 s three times on ice at 50% amplitude using the Vibra-Cell VC130 Ultrasonic Processor (Sonics). Lysates were cleared by centrifugation at 14,000× g for 30 min at 4 °C. Protein concentration was determined by BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. For CFTR analysis, 40 μg of total protein extracts were heated in Blue Loading Buffer by adding 50 nM of dithiothreithol (DTT) (Cell Signaling Technology, Danvers, MA, USA) at 37 °C for 10 min and loaded onto a 7% SDS-polyacrylamide gel. Then, the gel proteins were transferred to nitrocellulose membrane (Thermo Fischer Scientific, Waltham, MA, USA) by using Trans-Blot Turbo (Bio-Rad Laboratories, Hercules, CA, USA) and gross loading errors were excluded by staining of the gels with Ponceau S solution, as depicted in Figure S1 (bottom panel). Membranes was processed for Western blotting by using mouse monoclonal anti-body, clone 596, against NBD2 domain of CFTR (University of North Carolina, Cystic Fibrosis Center, Chapel Hill, NC, USA) at a dilution of 1:2,500 by overnight incubation at 4 °C. After washes, membranes were incubated with horseradish peroxidase-coupled anti-mouse immunoglobulin (R&D System, Minneapolis, MN, USA) at room temperature for 1 h and, after washes, the signal was developed by enhanced chemiluminescence (LumiGlo Reagent and Peroxide, Cell Signaling Technology, Danvers, MA, USA). The acquisition of the membrane images and the densitometric values of the bands were obtained using the ChemiDoc Imaging Systems (Bio-Rad Laboratories, Hercules, CA, USA). After membrane stripping, Na+/K+-ATPase α1 monoclonal antibody (sc-514614, Santa Cruz Biotechnology, Dallas, TX, USA) was used to compare the CFTR content with that of an internal control (Na+/K+-ATPase), a widely accepted strategy to assess changes of gene expression in treated cells by Western blotting [72]. The relative amount of CFTR protein was calculated after normalization of the densitometric values of CFTR bands with the densitometric values obtained using the anti-Na+/K+-ATPase antibody. In particular, for the sample loading normalization, the CFTR protein signal in each lane is divided (normalized) by the signal value for the Na+/K+-ATPase in that lane, and then relative levels of CFTR protein were compared across the blot using untreated cells (−) as a reference sample [72]. Na+/K+-ATPase was selected as an internal control for two main reasons: (a) its molecular weight is in the range of CFTR (100 kDa for Na+/K+-ATPase α1; 170–180 kDa for CFTR band C), allowing the use of the same Western blotting filter; (b) its protein extraction, purification, and pre-loading denaturation protocol are very similar to that employed for CFTR. Of course, as suggested elsewhere [72], our conclusion might be further supported by the use of additional internal protein controls (GAPDH, β-actin and others).
4.7. Statistical Analysis
Results are expressed as mean ± standard error of the mean (SEM). Comparisons between groups were made by using one-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001).
Acknowledgments
The Association “Tutti per Chiara” is acknowledged for supporting C.P. and M.Z. with a research fellowship.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23169348/s1.
Author Contributions
Conceptualization, R.G., R.C., G.C. and A.F.; methodology, C.P., J.G., M.Z. and A.M.; software, C.P. and A.M.; validation, C.P., R.G. and A.F.; formal analysis, C.P., A.F. and R.G.; investigation, C.P. and A.F.; resources, R.G., R.C. and G.C.; data curation, R.G. and A.F.; writing—original draft preparation, R.G. and C.P.; writing—review and editing, C.P., J.G., M.Z., A.M., R.G., A.F., R.C. and G.C.; supervision, A.F.; funding acquisition, R.G. and A.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets and materials generated and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This work was supported by the Fondazione Fibrosi Cistica (FFC), Project “Revealing the microRNAs-transcription factors network in cystic fibrosis: from microRNA therapeutics to precision medicine (CF-miRNA-THER)”, FFC#7/2018. This work has benefited from the equipment and framework of the COMP-HUB Initiative, funded by the ‘Departments of Excellence’ program of the Italian Ministry for Education, University and Research (MIUR, 2018–2022) for the Department of Chemistry, Life Sciences and Environmental Sustainability of the University of Parma. A.F. was funded by FAR (University Fund for Scientific Research, FAR-AF-Unife-2018 and FAR-AF-Unife-2019).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sontheimer E.J., Carthew R.W. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W., Jaskiewicz L., Kolb F.A., Pillai R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Garcia I., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 4.He L., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Kowdley K.V. MicroRNAs in common human diseases. Genom. Proteom. Bioinform. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu M., Zhang Q., Deng M., Miao J., Guo Y., Gao W., Cui Q. An analysis of human microRNA and disease associations. PLoS ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das J., Podder S., Ghosh T.C. Insights into the miRNA regulations in human disease genes. BMC Genom. 2014;15:1010. doi: 10.1186/1471-2164-15-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H., Xu J., Zhang G., Xu L., Li C., Wang L., Zhao Z., Jiang W., Guo Z., Li X. Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Syst. Biol. 2013;7:101. doi: 10.1186/1752-0509-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupaimoole R., Slack F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 12.Fabbri E., Brognara E., Borgatti M., Lampronti I., Finotti A., Bianchi N., Sforza S., Tedeschi T., Manicardi A., Marchelli R., et al. miRNA therapeutics: Delivery and biological activity of peptide nucleic acids targeting miRNAs. Epigenomics. 2011;3:733–745. doi: 10.2217/epi.11.90. [DOI] [PubMed] [Google Scholar]
- 13.Hanna J., Hossain G.S., Kocerha J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christopher A.F., Kaur R.P., Kaur G., Kaur A., Gupta V., Bansal P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016;7:68–74. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakraborty C., Sharma A.R., Sharma G., Lee S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2020;28:127–138. doi: 10.1016/j.jare.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Ree M.H., de Vree J.M., Stelma F., Willemse S., van der Valk M., Rietdijk S., Molenkamp R., Schinkel J., van Nuenen A.C., Beuers U., et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: A phase 1B, double-blind, randomised controlled trial. Lancet. 2017;389:709–717. doi: 10.1016/S0140-6736(16)31715-9. [DOI] [PubMed] [Google Scholar]
- 17.Ottosen S., Parsley T.B., Yang L., Zeh K., van Doorn L.J., van der Veer E., Raney A.K., Hodges M.R., Patick A.K. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob. Agents Chemother. 2015;59:599–608. doi: 10.1128/AAC.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyagi A.C., Sen U., Mishra P.K. Synergy of microRNA and stem cell: A novel therapeutic approach for diabetes mellitus and cardiovascular diseases. Curr. Diabetes Rev. 2011;7:367–376. doi: 10.2174/157339911797579179. [DOI] [PubMed] [Google Scholar]
- 19.Kohnken R., Mishra A. MicroRNAs in Cutaneous T-Cell Lymphoma: The Future of Therapy. J. Investig. Dermatol. 2019;139:528–534. doi: 10.1016/j.jid.2018.10.035. [DOI] [PubMed] [Google Scholar]
- 20.Xu W., Hui C., Yu S.S., Jing C., Chan H.C. MicroRNAs and cystic fibrosis-an epigenetic perspective. Cell Biol. Int. 2011;35:463–466. doi: 10.1042/CBI20100664. [DOI] [PubMed] [Google Scholar]
- 21.Greene C.M. MicroRNA Expression in Cystic Fibrosis Airway Epithelium. Biomolecules. 2013;3:157–167. doi: 10.3390/biom3010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKiernan P.J., Greene C.M. MicroRNA Dysregulation in Cystic Fibrosis. Mediat. Inflamm. 2015;2015:529642. doi: 10.1155/2015/529642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonneville F., Ruffin M., Guillot L., Rousselet N., Le Rouzic P., Corvol H., Tabary O. New insights about miRNAs in cystic fibrosis. Am. J. Pathol. 2015;185:897–908. doi: 10.1016/j.ajpath.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Oglesby I.K., McKiernan P.J. MiRNA Expression in Cystic Fibrosis Bronchial Epithelial Cells. Methods Mol. Biol. 2017;1509:57–69. doi: 10.1007/978-1-4939-6524-3_7. [DOI] [PubMed] [Google Scholar]
- 25.Bardin P., Sonneville F., Corvol H., Tabary O. Emerging microRNA Therapeutic Approaches for Cystic Fibrosis. Front. Pharmacol. 2018;9:1113. doi: 10.3389/fphar.2018.01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glasgow A.M.A., De Santi C., Greene C.M. Non-coding RNA in cystic fibrosis. Biochem. Soc. Trans. 2018;46:619–630. doi: 10.1042/BST20170469. [DOI] [PubMed] [Google Scholar]
- 27.De Santi C., Greene C.M. Challenges facing microRNA therapeutics for cystic fibrosis lung disease. Epigenomics. 2020;12:179–181. doi: 10.2217/epi-2019-0395. [DOI] [PubMed] [Google Scholar]
- 28.De Palma F.D.E., Raia V., Kroemer G., Maiuri M.C. The Multifaceted Roles of MicroRNAs in Cystic Fibrosis. Diagnostics. 2020;10:1102. doi: 10.3390/diagnostics10121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riordan J.R., Rommens J.M., Kerem B.S., Alon N.O.A., Rozmahel R., Grzelczak Z., Zielenski J., Lok S.I., Plavsic N., Chou J.L., et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 30.Fanen P., Wohlhuter-Haddad A., Hinzpeter A. Genetics of cystic fibrosis: CFTR mutation classifications toward genotype-based CF therapies. Int. J. Biochem. Cell Biol. 2014;52:94–102. doi: 10.1016/j.biocel.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 31.Gillen A.E., Gosalia N., Leir S.H., Harris A. MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 2011;438:25–32. doi: 10.1042/BJ20110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Megiorni F., Cialfi S., Dominici C., Quattrucci S., Pizzuti A. Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS ONE. 2011;6:e26601. doi: 10.1371/journal.pone.0026601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan F., Nuovo G.J., Crawford M., Boyaka P.N., Kirkby S., Nana-Sinkam S.P., Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS ONE. 2012;7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran S., Karp P.H., Jiang P., Ostedgaard L.S., Walz A.E., Fisher J.T., Keshavjee S., Lennox K.A., Jacobi A.M., Rose S.D., et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. USA. 2012;109:13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramachandran S., Karp P.H., Osterhaus S.R., Jiang P., Wohlford-Lenane C., Lennox K.A., Jacobi A.M., Praekh K., Rose S.D., Behlke M.A., et al. Post-transcriptional regulation of cystic fibrosis transmembrane conductance regulator expression and function by microRNAs. Am. J. Respir. Cell Mol. Biol. 2013;49:544–551. doi: 10.1165/rcmb.2012-0430OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oglesby I.K., Chotirmall S.H., McElvaney N.G., Greene C.M. Regulation of cystic fibrosis transmembrane conductance regulator by microRNA-145, -223, and -494 is altered in ΔF508 cystic fibrosis airway epithelium. J. Immunol. 2013;190:3354–3362. doi: 10.4049/jimmunol.1202960. [DOI] [PubMed] [Google Scholar]
- 37.Amato F., Tomaiuolo R., Nici F., Borbone N., Elce A., Catalanotti B., D’Errico S., Morgillo C.M., De Rosa G., Mayol L., et al. Exploitation of a very small peptide nucleic acid as a new inhibitor of miR-509-3p involved in the regulation of cystic fibrosis disease-gene expression. Biomed. Res. Int. 2014;2014:610718. doi: 10.1155/2014/610718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabbri E., Tamanini A., Jakova T., Gasparello J., Manicardi A., Corradini R., Sabbioni G., Finotti A., Borgatti M., Lampronti I., et al. A Peptide Nucleic Acid against MicroRNA miR-145-5p Enhances the Expression of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) in Calu-3 Cells. Molecules. 2017;23:71. doi: 10.3390/molecules23010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gambari R., Gasparello J., Fabbri E., Borgatti M., Tamanini A., Finotti A. Peptide Nucleic Acids for MicroRNA Targeting. Methods Mol. Biol. 2020;2105:199–215. doi: 10.1007/978-1-0716-0243-0_12. [DOI] [PubMed] [Google Scholar]
- 40.Bartoszewska S., Kamysz W., Jakiela B., Sanak M., Króliczewski J., Bebok Z., Bartoszewski R., Collawn J.F. miR-200b downregulates CFTR during hypoxia in human lung epithelial cells. Cell Mol. Biol. Lett. 2017;22:23. doi: 10.1186/s11658-017-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutful Kabir F., Ambalavanan N., Liu G., Li P., Solomon G.M., Lal C.V., Mazur M., Halloran B., Szul T., Gerthoffer W.T., et al. MicroRNA-145 Antagonism Reverses TGF-beta Inhibition of F508del CFTR Correction in Airway Epithelia. Am. J. Respir. Crit. Care Med. 2018;197:632–643. doi: 10.1164/rccm.201704-0732OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Santi C., Gadi S., Swiatecka-Urban A., Greene C.M. Identification of a novel functional miR-143-5p recognition element in the Cystic Fibrosis Transmembrane Conductance Regulator 3′UTR. AIMS Genet. 2018;5:53–62. doi: 10.3934/genet.2018.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitash N., Mu F., Donovan J.E., Myerburg M.M., Ranganathan S., Greene C.M., Swiatecka-Urban A. Transforming Growth Factor-beta1 Selectively Recruits microRNAs to the RNA-Induced Silencing Complex and Degrades CFTR mRNA under Permissive Conditions in Human Bronchial Epithelial Cells. Int. J. Mol. Sci. 2019;20:4933. doi: 10.3390/ijms20194933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dutta R.K., Chinnapaiyan S., Rasmussen L., Raju S.V., Unwalla H.J. A Neutralizing Aptamer to TGFBR2 and miR-145 Antagonism Rescue Cigarette Smoke- and TGF-beta-Mediated CFTR Expression. Mol. Ther. 2019;27:442–455. doi: 10.1016/j.ymthe.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finotti A., Gasparello J., Fabbri E., Tamanini A., Corradini R., Dechecchi M.C., Cabrini G., Gambari R. Enhancing the Expression of CFTR Using Antisense Molecules against MicroRNA miR-145-5p. Am. J. Respir. Crit. Care Med. 2019;199:1443–1444. doi: 10.1164/rccm.201901-0019LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabbri E., Tamanini A., Jakova T., Gasparello J., Manicardi A., Corradini R., Finotti A., Borgatti M., Lampronti I., Munari S., et al. Treatment of human airway epithelial Calu-3 cells with a Peptide-Nucleic Acid (PNA) targeting the microRNA miR-101-3p is associated with increased expression of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene. Eur. J. Med. Chem. 2021;209:112876. doi: 10.1016/j.ejmech.2020.112876. [DOI] [PubMed] [Google Scholar]
- 47.Sultan S., Rozzi A., Gasparello J., Manicardi A., Corradini R., Papi C., Finotti A., Lampronti I., Reali E., Cabrini G., et al. A Peptide Nucleic Acid (PNA) Masking the miR-145-5p Binding Site of the 3′UTR of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) mRNA Enhances CFTR Expression in Calu-3 Cells. Molecules. 2020;25:1677. doi: 10.3390/molecules25071677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Santi C., Fernández E.F., Gaul R., Vencken S., Glasgow A., Oglesby I.K., Hurley K., Hawkins F., Mitash N., Mu F., et al. Precise Targeting of miRNA Sites Restores CFTR Activity in CF Bronchial Epithelial Cells. Mol. Ther. 2020;28:1190–1199. doi: 10.1016/j.ymthe.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gasparello J., Fabbri E., Gambari R., Finotti A. Differential effects on the miRNome of the treatment of human airway epithelial Calu-3 cells with peptide-nucleic acids (PNAs) targeting microRNAs miR-101-3p and miR-145-5p: Next generation sequencing datasets. Data Brief. 2021;35:106718. doi: 10.1016/j.dib.2021.106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamanini A., Fabbri E., Jakova T., Gasparello J., Manicardi A., Corradini R., Finotti A., Borgatti M., Lampronti I., Munari S., et al. A Peptide-Nucleic Acid Targeting miR-335-5p Enhances Expression of Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Gene with the Possible Involvement of the CFTR Scaffolding Protein NHERF1. Biomedicines. 2021;9:117. doi: 10.3390/biomedicines9020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen P.E., Egholm M., Berg R.H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen P.E. Targeting double stranded DNA with peptide nucleic acid (PNA) Curr. Med. Chem. 2001;8:545–550. doi: 10.2174/0929867003373373. [DOI] [PubMed] [Google Scholar]
- 53.Egholm M., Buchardt O., Christensen L., Behrens C., Freier S.M., Driver D.A., Berg R.H., Kim S.K., Norden B., Nielsen P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 54.Nielsen P.E. Gene targeting and expression modulation by peptide nucleic acids (PNA) Curr. Pharm. Des. 2010;16:3118–3123. doi: 10.2174/138161210793292546. [DOI] [PubMed] [Google Scholar]
- 55.Shiraishi T., Hamzavi R., Nielsen P.E. Subnanomolar antisense activity of phosphonate-peptide nucleic acid (PNA) conjugates delivered by cationic lipids to HeLa cells. Nucleic Acids Res. 2008;36:4424–4432. doi: 10.1093/nar/gkn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nekhotiaeva A., Awasthi S.K., Nielsen P.E., Good L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 2004;10:652–659. doi: 10.1016/j.ymthe.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 57.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H., et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2017;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brognara E., Fabbri E., Bazzoli E., Montagner G., Ghimenton C., Eccher A., Cantù C., Manicardi A., Bianchi N., Finotti A., et al. Uptake by human glioma cell lines and biological effects of a peptide-nucleic acids targeting miR-221. J. Neurooncol. 2014;118:19–28. doi: 10.1007/s11060-014-1405-6. [DOI] [PubMed] [Google Scholar]
- 59.Volpi S., Cancelli U., Neri M., Corradini R. Multifunctional Delivery Systems for Peptide Nucleic Acids. Pharmaceuticals. 2020;14:14. doi: 10.3390/ph14010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chou T.C., Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 61.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 62.Kerem E. ELX-02: An investigational read-through agent for the treatment of nonsense mutation-related genetic disease. Expert Opin. Investig. Drugs. 2020;29:1347–1354. doi: 10.1080/13543784.2020.1828862. [DOI] [PubMed] [Google Scholar]
- 63.Zainal Abidin N., Haq I.J., Gardner A.I., Brodlie M. Ataluren in cystic fibrosis: Development, clinical studies and where are we now? Expert Opin. Pharmacother. 2017;18:1363–1371. doi: 10.1080/14656566.2017.1359255. [DOI] [PubMed] [Google Scholar]
- 64.Cabrini G. Innovative Therapies for Cystic Fibrosis: The Road from Treatment to Cure. Mol. Diagn. Ther. 2019;23:263–279. doi: 10.1007/s40291-018-0372-6. [DOI] [PubMed] [Google Scholar]
- 65.Lopes-Pacheco M., Pedemonte N., Veit G. Discovery of CFTR modulators for the treatment of cystic fibrosis. Expert Opin. Drug Discov. 2021;16:897–913. doi: 10.1080/17460441.2021.1912732. [DOI] [PubMed] [Google Scholar]
- 66.Bardin E., Pastor A., Semeraro M., Golec A., Hayes K., Chevalier B., Berhal F., Prestat G., Hinzpeter A., Gravier-Pelletier C., et al. Modulators of CFTR. Updates on clinical development and future directions. Eur. J. Med. Chem. 2021;213:113195. doi: 10.1016/j.ejmech.2021.113195. [DOI] [PubMed] [Google Scholar]
- 67.Dechecchi M.C., Tamanini A., Cabrini G. Molecular basis of cystic fibrosis: From bench to bedside. Ann. Transl. Med. 2018;6:334. doi: 10.21037/atm.2018.06.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zarrilli F., Amato F., Morgillo C.M., Pinto B., Santarpia G., Borbone N., D’Errico S., Catalanotti B., Piccialli G., Castaldo G., et al. Peptide Nucleic Acids as miRNA Target Protectors for the Treatment of Cystic Fibrosis. Molecules. 2017;22:1144. doi: 10.3390/molecules22071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith E.S., Whitty E., Yoo B., Moore A., Sempere L.F., Medarova Z. Clinical Applications of Short Non-Coding RNA-Based Therapies in the Era of Precision Medicine. Cancers. 2022;14:1588. doi: 10.3390/cancers14061588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diener C., Keller A., Meese E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022;38:613–626. doi: 10.1016/j.tig.2022.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Kreft M.E., Jerman U.D., Lasič E., Hevir-Kene N., Rižner T.L., Peternel L., Kristan K. The characterization of the human cell line Calu-3 under different culture conditions and its use as an optimized in vitro model to investigate bronchial epithelial function. Eur. J. Pharm. Sci. 2015;69:1–9. doi: 10.1016/j.ejps.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 72.Pillai-Kastoori L., Schutz-Geschwender A.R., Harford J.A. A systematic approach to quantitative Western blot analysis. Anal. Biochem. 2020;593:113608. doi: 10.1016/j.ab.2020.113608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and materials generated and/or analyzed during the present study are available from the corresponding author upon reasonable request.