Abstract
The trp RNA-binding attenuation protein (TRAP) regulates expression of the Bacillus subtilis trpEDCFBA operon by transcription attenuation. Tryptophan-activated TRAP binds to the nascent trp leader transcript by interacting with 11 (G/U)AG repeats. TRAP binding prevents formation of an antiterminator structure, thereby promoting formation of an overlapping terminator, and hence transcription is terminated before RNA polymerase can reach the trp structural genes. In addition to the antiterminator and terminator, a stem-loop structure is predicted to form at the 5′ end of the trp leader transcript. Deletion of this structure resulted in a dramatic increase in expression of a trpE′-′lacZ translational fusion and a reduced ability to regulate expression in response to tryptophan. By introducing a series of point mutations in the 5′ stem-loop, we found that both the sequence and the structure of the hairpin are important for its regulatory function and that compensatory changes that restored base pairing partially restored wild-type-like expression levels. Our results indicate that the 5′ stem-loop functions primarily through the TRAP-dependent regulatory pathway. Gel shift results demonstrate that the 5′ stem-loop increases the affinity of TRAP for trp leader RNA four- to fivefold, suggesting that the 5′ structure interacts with TRAP. In vitro transcription results indicate that this 5′ structure functions in the attenuation mechanism, since deletion of the stem-loop caused an increase in transcription readthrough. An oligonucleotide complementary to a segment of the 5′ stem-loop was used to demonstrate that formation of the 5′ structure is required for proper attenuation control of this operon.
Expression of the Bacillus subtilis tryptophan biosynthetic genes is regulated in response to changes in the intracellular level of tryptophan by TRAP, the trp RNA-binding attenuation protein (5, 9, 17, 24, 30, 35), which is the product of the mtrB gene (17). (For a recent review, see reference 4.) Examination of the crystal structure of TRAP has revealed that it consists of 11 identical subunits arranged in a single ring structure termed the β-wheel (2, 3). Cooperative TRAP activation occurs by binding of one tryptophan molecule between every two adjacent subunits (3, 6).
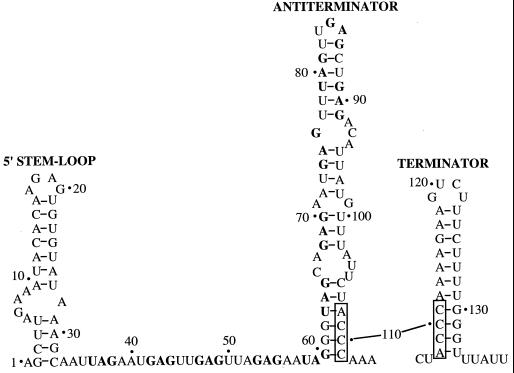
The 203-nucleotide (nt) untranslated trpEDCFBA operon leader transcript contains inverted repeats that allow folding of the transcript to form three RNA secondary structures (Fig. 1). Two of these structures, the antiterminator and an intrinsic terminator, overlap by 4 nt and therefore are mutually exclusive (Fig. 1). The TRAP binding target in the trp leader transcript consists of 11 closely spaced (G/U)AG repeats, 6 of which are present within the antiterminator (3, 7). When activated by tryptophan, 11 KKR motifs that outline the periphery of the TRAP complex interact with the 11 triplet repeats, presumably due to extensive hydrogen bond formation (42). Thus, TRAP binding to the nascent trp leader transcript blocks formation of the antiterminator structure, thereby allowing formation of the overlapping terminator, and hence promoting transcription termination upstream of the trp structural genes. In the absence of TRAP binding, the antiterminator structure forms, which results in transcription of the entire operon (5, 35).
FIG. 1.
Nucleotide sequence of the B. subtilis trp leader transcript showing the 5′ stem-loop and the mutually exclusive antiterminator and terminator structures. Boxed nucleotides mark overlapping segments of the competing secondary structures. The (G/U)AG repeats known to be involved in TRAP-RNA recognition are boldfaced. Numbering is from the start of transcription. RNA secondary-structure predictions were performed by using MFOLD (40, 43).
In addition to the transcription attenuation mechanism described above, TRAP also regulates translation of trpE and the unlinked trpG gene. TRAP-mediated translational control of trpE can occur by a novel RNA conformational switch mechanism (15). TRAP binding to trp operon readthrough transcripts promotes formation of an RNA hairpin that sequesters the trpE Shine-Dalgarno sequence (15, 24, 27). The trpG gene of B. subtilis is involved in the biosynthesis of both tryptophan and folic acid (23, 37). TRAP regulates TrpG synthesis by binding to nine trinucleotide repeats that surround and overlap the trpG Shine-Dalgarno sequence (7, 16, 41). Thus, TRAP binding directly blocks ribosome access to the trpG ribosome binding site (16).
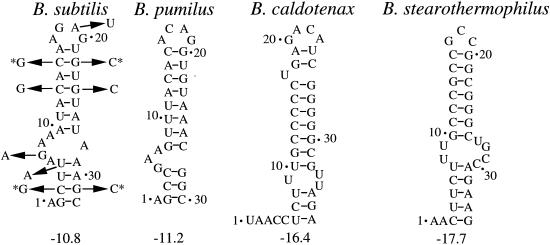
In addition to the antiterminator and terminator structures, an RNA hairpin is predicted to form at the extreme 5′ end of the B. subtilis trp leader transcript (Fig. 1). Similar 5′ stem-loop structures appear to be conserved in Bacillus pumilus, Bacillus caldotenax, and Bacillus stearothermophilus (Fig. 2). The trp operon leader transcripts of these bacilli also contain multiple triplet repeats, as well as overlapping antiterminator and terminator structures, suggesting that all four organisms control expression of the trp operon by similar transcription attenuation mechanisms (12, 20, 27).
FIG. 2.
Comparison of the 5′ stem-loop structures predicted to form in the trp operon leader transcripts of B. subtilis (18), B. pumilus (20), B. caldotenax (36), and B. stearothermophilus (12). Numbering is from the start of transcription. The number below each structure indicates the calculated free energy of formation (ΔG) in kilocalories per mole. Arrows associated with the B. subtilis structure indicate the single-nucleotide substitutions introduced into the 5′ stem-loop. Asterisks mark the double compensatory changes that restore the predicted secondary structure. The 5′ stem-loop deletion removed nt 3 to 32.
Since the 5′ stem-loop and the attenuation mechanisms appear to be evolutionarily conserved, we were interested in determining if the 5′ stem-loop affects expression of the B. subtilis trp operon. By examining the effect of a 5′ stem-loop deletion and several point mutations within the 5′ hairpin, we found that disruption of the structure resulted in a dramatic increase in trp operon expression and a substantial reduction in the ability of B. subtilis to regulate expression of this operon. Moreover, our results indicate that the 5′ stem-loop functions in the transcription attenuation mechanism by increasing the affinity of TRAP for trp leader RNA. Our results suggest that TRAP–5′ stem-loop interaction increases the probability that TRAP will bind to the (G/U)AG repeats before the antiterminator can form, thereby increasing the likelihood that transcription termination will occur before RNA polymerase can reach the trp operon structural genes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All of the B. subtilis strains used in this study are listed in Table 1. Plasmids pTZ18R and pTZ18U were obtained from United States Biochemical Corp. ptrpBGI-PLK (27) was used to construct all of the translational fusion integration plasmids for B. subtilis. This plasmid is a derivative of ptrpBGI (34) in which the EcoRI-HindIII fragment containing the B. subtilis trp promoter and leader region has been replaced by the polylinker of pTZ18R, leaving a promoterless lacZ gene. ptrpBGI-PLK is designed to allow integration of essentially any trp promoter and leader construct into the amyE locus of the B. subtilis chromosome such that lacZ expression is under the control of the trp leader construct. pHY300PLK is an Escherichia coli-B. subtilis shuttle vector (22). Plasmid pSI45 is a derivative of pHY300PLK that carries the mtrAB operon under the control of its natural promoter (17).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| W168 | Prototroph | BGSCb |
| BG4233 | ΔmtrB argC4 | 20 |
| PLBS44 | amyE::[trpP(−412 to +203)trpE′-′lacZ Cmr] | This study |
| PLBS104 | amyE::[trpP(−412 to +203) Δ(+3 to +32)trpE′-′lacZ Cmr] | This study |
| PLBS251 | ΔmtrB argC4 amyE::[trpP(−412 to +203)trpE′-′lacZ Cmr] | This study |
| PLBS252 | ΔmtrB argC4 amyE::[trpP(−412 to +203) Δ(+3 to +32)trpE′-′lacZ Cmr] | This study |
| PLBS253 | PLBS44/pSI45 (Tcr) | This study |
| PLBS254 | PLBS44/pHY300PLK (Tcr) | This study |
| PLBS255 | PLBS104/pSI45 (Tcr) | This study |
| PLBS256 | PLBS104/pHY300PLK (Tcr) | This study |
| PLBS257 | amyE::[trpP(−412 to +203) C3G trpE′-′lacZ Cmr] | This study |
| PLBS258 | amyE::[trpP(−412 to +203) T5A trpE′-′lacZ Cmr] | This study |
| PLBS259 | amyE::[trpP(−412 to +203) G7A trpE′-′lacZ Cmr] | This study |
| PLBS260 | amyE::[trpP(−412 to +203) C13G trpE′-′lacZ Cmr] | This study |
| PLBS261 | amyE::[trpP(−412 to +203) C15G trpE′-′lacZ Cmr] | This study |
| PLBS262 | amyE::[trpP(−412 to +203) A19T trpE′-′lacZ Cmr] | This study |
| PLBS263 | amyE::[trpP(−412 to +203) G22C trpE′-′lacZ Cmr] | This study |
| PLBS264 | amyE::[trpP(−412 to +203) G24C trpE′-′lacZ Cmr] | This study |
| PLBS265 | amyE::[trpP(−412 to +203) G31C trpE′-′lacZ Cmr] | This study |
| PLBS266 | amyE::[trpP(−412 to +203) C3G G31C trpE′-′lacZ Cmr] | This study |
| PLBS267 | amyE::[trpP(−412 to +203) C15G G22C trpE′-′lacZ Cmr] | This study |
Symbols: trpP, the trp promoter; prime, truncation of the gene; −412 to +203, the DNA fragment containing trpP and neighboring regions that was incorporated (numbered relative to the transcription start site); Δ(+3 to +32), the portion of the leader region that was deleted. C3G (and similar designations within genotypes for PLBS257 through PLBS267) indicates the position of a point mutation in the trp leader.
BGSC, Bacillus Genetic Stock Center, Ohio State University, Columbus.
Plasmid pPB22 (5) contains a 730-bp EcoRI-HindIII fragment with the wild-type trp promoter and leader region cloned into the polylinker of pTZ18R. Plasmid pPB77, containing nucleotides +1 to +111 of the trp leader region, has been described elsewhere (7). Plasmid pJY1 is a derivative of pPB22 in which nucleotides +3 to +32 relative to the start of trp operon transcription have been deleted. The deletion removes the DNA corresponding to the 5′ stem-loop present in the wild-type trp leader transcript (Fig. 1). pJY1 was constructed by PCR using the oligonucleotides 5′ CTGTTCTTATCGTATTACAATTC 3′ and 5′ ATTAGAATGAGTTGAGTTAGAG 3′ as primers and pPB22 as the template. Following PCR, the product was self-ligated, resulting in a plasmid, pJY1, containing a 700-bp EcoRI-HindIII fragment comprising the B. subtilis trp promoter and leader region with the 5′ stem-loop deletion. The precise deletion of the 30 nt was confirmed by DNA sequencing. pJY2 was constructed by subcloning the 700-bp EcoRI-HindIII fragment from pJY1 into the EcoRI-HindIII sites of the pTZ18U polylinker. The 700-bp EcoRI-HindIII fragment from pJY1 was also subcloned into the EcoRI-HindIII sites of the ptrpBG1-PLK polylinker to create the integration vector pMM3. Plasmid pPB310 contains nucleotides +32 to +111 of the B. subtilis trp leader region and was constructed by PCR using the oligonucleotides 5′ CCCGAATTCAATTAGAATGAGTTGAGTTAG 3′ and 5′ CCCGGATCCGGGTAGAATAAACATAATGTC 3′ as primers and plasmid pPB22 as the template.
Various point mutations that changed the primary and/or secondary structure of the 5′ stem-loop were generated. The MFOLD computer program (40, 43) was used to predict the secondary structure of the wild-type transcript and each mutant transcript. PCR-mediated mutagenesis using mutagenic DNA oligonucleotides was used to construct each of the point mutations (19, 29). Plasmid pPB22 was used as the template in each reaction. The resulting 730-bp EcoRI-HindIII fragments, each containing a point mutation, were subcloned into the EcoRI-HindIII sites of the pTZ18R polylinker. The 730-bp EcoRI-HindIII fragments containing the point mutations were then subcloned into the EcoRI-HindIII sites of the ptrpBGI-PLK polylinker to generate a series of integration plasmids. We found that several of the mutant B. subtilis trp leaders accumulated additional mutations when grown in E. coli. Therefore, the trp promoter and leader region was sequenced after each step of the plasmid constructions to ensure that secondary changes had not been introduced. Plasmids derived from ptrpBGI-PLK that carried trpE′-′lacZ translational fusions under the control of the wild-type trp promoter and wild-type or mutant trp leaders were linearized with PstI and subsequently integrated into the B. subtilis W168 amyE locus by homologous recombination (Table 1) (27, 34). Integration of the various translational fusions into amyE was confirmed by the starch iodine test (33). Finally, to confirm that no undesirable secondary mutations had been introduced, PCR products were generated from the chromosomal DNA of the transformed B. subtilis strains and subsequently sequenced.
B. subtilis BG4233 contains a deletion of mtrB (20). PLBS251 and PLBS252 were constructed by transforming BG4233 with chromosomal DNA purified from PLBS44 (wild-type trp leader; trpE′-′lacZ in amyE) or PLBS104 (trp leader with 5′ stem-loop deletion [Δ5′S-L]; trpE′-′lacZ in amyE), respectively. Selection was for chloramphenicol resistance. Transformants were screened for amylase deficiency (AmyE−) by the starch iodine test. The absence of TRAP (ΔmtrB) was confirmed by screening for 5-fluorotryptophan resistance (growth in the presence of 200 μg/ml).
Bacterial growth, transformation, and DNA isolation.
B. subtilis and E. coli strains were routinely grown on L agar, L broth, or minimal acid casein hydrolysate (ACH) medium (38). Plasmid (11) and chromosomal (25) DNA was isolated by standard procedures. E. coli (32) and B. subtilis (1) transformations were performed as described previously. Appropriate antibiotics were added, as needed, to the following concentrations: ampicillin, 100 μg/ml; tetracycline, 10 μg/ml; chloramphenicol, 5 μg/ml.
β-Galactosidase assays.
Bacterial cultures were grown in minimal ACH medium in the presence or absence of 50 μg of l-tryptophan/ml supplemented with the appropriate antibiotics. Cells were harvested during late-exponential growth (110 Klett units, filter no. 54; Klett Manufacturing Co., Inc.). Aliquots were then assayed for β-galactosidase activity by the method of Miller (28).
Gel mobility shift assay.
TRAP purification was performed as described previously (5). Gel-purified transcripts used in this analysis were synthesized by using the Ambion MEGAscript in vitro transcription kit. 5′-end-labeled RNAs were generated by treating in vitro-generated transcripts with calf intestinal phosphatase and subsequently with polynucleotide kinase and [γ-32P]ATP. The unlabeled and labeled RNA was gel purified as described previously (15). The binding affinity between TRAP and trp leader RNA was estimated by using gel mobility shift assays designed by modifying a published procedure (30). The binding data was fit to a nonlinear least-squares algorithm. Transcripts used in the analysis were generated from pPB77 (wild type) or pPB310 (5′ stem-loop deletion) that had been linearized with BamHI. Binding reaction mixtures (40 μl) containing 0.2 nM 5′-end-labeled RNA, various concentrations of TRAP (TRAP excess), and 1 mM l-tryptophan in TKM buffer (40 mM Tris-HCl [pH 8.0], 250 mM KCl, 4 mM MgCl2) were incubated at 25°C for 20 min. Aliquots of reaction mixtures were fractionated through 6% native polyacrylamide gels. Electrophoresis was performed at 4°C and in 0.5× Tris-borate-EDTA. Gels were dried, and the bound and free RNA bands were quantified by using a Phosphorimager (Molecular Dynamics) and the ImageQuant software package. Modifications of the standard reaction are described in the text or the appropriate figure legend.
In vitro transcription attenuation assay.
In vitro transcription attenuation assays followed a previously published procedure (5). Briefly, transcription reaction mixtures contained B. subtilis vegetative (ςA) RNA polymerase, 20 nM DNA template, TRAP (various concentrations), 1 mM l-tryptophan, and ribonucleoside triphosphates (2.7 mM ATP, 0.7 mM CTP, 1.1 mM GTP, 1.4 mM UTP) of which UTP was radioactively labeled. EcoRI-HindIII restriction fragments that contained the B. subtilis trp promoter and leader region from plasmids pPB22 and pJY2 were used as DNA templates in transcription reactions. Reactions were carried out at 30°C for 30 min. Samples were fractionated through 6% polyacrylamide gel electrophoresis (PAGE) gels containing 7 M urea. Radiolabeled RNA bands were quantified with a Phosphorimager (Molecular Dynamics, Inc.) and the ImageQuant software package. Modifications of the standard assay are described in the text or figure legends.
RESULTS
The 5′ stem-loop is involved in regulating expression of the trpEDCFBA operon.
A stem-loop structure is predicted to form at the 5′ end of trp operon transcripts in B. subtilis, B. pumilus, B. caldotenax, and B. stearothermophilus (Fig. 2). Since the trp operon transcription attenuation mechanism appears to be conserved in all four of these organisms (12, 20, 27), we were interested in determining if the 5′ stem-loop is involved in regulating expression of the B. subtilis trp operon. Accordingly, we deleted the DNA corresponding to the 5′ stem-loop from the trp operon leader. We constructed B. subtilis strains containing trpE′-′lacZ translational fusions that were controlled by the wild-type (trpLWT) or Δ5′S-L trp leader and analyzed β-galactosidase expression when each strain was grown in the presence and absence of exogenous tryptophan. We observed minimal expression in the trpLWT strain PLBS44 grown in the presence of tryptophan (Table 2). The effect of exogenous tryptophan on expression of the trpLWT trpE′-′lacZ can be assessed from the ratio of expression in the absence of tryptophan to expression in the presence of tryptophan (−Trp/+Trp ratio), which was 140. Comparable experiments were performed with the Δ5′S-L strain PLBS104. In this case the −Trp/+Trp ratio was only 8.4, significantly lower than that observed for the trpLWT strain. The 17-fold (140/8.4) reduction in the ability of the Δ5′S-L strain to regulate expression was primarily due to the dramatic increase in expression found to occur when this strain was grown in the presence of tryptophan. When cultures were grown in the presence of tryptophan, β-galactosidase activity was approximately 200-fold higher in the deletion strain than in the wild type, while β-galactosidase activity was only 13-fold higher in the deletion strain when cultures were grown without tryptophan (Table 2). The dramatic increase in expression, as well as the 17-fold reduction in regulation observed for the Δ5′S-L strain, demonstrated that the 5′ stem-loop influences regulation of expression of the B. subtilis trp operon.
TABLE 2.
Effect of 5′ stem-loop mutations on trp operon expression
| Strain | Relevant genotype | Sequence and/or structural changea | β-Gal activityb
|
β-Gal ratio (−Trp/+Trp) | |
|---|---|---|---|---|---|
| +Trp | −Trp | ||||
| PLBS44 | Wild type | Wild type | 0.2 ± 0.1 | 28 ± 4.3 | 140 |
| PLBS104 | Δ5′S-L | Deletion | 44 ± 2.4 | 371 ± 49 | 8.4 |
| PLBS251 | ΔmtrB | Wild type | 509 ± 52 | 407 ± 32 | 0.8 |
| PLBS252 | Δ5′S-L ΔmtrB | Deletion | 1,110 ± 235 | 1,150 ± 317 | 1.0 |
| PLBS258 | T5A | Structure | 8 ± 2.7 | 178 ± 37 | 22 |
| PLBS259 | G7A | Sequence | 31 ± 6.4 | 350 ± 19 | 11 |
| PLBS260 | C13G | Structure | 13 ± 1 | 334 ± 84 | 26 |
| PLBS262 | A19T | Sequence | 0.9 ± 0.2 | 70 ± 9 | 78 |
| PLBS264 | G24C | Structure | 5 ± 1 | 69 ± 22 | 14 |
| PLBS257 | C3G | Structure | 22 ± 4.3 | 348 ± 76 | 16 |
| PLBS265 | G31C | Structure | 12 ± 2.5 | 260 ± 37 | 22 |
| PLBS266 | C3G G31C | Compensatoryc | 2.9 ± 1.2 | 261 ± 20 | 90 |
| PLBS261 | C15G | Structure | 111 ± 30 | 340 ± 2.3 | 3.1 |
| PLBS263 | G22C | Structure | 100 ± 16 | 376 ± 9.2 | 3.8 |
| PLBS267 | C15G G22C | Compensatory | 2.0 ± 0.7 | 237 ± 9.3 | 119 |
The mutation in the 5′ stem-loop alters either the sequence alone or both the sequence and the predicted secondary structure.
The β-Galactosidase (β-Gal) activity expressed from the trpE′-′lacZ fusion is given in Miller units (28). Values are the averages from at least four independent experiments ± standard deviations. +, in the presence of; −, in the absence of.
Double mutations that are predicted to restore the secondary structure of the 5′ stem-loop.
Effect of various 5′ stem-loop point mutations on trp operon expression.
Since deletion of the entire 5′ stem-loop had such a dramatic effect on trp operon expression, we wanted to determine if the primary sequence, the secondary structure, or both were responsible for 5′ stem-loop function. We examined the effects of several point mutations that altered the primary sequence and/or the predicted secondary structure of the 5′ stem-loop (Fig. 2). Leader regions bearing these 5′ stem-loop mutations were integrated as a single copy into the B. subtilis amyE locus as trpE′-′lacZ translational fusions. The effect of the point mutations in the 5′ stem-loop of the trp leader on trp operon expression was assessed by comparing β-galactosidase expression levels of these mutant strains to those of the wild-type and Δ5′S-L strains (Table 2). A substantial increase in β-galactosidase activity was observed with all of the mutations predicted to disrupt the 5′ stem-loop (C3G, U5A, C13G, C15G, G22C, G24C, and G31C). Furthermore, compared to that in the wild-type strain, the fold change in β-galactosidase activity when cultures were grown in the absence versus the presence of tryptophan was reduced considerably in these 5′ stem-loop mutants (Table 2). Interestingly, each of these single-nucleotide substitutions had an effect similar to that of the 5′ stem-loop deletion.
Compensatory double mutations that were predicted to restore the secondary structure of the 5′ stem-loop resulted in partial restoration of wild-type expression levels (Table 2). This effect was most pronounced when cultures were grown in the presence of l-tryptophan (compare C3G and G31C with C3G G31C; compare C15G and G22C with C15G G22C). Note that while the β-galactosidase expression levels of the compensatory mutants were approximately 10-fold higher than that observed for the wild-type strain, the −Trp/+Trp ratios were similar.
Two other nucleotide substitutions were tested; these changes were not predicted to alter the predicted secondary structure. Instead, they altered a possible UAG (G7A) or a GAG (A19U) located in the 5′ portion of the stem or in the loop, respectively (Fig. 2). The G7A mutation had an effect similar to that of the Δ5′S-L mutation, whereas A19U had only a modest effect on trp operon expression (Table 2). Taken together, the trp leader point mutation studies indicate that both the primary sequence and the structure of the 5′ stem-loop are important for proper regulation of the trp operon.
The 5′ stem-loop functions in TRAP-dependent regulation of the trp operon.
Since it is well established that TRAP is responsible for regulating expression of the B. subtilis trp operon, we expected that the 5′ stem-loop would function in the TRAP-dependent regulatory pathway. To test this hypothesis, we constructed two new strains by integrating the wild-type or Δ5′S-L trpE′-′lacZ fusion into a TRAP-deficient (ΔmtrB) background. As expected, both strains lost the ability to regulate trp operon expression in response to tryptophan (Table 2). Interestingly, we observed a reproducible two- to threefold increase in expression in the Δ5′S-L ΔmtrB strain (PLBS252) relative to the ΔmtrB control (PLBS251). Thus, while it appears that the 5′ stem-loop functions primarily through the TRAP-dependent regulatory mechanism, these results indicate that the 5′ hairpin has a small effect on trp operon expression that is independent of TRAP.
Overexpression of mtrB partially suppresses the defect associated with the 5′ stem-loop deletion.
The results described above indicate that the 5′ stem-loop primarily functions in TRAP-dependent regulation of the B. subtilis trp operon. One possible explanation for these results is that the 5′ stem-loop participates in TRAP-RNA recognition. To test this possibility, we examined the effect of overexpressing mtrB (TRAP) in the 5′ stem-loop deletion strain by measuring β-galactosidase activity when cells were grown in the presence and absence of tryptophan. We found that mtrB overexpression largely suppressed the defect associated with the 5′ stem-loop deletion (Table 3). Compare the expression levels of PLBS256 [trpLΔ5′S-L trpE′-′lacZ/pHY300PLK (vector)] and PLBS255 [trpLΔ5′S-L trpE′-′lacZ/pSI45 (mtrB+)] (Table 3) with that of PLBS44 (trpLWT trpE′-′lacZ) (Table 2). These results suggest that the 5′ stem-loop assists TRAP in binding to the (G/U)AG repeats present in the trp leader transcript by increasing the affinity of TRAP for trp leader RNA.
TABLE 3.
Effect of mtrB overexpression in strains containing a 5′ stem-loop deletion
| Strain | Relevant genotype | Plasmid | β-Gal activitya
|
β-Gal ratio (−Trp/+Trp) | |
|---|---|---|---|---|---|
| +Trp | −Trp | ||||
| PLBS254 | Wild type | pHY300PLK | 0.1 ± 0.03 | 44 ± 12 | 440 |
| PLBS253 | Wild type | pSI45 (mtrB+) | 0.1 ± 0.02 | 18 ± 5.9 | 180 |
| PLBS256 | Δ5′S-L | pHY300PLK | 23 ± 6.7 | 391 ± 73 | 17 |
| PLBS255 | Δ5′S-L | pSI45 (mtrB+) | 0.3 ± 0.08 | 143 ± 23 | 477 |
The β-Galactosidase (β-Gal) activity expressed from the trpE′-′lacZ fusion is given in Miller units (28). Values are averages from at least four independent experiments ± standard deviations. +Trp, in the presence of Trp; −Trp, in the absence of Trp.
The 5′ stem-loop structure increases the affinity of TRAP for trp leader RNA.
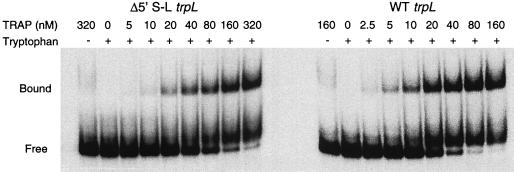
The results described above suggested that the 5′ stem-loop increases the affinity of TRAP for trp leader RNA. We performed TRAP-trp leader RNA gel mobility shift experiments to determine if the 5′ stem-loop does in fact increase the affinity of TRAP for the trp leader transcript. We used in vitro-generated transcripts that contained nucleotides +1 to +111 of the trp leader (wild type) or a similar transcript containing nucleotides +32 to +111 in which the 5′ stem-loop had been deleted (Δ5′S-L). Note that both of these transcripts contained the 11 (G/U)AG repeats that are known to interact with TRAP (Fig. 1). Binding to the wild-type trp leader transcript was detectable at 2.5 nM TRAP and saturated at approximately 80 nM TRAP (Fig. 3). With the Δ5′S-L transcript, binding was detected at 10 nM TRAP and saturated at approximately 320 nM TRAP (Fig. 3). Note that the relative intensity of the TRAP-dependent band that is visible just above the free RNA band fluctuated from experiment to experiment. Since this species was found to increase with increasing TRAP concentrations and never saturated, and the shift was too small for it to represent a distinct TRAP-RNA complex, it is likely that this band resulted from complex dissociation soon after gel loading. Nonlinear least-squares analysis of these data yielded estimated Kd values of 17 ± 5.3 nM TRAP for the wild-type transcript and 79 ± 9.2 nM TRAP for the Δ5′S-L transcript. Our in vitro binding data indicate that deletion of the 5′ stem-loop results in a four- to fivefold decrease in the affinity of TRAP for trp leader RNA. This result is consistent with the previous finding that overexpression of TRAP partially suppressed the defect associated with a 5′ stem-loop deletion in vivo (Table 3).
FIG. 3.
Gel mobility shift analysis of TRAP complexed with wild-type or 5′ stem-loop deletion trp leader transcripts. 5′-32P-labeled wild-type (WT) or 5′ stem-loop deletion (Δ5′S-L) trp leader transcripts (0.2 nM) were incubated with 1 mM tryptophan and TRAP at the concentrations indicated above the gel. Each transcript contained the 11 (G/U)AG repeats between +36 and +91. Control reactions without tryptophan were performed with each RNA at the highest TRAP concentration used. Bands corresponding to free RNA (Free) and shifted TRAP-RNA complexes (Bound) are indicated at the left.
The 5′ stem-loop participates in the trp operon transcription attenuation mechanism.
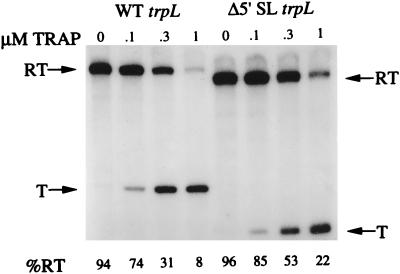
Since TRAP binding to trp leader RNA is responsible for controlling trp operon expression by transcription attenuation and translational control mechanisms, we were interested in determining if the 5′ stem-loop participates in the transcription attenuation mechanism. To test this possibility, we performed in vitro transcription attenuation assays using wild-type (pPB22) and 5′ stem-loop deletion (pJY2) templates in a reaction mixture containing B. subtilis vegetative (ςA) RNA polymerase, TRAP, tryptophan, and the four ribonucleoside triphosphates. The major transcript produced from the wild-type template in the absence of TRAP was the 320-nt readthrough transcript (Fig. 4). Essentially identical results were observed when TRAP was present but tryptophan was omitted from the reaction mixture (data not shown). In the presence of increasing concentrations of TRAP (0.1, 0.3, and 1 μM), we observed a decrease in the readthrough transcript and an increase in the 139-nt terminated transcript (Fig. 4). Thus, as demonstrated previously, transcription termination in the wild-type trp leader in vitro is dependent on the presence of TRAP and tryptophan (5, 27). When the 5′ stem-loop deletion template was used in the reaction in the absence of TRAP, the major transcript was the 290-nt readthrough transcript (Fig. 4). In the presence of increasing concentrations of TRAP (0.1, 0.3, and 1 μM), we observed a decrease in the readthrough transcript and an increase in the 109-nt terminated transcript (Fig. 4). At each concentration of TRAP tested, we observed a higher percentage of readthrough transcripts with the 5′ stem-loop deletion template than with the wild-type template. The effect of the deletion was most pronounced at the highest concentration of TRAP used. In this case, the percentage of readthrough transcripts was approximately threefold higher than that observed with the wild-type template (Fig. 4). While the observed difference in readthrough efficiency is modest compared to the effect observed in vivo, these results demonstrate that deletion of the 5′ stem-loop leads to an increase in transcriptional readthrough, consistent with the expression and gel shift results described above. Moreover, these results demonstrate that the 5′ stem-loop influences the efficiency of the transcription attenuation mechanism.
FIG. 4.
Deletion of the 5′ stem-loop increases transcription readthrough in vitro. The concentration of TRAP used in each reaction is indicated. Arrows on the left indicate the positions of the 320-nt readthrough transcript (RT) and the 139-nt terminated transcript (T) generated from the wild-type template. Arrows on the right indicate the positions of the 290-nt readthrough and the 109-nt terminated transcripts generated from the 5′ stem-loop deletion template. The molar percentage of readthrough transcripts is shown below each lane.
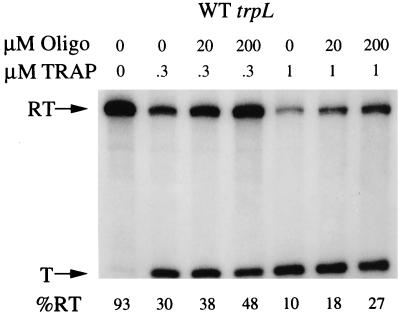
The in vivo and in vitro analyses with the 5′ stem-loop deletion described above suggested that this 5′ structure plays a role in the transcription attenuation mechanism of the trp operon. To provide additional support for this interpretation, we performed in vitro transcription attenuation assays in the presence of an oligonucleotide complementary to a portion of the hairpin structure. This approach was used previously to demonstrate that the antiterminator and terminator structures in the B. subtilis trp leader function as predicted in vitro (5, 27). Transcription was carried out with a wild-type DNA template in a reaction mixture containing B. subtilis vegetative (ςA) RNA polymerase, TRAP, tryptophan, and the four ribonucleoside triphosphates in the presence or absence of an oligonucleotide complementary to nt +2 through +16 of the 5′ stem-loop structure (Fig. 1). We expected that the oligonucleotide would base pair with the 5′ half of the stem, which would prevent formation of the structure, thereby mimicking the effect of the 5′ stem-loop deletion. The oligonucleotide concentration used was in 102- or 103-fold molar excess over the template DNA concentration. In Fig. 5, it can be seen that in the absence of TRAP, the majority of the transcripts were full length. The addition of TRAP decreased the percentage of readthrough transcripts (Fig. 5). As predicted, the addition of increasing amounts of the oligonucleotide to the transcription reaction increased the percentage of readthrough transcripts (Fig. 5). By contrast, a control oligonucleotide that was not complementary to any portion of the trp leader had no effect (data not shown). Note that the effect of the complementary oligonucleotide was most pronounced at the higher concentration of TRAP tested (approximately threefold) and was similar to the results with the deletion template (compare Fig. 4, trpLWT and trpLΔ5′S-L lanes with 1 μM TRAP, with Fig. 5, lanes with 1 μM TRAP and 0 or 200 mM oligonucleotide. These results support the conclusion that formation of the 5′ stem-loop structure plays a role in the transcription attenuation mechanism of the B. subtilis trp operon by increasing the affinity of TRAP for trp leader RNA.
FIG. 5.
Oligonucleotide competition in vitro transcription assay. The DNA oligonucleotide was complementary to nt 2 to 16 of the trp leader transcript. The concentrations of TRAP and the oligonucleotide added to each reaction mixture are given above the gel. Arrows indicate the positions of the 320-nt readthrough (RT) and the 139-nt terminated (T) transcripts. The molar percentage of readthrough transcripts is shown below each lane.
DISCUSSION
The B. subtilis trp operon is regulated by transcription attenuation in response to changes in the intracellular level of tryptophan by the mtrB gene product, TRAP. TRAP interaction with the 11 (G/U)AG repeats present within the nascent trp leader transcript promotes formation of the intrinsic terminator by blocking formation of the overlapping antiterminator structure (Fig. 1). An essentially identical transcription attenuation mechanism was shown to regulate expression of the B. pumilus trp operon (20). In addition, the trp operon leaders of B. caldotenax and B. stearothermophilus contain multiple triplet repeats, as well as overlapping antiterminator and terminator structures. Thus, it appears that all four of these bacilli regulate trp operon expression by a conserved attenuation mechanism.
In this study, the function of an RNA secondary structure predicted to form at the 5′ end of the B. subtilis trp leader transcript was examined. The conservation of similar structures in the trp operon leaders of B. pumilus, B. caldotenax, and B. stearothermophilus suggested that it may have a regulatory role in trp operon expression (Fig. 2). We found that deletion of the B. subtilis 5′ stem-loop resulted in a dramatic increase in expression of a trpE′-′lacZ translational fusion (PLBS104) compared to a similar fusion containing a wild-type trp leader (PLBS44). The effect of the deletion was most pronounced when cells were grown in the presence of tryptophan in the growth medium (Table 2). The −Trp/+Trp β-galactosidase ratios of these two strains demonstrate that the absence of the 5′ stem-loop results in a partial loss of trp operon regulation in response to tryptophan.
To determine the features of the 5′ stem-loop involved in trp operon regulation, we examined the effects of single point mutations that altered its sequence, the predicted secondary structure, or both. All of the point mutations that were predicted to disrupt the secondary structure (C3G, U5A, C13G, C15G, G22C, G24C, and G31C) had effects on trp operon expression that were similar to deletion of the entire hairpin. In fact, C15G and G22C were more deleterious than the deletion itself (Table 2). The finding that the 5′ stem-loop increases the affinity of TRAP for trp leader RNA four- to fivefold (Fig. 3) suggests that TRAP interacts with the 5′ stem-loop. Thus, disruption of the 5′ structure by various point mutations probably prevents this interaction, resulting in higher expression levels. However, it should be pointed out that in addition to disruption of the 5′ structure, the optimal and/or suboptimal structures predicted to form in several of these mutant transcripts (C3G, C13G, C15G, G22C, and G31C) sequester several of the normally single-stranded (G/U)AG repeats previously shown to function in TRAP binding (data not shown). Since it is known that RNA secondary structures that sequester (G/U)AG repeats inhibit TRAP binding (8), it is possible that these optimal and/or suboptimal structures contribute to the regulatory defects associated with these mutations. Despite this, the finding that the C3G G31C and C15G G22C compensatory changes resulted in the partial restoration of wild-type-like expression levels, especially when cultures were grown in the presence of tryptophan, clearly indicates that the structure of the hairpin is important for proper regulation of the trp operon (Table 2). Interestingly, while the expression levels of the compensatory mutants are approximately 10-fold higher than those of the wild-type strain in both the presence and the absence of tryptophan in the growth medium, the fold regulation (−Trp/+Trp) was similar to that of the wild type in both cases. Results from in vitro transcription attenuation experiments are consistent with the in vivo trp leader mutation studies. Deletion of the 5′ stem-loop or disruption of hairpin formation with a complementary oligonucleotide resulted in an increase of approximately threefold in transcriptional readthrough (Fig. 4 and 5).
The two point mutations that were predicted to alter only the primary sequence of the 5′ stem-loop (G7A and A19U) had very different effects on trp operon expression. The free energies of the structures containing the G7A or A19U mutations are predicted to be the same as that of the wild-type hairpin. Both of these mutations alter potential UAG (G7A) or GAG (A19U) TRAP recognition sites. The A19U mutation had a minimal effect on trp operon regulation, suggesting that a KKR motif interaction with this GAG sequence is not involved in TRAP-trp leader RNA interaction. In contrast, the G7A mutation led to a substantial loss of regulation, comparable to that with the 5′ stem-loop deletion. Thus, it is possible that TRAP recognizes this UAG sequence with a KKR motif. An alternative explanation is that this residue contributes to TRAP-RNA recognition by a non-KKR motif interaction. For example, G7 is part of a GAAA sequence that could form a tetraloop, a structure consisting of non-Watson-Crick base pairs known to stabilize RNA secondary structures (39). Thus, it is possible that G7 contributes to the structure of the 5′ stem-loop.
Since TRAP is the only trans-acting factor known to regulate trp operon expression, we expected that the 5′ stem-loop would function in TRAP-dependent regulation of this operon. We found that the effect of the 5′ stem-loop deletion on trp operon expression was significantly reduced in a TRAP-deficient (ΔmtrB) background (Table 2). Moreover, the observation that overexpression of mtrB partially suppressed the effect of the 5′ stem-loop deletion (Table 3), combined with the finding that deletion of the 5′ structure reduced the affinity of TRAP for trp leader RNA four- to fivefold (Fig. 3), suggests that the 5′ structure increases the affinity of TRAP for trp leader RNA by a direct TRAP–5′ stem-loop interaction. The results of the in vitro transcription studies are consistent with this interpretation (Fig. 4 and 5). Thus, the 5′ stem-loop may tether TRAP to the nascent trp leader transcript in a manner not yet identified such that TRAP would be in position to bind to the (G/U)AG repeats as soon as they are transcribed. This multipartite binding mechanism may be important to increase the likelihood that tryptophan-activated TRAP would bind to the trp leader in time to block antiterminator formation.
In addition to TRAP-trp leader interaction, the 5′ stem-loop may play a role in transcript stability. This could provide an explanation for the consistent two- to threefold increase in expression levels that was observed for strain PLBS252 (Δ5′S-L ΔmtrB) compared to PLBS251 (ΔmtrB). The 5′ stem-loop may be a target for an endonuclease leading to the decay of the trp operon transcript. Various studies regarding mRNA decay in B. subtilis have pointed to the importance of the 5′ segment of mRNA in controlling the stability of the transcript under different growth conditions (10, 13, 26, 31). Some messages are thought to be degraded by a ribonucleolytic activity that begins at the 5′ end and degrades the message in a 5′-to-3′ direction (10), while processing of the thrS leader transcript plays a major role in the induction of thrS expression following threonine starvation in B. subtilis (13). Another study concludes that initiation of mRNA decay in B. subtilis generally occurs at or near the 5′ terminus (14). Studies have also revealed a sequence that specifies a 5′ stabilizer function which appears to be localized to a polypurine sequence that resembles a ribosome binding site in B. subtilis (14) and indicate that attack at the 5′ end is a principal mechanism for initiation of mRNA decay in B. subtilis (21). In our study of the trp leader 5′ stem-loop, loss of a potential endonucleolytic target would result in a two- to threefold increase in the stability of the trp operon transcript. It remains to be determined if the 5′ stem-loop does in fact serve as an mRNA instability determinant.
Deletion of the 5′ stem-loop affected regulation of the trp operon approximately 200-fold. Since only two- to threefold of this is TRAP independent, the remaining 67- to 100-fold effect of the deletion is due to a defect in TRAP-dependent regulation. While it is possible that the two- to threefold TRAP-independent effect is due to increased mRNA stability, the increase of approximately threefold in transcriptional readthrough observed in vitro cannot be attributed to mRNA stability, since the only proteins present in the transcription attenuation assay were RNA polymerase and TRAP. Taking into consideration the two- to threefold in vivo TRAP-independent and the approximately threefold in vitro TRAP-dependent effect of the 5′ stem-loop deletion, we are left with a 20- to 30-fold TRAP-dependent effect that is unaccounted for. Thus, it appears that some feature or factor involved in TRAP-dependent trp operon regulation is missing from the in vitro transcription attenuation system.
ACKNOWLEDGMENTS
We thank Jeanne Yealy and Behnam Bozorgnia for technical assistance, Paul Gollnick for sharing the B. stearothermophilus trp leader sequence prior to publication, and Phil Bevilacqua and Craig Cameron for thoughtful discussions. We also thank Charles Yanofsky, Paul Gollnick, and Phil Bevilacqua for critical reading of the manuscript.
This work was supported by grant GM52840 from the National Institutes of Health.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antson A A, Brzozowski A M, Dodson E J, Dauter Z, Wilson K S, Kurecki T, Otridge J, Gollnick P. 11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis. J Mol Biol. 1994;244:1–5. doi: 10.1006/jmbi.1994.1698. [DOI] [PubMed] [Google Scholar]
- 3.Antson A A, Otridge J, Brzozowski A M, Dodson E J, Dodson G G, Wilson K S, Smith T M, Yang M, Kurecki T, Gollnick P. The structure of the trp RNA attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 4.Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P, Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci USA. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babitzke P, Yanofsky C. Structural features of l-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 1995;270:12452–12456. doi: 10.1074/jbc.270.21.12452. [DOI] [PubMed] [Google Scholar]
- 7.Babitzke P, Stults J T, Shire S J, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994;269:16597–16604. [PubMed] [Google Scholar]
- 8.Babitzke P, Yealy J, Campanelli D. Interaction of the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis with RNA: effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J Bacteriol. 1996;178:5159–5163. doi: 10.1128/jb.178.17.5159-5163.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babitzke P, Gollnick P, Yanofsky C. The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of l-tryptophan biosynthesis. J Bacteriol. 1992;174:2059–2064. doi: 10.1128/jb.174.7.2059-2064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bechhofer D H, Wang W. Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J Bacteriol. 1998;180:5968–5977. doi: 10.1128/jb.180.22.5968-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X-P, Antson A A, Yang M, Li P, Baumann C, Dodson E J, Dodson G G, Gollnick P. Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus. J Mol Biol. 1999;289:1003–1016. doi: 10.1006/jmbi.1999.2834. [DOI] [PubMed] [Google Scholar]
- 13.Condon C, Putzer H, Grunberg-Manago M. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:6992–6997. doi: 10.1073/pnas.93.14.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMari J F, Bechhofer D H. Initiation of mRNA decay in Bacillus subtilis. Mol Microbiol. 1993;7:705–717. doi: 10.1111/j.1365-2958.1993.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 15.Du H, Babitzke P. trp-RNA binding attenuation protein-mediated long-distance RNA refolding regulates translation of trpE in Bacillus subtilis. J Biol Chem. 1998;273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- 16.Du H, Tarpey R, Babitzke P. The trp-RNA binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J Bacteriol. 1997;179:2582–2586. doi: 10.1128/jb.179.8.2582-2586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gollnick P, Ishino S, Kuroda M I, Henner D J, Yanofsky C. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:8726–8730. doi: 10.1073/pnas.87.22.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henner D, Band L, Shimotsu H. Nucleotide sequence of the Bacillus subtilis tryptophan operon. Gene. 1984;34:169–177. doi: 10.1016/0378-1119(85)90125-8. [DOI] [PubMed] [Google Scholar]
- 19.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman R J, Gollnick P. The mtrB gene of Bacillus pumilus encodes a protein with sequence and functional homology to the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis. J Bacteriol. 1995;177:839–842. doi: 10.1128/jb.177.3.839-842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hue K K, Cohen S D, Bechhofer D H. A polypurine sequence that acts as a 5′ mRNA stabilizer in Bacillus subtilis. J Bacteriol. 1995;177:3465–3471. doi: 10.1128/jb.177.12.3465-3471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishiwa H, Shibahara H. New shuttle vectors for Escherichia coli and Bacillus subtilis. II. Plasmid pHY300PLK, a multipurpose cloning vector with a polylinker, derived from pHY460. Jpn J Genet. 1985;60:235–243. [Google Scholar]
- 23.Kane J F. Regulation of a common amidotransferase subunit. J Bacteriol. 1977;132:419–425. doi: 10.1128/jb.132.2.419-425.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda M I, Henner D, Yanofsky C. cis-acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J Bacteriol. 1988;170:3080–3088. doi: 10.1128/jb.170.7.3080-3088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovett P S, Keggins K M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- 26.Melin L, Friden H, Dehlin E, Rutberg L, von Gabain A. The importance of the 5′-region in regulating the stability of sdh mRNA in Bacillus subtilis. Mol Microbiol. 1990;4:1881–1889. doi: 10.1111/j.1365-2958.1990.tb02037.x. [DOI] [PubMed] [Google Scholar]
- 27.Merino E, Babitzke P, Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 29.Morrison H G, Desrosiers R C. A PCR-based strategy for extensive mutagenesis of a target DNA sequence. BioTechniques. 1993;14:454–457. [PubMed] [Google Scholar]
- 30.Otridge J, Gollnick P. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc Natl Acad Sci USA. 1993;90:128–132. doi: 10.1073/pnas.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resnekov O, Rutberg L, von Gabain A. Changes in the stability of specific mRNA species in response to growth stage in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:8355–8359. doi: 10.1073/pnas.87.21.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Sekiguchi J, Takada N, Okada H. Genes affecting the productivity of α-amylase in Bacillus subtilis Marburg. J Bacteriol. 1975;121:688–694. doi: 10.1128/jb.121.2.688-694.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimotsu H, Henner D J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43:85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- 35.Shimotsu H, Kuroda M I, Yanofsky C, Henner D J. Novel form of transcription attenuation regulates expression of the Bacillus subtilis tryptophan operon. J Bacteriol. 1986;166:461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiratsuchi A, Sato S. Nucleotide sequence of trpE, anthranilate synthase I gene, of Bacillus caldotenax. Biochim Biophys Acta. 1991;1090:348–350. doi: 10.1016/0167-4781(91)90201-v. [DOI] [PubMed] [Google Scholar]
- 37.Slock J, Stahly D P, Han C-Y, Six E W, Crawford I P. An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. J Bacteriol. 1990;172:7211–7226. doi: 10.1128/jb.172.12.7211-7226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varani G. Exceptionally stable nucleic acid hairpins. Annu Rev Biophys Biomol Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 40.Walter A E, Turner D H, Kim J, Lyttle M H, Mueller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M, de Saizieu A, van Loon A P G M, Gollnick P. Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP) J Bacteriol. 1995;177:4272–4278. doi: 10.1128/jb.177.15.4272-4278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M, Chen X-P, Militello K, Hoffman R, Fernandez B, Baumann C, Gollnick P. Alanine-scanning mutagenesis of Bacillus subtilis trp-RNA binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J Mol Biol. 1997;270:696–710. doi: 10.1006/jmbi.1997.1149. [DOI] [PubMed] [Google Scholar]
- 43.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]