Abstract
The methylotrophic proteobacterium Methylobacterium extorquens AM1 possesses tetrahydromethanopterin (H4MPT)-dependent enzymes, which are otherwise specific to methanogenic and sulfate-reducing archaea and which have been suggested to be involved in formaldehyde oxidation to CO2 in M. extorquens AM1. The distribution of H4MPT-dependent enzyme activities in cell extracts of methylotrophic bacteria from 13 different genera are reported. H4MPT-dependent activities were detected in all of the methylotrophic and methanotrophic proteobacteria tested that assimilate formaldehyde by the serine or ribulose monophosphate pathway. H4MPT-dependent activities were also found in autotrophic Xanthobacter strains. However, no H4MPT-dependent enzyme activities could be detected in other autotrophic α-proteobacteria or in gram-positive methylotrophic bacteria. Genes encoding methenyl H4MPT cyclohydrolase (mch genes) were cloned and sequenced from several proteobacteria. Bacterial and archaeal Mch sequences have roughly 35% amino acid identity and form distinct groups in phylogenetic analysis.
Aerobic methylotrophs are gram-negative or gram-positive bacteria capable of growth on reduced C1 compounds such as methanol or methane as their carbon and energy source (24). Formaldehyde is formed as a central intermediate in methylotrophic metabolism. At least four different enzyme systems have been proposed as C1 pathways for the oxidation of formaldehyde to CO2 in methylotrophic bacteria (see also 13). The first of these is a linear pathway involving thiol-linked formaldehyde dehydrogenase, a hydrolase and formate dehydrogenase. This pathway is found in the autotrophic α-proteobacterium Paracoccus denitrificans and the nonsulfur purple bacteria Rhodopseudomonas acidophila and Rhodobacter sphaeroides, which possess a glutathione-dependent formaldehyde dehydrogenase (2, 3, 20, 32, 36). Similarly, the gram-positive methylotroph Amycolatopsis methanolica that contains the ribulose monophosphate cycle was shown to contain a mycothiol-dependent formaldehyde dehydrogenase (27, 28), which was previously called NAD factor-dependent formaldehyde dehydrogenase (15, 30). As a second route, tetrahydrofolate (H4F)-dependent enzymes have been discussed as being involved in formaldehyde oxidation in serine cycle methylotrophs (10, 26). A third proposed route for formaldehyde oxidation entails the cyclic ribulose monophosphate pathway involving 6-phosphogluconate dehydrogenase (1, 4, 18). This pathway has been proposed for β-proteobacterial methylotrophs that contain the ribulose monophosphate cycle. However, only low activities of this pathway could be detected in the other group containing the ribulose monophosphate cycle, the obligate methanotrophic bacteria that belong to the γ-proteobacteria class (48). Therefore, the formaldehyde oxidation pathway in these methanotrophs has been uncertain.
Recently, we described a fourth pathway for formaldehyde oxidation, involving tetrahydromethanopterin (H4MPT)-dependent enzymes in the α-proteobacterium Methylobacterium extorquens AM1, a serine cycle methylotroph (11, 31, 47). These enzymes were previously thought to be restricted to methanogenic and sulfate-reducing archaea (for a review, see reference 43). Three H4MPT-dependent and H4F-dependent enzymes from M. extorquens AM1 have been purified and characterized (31, 47): a methenyl H4MPT cyclohydrolase, a methenyl H4F cyclohydrolase, and an NADP-dependent methylene H4MPT dehydrogenase, which also catalyzes the dehydrogenation of methylene H4F (47). Both pterin cofactors have been shown to be present in similar amounts (47). It therefore appears that in Methylobacterium extorquens AM1, two independent C1 transfer pathways are simultaneously operative, one involving H4MPT (in a dephospho form) and one involving H4F (11, 47). On the basis of mutant and enzyme studies, the H4MPT-dependent pathway was proposed to be the main pathway for oxidation of formaldehyde to CO2 (11, 31, 47).
In this study, we investigated the distribution of H4MPT-dependent enzyme activities in methylotrophic representatives from different phylogenetic and physiological groups. For those bacteria that showed H4MPT-dependent enzyme activities, we cloned one of the encoding genes to obtain sequence data and gain insights into the origin and phylogeny of H4MPT-dependent enzymes. We concentrated on methenyl H4MPT cyclohydrolase, which is also present in methanogenic archaea, rather than NAD- and NADP-dependent methylene H4MPT dehydrogenases, which are novel enzymes not found in archaea (47). The gene encoding methenyl H4MPT cyclohydrolase (mch) from M. extorquens AM1 has been identified by N-terminal sequencing of the purified protein and shown to consist of only one type of subunit of molecular mass 33 kDa (31). Mch from M. extorquens AM1 possesses about 36% amino acid sequence identity to homologues from methanogenic archaea and Archaeoglobus fulgidus (11).
MATERIALS AND METHODS
Organisms.
The following are laboratory strains (University of Washington, Seattle) that were used in this study: Methylobacterium extorquens AM1 (29), Methylosinus trichosporium OB3b (5), Xanthobacter flavus H4-14 (33), Methylococcus capsulatus Bath (5), Methylococcus thermophilus IIIp (25), Methylomicrobium album BG8 (5), Methylomonas rubra 15sh (35), and Shewanella putrefaciens MR-1 (34). Methylobacterium organophilum XX (DSM 760), Hyphomicrobium methylovorum GM2 (DSM 5458), Xanthobacter autotrophicus (DSM 432), Paracoccus denitrificans (DSM 413), Rhodopseudomonas acidophila (DSM 145), Methylobacillus flagellatum KT (DSM 6875), Methylophilus methylotrophus AS1 (DSM 46235), Amycolatopsis methanolica (DSM 44096), and Methanobacterium thermoautotrophicum Marburg (DSM 2133) were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). Rhodobacter sphaeroides 2.4.1 (44) was obtained from S. Kaplan (University of Texas). Cell mass of Bacillus methanolicus MGA3 (38) was kindly provided by S. Pluschkell, University of Minnesota.
Growth of bacteria.
Methylobacterium extorquens AM1, Methylobacterium organophilum XX, H. methylovorum GM2, Methylobacillus flagellatum KT, and Methylophilus methylotrophus AS1 were grown on methanol on minimal medium as described previously or on succinate (0.5%) on the same medium (17). The methanotrophic strains Methylosinus trichosporium OB3b, Methylococcus capsulatus Bath, Methylococcus thermophilus IIIp, Methylomicrobium album BG8, and Methylomonas rubra 15sh were grown on methane as described previously (19). The following bacteria were grown on the media described by the DSMZ (12a). X. autotrophicus and X. flavus H4-14 were grown on nutrient broth medium plus methanol (100 mM) or on methanol (100 mM) in minimal medium 81 supplemented with 0.01% yeast extract, P. denitrificans was grown on methanol (100 mM) in minimal medium 81 supplemented with 0.01% yeast extract, and Rhodopseudomonas acidophila was grown on medium 26 plus methanol (100 mM) under anoxic conditions in the light. Amycolatopsis methanolica was also grown on methanol (100 mM) or glucose (0.4%) in the minimal medium described previously (21), and R. sphaeroides 2.4.1 was grown on Luria-Bertani medium (37) containing 5 g (instead of 10 g) NaCl per liter.
Rhodopseudomonas acidophila was grown at room temperature, M. thermophilus IIIp was grown at 42°C, and all other bacteria were grown at 30°C.
Coenzymes.
Tetrahydromethanopterin (H4MPT) and methenyl H4MPT were purified from Methanobacterium thermoautotrophicum Marburg (7). H4F was purchased from Sigma.
Preparation of cell extracts.
Frozen cells (0.5 g) were resuspended in 1 ml of 50 mM morpholinepropanesulfonic acid (MOPS)-KOH (pH 7.0) at 4°C and passed two times through a French pressure cell at 1.2 × 108 Pa. Cell debris and unbroken cells were removed by centrifugation in an Eppendorf microcentrifuge. The resulting supernatant is referred to as cell extract.
Protein concentration was determined by the Bradford assay (6) by using the Bio-Rad reagent with bovine serum albumin as the standard.
Determination of enzyme activities.
The assays were routinely performed at room temperature in 1-ml cuvettes (diameter, 1 cm) in a total volume of 0.7 ml.
Methenyl H4MPT cyclohydrolase activity was determined photometrically at 335 nm as described before (31). NADP- and NAD-dependent methylene H4MPT dehydrogenase and NADP-dependent methylene H4F dehydrogenase activities were monitored at 340 nm (47). Glutathione-linked formaldehyde dehydrogenase was measured at 340 nm as described before (32).
Standard molecular biology techniques.
All standard procedures were performed as described by Sambrook et al. (37). Enzymes were used in accordance with recommendations of the manufacturers (Boehringer Mannheim and New England Biolabs).
PCR amplification.
The heterologous primers mch-1 (5′-TTCTTCGCCMTGGGKTCKGGKCC), mch-2a (5′-TGCCTCGGCTCKCAATATGCYGGBTGG), mch-2b (5′-TGCCTCGGCAGCCAGWAYGCSGGCTGG), mch-3 (5′-GCGTCGTTKGTKCKBCCCAT), and mch-4 (5′-AACAGCGCTCCGTCKATYTTGTARAARTC) were derived from four sequence parts of Mch conserved in the sequences from Methylobacterium extorquens AM1, methanogenic archaea, and Archaeoglobus fulgidus. The primer combination mch-2a–mch-3 was used to amplify a PCR product of approximately 420 nucleotides with chromosomal DNA from Methylococcus capsulatus Bath, Methylomonas rubra 15sh, Methylococcus thermophilus IIIp, and Methylobacillus flagellatum KT. From the newly obtained Mch sequences, additional heterologous primers were designed, namely, mch-5 (5′-GAGGAGCTSTACAAGGAGCTSGGBTA) and mch-6 (5′-TGSGCCTTGTGYAGYGCBACYTC). PCR products were obtained for Methylophilus methylotrophus AS1 (primer combination mch-4–mch-5, 489-nucleotide product) and H. methylovorum GM2 (primer combination mch-1–mch-6, 264-nucleotide product). Homologous primers were derived from part of mch from both organisms: m1 (5′-GGCCTTGAACACTTCAGC) and m2 (5′-CAAAGACAGTGCTGATGC), h1 (5′-GGCGATCTGCACGGTGCC) and h2 (5′-GTGGAAGACCTCTTCAAGG). With template DNA from Methylophilus methylotrophus AS1, m1 and mch-2a were used to amplify an additional PCR fragment. With template DNA from H. methylovorum GM2, two additional PCR fragments were cloned, generated with primers h1 and mch-4 as well as h2 and mch-2b.
PCR amplifications were carried out at annealing temperatures of 35, 38, and 42°C by using Taq polymerase (Boehringer Mannheim). PCR products were cloned into the pCR2.1 vector with the Invitrogen TA cloning kit and identified by sequencing.
Cloning of mch genes.
Southern blots indicated the presence of the mch gene in chromosomal digests of Methylobacterium organophilum XX DNA by using mch-2 as a heterologous oligonucleotide probe, the presence of the mch gene in chromosomal digests of X. autotrophicus DNA by using mch-6 as a heterologous oligonucleotide probe, and the presence of the mch gene in chromosomal digests of Methylosinus trichosporium OB3b by using mch-3 as a heterologous oligonucleotide probe. Hybridizations were carried out at 55°C, and washing steps were performed at 55°C and with 5× SSC (0.75 M NaCl plus 0.075 M sodium citrate). For cloning of the mch gene from Methylobacterium organophilum XX, chromosomal DNA was digested by SalI, eluted from the gel at approximately 2.1 kb (Qiagen gel extraction kit), and ligated to pAYC63 (9) predigested with SalI. For cloning of the mch gene from X. autotrophicus, an SphI digest was used and DNA at approximately 4 kb was ligated to pAYC63 (9) predigested with the same enzyme. For cloning of the mch gene from Methylosinus trichosporium OB3b, a SalI digest was used and DNA at approximately 0.9 kb was ligated to pAYC63 (9) predigested with the same enzyme. Positive clones were obtained from the partial clone banks, which were identified by colony hybridizations (37) by using the probes mentioned and by sequencing. The positive clone obtained for Methylosinus trichosporium OB3b contained only the 3′ region of the mch gene. To obtain the sequence further upstream, the oligonucleotides ob3b-6 (5′-GGAATAGATGATCGCGTC) and ob3b-7 (5′-CAGGCGAGACGTTCCGTGC) were used for inverted PCR. Chromosomal DNA of Methylosinus trichosporium OB3b was digested with BglII and self-ligated. This DNA was used as a template for amplification employing the oligonucleotides ob3b-6 and ob3b-7. The band of interest (approximately 1.8 kb) was identified by Southern hybridization using the oligonucleotide ob3b-4 (5′-GTCGACGGCATCGCCACG) as a probe and cloned into pCR2.1 (Invitrogen) and sequenced. The insert was shown to contain 449 nucleotides of new sequence.
The complete mch gene from Methylococcus capsulatus Bath was cloned by using an oligonucleotide probe (MC-1 5′-CCACCAAGATCAAGGACGGCAACG) derived from the sequence of the PCR product obtained as described above.
Chromosomal DNA was digested by EcoRI, eluted at approximately 1.6 kb, and ligated to pAYC63 (9) predigested with EcoRI. The blots were washed at 50°C and with 2× SSC. Positive clones were obtained from the partial EcoRI clone bank and identified by colony hybridizations and sequencing. The complete mch gene from Methylobacillus flagellatum KT was cloned in a similar fashion, from a partial chromosomal library in pAYC63 (9) containing SacI inserts of approximately 2.7 kb. The PCR product described above was used as a probe.
Oligonucleotide labeling, hybridizations, and detections were done with the digoxigenin oligonucleotide tailing kit and by following the protocol provided by Boehringer Mannheim.
DNA sequencing and analysis.
DNA sequencing was performed at the sequencing facility of the University of Washington. Nucleotides and inferred peptide sequences were aligned by the Clustal method (DNAStar package) and manually. Dendrograms were constructed by using PUZZLE version 4.0 (42) and the programs from the PHYLIP version 3.5c package (16).
Nucleotide sequence accession numbers.
The gene sequences have been deposited in the GenBank database under accession no. AF139592, AF139593, AF142649 to AF142655, and AF162786.
RESULTS
Occurrence of H4MPT-dependent enzyme activities in cell extracts of methylotrophic bacteria.
Cell extracts from different methylotrophic bacteria were tested for the presence of methenyl H4MPT cyclohydrolase, NADP-dependent methylene H4MPT dehydrogenase, and NAD-dependent methylene H4MPT dehydrogenase activities, which were shown to involve three different enzymes in Methylobacterium extorquens AM1 (31, 47). The results are shown in Table 1. Cells were grown on the C1 substrates indicated. Additionally, most facultative methylotrophic bacteria were grown under a nonmethylotrophic growth condition for induction studies. All of the methanotrophic bacteria tested were found to exhibit H4MPT-dependent enzyme activities in the range of 0.1 to 1.7 U/mg (Table 1). The methanotrophs are either α-proteobacteria containing the serine pathway for assimilation (Methylosinus trichosporium OB3b) or γ-proteobacteria containing the ribulose monophosphate pathway (Methylococcus capsulatus Bath, Methylococcus thermophilus IIIp, Methylomicrobium album BG8, and Methylomonas rubra 15sh). The methanotrophic bacteria are all obligate methylotrophic bacteria growing only on methane or, in some cases, also on methanol. Methylomicrobium album BG8 also exhibited a comparable activity of methenyl H4MPT cyclohydrolase when growing in the presence of methanol (data not shown). No significant activities (<0.01 U/mg) of a pyridine-dependent methylene H4F dehydrogenase could be detected in methanotrophic bacteria.
TABLE 1.
Activities of methenyl H4MPT cyclohydrolase, NAD-dependent methylene H4MPT dehydrogenase, and NADP-dependent methylene H4MPT dehydrogenase in cell extracts of different methylotrophic bacteria
| Organism | Major assimilation pathwaya | Grown on: | Activity (U/mg)b
|
||
|---|---|---|---|---|---|
| Methenyl H4MPT CH | Methylene H4MPT DH (NAD) | Methylene H4MPT DH (NADP) | |||
| α-Proteobacteria: | |||||
| Methylobacterium extorquens AM1 | Serine | Methanol | 0.7 | 0.6 | 5.1 |
| Succinate | 0.3 | 0.5 | 1.0 | ||
| Methylobacterium organophilum XX | Serine | Methanol | 0.3 | 0.1 | 1.3 |
| Succinate | 0.1 | 0.1 | 0.2 | ||
| Hyphomicrobium methylovorum GM2 | Serine | Methanol | 0.4 | 0.1 | 1.3 |
| Methylosinus trichosporium OB3b | Serine | Methane | 0.7 | 0.7 | 1.0 |
| Xanthobacter autotrophicus | CBB | Methanol | 0.6 | 0.1 | 0.2 |
| Nh + Methanol | 0.3 | 0.03 | 0.2 | ||
| Xanthobacter flavus H4-14 | CBB | N + Methanol | 0.2 | 0.2 | 0.4 |
| Paracoccus denitrificansc | CBB | Methanol | <0.01 | <0.01 | <0.01 |
| Rhodopseudomonas acidophilad,e | CBB | Succinate + Methanol | <0.01 | <0.01 | <0.01 |
| Rhodobacter sphaeroides 2.4.1f | CBB | N | <0.01 | <0.01 | <0.01 |
| β-Proteobacteria: | |||||
| Methylobacillus flagellatum KT | RuMP | Methanol | 1.0 | 1.1 | 1.8 |
| Methylophilus methylotrophus AS1 | RuMP | Methanol | 0.6 | 0.2 | 0.4 |
| γ-Proteobacteria: | |||||
| Methylococcus capsulatus Bath | RuMP | Methane | 0.7 | 0.5 | 1.7 |
| Methylococcus thermophilus IIIp | RuMP | Methane | 0.2 | 0.1 | 0.3 |
| Methylomicrobium album BG8 | RuMP | Methane | 0.4 | 0.3 | 0.7 |
| Methylomonas rubra 15sh | RuMP | Methane | 0.6 | 0.4 | 0.6 |
| Gram-positive bacteria: | |||||
| Bacillus methanolicus MGA3 | RuMP | Methanol | <0.01 | <0.01 | <0.01 |
| Amycolatopsis methanolicag | RuMP | Methanol | <0.01 | <0.01 | <0.01 |
| Glucose | <0.01 | <0.01 | <0.01 | ||
CBB, Calvin-Benson-Bassham; RuMP, ribulose monophosphate.
Enzyme activities were measured under standard assay conditions at room temperature. CH, cyclohydrolase; DH, dehydrogenase.
Contains a glutathione-linked formaldehyde dehydrogenase (32).
Contains a glutathione-linked formaldehyde dehydrogenase (36).
Cells were grown anaerobically in the light.
N, nutrient agar.
The ratio of NADP- to NAD-dependent H4MPT dehydrogenase activity in methanotrophic bacteria appears to be more balanced than that in other methylotrophic bacteria. This might reflect the higher demand for NADH for methane monooxygenase, which uses NADH rather than NADPH (12).
Hyphomicrobium methylovorum GM2 and the facultative methylotroph Methylobacterium organophilum XX contain the serine pathway for assimilation. Cell extracts of both α-proteobacteria were found to exhibit H4MPT-dependent enzyme activities. Like Methylobacterium extorquens AM1, Methylobacterium organophilum XX showed C1-inducible activities of methenyl H4MPT cyclohydrolase and NADP-dependent methylene H4MPT dehydrogenase and no distinct induction of NAD-dependent methylene H4MPT dehydrogenase (Table 1). The NADP-dependent methylene H4F dehydrogenase activity in cell extracts of H. methylovorum GM2 (0.13 U/mg) and Methylobacterium organophilum XX (0.03 U/mg) was found to be very low (see also reference 26).
The β-proteobacteria Methylobacillus flagellatum KT and Methylophilus methylotrophus AS1 contain the ribulose monophosphate pathway for formaldehyde assimilation and were found to exhibit rather high activities of H4MPT-dependent enzymes of 0.2 to 1.8 U/mg.
In contrast to these proteobacteria, no H4MPT-dependent enzyme activities could be detected in the gram-positive methylotrophic bacteria Amycolatopsis methanolica and Bacillus methanolicus MGA3.
Five autotrophic bacteria were tested for H4MPT-dependent cyclohydrolase and dehydrogenase activities, Xanthobacter autotrophicus and Xanthobacter flavus H4-14, Paracoccus denitrificans, Rhodobacter sphaeroides 2.4.1, and Rhodopseudomonas acidophila. The latter organism was grown on methanol anaerobically in the light, while R. sphaeroides was grown aerobically on rich medium. Interestingly, it was found that cell extracts of the Xanthobacter strains contain H4MPT-dependent enzyme activities. However, in cell extracts of the other autotrophic bacterial strains tested, these enzyme activities could not be detected. P. denitrificans, Rhodopseudomonas acidophila, and Rhodobacter sphaeroides were previously shown to contain glutathione-dependent formaldehyde dehydrogenase (2, 3, 32, 36), which was measured as a control enzyme (data not shown).
Besides the organisms shown in Table 1, cell extracts of Shewanella putrefaciens MR1 were assayed for H4MPT-dependent enzyme activities. S. putrefaciens is a γ-proteobacterium that can grow anaerobically on formate as the sole carbon source and has been shown to contain hydroxypyruvate reductase activity, the key enzyme of the serine pathway (39). The bacterium was not grown on C1 substrates but on minimal medium supplemented with glucose or in rich media. In cell extracts of S. putrefaciens, no H4MPT-dependent enzyme activities could be detected under these growth conditions and it was not investigated further.
Cloning of methenyl H4MPT cyclohydrolase genes (mch).
The known sequences of methenyl H4MPT cyclohydrolases (Mch) from Methylobacterium extorquens AM1 (11), methanogenic archaea (8, 41, 45, 46), and Archaeoglobus fulgidus (23) were used to deduce heterologous primers for PCR (for an alignment, see reference 11). PCR products of approximately 430 nucleotides (corresponding to bp 279 to 706 of the mch gene from Methylobacterium extorquens AM1) were obtained by using template DNA from Methylococcus capsulatus Bath, Methylomonas rubra 15sh, Methylococcus thermophilus IIIp, and Methylobacillus flagellatum KT. The PCR products were cloned and sequenced. They showed sequence identities to other Mch proteins, with highest identity to Mch from Methylobacterium extorquens AM1. From the newly obtained bacterial Mch sequences, additional heterologous primers were designed and used for PCR. Using these new primers, positive PCR products were obtained for Methylophilus methylotrophus AS1 and H. methylovorum GM2 (see Materials and Methods).
The mch genes of Methylobacterium organophilum XX and X. autotrophicus were cloned directly from the chromosome as complete genes by using heterologous oligonucleotide probes for Southern hybridization and constructing partial clone banks. The mch gene of Methylosinus trichosporium OB3b was cloned in part directly from the chromosome by using a heterologous oligonucleotide probe and in part by inverted PCR. For Methylococcus capsulatus Bath and Methylobacillus flagellatum KT, the complete mch genes were cloned and sequenced after the partial sequence was obtained by PCR (see Materials and Methods).
Phylogenetic analysis of Mch sequences.
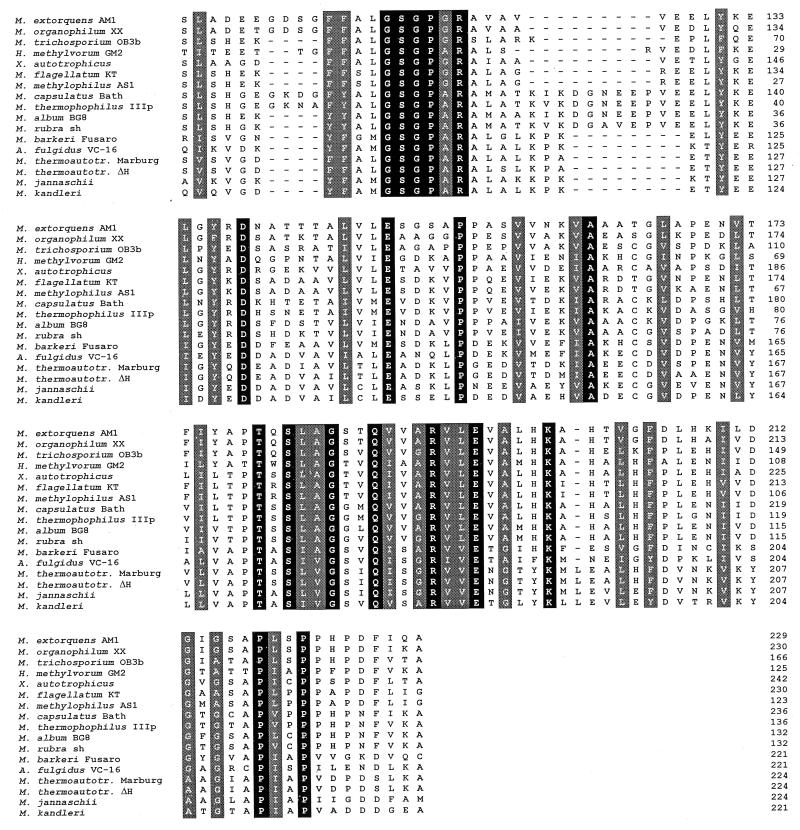
In Fig. 1, an alignment of partial Mch fragments from bacteria and archaea is shown. Approximately 15% of the amino acids are conserved in the region studied for all bacterial and archaeal fragments, while approximately 30% of the amino acids are conserved in all bacterial fragments.
FIG. 1.
Alignment of predicted amino acid sequences corresponding to partial mch genes from bacteria and archaea by the Clustal method (DNAStar). Sequences were obtained by cloning the gene directly from the chromosome from Methylobacterium organophilum XX, Xanthobacter autotrophicus, Methylobacillus flagellatum KT, and Methylococcus capsulatus Bath. Sequences were retrieved by PCR from Hyphomicrobium methylovorum GM2, Methylophilus methylotrophus AS1, Methylococcus thermophilus IIIp, Methylomicrobium album BG8, and Methylomonas rubra 15sh. Sequences from Methylobacterium exotorquens AM1 (11), Methanosarcina barkeri Fusaro (Vaupel and Thauer, EMBL accession no. Y08843), Archaeoglobus fulgidus VC-16 (23), Methanobacterium thermoautotrophicum Marburg (45), Methanobacterium thermoautotrophicum ΔH (41), Methanococcus jannaschii (8), and Methanopyrus kandleri (46) were known before. Amino acid positions conserved in all bacterial and archaeal sequences are shown in black boxes; positions where chemically similar amino acids were found are shown in grey boxes.
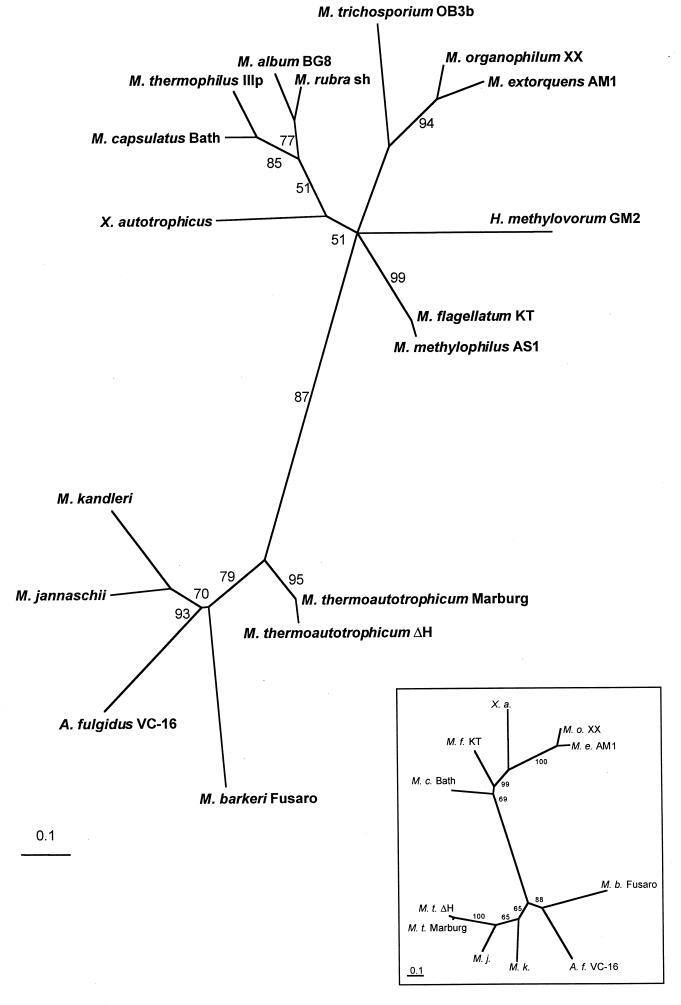
In Fig. 2, the result of a phylogenetic analysis by PUZZLE (42) of Mch fragments, based on the alignment shown in Fig. 1, is given. Phylip (16) was also used for analysis, and it resulted in a similar phylogenetic tree (data not shown). The exact positions of H. methylovorum GM2 and X. autotrophicus are uncertain according to the programs used. An alignment of the known complete Mch sequences including the five new bacterial sequences reported here is shown in the inset to Fig. 1 and was based on an alignment by the Clustal method (DNAStar). The alignment of Mch sequences as well as the phylogenetic tree shows that all the sequences are related to one another, with a strict separation between the bacterial and archaeal sequences.
FIG. 2.
Phylogenetic analysis of the partial Mch amino acid sequences from bacteria and archaea based on the alignment shown in Fig. 2. The tree shows the result of an analysis with PUZZLE (42); the BOOTSTRAP values above 50% from 1,000 replicates are shown. The bar scale stands for numbers of substitutions per site. The inset shows a phylogenetic analysis from Mch sequences which are known completely.
The highest degree of divergence within the groups of bacteria analyzed is found among the α-proteobacteria. Although the fragments from the two Methylobacterium species show 84% identity, the other pairwise comparisons in this group show only 48 to 63% identity. In contrast, the identities between the fragments for the two β-proteobacteria are 97% and those for the four γ-proteobacteria are 71 to 86%. The lowest sequence identity between different proteobacteria was found to be 48% between an α-proteobacterium (Methylosinus trichosporium OB3b) and a γ-proteobacterium (Methylococcus thermophilus IIIP) as well as two α-proteobacteria (Methylosinus trichosporium OB3b and H. methylovorum GM2). Positioning of some α-proteobacterial sequences turned out to be uncertain, consistent with the greater divergence of sequence identities in this group. However, the sequences of the β- and γ-proteobacteria form distinct clusters within the bacterial sequences. Within the group of archaea, sequence identities from 52 to 73% between different species were found. The identity between the bacterial and the archaeal Mch sequences is in the 26 to 42% range.
For those mch genes which were cloned from the chromosome, the genomic arrangement was determined and found to be conserved with respect to Methylobacterium extorquens AM1 for M. organophilum XX and the γ-proteobacterium Methylococcus capsulatus Bath. Upstream of mch, an as-yet-unidentified but conserved open reading frame designated orfY (11) was found with identities only to open reading frames from methanogenic archaea. Downstream of mch, an open reading frame (orf5) with similarity to that coding for the ribosomal S6 modification protein was found. The gene arrangement is different in X. autotrophicus in that orf5 overlaps mch by 64 nucleotides, while upstream of mch a gene similar to mtdA (10, 47) was found. In M. extorquens AM1, mtdA was shown to encode the NADP-dependent methylene H4MPT dehydrogenase and is located approximately 35 kb upstream of mch. These results show that the genomic arrangement of genes for H4MPT-dependent enzymes is not strictly conserved among all methylotrophic proteobacteria. In contrast to the clustered occurrence of genes that are proposed to be involved in a H4MPT-dependent formaldehyde oxidation pathway in bacteria, in methanogenic archaea and the sulfate-reducing archaeon Archaeoglobus fulgidus, those genes are widely scattered over the chromosome.
DISCUSSION
In this report, we demonstrate that the H4MPT-dependent enzymes involved in formaldehyde oxidation are not restricted to serine cycle methylotrophs like Methylobacterium species. These enzymes were also found in the methylotrophic proteobacteria assimilating formaldehyde by the ribulose monophosphate cycle. Even autotrophic Xanthobacter strains were found to contain H4MPT-dependent enzyme activities and the respective genes.
The methylotrophs tested in this study that are known to contain thiol-dependent formaldehyde oxidation systems did not contain detectable levels of H4MPT-dependent enzyme activities. These include Rhodopseudomonas acidophila, Rhodobacter sphaeroides, and Paracoccus denitrificans, all α-proteobacterial methylotrophs that grow autotrophically on methanol via the Calvin-Benson-Bassham cycle and that contain a glutathione-dependent formaldehyde dehydrogenase (3, 32, 36). In the latter two cases, this enzyme has been shown to be required for methylotrophic growth (3, 32). In addition, the gram-positive methylotroph Amycolatopsis methanolica contains a mycothiol-dependent formaldehyde dehydrogenase (27, 28). The other methylotrophs in which H4MPT-dependent enzyme activities could not be detected have not yet been tested for thiol-dependent formaldehyde oxidation systems, and so the extent of this correlation is not yet known.
Genes similar to the H4MPT-dependent enzymes were not present in other nonarchaeal microbial genome sequences that are currently available, which include a yeast genome and representatives from 10 of the major branches of the bacteria. Therefore, a H4MPT-dependent C1 pathway does not appear to be widespread in bacteria as a whole but is present in most of the proteobacterial methylotrophs tested. The broad distribution of H4MPT-dependent enzymes in methylotrophic proteobacteria leads to speculation on the potential function of this pathway. In Methylobacterium extorquens AM1, the H4MPT-dependent formaldehyde oxidation system appears to be the main dissimilatory pathway during growth on C1 substrates (11, 47). This pathway is indispensable for methylotrophic growth, and the enzymes are present at high activities in methylotrophically grown cells as compared to cells grown on succinate (11). Methylobacterium extorquens AM1 also contains a functionally analogous H4F-linked pathway, albeit with much lower enzyme activities (11). Mutant evidence suggests that in Methylobacterium extorquens AM1, both pathways are involved in methylotrophy (unpublished data), and so the H4F-linked pathway may also contribute to formaldehyde dissimilation or detoxification. It seems likely that the H4MPT-dependent formaldehyde oxidation system plays a similar role in other serine cycle methylotrophs and in the Xanthobacter strains, since they do not contain sufficiently high activities of enzymes from other potential formaldehyde oxidation pathways to account for their growth rate on C1 substrates. A similar argument follows for the methanotrophic bacteria containing the ribulose monophosphate cycle, since they also lack sufficient enzyme activities of alternative oxidative pathways (48). However, the presence of the H4MPT-dependent formaldehyde oxidation system in the β-proteobacteria is surprising, since these bacteria have been proposed to employ a cyclic route of formaldehyde oxidation (1, 4). The results reported here show that like the serine cycle methylotrophs, these ribulose monophosphate cycle methylotrophs contain two possible formaldehyde oxidation pathways. Mutagenesis studies will be necessary to address the role of these two pathways in the β-proteobacterial methylotrophs. The potential role of the H4MPT-dependent oxidation pathway in formaldehyde dissimilation in methylotrophs is consistent with the apparent absence of this pathway in methylotrophs that contain a thiol-linked formaldehyde oxidation system. These results suggest that methylotrophic bacteria have either one or the other of these formaldehyde oxidation pathways but may also have a second pathway as in the β-proteobacterial methylotrophs (cyclic oxidation pathway) and the serine cycle nonmethanotrophs (H4F-linked pathway). However, we cannot rule out completely the possibility that the absence of H4MPT-dependent enzyme activities might be due to the presence of a modified pterin in these methylotrophic bacteria that is not functional with the H4MPT that was used in this study, which was isolated from the methanogenic archaeon Methanobacterium thermoautotrophicum. Methylobacterium extorquens AM1 was shown to contain a modified H4MPT that lacks the α-hydroxyglutaryl phosphate unit, although enzyme activity was detected with both pterins (11, 47).
The data presented here show the presence of functional H4MPT-dependent enzymes in three branches of the proteobacteria. Phylogenetic analysis revealed that the bacterial and archaeal branches in the tree of mch genes are clearly separated but that bacterial and archaeal mch genes are clearly related. H4MPT and H4MPT-dependent enzymes were previously thought to be a specific attribute of archaea. Their sparse occurrence among the bacteria in general and the proteobacteria in particular raises the question of the evolutionary process underlying this phylogenetic distribution. In principle, it is possible that the common ancestor of archaea and bacteria possessed H4MPT, Mch, and other H4MPT-dependent enzymes and that countless parallel losses have occurred in independent lineages. This would mean that these enzymes and their corresponding cofactors were retained throughout the trunk of bacterial and archaeal evolution but have only been retained in selected lineages that use these pathways for C1 metabolism today.
Alternatively, it is possible that the genes for these enzymes and cofactors have been transferred between bacteria and archaea by horizontal gene transfer (14, 22). The information available at this time is not sufficient to distinguish between these possibilities. However, it is noteworthy that Mch and H4MPT are ubiquitous among methanogenic archaea, where they are integral to the ecologically specialized energy metabolism of this group, while in contrast, results to date suggest that only some proteobacteria possess this gene and the cofactor. This finding led to the suggestion that proteobacteria may have acquired Mch and functionally related genes of typically methanogenic C1 metabolism through lateral transfer from archaeal donors (11). If so, the small sample of sequences studied here would be more supportive of a single rather than multiple transfer events, because the bacterial and archaeal genes are distinct rather than interleaved in the tree. It was previously noted that bacterial Mch is activated 20-fold by 1.2 M potassium phosphate (31). This is an unusual property for α-proteobacterial enzymes but is a property of some enzymes from methanogenic archaea, which require such salt concentrations for activity (40). Activation of Mch by 1.2 M potassium phosphate would be consistent with the hypothesis of lateral transfer from methanogens to proteobacteria. However, at this time we can state with confidence only that the two major lineages of these genes, bacterial and archaeal, show separate evolution.
ACKNOWLEDGMENTS
We thank Stefanie Pluschkell, University of Minnesota, for providing cell mass from methanol-grown Bacillus methanolicus. We thank William Martin, Technische Universität Braunschweig, for many helpful discussions.
This work was supported by a grant from NIH (GM36296) and the Max-Planck-Gesellschaft. J.A.V. was the recipient of a fellowship from the Max-Planck-Gesellschaft.
REFERENCES
- 1.Anthony C. The biochemistry of methylotrophs. London, United Kingdom: Academic Press Ltd.; 1982. [Google Scholar]
- 2.Barber R D, Rott M A, Donohue T J. Characterization of a glutathione-dependent formaldehyde dehydrogenase from Rhodobacter sphaeroides. J Bacteriol. 1996;178:1386–1393. doi: 10.1128/jb.178.5.1386-1393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber R D, Donohue T J. Function of a glutathione-dependent formaldehyde dehydrogenase in Rhodobacter sphaeroides formaldehyde oxidation and assimilation. Biochemistry. 1998;37:530–537. doi: 10.1021/bi971463t. [DOI] [PubMed] [Google Scholar]
- 4.Beardsmore A J, Aperghis P N G, Quayle J R. Characterization of the assimilatory and dissimilatory pathways of carbon metabolism during growth of Methylophilus methylotrophus on methanol. J Gen Microbiol. 1982;128:1423–1439. [Google Scholar]
- 5.Bowman J P, Sly L I, Stackebrandt E. The phylogenetic position of the family Methylococcaceae. Int J Syst Bacteriol. 1995;45:182–185. doi: 10.1099/00207713-45-1-182. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 7.Breitung J, Börner G, Scholz S, Linder D, Stetter K O, Thauer R K. Salt dependence, kinetic properties and catalytic mechanism of N-formylmethanofuran:tetrahydromethanopterin formyltransferase from the extreme thermophile Methanopyrus kandleri. Eur J Biochem. 1992;210:971–981. doi: 10.1111/j.1432-1033.1992.tb17502.x. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrik J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Chistoserdov A Y, Boyd J, Mathews F S, Lidstrom M E. The genetic organization of the mau gene cluster of the facultative autotroph Paracoccus denitrificans. Biochem Biophys Res Commun. 1992;184:1226–1234. doi: 10.1016/s0006-291x(05)80007-5. [DOI] [PubMed] [Google Scholar]
- 10.Chistoserdova L V, Lidstrom M E. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification of sgaA and mtdA and sequences of sgaA, hprA, and mtdA. J Bacteriol. 1994;176:1957–1968. doi: 10.1128/jb.176.7.1957-1968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. C1 transfer enzymes and coenzymes linking methytrophic bacteria and methanogenic archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 12.Dalton H. Methane oxidation by methanotrophs. In: Murrell J C, Dalton H, editors. Biotechnology handbooks: Methane and methanol utilizers. New York, N.Y: Plenum Press; 1992. pp. 85–114. [Google Scholar]
- 12a.Deutsche Sammlung von Mihroorganismen und Zellkulturen CombH Website. July 1999, revision date. Media list [Online.] http://www.dsmz.de/media/media.htm. [17 March 1999, last date accessed.]
- 13.Dijkhuizen L, Levering P R, de Vries G E. The physiology and biochemistry of aerobic methanol-utilizing Gram-negative and Gram-positive bacteria. In: Murrell J C, Dalton H, editors. Biotechnology handbooks: methane and methanol utilizers. New York, N.Y: Plenum Press; 1992. pp. 149–181. [Google Scholar]
- 14.Doolittle W F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 15.Eggeling L, Sahm H. The formaldehyde dehydrogenase of Rhodococcus erythropolis, a trimeric enzyme requiring a cofactor and active with alcohols. Eur J Biochem. 1985;150:129–134. doi: 10.1111/j.1432-1033.1985.tb08997.x. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 17.Fulton G L, Nunn D N, Lidstrom M E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984;160:718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gründig M W, Babel W. The linear oxidation of formaldehyde to CO2 as the proper energy generating sequence for the assimilation of methanol in Acetobacter methanolicus MB58. Arch Microbiol. 1987;149:149–155. [Google Scholar]
- 19.Hanson R S, Netrusov A I, Tsuji K. The obligate methanotrophic bacteria Methylococcus, Methylomonas, and Methylosinus. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 2350–2364. [Google Scholar]
- 20.Harms N, Ras J, Reijnders W N M, van Spanning R J M, Stouthamer A H. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway of formaldehyde detoxification? J Bacteriol. 1996;178:6296–6299. doi: 10.1128/jb.178.21.6296-6299.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazeu W, de Bruyn J C, Dijken J P. Nocardia sp. 239, a facultative methanol utilizer with the ribulose monophosphate pathway of formaldehyde fixation. Arch Microbiol. 1983;135:205–210. [Google Scholar]
- 22.Jain R, Rivera M C, Lake J A. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 24.Lidstrom M E. The aerobic methylotrophic bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 431–445. [Google Scholar]
- 25.Malashenko I R, Romanovskaia V A, Bogachenko V N, Shved A D. Thermophilic and thermotolerant bacteria that assimilate methane. Mikrobiologiya. 1975;44:855–862. [PubMed] [Google Scholar]
- 26.Marison I W, Attwood M M. A possible alternative mechanism for the oxidation of formaldehyde to formate. J Gen Microbiol. 1982;128:1441–1446. [Google Scholar]
- 27.Misset-Smits M, van Ophem P W, Sakuda S, Duine J A. Mycothiol, 1-O-(2′-[N-acetyl-l-cysteinyl]amido-2′-deoxy-α-d-glucopyranosyl)-d-myo-inositol, is the factor of NAD/factor-dependent formaldehyde dehydrogenase. FEBS Lett. 1997;409:221–222. doi: 10.1016/s0014-5793(97)00510-3. [DOI] [PubMed] [Google Scholar]
- 28.Norin A, van Ophem P W, Piersma S R, Persson B, Duine J A, Jörnvall H. Mycothiol-dependent formaldehyde dehydrogenase, a prokaryotic medium-chain dehydrogenase/reductase, phylogenetically links different eukaryotic alcohol dehydrogenases. Primary structure, conformational modelling and functional correlations. Eur J Biochem. 1997;248:282–289. doi: 10.1111/j.1432-1033.1997.00282.x. [DOI] [PubMed] [Google Scholar]
- 29.Nunn D N, Lidstrom M E. Isolation and complementation analysis of ten methanol oxidation (Mox) mutant classes and the identification of the methanol dehydrogenase structural gene of Methylobacterium AM1. J Bacteriol. 1986;166:581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ophem P W van, van Beeumen J, Duine J A. NAD-linked, factor-dependent formaldehyde dehydrogenase or trimeric, zinc-containing, long-chain alcohol dehydrogenase from Amycolatopsis methanolica. Eur J Biochem. 1992;206:511–518. doi: 10.1111/j.1432-1033.1992.tb16954.x. [DOI] [PubMed] [Google Scholar]
- 31.Pomper B K, Vorholt J A, Chistoserdova L, Lidstrom M E, Thauer R K. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur J Biochem. 1999;261:475–480. doi: 10.1046/j.1432-1327.1999.00291.x. [DOI] [PubMed] [Google Scholar]
- 32.Ras J, van Ophem P W, Reijnders W N M, van Spanning R J M, Duine J A, Stouthamer A H, Harms N. Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J Bacteriol. 1995;177:247–251. doi: 10.1128/jb.177.1.247-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reding H K, Croes G L, Dijkhuizen L, Wiegel J. Emendation of Xanthobacter flavus as a motile species. Int J Syst Bacteriol. 1992;42:309–311. doi: 10.1099/00207713-42-2-309. [DOI] [PubMed] [Google Scholar]
- 34.Reid G A, Gordon E H. Phylogeny of marine and freshwater Shewanella: reclassification of Shewanella putrefaciens NCIMB 400 as Shewanella frigidimarina. Int J Syst Bacteriol. 1999;49:189–191. doi: 10.1099/00207713-49-1-189. [DOI] [PubMed] [Google Scholar]
- 35.Romanovskaia V A, Malashenko I R, Bogachenko V N. Refinement of the diagnosis of the genera and species of methane-using bacteria. Mikrobiologiya. 1978;47:120–130. [PubMed] [Google Scholar]
- 36.Sahm H, Cox R B, Quayle J R. Metabolism of methanol by Rhodopseudomonas acidophila. J Gen Microbiol. 1976;94:313–322. doi: 10.1099/00221287-94-2-313. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schendel F J, Bremmon C E, Flickinger M C, Guettler M, Hanson R S. l-Lysine production at 50°C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl Environ Microbiol. 1990;56:963–970. doi: 10.1128/aem.56.4.963-970.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott J H, Nealson K H. A biochemical study of the intermediary carbon metabolism of Shewanella putrefaciens. J Bacteriol. 1994;176:3408–3411. doi: 10.1128/jb.176.11.3408-3411.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shima S, Hérault D A, Berkessel A, Thauer R K. Activation and thermostabilization effects of cyclic 2,3-diphosphoglycerate on enzymes from the hyperthermophilic Methanopyrus kandleri. Arch Microbiol. 1998;170:469–472. doi: 10.1007/s002030050669. [DOI] [PubMed] [Google Scholar]
- 41.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thauer R K. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 44.Van Neil C B. The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol Rev. 1944;8:1–118. doi: 10.1128/br.8.1.1-118.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaupel M, Dietz H, Linder D, Thauer R K. Primary structure of cyclohydrolase (Mch) from Methanobacterium thermoautotrophicum (strain Marburg) and functional expression of the mch gene in Escherichia coli. Eur J Biochem. 1996;236:294–300. doi: 10.1111/j.1432-1033.1996.00294.x. [DOI] [PubMed] [Google Scholar]
- 46.Vaupel M, Vorholt J A, Thauer R K. Overproduction and one-step purification of the N5,N10-methenyltetrahydromethanopterin cyclohydrolase (Mch) from the hyperthermophilic Methanopyrus kandleri. Extremophiles. 1998;2:15–22. doi: 10.1007/s007920050038. [DOI] [PubMed] [Google Scholar]
- 47.Vorholt J A, Chistoserdova L, Lidstrom M E, Thauer R K. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol. 1998;180:5351–5356. doi: 10.1128/jb.180.20.5351-5356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zatman L J. A search for patterns in methylotrophic pathways. In: Dalton H, editor. Microbial growth on C1 compounds. 1981. pp. 42–54. Heyden, London, United Kingdom. [Google Scholar]