Abstract
Messenger ribonucleic acid (mRNA) vaccines made their successful public debut in the effort against the COVID‐19 outbreak starting in late 2019, although the history of mRNA vaccines can be traced back decades. This review provides an overview to discuss the historical course and present situation of mRNA vaccine development in addition to some basic concepts that underly mRNA vaccines. We discuss the general preparation and manufacturing of mRNA vaccines and also discuss the scientific advances in the in vivo delivery system and evaluate popular approaches (i.e., lipid nanoparticle and protamine) in detail. Next, we highlight the clinical value of mRNA vaccines as potent candidates for therapeutic treatment and discuss clinical progress in the treatment of cancer and coronavirus disease 2019. Data suggest that mRNA vaccines, with several prominent advantages, have achieved encouraging results and increasing attention due to tremendous potential in disease management. Finally, we suggest some potential directions worthy of further investigation and optimization. In addition to basic research, studies that help to facilitate storage and transportation will be indispensable for practical applications.
Keywords: cancer, infectious diseases, lipid nanoparticles, mRNA delivery, mRNA vaccine
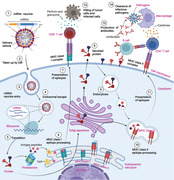
Mechanisms of mRNA vaccines for infectious diseases and cancers. mRNA molecules encoding tumor antigens are injected into body (either with or without delivery vehicles). The mRNA molecules are taken up and translated into protein antigens by antigen‐presenting cells (APCs). After proteasomal processing of proteins, antigen peptides associate with MHC Class I molecule in the endoplasmic reticulum and are transferred to the APC surface, activating CD8+ T cells for a specific cellular immune response. Protein antigens, which are sorted for the endosome route, can activate CD4+ T cells via the MHC Class II presentation pathway. The secretory protein antigen or membrane antigen encoded by mRNA can stimulate B cells to produce neutralizing antibodies and activate phagocytes such as macrophages to secrete inflammatory cytokines, facilitating the clearance of circulating infectious pathogens and tumor cells.
1. INTRODUCTION
Messenger ribonucleic acid (mRNA) vaccines are now regarded as an attractive and promising alternative to conventional vaccines. mRNA vaccines show several prominent advantages compared to live attenuated and subunit‐based vaccines. 1 , 2 First, they exhibit reliable potency in the induction of immune responses. 3 mRNA vaccines are able to drive both humoral and cellular immunity. 4 , 5 In addition, RNA interacts directly with pattern recognition receptors (PRRs), driving a robust innate immune response without the need to involve additional adjuvants. However, innate antiviral responses may be a double‐edged sword, as they may reduce vaccine effectiveness by downgrading mRNA expression. Second, mRNA vaccines are safe, avoiding many risks of conventional vaccines, such as reversion to virulence or insertional mutagenesis. 6 , 7 Furthermore, mRNA, as a minimal genetic unit, does not stimulate any antivector immunity, making recurrent administration of mRNA vaccine safe and easy. 8 , 9 Third, mRNA vaccines are rapid and inexpensive to manufacture, because the synthesis of mRNA vaccines mainly uses in vitro transcription (IVT), 10 which bypasses the time‐consuming steps of in vivo protein expression (i.e., cloning and cell culture). 11 , 12 Lastly, mRNA is a manageable transient modulator of physiologic processes.
The global COVID‐19 pandemic beginning in 2020 brought mRNA vaccines into the spotlight and has greatly accelerated their deployment. Recently, the Pfizer‐BioNTech COVID‐19 vaccine became the first‐ever approved mRNA‐based vaccine available to the public, although the idea of a nucleic acid vaccine was proposed decades ago. 13 For a long time, the susceptibility of naked RNA to extracellular RNase degradation was the major obstacle to RNA transfection; therefore, a delivery system that protected mRNA and mediated cellular internalization of mRNA was needed. The earliest solution arose as early as 1978, when scientists conducted a landmark experiment, which demonstrated that human cells could take up mRNA sequestered within the liposome and stably produce proteins from it. 14 However, the use of mRNA as a potential therapeutic agent still remained a concept with little practicality, until in 1984, when mRNA was successfully in vitro synthesized in the laboratory. 15 Since then, mRNA vaccine development has attracted more interest, and in 1993, Martinon and his team designed and tested the first mRNA vaccine encoding the influenza nucleoprotein (NP). They managed to elicit anti‐influenza cytotoxic T lymphocytes (CTL) responses in mice. 16 In later years, mRNA vaccines were verified to have versatility in giving effective protection to animal hosts against various diseases, including both infectious diseases (e.g., rabies, Zika virus, and others) 17 , 18 , 19 , 20 , 21 , 22 and noninfectious diseases (e.g., cancer). 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 In 2005, another revolutionary paper was published by Katalin Karikó and her colleagues, suggesting that different uridine modifications of RNA (e.g. m5C, m5U, pseudouridine Ψ, and others) could suppress the activation of different human Toll‐like receptors (TLRs), while the unmodified forms stimulated all TLRs such as TLR3, TLR7, and TLR8. 32 This finding made a profound impact on mRNA vaccine design and further advanced the use of mRNA (Figure 1).
FIGURE 1.
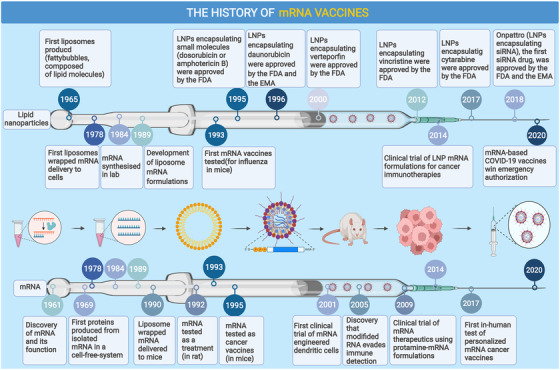
Timeline of major findings and breakthroughs in the development of messenger ribonucleic acid (mRNA) vaccines. EMA, European Medicines Agency; COVID‐19, coronavirus disease 2019; FDA, U.S. Food and Drug Administration; LNP, lipid nanoparticle
In this review, we provide a broad overview of the progress in mRNA vaccine technology (i.e., the engineering of mRNA sequence and in vivo delivery systems), current application and success of mRNA vaccines in the treatment of cancer, as well as a range of infectious diseases (e.g., COVID‐19).
2. BASICS OF mRNA VACCINES
2.1. mRNA biology
mRNA is single‐stranded RNA (ssRNA) encoding a genetic sequence that can be translated to a protein by ribosome. 33 There are three main types of mRNAs that mRNA vaccines can use: conventional mRNA, self‐amplifying RNA, and circular RNA. 34 , 35 Conventional mRNA is a nonreplicating linear mRNA and so far has been the focus of this field. Self‐amplifying RNA (saRNA) resembles conventional mRNA, albeit containing extra elements, including 5′ and 3′ conserved sequence elements, a large open reading frame (ORF) encoding four nonstructural proteins (nsP1‐4), which form alphavirus RNA‐dependent RNA polymerase (RdRP), and a subgenomic promoter dictating the expression of gene of interest as subgenomic RNA. saRNA can achieve same efficacy with a much lower dose. 36 , 37 , 38 However, reuse of saRNA may be limited by immunity against viral RdRP. 39 Circular RNA (circRNA) can be rendered translatable by inserting an internal ribosome entry site upstream of the coding sequence. The most viable approach for generating circRNA in vitro to date utilizes the group I intron self‐splicing ribozyme, which produces large circRNA using guanosine 5′‐phosphate (GTP) and magnesium ion as cofactors. 34 , 40 , 41 , 42 , 43 This method is also known as the permuted introns and exons method. 44
A typical mRNA consists of a 5′ cap, a 5′ untranslated region (UTR), an ORF, a 3′ UTR, and a poly‐A tail. In the eukaryotic system, translation initiation factor 4E (eIF4E) binds to the 5′ cap, and poly‐A binding protein (PABP) binds to the 3′ poly‐A tail. Through the interaction of eIF4E and PABP with the translation initiation factor eIF4G, the mRNA circularizes, which increases its structural stability 45 , 46 , 47 and allows enhanced translation. A poly‐A tail about 100 nucleotides long is preferred in mRNA design, because maintenance of a long poly(d(A/T)) stretch of DNA templates is usually difficult in the bacterial host. 48 , 49 , 50 Pfizer/BioNTech utilized a poly‐A tail 110 nucleotide long with a GCAUAUGACU linker placed between the poly(A)30 and poly(A)70 sequences. 51 Counterintuitively, recent insights into poly‐A tail biology suggested that mRNAs with shorter poly‐A tails (about 30 nt long) are generally better translated than those with longer poly‐A tails. Counteracting the roles of cytoplasmic poly(A) binding protein (PABPC) in protecting the poly‐A tail to sustain translation and in facilitating deadenylase activities to accelerate mRNA decay may be one explanation. 52
The UTR elements impact the translational efficiency of mRNA. 53 , 54 , 55 UTRs of customized mRNAs are usually adapted from highly expressed human genes. For example, UTRs of α‐globin and β‐globin were often used, and in the BNT162b2 vaccine, 56 3′ UTRs of hybrid AES‐mtRNR1 were adopted after screening by systematic evolution of ligands via the exponential enrichment method. 57 , 58 , 59 , 60 The ORF generally encodes a target protein antigen for the purpose of eliciting effective immune responses.
2.2. Innate immune responses: Something to exploit or to avoid?
Exogenous mRNAs are typically transferred to the cytosol through the endosomal pathway, along which the cargo mRNAs are readily recognized by PRRs, such as TLR‐7. The recognition triggers immediate induction of type I interferon that ultimately leads to both stimulation of innate immunity and inhibition of translation. 61 , 62 CureVac managed to solve this dilemma by introducing the RNActive technology, 63 in which a mixture of naked mRNAs and protamine‐complexed mRNAs work together to activate the TLR‐7‐dependent immune response, while maintaining an acceptable level of antigen expression. The two‐component mRNA vaccine system could thus elicit balanced humoral and T cell‐mediated responses, in particular the type 1 helper T (Th1)‐biased responses. 64 , 65 , 66 However, the more common practice is to avoid PRR recognition of the delivered mRNA, especially the uridine moiety, rather than to exploit it. CureVac also devised an mRNA sequence engineering method to select for the most germinal center (GC)‐rich codons, thereby reducing the presence of uridine and achieving significantly higher level of protein production. 67 Codon optimization, which could generate misfolded proteins or even novel proteins with unknown functions, should be scrutinized on a case by case basis. 68 , 69 , 70 The prevailing approach is to substitute the uridine in mRNA molecules with modified nucleosides, such as pseudouridine, N1‐methyl‐pseudouridine, or 5‐methoxyuridine. 71 , 72 , 73 However, the previously unexamined issue of modified nucleosides impeding translation elongation and termination deserves due attention. 74 , 75
2.3. Manufacture of mRNA vaccines
mRNA synthesis in vitro starts from bacteriophage (usually T7) RNA polymerase‐mediated transcription of a linear DNA template. A typical final product of in vitro mRNA manufacturing is Cap 1 mRNA. 76 The Cap 1 structure is derived from the Cap 0 structure, which has an N7‐methyl guanosine connected to the 5′ nucleotide by a 5′‐5′ triphosphate bridge. Further methylation at the 2′‐O position of Cap 0 gives the Cap 1 structure. For Cap 1 mRNA, the Cap 0 backbone enables high accessibility to the translational machinery, 77 while both 2′‐O methylation and elimination of 5′ triphosphate contribute to the evasion of translational inhibition followed by innate immune recognition.
Capping of RNA in vitro could be done in two ways: either via co‐transcription or via post‐transcriptional enzymatic reactions. Co‐transcriptional capping is a one‐step process, a typical example of which uses CleanCap technology. This method uses a proprietary 5′ capping solution, which has a capping efficiency of 94% without the yield being significantly compromised. Enzymatic capping is a conventional two‐step process, in which vaccinia capping enzyme catalyzes the formation of Cap 0, while 2′‐O‐methyltransferase converts Cap 0 to Cap 1 structure. Co‐transcriptional capping is simple and convenient. However, CleanCap not only requires the expensive cap analogue and achieves incomplete capping but also requires alkaline phosphatase processing of uncapped RNA to avoid innate immune recognition. In addition, the DNA template must be modified to replace the GG sequence, which often appears after the TATA box of the T7 promoter, with an AG sequence. As for enzymatic capping, it is highly efficient but requires a buffer exchange between steps, which is more complicated and time‐consuming. Furthermore, certain secondary structures at the 5′ end of RNA may hamper efficient enzyme‐mediated capping (Figure 2).
FIGURE 2.
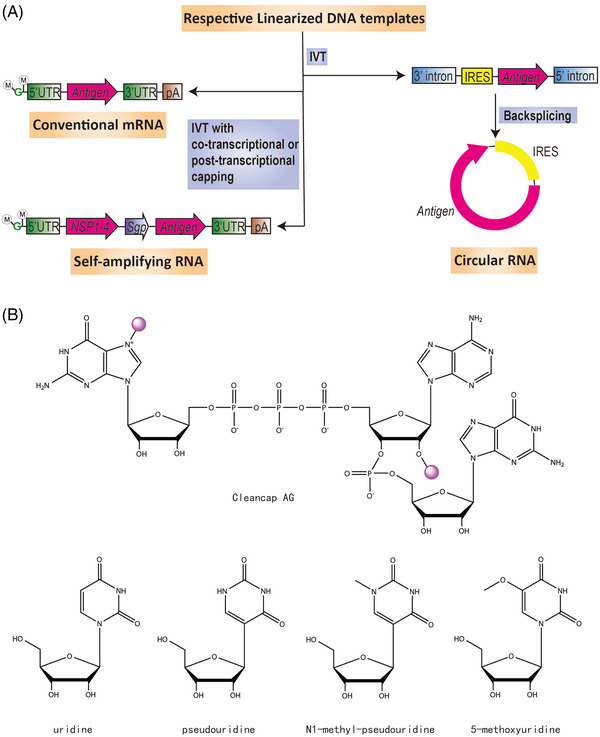
Three types of messenger ribonucleic acid (mRNA) and their production and modification. (A) Features of conventional mRNA, self‐amplifying RNA, and circular RNA, as well as their typical synthesis processes in vitro. M in circle, methyl group. G in green, guanylate. UTR, untranslated region. IVT, in vitro transcription. pA, poly (A) sequence. NSP1‐4, sequence encoding four nonstructural proteins, namely nsP1‐4, which together form RNA‐dependent RNA polymerase. Sgp, subgenomic promoter. IRES, internal ribosome entry site. (B) Structure of cap analogue Cleancap AG, as well as uridine and its modifications. Note that for Cleancap AG, the two methyl groups are highlighted with purple circles.
These in vitro reactions can be easily scaled up for bulk manufacture of mRNA. When the reactions are complete, impurities in the reaction systems (e.g., buffers, proteins, residue substrates, DNA templates, dsRNA byproducts, and short RNA fragments) must be removed to obtain pure mRNA products. mRNA purification involves degradation of DNA templates with DNase I digestion, removal of dsRNA via cellulose chromatography, removal of small‐molecule impurities with tangential flow filtration (TFF), and removal of particulate contaminants via a sterile filter. 78 This process is simple and convenient; however, remaining dsRNA byproducts could still trigger innate immune responses and interfere with vaccine efficacy. Strategies such as oligo dT affinity chromatography, 79 anion exchange chromatography, and hydrogen bond chromatography 80 can produce mRNA with even higher purity in combination with a subsequent polishing step. For oligo dT affinity chromatography, the poly‐A tail of mRNA basepairs with the oligo dT stretches under high ionic strength and dissociates under low ionic strength during elution. Other impurities that cannot bind to oligo dT are removed. 81 For anion exchange chromatography, the net contribution of hydrogen bonding is reduced with an anion exchanger with reduced hydrogen bonding potential, and impurities like dsRNA and DNA are washed off with 1 M NaCl and 10 mM EDTA. Subsequently, ssRNA can be eluted at ambient temperature by a pH gradient. For hydrogen bond chromatography, impurities like dsRNA and DNA can also be washed off with 1 M NaCl and with 10 mM EDTA, and ssRNA can be eluted by a pyrophosphate gradient. 82
Complexation of mRNA with lipid nanoparticles (LNPs) can be carried out with multiple microfluidics devices placed in parallel. 83 After purification by a TFF device and a sterile filter, the resulting mRNA‐LNPs are diluted with buffer to achieve pharmaceutical concentrations of interest (Figure 3).
FIGURE 3.
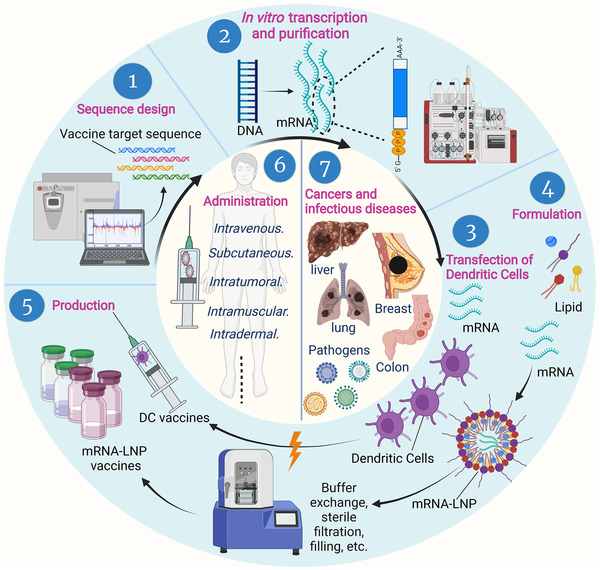
Preparation and application of messenger ribonucleic acid (mRNA) vaccines. Once pathogens or tumors are identified, sequences for the target antigens are determined by the combined efforts of sequencing, bioinformatics, and computational approaches. Target DNAs are synthesized and transcribed into mRNAs in vitro, and then mRNA transcripts are purified to remove contaminants and reactants. Purified mRNA is mixed with lipids in a microfluidic mixer to form lipid nanoparticle mRNA vaccines. Dendritic cells are loaded with candidate mRNA to form DC‐mRNA vaccines. Various vaccines are produced by scaling up, then quickly tested and stored in sterilized bottles to treat various cancers and infectious diseases through different administration methods. DC, dendritic cells; LNP, lipid nanoparticle
2.4. Storage of mRNA vaccines
The two approved mRNA vaccines, mRNA‐1273 and BNT162b2, both formulated in LNPs, have shelf lives of 6 months in a frozen state at −20°C and −80 to −60°C, 84 respectively. When thawed, they can be stored at 2–8°C for up to 1 month. 85 These strict storage conditions challenge the global distribution of those vaccines, and many studies have been dedicated to addressing this issue. 86 , 87 , 88 , 89 Lyophilized mRNA‐LNPs achieved a stable storage duration of 6 months at 4°C and 3 months at room temperature. 88
2.5. Quality control of mRNA vaccines
In 2021, the WHO released guidance on mRNA against infectious diseases, in which the drug substance (mRNA) and the drug product (final formulated vaccine) are somewhat divergent in quality control considerations. 90 For mRNA, the identity, purity and impurities, quantification and physical state, safety attributes, reference materials, as well as stability are points of concern, while for the final formulated vaccine, the identity, purity and impurities, content, strength or quantity, safety attributes, potency, reference materials, as well as stability testing, storage, and expiry date are the focus.
2.6. Adverse effects of mRNA vaccines
Data from mass vaccination efforts suggest that mRNA vaccines are safe, with adverse effects that are generally acceptable in frequency and severity. Nevertheless, rapid and robust local inflammation could be induced in mice‐administrated mRNA vaccines via intradermal, 91 intramuscular, 92 or intranasal routes. 93 This process was dependent on the ionizable lipid component of the LNP formulation. 94 Myocarditis is also an adverse effect of concern, which, however, could be attributed to inadvertent intravenous injection during intramuscular administration. 88 Furthermore, anaphylaxis could be induced by antibody response toward the polyethylene glycol (PEG) moiety, which is a rare, yet possibly fatal outcome. 95 , 96 , 97 These data warrant future study to optimize the delivery system of mRNA vaccines.
2.7. Administration routes of mRNA vaccines
Administration method is very important for the efficacy of mRNA vaccines and can affect the organ distribution, expression kinetics, induced immune response intensity, and side effects. For prevention vaccines, such as COVID‐19 mRNA vaccines, in order to induce a strong immune response, administration is typically intramuscularly and subcutaneously (Tables 1 and 2). 98 , 99 , 100 Therapeutic mRNA vaccines, such as a variety of tumor therapeutic mRNA vaccines, can be administered intravenously, intratumorally, 101 or subcutaneously (Table 3). 102 In addition, mRNA can be transfected into dendritic cells (DCs) in vitro to prepare a DCs cell vaccine, and the cell vaccine can be infused to prevent or treat diseases (Table 4). 103 , 104 , 105
TABLE 1.
Information on COVID19 mRNA vaccines approved for use
| Category | Pfizer‐BioNTech | Moderna |
|---|---|---|
| Name product | BNT162b2 | mRNA‐1273 |
| Lipid nanoparticle components and ratio | ALC‐0315/DSPC/Cholesterol/ALC‐0159 = 46.3:9.4:42.7:1.6 | SM‐102/DSPC/Cholesterol/PEG2000‐DMG = 50:10:38.5:1.5 |
| Ionizable nitrogen/phosphate molar raio | 6 | Estimated to be 6 |
| Excipients | KH2PO4; Na2HPO4; KCl; NaCl; Sucrose; Water for injection | Tris; sodium acetate; sucrose; water for injection |
| Ages recommended | 5+ years old | 18+ years old |
| mRNA dose; route of administration | 30 μg; intramuscular | 100 μg; intramuscular |
| Primary series | Two doses; given 3 weeks apart | two doses; given 4 weeks apart |
| Booster dose | Everyone aged 18 years and older should get a booster dose of either Pfizer‐BioNTech or Moderna (COVID‐19 vaccines) 5 months after the last dose in their primary series. | Everyone aged 18 years and older should get a booster dose of either Pfizer‐BioNTech or Moderna (COVID‐19 vaccines) 5 months after the last dose in their primary series. |
| Teens 12–17 years old should get a Pfizer‐BioNTech COVID‐19 Vaccine booster 5 months after the last dose in their primary series. | Null | |
| When fully vaccinated | 2 weeks after 2nd dose | 2 weeks after 2nd dose |
Note: The table's data adapted from https://www.sciencedirect.com/science/article/pii/S0378517321003914 and https://www.cdc.gov/coronavirus/2019‐ncov/vaccines/different‐vaccines.html.
Abbreviation: ALC‐0315, ([(4‐hydroxybutyl)azanediyl]di(hexane‐6,1‐diyl) bis(2‐hexyldecanoate)); ALC‐0159, 2‐[(polyethylene glycol)‐2000]‐N,N‐ditetradecylacetamide; COVID‐19, corona virus disease‐2019; DSPC, 1,2‐Distearoyl‐sn‐glycero‐3‐phosphocholine; mRNA, messenger ribonucleic acid; PEG, polyethylene glycol.
TABLE 2.
Clinical trials of mRNA vaccines targeting other infectious diseases
| Funding source | Vaccine name | Target | Route | Phase | NCT Number |
|---|---|---|---|---|---|
| Moderna | mRNA‐1189 | Epstein‐Barr Virus Infection | Intramuscular | Phase I | NCT05164094 |
| mRNA‐1345 | Respiratory Syncytial Virus (RSV) | Intramuscular | Phase II/III | NCT05127434 | |
| mRNA‐1647 | Cytomegalovirus Infection | Intramuscular | Phase III | NCT05085366 | |
| mRNA‐1010 | Seasonal Influenza | Intramuscular | Phase I/II | NCT04956575 | |
| mRNA‐1893 | Zika Virus | Intramuscular | Phase II | NCT04917861 | |
| mRNA‐1443 | CMV | Intramuscular | Phase I | NCT03382405 | |
| mRNA‐1325 | Zika | Intramuscular | Phase I | NCT03014089 | |
| mRNA‐1653 | hMPV/PIV3 | Intramuscular | Phase I | NCT04144348; NCT03392389 | |
| mRNA‐1851 (VAL‐339851) | Influenza A (H7N9) | Intramuscular | Phase 1 | NCT03345043 | |
| mRNA‐1440 (VAL‐506440) | Influenza A (H10N8) | Intramuscular | Phase 1 | NCT03076385 | |
| mRNA‐1010 | Influenza A (H1N1, H3N2), influenza B (Yamagata lineage, Victoria lineage) | Intramuscular | Phase I/II | NCT04956575 | |
| mRNA‐1944 | Chikungunya | Intramuscular | Phase I | NCT03829384 | |
| mRNA‐1388 (VAL‐181388) | Chikungunya | Intramuscular | Phase I | NCT03325075 | |
| CureVac | CV7201 | Rabies | Intradermal, intramuscular | Phase I | NCT02241135 |
| CV7202 | Rabies | Intramuscular | Phase I | NCT03713086 | |
| CureVac AG | CVSQIV | Influenza | Intramuscular | Phase I | NCT05252338 |
| GSK | GSK3903133A | Rabies | Intramuscular | Phase I | NCT04062669 |
| NIAID | BG505 MD39.3 mRNA; BG505 MD39.3 gp151 mRNA; BG505 MD39.3 gp151 CD4KO mRNA | HIV Infections | Intramuscular | Phase I | NCT05217641 |
Note: The table summarizes the clinical trials of mRNA vaccines targeting other infectious diseases registered at Clinical Trials.gov.
Abbreviations: CMV, cytomegalo virus; GSC, Glaxosmithkline Plc; mRNA, messenger ribonucleic acid; NIAID, National Institute of Allergy and Infectious Diseases.
TABLE 3.
Currently clinical trials with mRNA tumor vaccines
| Vaccine type | NCT Number | Cancer type | Status | Phases | mRNA | Combo | Route | Completion |
|---|---|---|---|---|---|---|---|---|
| mRNA encoding TAAs | NCT02410733 | Melanoma | Active, not recruiting | I | TAAs: NY‐ESO‐1/MAGE C3/ tyrosinase/TPTE; Lipo‐MERIT | Null | i.v. | 2023/5 |
| NCT01890213 | Stage III colon cancer | Completed | I | An alphavirus replicon (VRP) encoding the protein (CEA); AVX701 | Null | i.m. | 2019/7 | |
| NCT04534205 | Unresectable/Metastatic/Recurrent) HNSCC | Recruiting | II | TAAs:E6/E7; BNT113 | Pembrolizumab (i.v. infusion) | i.v. | 2025/5 | |
| NCT04526899 | Melanoma stage III, IV; unresectable melanoma | Recruiting | II | TAAs:NY‐ESO‐1, MAGE‐A3, tyrosinase, TPTE; BNT111 | Cemiplimab (i.v. infusion) | i.v. | 2023/12 | |
| NCT04382898 | Prostate cancer | Recruiting | I/II | TAAs: RBL038/RBL039/RBL‐040/RBL‐041/RBL‐045; BNT‐112 | Cemiplimab (i.v. infusion) | i.v. | 2023/7 | |
| NCT00204516 | Malignant melanoma | Completed | I/II | TAAs: Melan‐A/ Mage‐A1/Mage‐A3/Survivin/GP100 /Tyrosinase; mRNA coding for melanoma‐associated antigens | GM‐CSF (s.c.) | s.c. | 2012/7 | |
| NCT03164772 | Metastatic NSCLC; NSCLC | Completed | I/II | TAAs:NY‐ESO‐1/MAGE‐C2/MAGE‐C1/survivin/5T4/MUC1; BI 1361849 | Durvalumab + tremelimumab | Null | 2021/10/29 | |
| mRNA encoding neoantigens | NCT03289962 | Melanoma; NSCLC; bladder cancer; COAD; TNBC | Active, not recruiting | I | Neo‐Ag (mRNA); autogene cevumeran | Atezolizumab (i.v. infusion) | i.v. | 2024/2/1 |
| NCT03313778 | Solid tumors | Recruiting | I | Neo‐Ag (mRNA); mRNA‐4157 | Pembrolizumab (i.m. injection) | i.m. | 2022/6/30 | |
| NCT04161755 | Pancreatic cancer | Active, not recruiting | I | Neo‐Ag (mRNA); RO7198457 | Atezolizumab + mFOLFIRINOX | Null | 2023/11/11 | |
| NCT03948763 | Pancreatic neoplasms; colorectal neoplasms; carcinoma, NSCLC | Active, not recruiting | I | KRAS mutations: G12D/G12V/G13D/G12C; mRNA‐5671/V941 | Pembrolizumab (i.v. infusion) | i.m. | 2022/8/12 | |
| NCT04397926 | NSCLC | Recruiting | I | Neo‐Ag (mRNA) | Null | s.c. | 2022/5 | |
| NCT04487093 | NSCLC | Recruiting | I | Neo‐Ag (mRNA) | EGFR‐TKI+ anti‐angioge | s.c. | 2022/12/1 | |
| NCT02035956 | Melanoma | Completed | I | Neo‐Ag (mRNA):IVAC MUTANOME; NY‐ESO‐1/tyrosinase/personalized neoantigen petide | RBL001/RBL002 | i.n. | 2019/10 | |
| NCT03597282 | Metastatic Melanoma | Terminated | I | Neo‐Ag (mRNA):NEO‐PV‐01 | Nivolumab (i.v.) + Adjuvant + APX005M + ipilimumab | s.c. | 2020/8/11 | |
| NCT03166254 | NSCLC | Withdrawn | I | Neo‐Ag (mRNA):NEO‐PV‐01 | Pembrolizumab + Poly ICLC | s.c. | 2027/5 | |
| NCT03468244 | Advanced ESC; GAC; PAAD; COAD | Recruiting | I | Neo‐Ag (mRNA) | Null | s.c. | 2021/12/31 | |
| NCT02316457 | TNBC | Active, not yet recruiting | I | IVAC_W_bre1_uID/IVAC_M_uID | Null | Null | 2023/12 | |
| mRNA encoding neoantigens | NCT04163094 | Ovarian Cancer | Recruiting | I | W_ova1 | (Neo‐)Adjuvant Chemotherapy | i.v. | 2023/12 |
| NCT03380871 | Carcinoma, NSCLC; Nonsquamous NSCLC | Completed | I | Neo‐Ag (mRNA):NEO‐PV‐01 | Pembrolizumab + Adjuvant + Carboplatin + Pemetrexed | s.c. | 2021/2/5 | |
| NCT02897765 | UBC; Bladder Tumors; TCC of the Bladder; Melanoma; Skin Cancer; NSCLC | Completed | I | Neo‐Ag (mRNA):NEO‐PV‐01 | Adjuvant + Nivolumab (i.v.) | s.c. | 2020/5 | |
| NCT03815058 | Advanced Melanoma | Active, not recruiting | II | Neo‐Ag (mRNA); RO7198457 | Pembrolizumab (i.v. infusion) | i.v. | 2022/9/1 | |
| NCT03897881 | Melanoma | Active, not recruiting | II | Neo‐Ag (mRNA); mRNA‐4157 | Pembrolizumab (i.v. infusion) | Null | 2024/6/30 | |
| NCT04267237 | NSCLC | Withdrawn | II | Neo‐Ag (mRNA); RO7198457 | Atezolizumab | i.v. | 2025/9/30 | |
| NCT04486378 | Colorectal Cancer Stage II/III | Recruiting | II | Neo‐Ag (mRNA); RO7198457 | Null | i.v. | 2027/7 | |
| NCT03480152 | Melanoma; COAD; GAC; genitourinary cancer; LIHC | Terminated | I/II | Neo‐Ag (mRNA) | Null | i.m. | 2019/11/5 | |
| NCT03908671 | Esophageal cancer; NSCLC | Not yet recruiting | NA | Neo‐Ag (mRNA) | Null | s.c. | 2022/9 | |
| mRNA encoding immunostimulants | NCT03788083 | Breast cancer; early‐stage breast cancer | Recruiting | I | Trimix mRNA; mRNA encoding CD40L, CD70, acTLR4 | Null | i.t. | 2022/12/30 |
| NCT03394937 | Melanoma | Recruiting | I | ECI‐006;mRNA encoding tyrosinase, gp100, MAGE‐A3, MAGE‐C2, PRAME | Null | i.n. | 2021/1/29 | |
| NCT03291002 | Melanoma (Skin); SCC; HNSCC; ADCC | Active, not yet recruiting | I | CV8102; mRNA encoding TLR‐7,8 and RIG‐1 | anti‐PD‐1 therapy | Null | 2023/2 | |
| NCT03946800 | Solid Tumors | Recruiting | I | MEDI1191; mRNA encoding IL‐12 | Durvalumab (i.v.) | i.t. | 2027/1/15 | |
| NCT03739931 | TNBC; HNSCC; NHL; Melanoma; NSCLC | Recruiting | I | mRNA‐2752; mRNA encoding OX40L, IL‐23, IL36γ | Durvalumab | i.t. | 2023/1/30 | |
| NCT03871348 | Metastatic neoplasm | Recruiting | I | SAR441000; mRNA encoding IL‐12sc, IL‐15sushi, IFN α, GM‐CSF | Cemiplimab REGN2810 (i.v.) | i.t. | 2024/4 | |
| NCT03323398 | Solid tumor; ovarian cancer | Recruiting | I/II | mRNA‐2416; mRNA encoding OX40L | Durvalumab (i.v.) | i.t. | 2022/9/20 | |
| NCT04455620 | Solid tumor | Recruiting | I/II | BNT151; mRNA encoding IL‐2 | Null | i.v. | 2025/1/1 |
Note: The table summarizes the currently clinical trials with mRNA tumor vaccines registered at Clinical Trials.gov.
Abbreviations: ADCC, adenoid cystic arcinoma; CD40L, CD40 ligand; CEA, carcinoembryonic antigen; COAD, colon adenocarcinoma; ESC, esophageal squamous carcinoma; GAC, gastric adenocarcinoma; HNSCC, head and neck squamous cell carcinoma; i.m., intramuscular injection; i.n., intranodal injections; i.t., intratumoral injection; i.v., intravenous injection; LIHC, liver hepatocellular carcinoma; LNP, Lipid nanoparticles; mRNA, messenger ribonucleic acid; NA, not available; Neo‐Ag, neoantigen; NHL, non‐Hodgkin's lymphoma; NSCLC, non‐small cell lung cancer; OX40L, OX40 ligand; PAAD, pancreatic adenocarcinoma; PD‐L1, programmed death ligand‐1; RIG‐1, retinoic acid induced genes 1; s.c.,subcutaneous injection; SCC, squamous cell carcinoma; STES, stomach and esophageal carcinoma; TAAs, tumor‐associated antigens; TCC, transitional cell carcinoma of the bladder; TNBC, triple negative breast cancer; UBC, urinary bladder cancer.
TABLE 4.
Currently clinical trials with mRNA transfected dendritic cells tumor vaccines
| NCT number | Cancer type | Phases | mRNA encoding for | Interventions | Status | Route | Completion |
|---|---|---|---|---|---|---|---|
| NCT00639639 | Malignant neoplasms of brain | I/II | cmvpp65 | Tetanus toxoid; therapeutic autologous dendritic cells; herapeutic autologous lymphocytes | Active, not recruiting | i.d. | 2019/12 |
| NCT03615404 | Glioblastoma; malignant glioma; medulloblastoma recurrent; pediatric glioblastoma multiforme; pediatric brain tumor | I | pp65 | CMV‐DCs with GM‐CSF; Td (tetanus toxoid) | Completed | Null | 2020/7/2 |
| NCT02709616 | Glioblastoma | I | Multiple TAAs | Personalized cellular vaccine | Completed | i.d./i.v. | 2020/6 |
| NCT02808416 | Brain Cancer; Neoplasm Metastases | I | Multiple TAAs | Personalized cellular vaccine | Completed | Null | 2020/9/1 |
| NCT03334305 | Malignant glioma; high grade glioma | I | Autologous tumor‐mRNA | TTRNA‐DC vaccines with GM‐CSF; Dose‐intensified TMZ; autologous hematopoietic stem cells (HSCs); TTRNA‐xALT; Td vaccine | Recruiting | i.d. | 2026/5 |
| NCT03396575 | Diffuse intrinsic pontine glioma (DIPG); brain stem glioma | I | Autologous tumor‐mRNA | TTRNA‐DC vaccines with GM‐CSF; TTRNA‐xALT; cyclophosphamide + fludarabine lymphodepletive conditioning; dose‐intensified TMZ; Td vaccine; autologous hematopoietic stem cells (HSC) | Recruiting | i.d. | 2024/6 |
| NCT01456104 | Melanoma | I | tyrosinase‐related peptide 2 (TRP2) | Langerhans‐type dendritic cells (a.k.a. Langerhans cells or LCs) | Active, not recruiting | i.d. | 2019/10/1 |
| NCT01995708 | Multiple myeloma | I | CT7; MAGE‐A3; WT1 | CT7, MAGE‐A3, and WT1 mRNA‐electroporated langerhans cells (LCs)/Standard of care | Active, not recruiting | i.d. | 2019/11/1 |
| NCT03927222 | Glioblastoma | II | cmvpp65 | Human CMV pp65‐LAMP mRNA‐pulsed autologous DCs containing GM‐CSF; temozolomide; tetanus‐diphtheria toxoid (Td); GM‐CSF; 111‐Indium‐labeling of cells for in vivo trafficking studies | Suspended | i.d. | 2023/12/1 |
| NCT03688178 | Glioblastoma | II | cmvpp65 | Human CMV pp65‐LAMP mRNA‐pulsed autologous DCs; temozolomide; varlilumab; Td; 111In‐labeled DCs; unpulsed DCs; HIV‐Gag mRNA‐pulsed autologous DCs | Recruiting | i.d. | 2025/3/1 |
| NCT02465268 | Glioblastoma multiforme; malignant glioma; astrocytoma, grade IV | II | pp65 | pp65‐shLAMP DC with GM‐CSF; unpulsed PBMC and saline; Td; saline; pp65‐flLAMP DC with GM‐CSF | Recruiting | i.d. | 2024/6 |
| NCT02692976 | Prostatic Neoplasms | II | NY‐ESO‐1; MUC1; PepTivator | mDC vaccination; pDC vaccination; mDC and pDC vaccination | Completed | i.t. | 2019/3/6 |
| NCT03083054 | Myelodysplastic syndromes; AML | I/II | WT1 | Autologous dendritic cells electroporated with WT1 mRNA | Active, not recruiting | Null | 2020/7/1 |
| NCT02649829 | Malignant pleural; mesothelioma | I/II | WT1 | Dendritic cell vaccination plus chemotherapy | Recruiting | i.d. | 2021/11 |
| NCT04911621 | Glioma; diffuse intrinsic pontine glioma | I/II | WT1 | Dendritic cell vaccination + temozolomide‐based chemoradiation; Dendritic cell vaccination ± conventional next‐line treatment | Recruiting | i.d. | 2027/6 |
| NCT02649582 | Glioblastoma multiforme of brain | I/II | WT1 | Dendritic cell vaccine plus temozolomide chemotherapy | Recruiting | i.d. | 2022/12 |
| NCT01885702 | Colorectal cancer | I/II | CEA | DC vaccination | Active, not recruiting | Null | 2019/6 |
| NCT04567069 | Gastric cancer | I/II | MG‐7 | MG‐7‐DC vaccine; CTL; Sintilimab Injection | Recruiting | s.c. | 2022/6 |
| NCT02528682 | Hematological malignancies | I/II | minor histocompatibility antigens (MiHA) | MiHA‐loaded PD‐L‐silenced DC vaccination | Completed | i.v. | 2019/7/1 |
| NCT01334047 | Recurrent epithelial; ovarian cancer | I/II | survivin;hTERT; cancer stem cell mRNA | DC‐006 vaccine | Terminated | Null | 2022/4 |
| NCT01197625 | Prostate cancer | I/II | survivin; hTERT; cancer stem cell mRNA | Dendritic cell vaccine | Active, not recruiting | Null | 2025/9 |
| NCT03548571 | Glioblastoma | II/III | survivin;hTERT; cancer stem cell mRNA | Dendritic cell immunization; adjuvant temozolomide | Recruiting | i.d. | 2023/5 |
| NCT05000801 | Acute myeloid leukemia | NA | WT1; hTERT; survivin | DC vaccine (dendritic cells loaded with tri‐antigens (WT1/hTERT/survivin) | Recruiting | Null | 2026/7 |
Note: The table summarizes the currently clinical trials with mRNA transfected dendritic cells tumor vaccines registered at Clinical Trials.gov.
Abbreviations: AML, acute myeloid leukemia; CEA, carcinoembryonic antigen; CMV,cytomegalovirus; CTL, cytotoxic T lymphocyte; DC, dendritic cell; GM‐CSF, granulocyte‐macrophage colony stimulating factor; i.d., intradermal injection; i.t., intradermal injection; i.v., intravenous injection; MG‐7, Ag monoclonal gastric cancer 7 antigen; mRNA, messenger ribonucleic acid; NA, not available; s.c.,subcutaneous injection; TTRNA‐DC: total tumor RNA‐dendritic cell.
3. IN VIVO DELIVERY SYSTEMS FOR mRNA VACCINES
The selection of an optimal delivery system for mRNA vaccines remains one of the greatest challenges in their development. Intratumoral injection of naked mRNA can be used for cancer treatment while showing an appropriate adjuvant effect. 106 However, due to the extremely low uptake of naked mRNA, this drug delivery system is often supplemented to reinforce the stability and efficiency of mRNA in vivo delivery. In addition, mRNA molecules are large, hydrophilic, and carry negative charges, and electrostatic repulsion makes them thermodynamically unfavorable to diffuse across the plasma membrane, which is also negatively charged. 107 , 108 , 109 Once entering the body, mRNA will be further challenged by endogenous nucleases for degradation. Therefore, having a nontoxic and efficient delivery system is of critical importance to the success of mRNA vaccines. An ideal mRNA delivery system should not only enhance cellular uptake and expression of mRNA by target cells, but also protect them from nuclease degradation. 99 , 100 , 110
At present, there is a large number of mRNA delivery systems being reported, many of which have already entered clinical trials. Mature delivery systems include LNP 84 , 111 , 112 and protamine carriers, 113 , 114 , 115 while other less popular attempts include polyethylenimine, 116 cell‐penetrating peptides, 117 lipopolyplexes, 118 cationic nanoemulsions, 119 and exosomes. 120 , 121 LNPs and other synthetic nanoparticles are foreign substances, which can cause related side effects ranging from mild to severe. Exosomes are naturally occurring, nano‐sized extracellular vesicles with low immunogenicity and high safety. In addition, exosomes can penetrate physiological barriers that synthetic nanoparticle carriers cannot. These characteristics make exosomes an ideal delivery carrier for mRNA. 122 The challenges of using exosomes as carriers of mRNA delivery currently lie in large‐scale GMP‐grade production and the encapsulation efficiency of exosomes. 8 , 123 The three large‐scale developers of mRNA vaccines—Moderna, CureVac, and BioNTech—invested heavily in the LNP delivery system in the development of mRNA vaccines, rendering LNP the most prevalent nonviral delivery system for nucleic acid drugs.
3.1. LNP
LNP is a nonviral carrier that is regarded as nontoxic and safe. The main components of LNP include PEGylated lipids, cholesterol, neutral helper lipids, and ionizable cationic lipids (Figure 4). 108 , 124 PEG locates on the surface of LNP. It prevents the interaction of LNP with other lipid particles or serum components, 125 which blocks LNP from aggregating and being phagocytosed by immune cells. Cholesterol allows strong membrane fusion, which facilitates the entry of mRNA into the cytoplasm. 126 The neutral helper lipids are, in general, saturated phospholipids. 127 They increase the phase transition temperature of cationic liposomes, which support the formation of a lamellar lipid bilayer and stabilize its structure. 128 The ionizable cationic phospholipids, the most critical excipient in LNP composition, determine the efficiency of mRNA delivery and transfection. 129 They play a critical role in the intellectual property protection of the current LNP delivery systems. 130 For example, the ionizable cationic lipids used in the Pfizer‐BioNTech and Moderna COVID‐19 mRNA vaccines (BNT162b2 and mRNA‐1273) are [(4‐hydroxybutyl)azanediyl]di(hexane‐6,1‐diyl) bis(2‐hexyldecanoate) (ALC‐0315) and SM‐102, 51 respectively; the siRNA drug Onpattro uses Dlin‐MC3‐DMA. 131 Microfluidic technology based on the organic solvent injection method to prepare LNPs can accurately control the size of LNPs, and this represents the most suitable preparation technology for LNPs at present. 132
FIGURE 4.
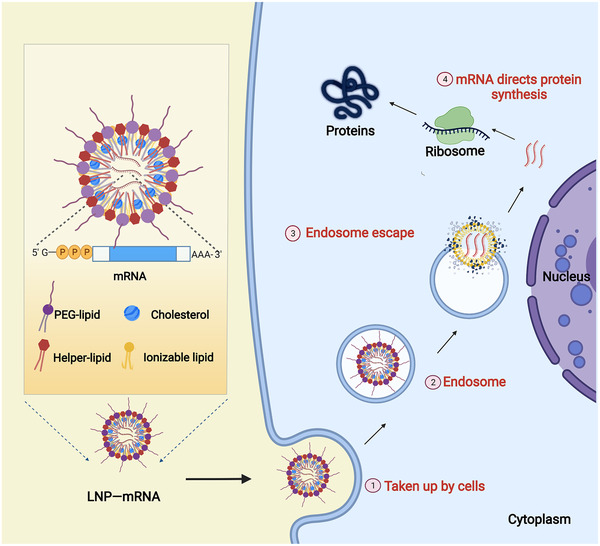
Schematic diagram of the typical structure of messenger ribonucleic acid (mRNA)‐LNP and in vivo delivery. In an acidic environment, the cationic LNP can form a complex with nucleic acids via electrostatic interaction. In the neutral environment, the formula becomes neutrally charged and thereby interacts less with serum components. Once mRNA‐LNP reaches the cell membrane, cationic phospholipids fuse with and destabilize the cell membrane, promoting the delivery of mRNA molecules. After being internalized into the cell, the mRNA‐LNP is engulfed by the endosome. The endosomal environment acidifies the ionizable phospholipids, allowing fusion with the negatively charged primary lysosomal membrane. LNP integrity is disrupted by this interaction, and therefore mRNA is released. Membrane fusions and structural changes in LNPs are thought to be the main causes of endosomal membrane destabilization and mRNA escape.
Although mRNA‐LNP vaccines have shown unprecedented potential, LNP delivery technology still has potential limitations, which hinder practical application, such as induction of allergic reactions, easy oxidative degradation, and poor preparation reproducibility. 94 , 112 , 133 , 134 , 135 , 136 The biggest concern is that LNP constituents may increase the risk of allergic reactions. A series of LNP‐triggered allergic reactions is currently considered to be related to several factors. First among them is the strong immunogenic nature of ionizable cationic phospholipids. 94 , 137 For example, after injection of the Pfizer‐BioNTech or Moderna COVID‐19 mRNA vaccines, various symptoms including swelling, pain, chills, and fever have been reported. 138 , 139 , 140 These are suspected to be the consequence of the production of pro‐inflammatory cytokines such as IL‐1β and IL‐6. 141 , 142 , 143 Second, an increasing number of studies has verified the immunogenicity of PEG; therefore, repeated administration of PEG can induce allergic reactions. 143 , 144 In addition, the hypersensitivity reactions observed after mRNA‐1273 and BNT162b2 administration are likely to be attributed to the formation of anti‐PEG antibodies after the first vaccination. 145 , 146 Fortunately, it has been reported that alternative lipids can be involved to prevent aggregate formation. For example, polysarcosine‐modified lipids can stabilize the LNP delivery system and prevent aggregate formation with no allergic reactions yet observed. 147
3.2. Protamine
Protamine is a natural cationic polypeptide mixture and exhibits two main advantages as an mRNA delivery vehicle: first, due to its high positive charge, 114 it can spontaneously complex with negatively charged mRNA, thereby protecting mRNA from serum nuclease degradation. Second, protamine also acts as an adjuvant. 66 , 148 , 149 The protamine‐mRNA complex is recognized by immune cells via the TLR‐7/TLR‐8 pathway and activates a strong immune response to secrete type I interferon, TNF‐α, IL‐12, and other cytokines. 115 , 150 , 151 , 152 , 153 , 154 , 155 Protamine‐mRNA nanoparticles of different sizes have different stimulatory effects on immune cells. For example, larger particles mainly activate monocytes and promote the production of TNF‐α, while particles smaller than 450 nm effectively stimulate plasmacytoid DCs to release IFN‐α. 152 , 156 To meet varying research needs, it was discovered that by adjusting the protamine to mRNA ratio and the salt concentration of the dilution solution, protamine‐mRNA can be prepared into particles with average diameters ranging from 50 to 1000 nm. 157
CureVac rabies vaccine (CV7201) uses protamine as a delivery vehicle. CV7201 is made up of protamine and mRNA that encodes rabies virus glycoprotein (RABV‐G). 158 In addition, CureVac also established an RNActive platform containing a protamine delivery vector. Based on RNActive, 159 CureVac carried out clinical trials for melanoma, prostate cancer (CV9104), and nonsmall cell lung cancer (NSCLC) (CV9201 and CV9202). 160 , 161 In the phase IIb clinical trial of the CV9104 vaccine (NCT01817738), the overall survival (OS) of patients with metastatic castration‐resistant prostate cancer was not significantly improved compared with the placebo control group. 162 Clinical trials for CV9201 and CV9202 also failed to show significant efficacy, which may be a result of tumor‐induced immunosuppression. Therefore, it has been proposed to combine mRNA vaccines with immune checkpoint inhibitors in future studies. Based on these results, CV9202 is undergoing a phase 1/2 study of combination immunotherapy and mRNA vaccine in subjects with NSCLC. The vaccine, administrated by intradermal injection using a needle‐free jet injector, is used in combination with the anti‐PD‐L1 antibody (Durvalumab) and/or the anti‐CTLA4 antibody (Tremlimumab) (NCT03164772). 66
Currently, protamine‐formulated mRNA delivery systems result in limited mRNA translation efficiency and immunity strength. Compared with LNP, lower transfection efficiency of protamine may arise from its hydrophilicity, which makes it difficult to permeate the cell membrane and escape from endosome. 114 , 163 , 164 Adding endosome destabilizing agents or adjusting the protamine to DOTAP (1,2‐Dioleoyl‐3‐trimethylammonium‐propane, a cationic lipids) ratio is expected to improve the transfection efficiency of protamine‐mRNA complexes. 165
3.3. Tissue and cell targeted delivery systems
For mRNA therapy, it is absolutely critical that the mRNA is delivered to the right place in the body to have the desired therapeutic effect. This not only enhances the curative effect but also reduces off‐target effects. However, the liver has been found to be a natural site for accumulation upon systemic administration of mRNA‐LNP. 166 , 167 In order to promote the efficacy of mRNA, the delivery of mRNA to specific organs, tissues, and cells needs to be resolved.
Targeted delivery of mRNA to the spleen is beneficial for vaccines and immunotherapy because there are a large number of immune cells in this organ. It was previously reported that using intravenously‐administered positively charged RNA‐lipoplexes (RNA‐LPX) (lipid to RNA charge ratio of 5:1) targeted Luc expression predominantly in the lungs of mice. After optimally adjusting the RNA‐LPX formulation with a charge ratio of 1.3:2, which effectively targeted RNA to the spleen, LPX mediated mRNA efficient uptake and expression of the encoded antigen by DC populations and macrophages. 168
Selective organ targeting (SORT) nanoparticles are in the development for mRNA delivery to nonliver tissues. LNPs are usually composed of ionizable cationic lipids, amphiphilic phospholipids, cholesterol, and poly(ethylene glycol) lipids. SORT molecules are the fifth component added to the traditional four‐component LNPs, with the added SORT molecules controlling biodistribution, global/apparent pKa, and serum protein interactions of the SORT nanoparticles. With the help of SORT molecules, LNPs can specifically target the lungs, the spleen, and the liver, and effect mRNA delivery and gene editing of related cells, such as epithelial cells, endothelial cells, B cells, T cells, and hepatocytes. 41 , 169
4. mRNA VACCINES FOR PREVENTION OF INFECTIOUS DISEASES
Infectious diseases are transmissible diseases caused by infection by pathogens, such as bacteria and viruses. The first active defense of host against those pathogens is innate immunity, which is usually evoked by PRR recognition of pathogen‐associated molecular patterns (PAMPs). PAMPs are a series of conserved and highly abundant molecular motifs within a class of pathogens. Innate immunity utilizes various mechanisms to combat invading pathogens, such as inflammation, engulfment, and the complement system. Later, adaptive immunity, elicited by antigen presentation in an inflammatory milieu, reacts specifically to antigens of pathogens by secreting antibodies and activating cytotoxic lymphocytes. Effective innate immunity creates the optimal microenvironment to foster a potent adaptive immunity, or it will otherwise be too weak and too late to prevail over pathogens in cases of serious infections. However, prophylactic vaccination is able to boost a strong adaptive defense and adaptive memory before infection takes place, achieving enduring immunity against relevant pathogens. mRNA vaccines encoding pathogen antigens are an active family of vaccines for controlling infectious diseases. 1 , 170
4.1. COVID‐19 mRNA vaccines
The COVID‐19 pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has more than 447 million cases and six million deaths globally as of March 8, 2022. It has devastating effects on health, as well as social and economic situations. Two mRNA vaccines were released after a short period of development. BNT162b2, under the brand name Comirnaty, and mRNA‐1273, under the brand name Spikevax, were developed by Pfizer–BioNTech and Moderna, respectively. They were authorized for emergency use in December 2020 and have since made considerable contributions to infection control.
4.1.1. Design of the COVID‐19 mRNA vaccines
The two approved COVID‐19 mRNA vaccines, BNT162b2 and mRNA‐1273 (see Table 1), 171 both contain mRNAs encoding the SARS‐CoV‐2 spike (S) protein, a 1273 amino acid long type I transmembrane glycoprotein that dictates the infectivity of the virus. One common feature is that they both harbor two substitutions of amino acids, namely K986P and V987P, in the peptide chain to stabilize the prefusion conformation of the S protein. 112 , 172 In addition, they both use N1‐methyl‐pseudouridine, instead of uridine, as a substrate to limit innate immune responses to mRNAs. However, the capping strategies differ. Capping for BNT162b2 is fulfilled co‐transcriptionally with the previously described CleanCap cap analogues, 84 , 173 while capping for mRNA‐1273 is achieved by post‐transcription reactions catalyzed by vaccinia capping enzyme and 2′ O‐methyltransferase as discussed above. 84 , 174
Other modifications of the S antigen were also investigated. In one study, wild‐type S antigen versions with two proline substitutions (2P), four additional proline substitutions (6P), furin cleavage site elimination, and endoplasmic reticulum retention signal elimination were compared for their capacity to induce neutralizing antibodies. The S antigen with both 2P and furin cleavage site elimination outperformed the others, inducing Th1‐biased responses in both the mouse and nonhuman primate models. 175 In another study, glycosylation site depletion in the receptor‐binding domain or the S2 domain of the S antigen exposed more conserved epitopes, and the mRNA vaccine encoding this type of S protein conferred broad protection against wild‐type virus and different variants of concern. The resulting misfolded protein product induced cell apoptosis and potent T cell responses. 176
Although the S antigen is the predominant target for COVID‐19 mRNA vaccines, other structural and nonstructural genes have been proposed as targets. 177 , 178 , 179 , 180 , 181 , 182 , 183 , 184 These genes are not under constant high selection pressure and are thus more conserved. Structural gene encoding NP (N), which is indispensable in the viral life cycle, was shown to elicit protective immune responses. 177 , 180 Nonstructural genes, although not present in viral particles, can also be exploited to mark the infected cells for destruction. Studies have suggested that nonstructural protein 3 (Nsp3), Nsp8, and Proteinase 3CL‐PRO could serve as potential targets. 181 , 182 , 183
4.1.2. Mechanisms of action of COVID‐19 mRNA vaccines in humans
mRNA vaccines can induce adaptive immunity against specific antigens. In the case of COVID‐19 mRNA vaccines, while the induction of neutralizing antibodies, which inactivate the live viruses, is currently a major concern, the roles of T cell immunity in the process are becoming more appreciated. 185
Analyses on vaccinated individuals revealed that, after the first dose, COVID‐19 mRNA vaccines elicited rapid Th1 and follicular helper T (Tfh) cell responses, which positively correlated with the level of follow‐up CD8+ T cells and neutralizing antibodies after the second dose. Importantly, CD4+ and CD8+ T cells generated by mRNA vaccination exhibited memory characteristics, which could be pivotal to recall responses for future infection. 4 , 186 In another study, by directly probing GC responses in the lymph nodes (LNs) of vaccinees, a connection between GC formation and the generation of neutralizing antibodies and memory B cells after COVID‐19 mRNA vaccination was proposed. 187 It was demonstrated in a recent study that, after COVID‐19 mRNA vaccination, GCs in LNs were robustly developed, and vaccine mRNA and the resulting S antigen were present in the GCs for a prolonged period of time. Nine months after vaccination, the titre of spike‐specific IgG dropped to about 1/20 of the peak level; however, a third‐dose booster managed to generate another new peak of antibody titer within 1 week. 188 Several other studies also corroborated a waning of immune responses against COVID‐19 over time, arguing for the necessity of further boosters to consolidate long‐term immunity. 19 , 189 , 190 , 191 , 192 , 193
Mucosal immunity, which directly responds to viral challenges, is also a concern. Disappointingly, in contrast to systemic immunity, mucosal immunity was poorly activated following standard intramuscular administration. 194 In light of this, a mucosal booster strategy was proposed to elevate mucosal immunity against COVID‐19. 195
4.1.3. Real‐world efficacy and strategies of COVID‐19 mRNA vaccines
Data from real‐world observational studies have been accumulating, as mass vaccination programs have been implemented worldwide. 196 A general study on mRNA vaccines based on health care staff, first responders, and other frontline workers demonstrated vaccine effectiveness of 91% for those who received two doses, and 81% for those who had only 1 dose. Moreover, for those who contracted the virus, the vaccinated group benefited by experiencing a 40% reduction in viral load, a 58% lower risk of febrile symptoms, and 2.3 days less sickbed time. 197
In one study conducted by Israel's largest health care organization on the BNT162b2 vaccine, by exploring the data collected at days 14 through 20 after the first dose and at day 7 or more days after the second dose, the vaccine had effectiveness rates of 46% and 92% for documented infection, 57% and 94% for symptomatic COVID‐19, 74% and 87% for hospitalization, and 62% and 92% for severe disease. Importantly, estimated mortality avoidance rate was 72% at days 14 through 20 after the first dose. 198 Another study was carried out in a large health maintenance organization in Israel, with individuals who frequently underwent polymerase chain reaction testing for SARS‐CoV‐2 infection. For the prevention of asymptomatic SARS‐CoV‐2 infection, the BNT162b2 vaccine showed an estimated effectiveness of 89% 7 days after two doses, in contrast to 61% 2 weeks after 1 dose. 199 A third study involved health‐care workers in England, who also underwent regular asymptomatic testing. The effectiveness of BNT162b2 vaccine was 70% 21 days after the first dose and 85% 7 days after the second dose. 200 A more recent study confirmed the effectiveness of the BNT162b2 vaccine in a large health provider cohort in Israel, achieving a protection rate of 90% and 94% against SARS‐Cov‐2 infection and COVID‐19 after two doses, respectively. Results from immunosuppressed patients were also impressive, reaching 71% protection against infection. 201
Recently published results for the two‐dose mRNA‐1273 vaccines were also encouraging. mRNA‐1273 provided protection against COVID‐19 infection (87.4%), COVID‐19 hospitalization (95.8%), and hospital mortality (97.9 %). Effectiveness against symptomatic COVID‐19 was 88.3%, compared to 72.7% against asymptomatic cases. A moderate effectiveness of 8.2–33.6% was observed among individuals with COVID‐19 history, suggesting the need for vaccination of those who have recovered from the disease. 202
To date, there have been five SARS‐CoV‐2 variants of concern: alpha (B.1.1.7 and descendant lineages), beta (B.1.351), gamma (P.1), delta (B.1.617.2 and AY lineages), and omicron (B.1.1.529 and BA lineages), with each causing a surge in cases and mortalities. 203 Reduced neutralizing activity by vaccines against these variants was observed, most notably for the latest variants, Delta and Omicron. 204 , 205 , 206 , 207 Fortunately, strategies such as sequential immunization and additional boosters have shown promise for limiting the breakthrough infection, hospitalization, severe disease, and death caused by SARS‐CoV‐2. 26 , 208 , 209 , 210 , 211 In addition, there are also suggestions for alternative strategies, such as constantly updating vaccines with newly prevalent variant sequences, polyvalent vaccines, or pan‐coronavirus strategies. 212
A recent real‐world study in Qatar revealed that two doses of an mRNA vaccine achieved peak protection rates of 41.6% and 51.7% against symptomatic infection by Omicron subvariants BA.1 and BA.2 3 months after the last dose, before dropping to 10% or lower. 196 Similar waning protection was also observed for the third dose, where moderate peak protection rates of 59.9% and 43.7% against infection by the two subvariants, respectively, were achieved 1 month after the last dose. Encouragingly, two doses of an mRNA vaccine reduced COVID‐19 hospitalization and death by 70%–80%, and a third dose further reduced the rate by >90%. 196
These data establish that the COVID‐19 mRNA vaccines currently in use are substantially efficacious. Nevertheless, further improvements are needed to combat the constantly evolving virus.
4.2. mRNA vaccines for other infectious diseases
Clinical trials of multiple mRNA vaccines targeting other infectious diseases, such as influenza, 175 , 213 Zika virus, 18 , 22 , 214 and rabies 215 , 216 are underway. However, the development of these vaccines is not comparable to the rate of progress that development of the SARS‐CoV‐2 vaccine has achieved (Table 2).
5. mRNA CANCER VACCINES
Although the COVID‐19 pandemic has accelerated the application of mRNA vaccines against viral infectious diseases, mRNA cancer vaccines were the earliest direction assessed in exploratory clinical trials, and remain a field with the most intense competition and the most concentrated R&D pipelines. 217
There are two types of cancer vaccines with fundamentally different working principles: prevention vaccines and therapeutic vaccines (Figure 5). It is estimated that 15% of all human cancers are associated with viral infection (e.g., human papillomavirus, hepatitis B virus, and others). 218 , 219 , 220 , 221 , 222 Therefore, vaccines that protect against certain viruses serve as prophylactic measures against certain cancers. In contrast, therapeutic vaccines address existing cancers. The mRNA‐based vaccine allows the use of vaccines as an available immunotherapy for cancer and other nonviral diseases. Most cancer vaccines fall into the therapeutic category. In general, they target tumor‐associated antigens (TAA), which are preferentially expressed in cancerous cells. 8
FIGURE 5.
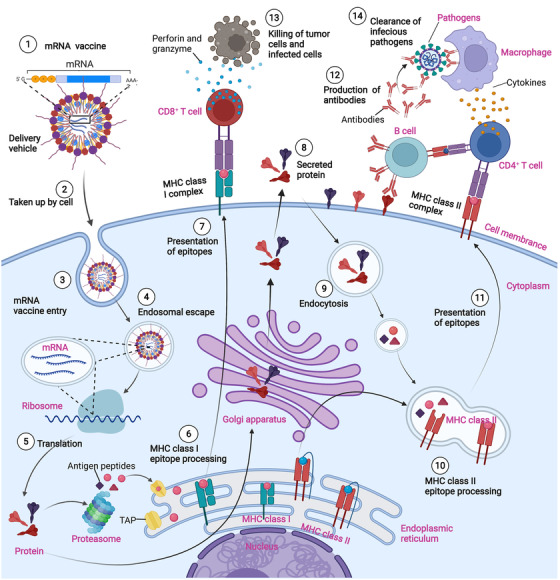
Mechanisms of messenger ribonucleic acid (mRNA) vaccines for infectious diseases and cancers. mRNA molecules encoding tumor antigens are injected into body (either with or without delivery vehicles). The mRNA molecules are taken up and translated into protein antigens by antigen presenting cells (APCs). After proteasomal processing of proteins, antigen peptides associate with major histocompatibility complex (MHC) Class I molecule in the endoplasmic reticulum and are transferred to the APC surface, activating CD8+ T cells for a specific cellular immune response. Protein antigens, which are sorted for the endosome route, can activate CD4+ T cells via the MHC Class II presentation pathway. The secretory protein antigen or membrane antigen encoded by mRNA can stimulate B cells to produce neutralizing antibodies, and activate phagocytes such as macrophages to secrete inflammatory cytokines, facilitating the clearance of circulating infectious pathogens and tumor cells
Currently, mRNA tumor vaccines are applied in two ways. One directly delivers mRNA into the body using delivery vehicles such as LNP or protamine, 223 , 224 , 225 and the other ex vivo transfects DCs with tumor antigen‐encoding mRNA to prepare a DC vaccine. 226 , 227 In addition, although mRNA encoded immunostimulants are not strictly part of tumor vaccines, their ability to turn cold tumors, with no or low immune response, into well‐responsive hot tumors has attracted attention. Consequently, many clinical trials have attempted to enhance antitumor efficacy 228 , 229 , 230 (Table 3).
The development of a therapeutic cancer vaccine is a research focus for many: using “cancer vaccine” as keywords in ClinicalTrials.gov results in more than 2000 clinical registered projects; however, this goal remains extremely challenging. After huge investments of human and financial resources, many blockbusting cancer vaccines failed in phase III trials, such as Stimuvax (Merck, USA), which targets the tumor antigen mucin (MUC1), 231 and the lead product of GlaxoSmithKline, GSK1572932A, 232 which targets melanoma‐related antigen 3 (MAGE‐A3). 233 , 234 These vaccines target a single TAA; however, for the development of mRNA tumor vaccines, simultaneous targeting of multiple TAAs or neoantigens is more promising (Table 3).
5.1. mRNA vaccines simultaneously encoding multiple TAAs
Simultaneous targeting of multiple TAAs reduces the chance of tumor antigen escape due to mutations and low expression of target antigens and thus increases the robustness and precision of specific antitumor responses. 235 , 236 , 237 The majority of ongoing clinical trials employ this strategy. More than 90% of melanoma patients express at least one of the four prevalent TAAs: NY‐ESO‐1, MAGE‐A3, Tyrosinase, and TPTE. 8 , 238 BNT111 is a lead candidate from the BioNTech FixVAC platform, which is experiencing the fastest research and developmental progress, and encodes all four TAAs of melanoma. According to a completed phase I clinical trial (NCT02410733), most melanoma patients given this vaccine that showed stable disease (SD) and partial responses have achieved ongoing survival. Some patients have survived for more than 2 years with TAA‐specific T cells still remaining at a detectable level in their body. 239 Based on the safety and preliminary efficacy of BNT111 demonstrated in that phase I trial, a phase II clinical trial of BNT111 in combination with Libtayo (Cemiplimab) was initiated in May 2021(NCT04526899), which targets patients with unresectable anti‐PD1‐refractory/relapsed melanoma. 240
More DNA mutations can produce more candidate peptides and lead to an increase in tumor antigens that can be presented. Several studies have demonstrated that tumor patients with a high tumor mutational burden are often associated with stronger T cell response and better clinical outcomes. 241 , 242 Therefore, multitargeting mRNA vaccines have shown promising prospects in the treatment of highly immunogenic melanomas but have not progressed as well in other tumor types. 243 , 244 , 245 For example, the mRNA vaccine BI‐1361849 (formerly called CV9202), developed by Curevac, is currently undergoing phase I/II clinical trials for treatment of NSCLC. BI‐1361849 encodes six TAAs (NY‐ESO‐1, MAGE‐C2, MAGE‐C1, survivin, 5T4, and MUC1). The completed phase I clinical trial (NCT01915524) showed that BI‐1361849 was well tolerated and immunogenic. 66 Furthermore, a phase I/II clinical trial (NCT03164772) of BI‐1361849 in combination with durvalumab and tremelimumab for NSCLC has also been completed. It is speculated that this may be related to poor clinical efficacy. CureVac terminated its cooperation agreement with Germany's Boehringer‐Ingelheim CV9202 project for the treatment of NSCLC in June 2021. 246
Although current studies focus on aberrantly or overexpressed autoantigens in tumors, several obstacles constrain the further clinical application of TAA‐targeted vaccines. First, only a limited number of TAAs have been identified for certain solid tumors, and extensive mutations in TAAs lead to immune escape and drug resistance. Second, TAAs exhibit poor immune specificity and immunogenicity, because they exist in normal tissues. Additionally, some TAAs used to be embryonic antigens, which have already induced an immune tolerance during the individual developmental processes. Lastly, TAA‐targeted therapy may trigger autoimmune responses can lead to serious safety concerns. 247 , 248 Therefore, developing cancer vaccines based on tumor‐specific antigens with high immunogenicity has been a hope of oncologists. mRNA tumor vaccines that encode neoantigens provide a new research path. 27 , 249 , 250 , 251
5.2. Personalized neoantigen‐encoding mRNA vaccines
Malignant cells are characterized by continuous and rapid proliferation, with the expansion of the tumor population often accompanied by mutations in a large number of genes, generating multiple neoantigens. Neoantigens originate from random somatic mutations in tumor cells and do not exist in normal cells. Therefore, neoantigens are recognized as nonself by the immune system and are presented to T cells by human leukocyte antigen (HLA), which makes them the most promising target for tumor vaccines. 252 , 253 , 254 , 255 , 256 , 257 , 258 , 259 Compared with a TAA‐targeted vaccine, a neoantigen‐based vaccine provides inspiring advantages: first, due to the constrained expression of neoantigens in tumor cells only, neoantigen vaccines elicit a true tumor‐specific T‐cell response, and avoid off‐target damage to nontumorous tissues. Next, a substantial fraction of nonsynonymous neoantigens causes strong immunogenicity, 260 this is perhaps due to the fact that somatic mutated neo‐epitopes manage to bypass the central tolerance. Lastly, prolonged T cell responses, along with immune memory, can be induced by neoantigens. They together prevent potential tumor recurrence and metastasis. 261 , 262 , 263 , 264
Each tumor patient has their own unique neoantigen profile, also known as the mutanome. With the wide application of next‐generation sequencing and the development of neoantigen prediction technology, each patient can benefit from individualized sequencing for quick and efficient neoantigen identification and selection. 265 , 266 Then, a monoepitope or, more commonly, polyepitope mRNA vaccine targeting neoantigens can be rapidly prepared. At present, the clinical results for several neoantigen‐targeting mRNA vaccines are encouraging. 23 , 25 , 267 , 268
In 2017, there were two significant medical breakthroughs of individualized vaccines for melanoma treatment. Both studies were based on whole‐exome sequencing of tumor tissues. In a study from Harvard Medical School, six melanoma patients received a neoantigen vaccine adjuvanted with an immunostimulant, poly‐ICLC. The results suggested that four patients showed no recurrence within a median of 25 months (from 20–32 months), and two other patients with lung metastases achieved complete remission after relapse by taking the immune checkpoint inhibitor Keytruda (NCT01970358). 253 At BioNTech, researchers prepared individualized mRNA neoantigen vaccines for each patient. After treatment, the recurrence and metastasis of melanoma were significantly reduced, and the progression‐free survival (PFS) of patients was significantly extended. 25
Inspired by these results, 21 clinical trials of mRNA tumor vaccines targeting neoantigens are currently under development, according to ClinicalTrials.gov (Table 3). Bioenterprises such as Moderna and BioNTech are actively exploiting customized tumor vaccine in competition with each other. The mRNA‐4157 vaccine developed by Moderna can encode up to 34 neoantigens and has completed a phase I clinical trial (NCT03313778) for the treatment of solid tumors. A combination therapy of the mRNA‐4157 vaccine and the immune checkpoint inhibitor pembrolizumab revealed antitumor potential against a variety of solid tumors. 269 The phase II clinical trial of mRNA‐4157 vaccine for melanoma has begun to recruit patients (NCT03897881). 229 Another completed phase I clinical trial (NCT03289962) is BioNTech's combination therapy for the treatment of locally advanced or metastatic solid tumors (e.g., NSCLC, colorectal cancer, melanoma, and triple negative breast cancer). This therapy combines personalized mRNA cancer vaccine BNT122 with the PD‐L1 inhibitor atezolizumab. Among 108 patients who received at least one postdose assessment, the objective response rate (ORR) was about 8% (nine patients), and the SD rate was 49% (53 patients). 270 NEO‐PV‐01, another personalized neoantigen vaccine for advanced solid tumors, is also being tested in combination with nivolumab. The phase Ib clinical trial (NCT02897765) suggested that, for vaccinated patients with melanoma, NSCLC, and bladder cancer, the corresponding ORRs were 59%, 39%, and 27%; the median PFS was 23.5 months, 8.5 months, and 5.8 months, respectively. Furthermore, the 1‐year OS rates were 96% for melanoma, 83% for NSCLC, and 67% for bladder cancer, all of which were superior to the historical data obtained from programmed death‐1 inhibitor monotherapy. 271 The existing clinical data suggest that in the treatment of tumor types with a high mutational burden (e.g., melanoma and NSCLC) and mismatch repair‐deficient colorectal cancer, neoantigen vaccines used in combination with immune checkpoint inhibitors should be the focus of future research, because they may provide synergistic effects to enable a higher response rate and PFS. 228 , 258 , 259 , 271
Although neoantigen vaccines have great prospects, the design of such vaccines depends heavily on the continuous optimization of algorithms, including HLA typing, binding strength of neoantigens and MHC, and T cell receptor (TCR) analysis. They are further limited by the median time from screening neoantigens to vaccine preparation of 103 days (ranging from 89 to 160 days), which is too long for patients with advanced tumors who urgently demand individualized treatment. 25 , 251 , 272 , 273 , 274 Despite these difficulties, mRNA vaccines targeting tumor neoantigens may enable in a new era of personalized treatment.
5.3. mRNA‐transfected DC vaccines
DC vaccines represent another type of popular cancer vaccine. DCs are ex vivo transfected with TAA‐encoding mRNA in various ways and are then adoptively transferred back. The world's first human clinical trial of mRNA‐transfected DC vaccines was carried out in 2001. 275 After two decades, there are now 23 clinical trials of DC tumor vaccines prepared by mRNA transfection (Table 4), many of which experience simultaneous transfection with mRNAs encoding multiple TAAs. 264 In 2010, the US Food and Drug Administration approved the world's first DC tumor vaccine, Sipuleucel‐T, for the treatment of advanced prostate cancer, starting a new chapter for antitumor vaccines. 276 , 277 , 278 , 279 Efficient loading of tumor antigens into DCs has been one of the core issues in the preparation of DC vaccines. Progress in the in vitro synthesis of mRNA considerably enhances the stability of mRNA, while reducing its immunogenicity by mRNA modification. Therefore, transfecting DCs with TAA‐encoded mRNA is a suitable way to load tumor antigens and has shown great application prospects. 280 , 281 , 282
The function of DCs is collectively determined by the balance of several factors, such as cell surface co‐stimulatory molecules, co‐inhibitory molecules, and activating and inhibitory cytokines. Therefore, DC vaccines designed to boost multiple aspects of DC function are likely to achieve optimal efficacy. The immunostimulatory activity of DC is elevated remarkably when it is electroplated with mRNAs that encode adjuvants, such as OX40L or 4‐1BBL. 283 , 284 Another representative approach is the TriMix platform developed by eTheRNA. TriMix includes three mRNA components that encode the adjuvants CD70 and CD40L, and constitutively active TLR4. 101 , 105 , 285 Co‐transfection of TriMix and mRNA encoding the HLA‐II targeting signal (DC‐LAMP) and the melanoma‐associated antigen fusion protein (MAGE‐A3, MAGE‐C2, tyrosinase or gp100) into autologous DC to prepare a DC vaccine (TriMixDC‐MEL) activation and promote DC maturation and function is underway (NCT01066390). 286 In a further clinical trial of TriMixDC‐MEL in combination with ipilimumab (TriMixDC‐MEL IPI), a 6‐month disease control rate of 51% was observed. After 5 years of clinical follow‐up, vaccine‐induced immune responses were evaluated in 15 patients (NCT01302496) and were sustained in 12 of the 15 patients. Stronger and more extensive immune responses were detected in patients with complete and partial responses compared to patients with stable and progressive disease. 105 , 287
The advantages of DC vaccines include controllable ex vivo culture conditions of DC cells, accurate tumor antigen loading by mRNA transfection, and high transfection efficiency. However, it also shares some problems common to all cell vaccines that require individualization, including a long production cycle, high labor costs, and poor tumor targeting. These issues need to be addressed before general application is feasible.
6. CONCLUSIONS AND PERSPECTIVES
mRNA vaccines have been developed at an unprecedented speed. As outlined in this review, although immuno‐oncologists were among the first to show early and wide acceptance of mRNA‐based vaccines, they can also be a generalized prophylactic measure for infectious diseases and have become the most promising approach to COVID‐19 prevention and control.
There are two key aspects that allow the rapid pace of mRNA vaccine development: the mRNA sequence design and in vivo delivery systems. 110 , 288 , 289 , 290 , 291 Because the therapeutic use of mRNA vaccine seems more feasible, owing to technical innovations that greatly improved the mRNA delivery efficiency and mRNA stability in the physiological environment, this potential billion‐dollar market has attracted funding from diverse sources, including governments, research project agencies, and pharmaceutical enterprises. Moderna Therapeutics, the “unicorn” set up in 2010, has already raised more than US $2 billion for the development and commercialization of mRNA‐based vaccines. 292
There are several aspects to mRNA vaccines that require further improvements. First, adverse effects should be harnessed, partly by introducing novel lipids that are more biocompatible, for example, ready to be fully catabolized into harmless substances in the body. 293 , 294 , 295 Current ionizable cationic lipid and PEGylated lipid components can be substituted. 26 Second, targeted delivery of mRNA vaccine into DCs should be put to real‐world use to enhance efficacy, reduce dosage, and limit adverse effects. Vehicles, such as LNPs, can be endowed with targeting ability by introducing functionalized components. 20 , 295 Novel intravital administration techniques can be developed. Third, novel adjuvants should be incorporated into the formulation to further boost the efficacy of the mRNA vaccines. 29 , 296 , 297 A delicate balance of several factors, for example, stimulation of immunity, exacerbated inflammation, and suppression of mRNA expression is required. Fourth, the issue raised by nucleoside modification and codon optimization of mRNA needs to be addressed to ensure efficient and accurate translation of full‐length polypeptides that are properly folded. 49 , 298 , 299 , 300 Fifth, mRNA cancer vaccines in combination with other immunotherapies should be investigated to determine which combinations exert synergetic effects in cancer treatment. 66 , 241 , 301 , 302 Lastly, it is important to determine whether mRNA vaccines are processed and function in the same way in animal models and in humans, because some mRNA vaccines, which showed immunogenicity in preclinical animal trials, have generated disappointing results in human trials. 28 , 91 , 175 , 303
CONFLICT OF INTEREST
The authors declare there is no conflict of interest.
AUTHOR CONTRIBUTIONS
X.Z. conceived the ideas of the manuscript. Y.Z.G., J.Y.D., N.Y. and Y.X.Y. searched and screened the literature, wrote the draft, and drew the figures and tables. Y.Z.G. and X.Z. critically revised the article, tables, and figures, supervised the coordination, and X.Z. was in charge of correspondence. All authors approved the final submitted version of this manuscript.
ETHICS STATEMENT
No ethical approval was required for this study.
ACKNOWLEDGEMENTS
This work was supported by the Postdoctoral Science Foundation of China (grant number: 2020T130086ZX), and funded by the Science and Technology Program of Guizhou Province (grant number: [2019]1263). Figures in this review were created with the aid of Adobe Illustrator, ChemDraw, Biorender software, and all images are original and no copyright issues.
Gu Y, Duan J, Yang N, Yang Y, Zhao X. mRNA vaccines in the prevention and treatment of diseases. MedComm. 2022;3:e167. 10.1002/mco2.167
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Maruggi G, Zhang C, Li J, et al. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol Ther. 2019;27(4):757‐772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kardani K, Bolhassani AJC. Vaccine development against SARS‐CoV‐2: from virology to vaccine clinical trials. Coronaviruses. 2021;2(2):159‐171. [Google Scholar]
- 3. Pardi N, Hogan MJ, Weissman D. Recent advances in mRNA vaccine technology. Curr Opin Immunol. 2020;65:14‐20. [DOI] [PubMed] [Google Scholar]
- 4. Painter MM, Mathew D, Goel RR, et al. Rapid induction of antigen‐specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS‐CoV‐2 mRNA vaccination. Immunity. 2021;54(9):2133‐2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gutiérrez‐Bautista JF, López‐Nevot M, Gómez‐Vicente E, et al. Study of humoral and cellular immunity in vaccinated with mRNA‐1273. Apmis. 2022;130(5):261‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Domazet‐Lošo T. mRNA vaccines: why is the biology of retroposition ignored?. Genes (Basel). 2022;13(5):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hajissa K, Mussa A. Positive aspects of the mRNA platform for SARS‐CoV‐2 vaccines. Hum Vaccin Immunother. 2021;17(8):2445‐2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu S, Yang K, Li R, et al. mRNA vaccine era‐mechanisms, drug platform and clinical prospection. Int J Mol Sci. 2020;21(18):6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sadarangani M, Marchant A, Kollmann TR. Immunological mechanisms of vaccine‐induced protection against COVID‐19 in humans. Nat Rev Immunol. 2021;21(8):475‐484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loomis KH, Lindsay KE, Zurla C, et al. In vitro transcribed mRNA vaccines with programmable stimulation of innate immunity. Bioconjug Chem. 2018;29(9):3072‐3083. [DOI] [PubMed] [Google Scholar]
- 11. Chen J, Chen J, Xu Q. Current developments and challenges of mRNA vaccines. Annu Rev Biomed Eng. 2022;24:85‐109. [DOI] [PubMed] [Google Scholar]
- 12. Noor R. mRNA vaccines as an efficient approach for the rapid and robust induction of host immunity against SARS‐CoV‐2. SN Compr Clin Med. 2022;4(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamb YN. BNT162b2 mRNA COVID‐19 vaccine: first approval. Drugs. 2021;81(4):495‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ostro MJ, Giacomoni D, Lavelle D, et al. Evidence for translation of rabbit globin mRNA after liposome‐mediated insertion into a human cell line. Nature. 1978;274(5674):921‐923. [DOI] [PubMed] [Google Scholar]
- 15. Melton DA, Krieg PA, Rebagliati MR, et al. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12(18):7035‐7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martinon F, Krishnan S, Lenzen G, et al. Induction of virus‐specific cytotoxic T lymphocytes in vivo by liposome‐entrapped mRNA. Eur J Immunol. 1993;23(7):1719‐1722. [DOI] [PubMed] [Google Scholar]
- 17. Schnee M, Vogel AB, Voss D, et al. An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl Trop Dis. 2016;10(6):e0004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richner JM, Himansu S, Dowd KA, et al. Modified mRNA vaccines protect against Zika virus infection. Cell. 1110;168(6):1114‐1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pegu A, O'Connell SE, Schmidt SD, et al. Durability of mRNA‐1273 vaccine‐induced antibodies against SARS‐CoV‐2 variants. Science. 2021;373(6561):1372‐1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liang F, Lindgren G, Lin A, et al. Efficient targeting and activation of antigen‐presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol Ther. 2017;25(12):2635‐2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. VanBlargan LA, Himansu S, Foreman BM, et al. An mRNA vaccine protects mice against multiple tick‐transmitted flavivirus infections. Cell Rep. 2018;25(12):3382‐3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhong Z, Catani JPP, Cafferty SM, et al. Immunogenicity and protection efficacy of a naked self‐replicating mRNA‐Based Zika virus vaccine. Vaccines (Basel). 2019;7(3):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Supabphol S, Li L, Goedegebuure SP, et al. Neoantigen vaccine platforms in clinical development: understanding the future of personalized immunotherapy. Expert Opin Investig Drugs. 2021;30(5):529‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rele S. COVID‐19 vaccine development during pandemic: gap analysis, opportunities, and impact on future emerging infectious disease development strategies. Hum Vaccin Immunother. 2021;17(4):1122‐1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly‐specific therapeutic immunity against cancer. Nature. 2017;547(7662):222‐226. [DOI] [PubMed] [Google Scholar]
- 26. Han X, Zhang H, Butowska K, et al. An ionizable lipid toolbox for RNA delivery. Nat Commun. 2021;12(1):7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cafri G, Gartner JJ, Zaks T, et al. mRNA vaccine‐induced neoantigen‐specific T cell immunity in patients with gastrointestinal cancer. J Clin Invest. 2020;130(11):5976‐5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu L, Wang Y, Miao L, et al. Combination immunotherapy of MUC1 mRNA nano‐vaccine and CTLA‐4 blockade effectively inhibits growth of triple negative breast cancer. Mol Ther. 2018;26(1):45‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Islam MA, Rice J, Reesor E, et al. Adjuvant‐pulsed mRNA vaccine nanoparticle for immunoprophylactic and therapeutic tumor suppression in mice. Biomaterials. 2021;266:120431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Zhang L, Xu Z, et al. mRNA vaccine with antigen‐specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol Ther. 2018;26(2):420‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang X, Zhang G, Tang T, et al. Identification of tumor antigens and immune subtypes of pancreatic adenocarcinoma for mRNA vaccine development. Mol Cancer. 2021;20(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karikó K, Buckstein M, Ni H, et al. Suppression of RNA recognition by Toll‐like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165‐175. [DOI] [PubMed] [Google Scholar]
- 33. Pardi N, Hogan MJ, Porter FW, et al. mRNA vaccines ‐ a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qu L, Yi Z, Shen Y, et al. Circular RNA vaccines against SARS‐CoV‐2 and emerging variants. Cell. 1716;185(10):1728‐1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rouf NZ, Biswas S, Tarannum N, et al. Demystifying mRNA vaccines: an emerging platform at the forefront of cryptic diseases. RNA Biol. 2022;19(1):386‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bloom K, van den Berg F, Arbuthnot P. Self‐amplifying RNA vaccines for infectious diseases. Gene Ther. 2021;28(3‐4):117‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brito LA, Kommareddy S, Maione D, et al. Self‐amplifying mRNA vaccines. Adv Genet. 2015;89:179‐233. [DOI] [PubMed] [Google Scholar]
- 38. Vogel AB, Lambert L, Kinnear E, et al. Self‐amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol Ther. 2018;26(2):446‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waheed Y, Safi SZ, Najmi MH, et al. Prediction of promiscuous T cell epitopes in RNA dependent RNA polymerase of Chikungunya virus. Asian Pac J Trop Med. 2017;10(8):760‐764. [DOI] [PubMed] [Google Scholar]
- 40. Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dilliard SA, Cheng Q, Siegwart DJ. On the mechanism of tissue‐specific mRNA delivery by selective organ targeting nanoparticles. Proc Natl Acad Sci U S A. 2021;118(52):e2109256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rausch JW, Heinz WF, Payea MJ, et al. Characterizing and circumventing sequence restrictions for synthesis of circular RNA in vitro. Nucleic Acids Res. 2021;49(6):e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wesselhoeft RA, Kowalski PS, Parker‐Hale FC, et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell. 2019;74(3):508‐520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Puttaraju M, Been MD. Group I permuted intron‐exon (PIE) sequences self‐splice to produce circular exons. Nucleic Acids Res. 1992;20(20):5357‐5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lim J, Ha M, Chang H, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159(6):1365‐1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kajjo S, Sharma S, Chen S, et al. PABP prevents the untimely decay of select mRNA populations in human cells. Embo j. 2022;41(6):e108650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Z, Kiledjian M. The poly(A)‐binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol. 2000;20(17):6334‐6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wells SE, Hillner PE, Vale RD, et al. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2(1):135‐140. [DOI] [PubMed] [Google Scholar]
- 49. Kim SC, Sekhon SS, Shin WR, et al. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol Cell Toxicol. 2022;18(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Passmore LA, Coller J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat Rev Mol Cell Biol. 2022;23(2):93‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Granados‐Riveron JT, Aquino‐Jarquin G. Engineering of the current nucleoside‐modified mRNA‐LNP vaccines against SARS‐CoV‐2. Biomed Pharmacother. 2021;142:111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9(4):337‐344. [DOI] [PubMed] [Google Scholar]
- 53. Szostak E, Gebauer F. Translational control by 3'‐UTR‐binding proteins. Brief Funct Genomics. 2013;12(1):58‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jia J, Yao P, Arif A, et al. Regulation and dysregulation of 3'UTR‐mediated translational control. Curr Opin Genet Dev. 2013;23(1):29‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5'‐untranslated regions of eukaryotic mRNAs. Science. 2016;352(6292):1413‐1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Linares‐Fernández S, Moreno J, Lambert E, et al. Combining an optimized mRNA template with a double purification process allows strong expression of in vitro transcribed mRNA. Mol Ther Nucleic Acids. 2021;26:945‐956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Orlandini von Niessen AG, Poleganov MA, Rechner C, et al. Improving mRNA‐based therapeutic gene delivery by expression‐augmenting 3' UTRs identified by cellular library screening. Mol Ther. 2019;27(4):824‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Verbeke R, Lentacker I, De Smedt SC, et al. The dawn of mRNA vaccines: the COVID‐19 case. J Control Release. 2021;333:511‐520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xia X. Detailed dissection and critical evaluation of the Pfizer/BioNTech and Moderna mRNA vaccines. Vaccines (Basel). 2021;9(7):734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Asrani KH, Farelli JD, Stahley MR, et al. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018;15(6):756‐762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sadler AJ, Williams BR. Interferon‐inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559‐568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sebastian M, Schröder A, Scheel B, et al. A phase I/IIa study of the mRNA‐based cancer immunotherapy CV9201 in patients with stage IIIB/IV non‐small cell lung cancer. Cancer Immunol Immunother. 2019;68(5):799‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fotin‐Mleczek M, Duchardt KM, Lorenz C, et al. Messenger RNA‐based vaccines with dual activity induce balanced TLR‐7 dependent adaptive immune responses and provide antitumor activity. J Immunother. 2011;34(1):1‐15. [DOI] [PubMed] [Google Scholar]
- 65. Rauch S, Roth N, Schwendt K, et al. mRNA‐based SARS‐CoV‐2 vaccine candidate CVnCoV induces high levels of virus‐neutralising antibodies and mediates protection in rodents. NPJ Vaccines. 2021;6(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Papachristofilou A, Hipp MM, Klinkhardt U, et al. Phase Ib evaluation of a self‐adjuvanted protamine formulated mRNA‐based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non‐small cell lung cancer. J Immunother Cancer. 2019;7(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thess A, Grund S, Mui BL, et al. Sequence‐engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther. 2015;23(9):1456‐1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mauro VP, Chappell SA. A critical analysis of codon optimization in human therapeutics. Trends Mol Med. 2014;20(11):604‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu CH, Dang Y, Zhou Z, et al. Codon usage influences the local rate of translation elongation to regulate co‐translational protein folding. Mol Cell. 2015;59(5):744‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lu J, Lu G, Tan S, et al. A COVID‐19 mRNA vaccine encoding SARS‐CoV‐2 virus‐like particles induces a strong antiviral‐like immune response in mice. Cell Res. 2020;30(10):936‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Anderson BR, Muramatsu H, Nallagatla SR, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38(17):5884‐5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Karikó K, Muramatsu H, Welsh FA, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16(11):1833‐1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li B, Luo X, Dong Y. Effects of chemically modified messenger RNA on protein expression. Bioconjug Chem. 2016;27(3):849‐853. [DOI] [PubMed] [Google Scholar]
- 74. Svitkin YV, Gingras AC, Sonenberg N. Membrane‐dependent relief of translation elongation arrest on pseudouridine‐ and N1‐methyl‐pseudouridine‐modified mRNAs. Nucleic Acids Res. 2021;50(13):7202‐7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Franco MK, Koutmou KS. Chemical modifications to mRNA nucleobases impact translation elongation and termination. Biophys Chem. 2022;285:106780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Henderson JM, Ujita A, Hill E, et al. Cap 1 messenger RNA synthesis with co‐transcriptional CleanCap(®) analog by in vitro transcription. Curr Protoc. 2021;1(2):e39. [DOI] [PubMed] [Google Scholar]
- 77. Banerjee AK. 5'‐terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44(2):175‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Labarta I, Hoffman S, Simpkins A. Manufacturing strategy for the production of 200 million sterile doses of an mRNA vaccine for COVID‐19. Accessed April 20, 2021. https://repository.upenn.edu/cbe_sdr/132/
- 79. Pemberton RE, Liberti P, Baglioni C. Isolation of messenger RNA from polysomes by chromatography on oligo(dT)‐cellulose. Anal Biochem. 1975;66(1):18‐28. [DOI] [PubMed] [Google Scholar]
- 80. Zhao X, Zhang S, Huang Q, et al. Regulation of hydrogen bond acidity and its effect on separation performances. J Chromatogr A. 2021;1657:462556. [DOI] [PubMed] [Google Scholar]
- 81. Korenč M, Mencin N, Puc J, et al. Chromatographic purification with CIMmultus™ Oligo dT increases mRNA stability. Cell and Gene Ther Insights. 2021;7(9):1207. [Google Scholar]
- 82. Gagnon P, Goričar B, Peršič S, et al. Two new capture options for improved purification of large mRNA. Cell Gene Ther Insights. 2020;6:7. [Google Scholar]
- 83. Shepherd SJ, Warzecha CC, Yadavali S, et al. Scalable mRNA and siRNA lipid nanoparticle production using a parallelized microfluidic device. Nano Lett. 2021;21(13):5671‐5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Corbett KS, Edwards DK, Leist SR, et al. SARS‐CoV‐2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Uddin MN, Roni MA. Challenges of storage and stability of mRNA‐based COVID‐19 vaccines. Vaccines (Basel). 2021;9(9):1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Crommelin DJA, Anchordoquy TJ, Volkin DB, et al. Addressing the cold reality of mRNA vaccine stability. J Pharm Sci. 2021;110(3):997‐1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Holm MR, Poland GA. Critical aspects of packaging, storage, preparation, and administration of mRNA and adenovirus‐vectored COVID‐19 vaccines for optimal efficacy. Vaccine. 2021;39(3):457‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Muramatsu H, Lam K, Bajusz C, et al. Lyophilization provides long‐term stability for a lipid nanoparticle‐formulated, nucleoside‐modified mRNA vaccine. Mol Ther. 2022;30(5):1941‐1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ramakanth D, Singh S, Maji PK, et al. Advanced packaging for distribution and storage of COVID‐19 vaccines: a review. Environ Chem Lett. 2021;19(5):3597‐3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. WHO . Evaluation of the quality, safety and efficacy of messenger RNA vaccines for the prevention of infectious diseases: regulatory considerations. Accessed December 7, 2021. https://www.who.int/publications/m/item/evaluation‐of‐the‐quality‐safety‐and‐efficacy‐of‐messenger‐rna‐vaccines‐for‐the‐prevention‐of‐infectious‐diseases‐regulatory‐considerations
- 91. Cagigi A, Loré K. Immune responses induced by mRNA vaccination in mice, monkeys and humans. Vaccines (Basel). 2021;9(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Maruggi G, Mallett CP, Westerbeck JW, et al. A self‐amplifying mRNA SARS‐CoV‐2 vaccine candidate induces safe and robust protective immunity in preclinical models. Mol Ther. 2022;30(5):1897‐1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dhama K, Dhawan M, Tiwari R, et al. COVID‐19 intranasal vaccines: current progress, advantages, prospects, and challenges. Hum Vaccin Immunother. 2022;18(5):2045853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ndeupen S, Qin Z, Jacobsen S, et al. The mRNA‐LNP platform's lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience. 2021;24(12):103479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kounis NG, Koniari I, de Gregorio C, et al. Allergic reactions to current available COVID‐19 vaccinations: pathophysiology, causality, and therapeutic considerations. Vaccines (Basel). 2021;9(3):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sellaturay P, Nasser S, Islam S, et al. Polyethylene glycol (PEG) is a cause of anaphylaxis to the Pfizer/BioNTech mRNA COVID‐19 vaccine. Clin Exp Allergy. 2021;51(6):861‐863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kadali RAK, Janagama R, Peruru S, et al. Side effects of BNT162b2 mRNA COVID‐19 vaccine: a randomized, cross‐sectional study with detailed self‐reported symptoms from healthcare workers. Int J Infect Dis. 2021;106:376‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Karer M, Stiasny K, Zeitlinger M, et al. Subcutaneous injection of mRNA vaccines against severe acute respiratory syndrome coronavirus 2: an option for severe bleeding disorders or anticoagulated patients?. Blood Coagul Fibrinolysis. 2021;32(6):423‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hou X, Zaks T, Langer R, et al. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6(12):1078‐1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen Q, Qi R, Chen X, et al. A targeted and stable polymeric nanoformulation enhances systemic delivery of mRNA to tumors. Mol Ther. 2017;25(1):92‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Van Lint S, Renmans D, Broos K, et al. Intratumoral delivery of TriMix mRNA results in T‐cell activation by cross‐presenting dendritic cells. Cancer Immunol Res. 2016;4(2):146‐156. [DOI] [PubMed] [Google Scholar]
- 102. Zeng C, Zhang C, Walker PG, et al. Formulation and delivery technologies for mRNA vaccines. Curr Top Microbiol Immunol. 2020. 10.1007/82_2020_217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wilgenhof S, Van Nuffel AMT, Benteyn D, et al. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann Oncol. 2013;24(10):2686‐2693. [DOI] [PubMed] [Google Scholar]
- 104. Chung DJ, Sharma S, Rangesa M, et al. Langerhans dendritic cell vaccine bearing mRNA‐encoded tumor antigens induces antimyeloma immunity after autotransplant. Blood Adv. 2022;6(5):1547‐1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. De Keersmaecker B, Claerhout S, Carrasco J, et al. TriMix and tumor antigen mRNA electroporated dendritic cell vaccination plus ipilimumab: link between T‐cell activation and clinical responses in advanced melanoma. J Immunother Cancer. 2020;8(1):e000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Weide B, Carralot JP, Reese A, et al. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother. 2008;31(2):180‐188. [DOI] [PubMed] [Google Scholar]
- 107. Wu Z, Li T. Nanoparticle‐mediated cytoplasmic delivery of messenger RNA vaccines: challenges and future perspectives. Pharm Res. 2021;38(3):473‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tenchov R, Bird R, Curtze AE, et al. Lipid nanoparticles‐from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano. 2021;15(11):16982‐17015. [DOI] [PubMed] [Google Scholar]
- 109. Iavarone C, O'Hagan DT, Yu D, et al. Mechanism of action of mRNA‐based vaccines. Expert Rev Vaccines. 2017;16(9):871‐881. [DOI] [PubMed] [Google Scholar]
- 110. Jackson NAC, Kester KE, Casimiro D, et al. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hassett KJ, Higgins J, Woods A, et al. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J Control Release. 2021;335:237‐246. [DOI] [PubMed] [Google Scholar]
- 112. Kon E, Elia U, Peer D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr Opin Biotechnol. 2022;73:329‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Jarzebska NT, Lauchli S, Iselin C, et al. Functional differences between protamine preparations for the transfection of mRNA. Drug Deliv. 2020;27(1):1231‐1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Jarzebska NT, Mellett M, Frei J, et al. Protamine‐based strategies for RNA transfection. Pharmaceutics. 2021;13(6):877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Weide B, Pascolo S, Scheel B, et al. Direct injection of protamine‐protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J Immunother. 2009;32(5):498‐507. [DOI] [PubMed] [Google Scholar]
- 116. Démoulins T, Milona P, Englezou PC, et al. Polyethylenimine‐based polyplex delivery of self‐replicating RNA vaccines. Nanomedicine. 2016;12(3):711‐722. [DOI] [PubMed] [Google Scholar]
- 117. Yokoo H, Oba M, Uchida S. Cell‐penetrating peptides: emerging tools for mRNA delivery. Pharmaceutics. 2021;14(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Persano S, Guevara ML, Li Z, et al. Lipopolyplex potentiates anti‐tumor immunity of mRNA‐based vaccination. Biomaterials. 2017;125:81‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Brito LA, Chan M, Shaw CA, et al. A cationic nanoemulsion for the delivery of next‐generation RNA vaccines. Mol Ther. 2014;22(12):2118‐2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Murphy DE, de Jong OG, Evers MJW, et al. Natural or synthetic RNA delivery: a stoichiometric comparison of extracellular vesicles and synthetic nanoparticles. Nano Lett. 2021;21(4):1888‐1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Amiri A, Bagherifar R, Dezfouli EA, Kiaie SH, Jafari R, Ramezani R. Exosomes as bio‐inspired nanocarriers for RNA delivery: preparation and applications. J Transl Med. 2022;20(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Tsai SJ, Atai NA, Cacciottolo M, et al. Exosome‐mediated mRNA delivery in vivo is safe and can be used to induce SARS‐CoV‐2 immunity. J Biol Chem. 2021;297(5):101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Aslan C, Kiaie SH, Zolbanin NM, et al. Exosomes for mRNA delivery: a novel biotherapeutic strategy with hurdles and hope. BMC Biotechnol. 2021;21(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kulkarni JA, Cullis PR, van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28(3):146‐157. [DOI] [PubMed] [Google Scholar]
- 125. Kalyanram P, Puri A, Gupta A. Thermotropic effects of PEGylated lipids on the stability of HPPH‐encapsulated lipid nanoparticles (LNP). J Therm Anal Calorim. 2022;147(11):6337‐6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Patel S, Ashwanikumar N, Robinson E, et al. Naturally‐occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat Commun. 2020;11(1):983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cheng X, Lee RJ. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv Drug Deliv Rev. 2016;99(Pt A):129‐137. [DOI] [PubMed] [Google Scholar]
- 128. García‐Pacios M, Collado MI, Busto JV, et al. Sphingosine‐1‐phosphate as an amphipathic metabolite: its properties in aqueous and membrane environments. Biophys J. 2009;97(5):1398‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Liu S, Cheng Q, Wei T, et al. Membrane‐destabilizing ionizable phospholipids for organ‐selective mRNA delivery and CRISPR‐Cas gene editing. Nat Mater. 2021;20(5):701‐710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Meyer RA, Hussmann GP, Peterson NC, et al. A scalable and robust cationic lipid/polymer hybrid nanoparticle platform for mRNA delivery. Int J Pharm. 2022;611:121314. [DOI] [PubMed] [Google Scholar]
- 131. Akinc A, Maier MA, Manoharan M, et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid‐based drugs. Nat Nanotechnol. 2019;14(12):1084‐1087. [DOI] [PubMed] [Google Scholar]
- 132. Kimura N, Maeki M, Sato Y, et al. Development of a microfluidic‐based post‐treatment process for size‐controlled lipid nanoparticles and application to siRNA delivery. ACS Appl Mater Interfaces. 2020;12(30):34011‐34020. [DOI] [PubMed] [Google Scholar]
- 133. Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA‐lipid nanoparticle COVID‐19 vaccines: structure and stability. Int J Pharm. 2021;601:120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Packer M, Gyawali D, Yerabolu R, et al. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat Commun. 2021;12(1):6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Szebeni J, Storm G, Ljubimova JY, et al. Applying lessons learned from nanomedicines to understand rare hypersensitivity reactions to mRNA‐based SARS‐CoV‐2 vaccines. Nat Nanotechnol. 2022;17(4):337‐346. [DOI] [PubMed] [Google Scholar]
- 136. Trougakos IP, Terpos E, Alexopoulos H, et al. Adverse effects of COVID‐19 mRNA vaccines: the spike hypothesis. Trends Mol Med. 2022;28(7):542‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Moghimi SM. Allergic reactions and anaphylaxis to LNP‐based COVID‐19 vaccines. Mol Ther. 2021;29(3):898‐900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Pilishvili T, Fleming‐Dutra KE, Farrar JL, et al. Interim estimates of vaccine effectiveness of Pfizer‐BioNTech and Moderna COVID‐19 vaccines among health care personnel ‐ 33 U.S. Sites, January‐March 2021. MMWR Morb Mortal Wkly Rep. 2021;70(20):753‐758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hamza MS, Tikamdas R, El Baghdady NS, et al. Safety and effectiveness of COVID‐19 vaccines: results from a cross‐sectional survey among staff, workers and students at an Egyptian university. Vaccines. 2022;10(6):846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jubishi D, Okamoto K, Hamada K, et al. The association between adverse reactions and immune response against SARS‐CoV‐2 spike protein after vaccination with BNT162b2 among healthcare workers in a single healthcare system: a prospective observational cohort study. Hum Vaccin Immunother. 2022;18(5):2048559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Walsh EE, Frenck RW, Falsey AR, et al. Safety and immunogenicity of two RNA‐based Covid‐19 vaccine candidates. N Engl J Med. 2020;383(25):2439‐2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Tanaka T, Narazaki M, Kishimoto T. IL‐6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6(10):a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kozma GT, Shimizu T, Ishida T, et al. Anti‐PEG antibodies: properties, formation, testing and role in adverse immune reactions to PEGylated nano‐biopharmaceuticals. Adv Drug Deliv Rev. 2020;154((155):163‐175. [DOI] [PubMed] [Google Scholar]
- 144. McSweeney MD, Mohan M, Commins SP, et al. Anaphylaxis to Pfizer/BioNTech mRNA COVID‐19 vaccine in a patient with clinically confirmed PEG allergy. Front Allergy. 2021;2:715844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Baden LR, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2021;384(5):403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;383(27):2603‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Nogueira SS, Schlegel A, Maxeiner K, et al. Polysarcosine‐functionalized lipid nanoparticles for therapeutic mRNA delivery. ACS Nano. 2020;3(11):10634‐10645. [Google Scholar]
- 148. González‐Aramundiz JV, Peleteiro M, González‐Fernández Á, et al. Protamine nanocapsules for the development of thermostable adjuvanted nanovaccines. Mol Pharm. 2018;15(12):5653‐5664. [DOI] [PubMed] [Google Scholar]
- 149. Thachil J. Protamine‐the journey from DNA to heparin neutralization to gene therapy. Semin Thromb Hemost. 2022;48(2):240‐243. [DOI] [PubMed] [Google Scholar]
- 150. Scheel B, Teufel R, Probst J, et al. Toll‐like receptor‐dependent activation of several human blood cell types by protamine‐condensed mRNA. Eur J Immunol. 2005;35(5):1557‐1566. [DOI] [PubMed] [Google Scholar]
- 151. Kowalczyk A, Doener F, Zanzinger K, et al. Self‐adjuvanted mRNA vaccines induce local innate immune responses that lead to a potent and boostable adaptive immunity. Vaccine. 2016;34(33):3882‐3893. [DOI] [PubMed] [Google Scholar]
- 152. Rettig L, Haen SP, Bittermann AG, et al. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010;115(22):4533‐4541. [DOI] [PubMed] [Google Scholar]
- 153. Zhao H, Wang TC, Li XF, et al. Long‐term stability and protection efficacy of the RBD‐targeting COVID‐19 mRNA vaccine in nonhuman primates. Signal Transduct Target Ther. 2021;6(1):438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sköld AE, van Beek JJ, Sittig SP, et al. Protamine‐stabilized RNA as an ex vivo stimulant of primary human dendritic cell subsets. Cancer Immunol Immunother. 2015;64(11):1461‐1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Higgins JP, Bernstein MB, Hodge JW. Enhancing immune responses to tumor‐associated antigens. Cancer Biol Ther. 2009;8(15):1440‐1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Scheel B, Aulwurm S, Probst J, et al. Therapeutic anti‐tumor immunity triggered by injections of immunostimulating single‐stranded RNA. Eur J Immunol. 2006;36(10):2807‐2816. [DOI] [PubMed] [Google Scholar]
- 157. Tusup M, Pascolo S. Generation of immunostimulating 130 nm protamine‐RNA nanoparticles. Methods Mol Biol. 2017;1499:155‐163. [DOI] [PubMed] [Google Scholar]
- 158. Armbruster N, Jasny E, Petsch B. Advances in RNA vaccines for preventive indications: a case study of a vaccine against rabies. Vaccines (Basel). 2019;7(4):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kallen KJ, Heidenreich R, Schnee M, et al. A novel, disruptive vaccination technology: self‐adjuvanted RNActive(®) vaccines. Hum Vaccin Immunother. 2013;9(10):2263‐2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Hong HS, Koch SD, Scheel B, et al. Distinct transcriptional changes in non‐small cell lung cancer patients associated with multi‐antigenic RNActive® CV9201 immunotherapy. Oncoimmunology. 2016;5(12):e1249560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Rausch S, Schwentner C, Stenzl A, et al. mRNA vaccine CV9103 and CV9104 for the treatment of prostate cancer. Hum Vaccin Immunother. 2014;10(11):3146‐3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Stenzl A, Feyerabend S, Syndikus I, et al. Results of the randomized, placebo‐controlled phase I/IIB trial of CV9104, an mRNA based cancer immunotherapy, in patients with metastatic castration‐resistant prostate cancer (mCRPC). Annals of Oncology. 2017;28:v408‐v409. [Google Scholar]
- 163. Liu J, Guo S, Li Z, et al. Synthesis and characterization of stearyl protamine and investigation of their complexes with DNA for gene delivery. Colloids Surf B Biointerfaces. 2009;73(1):36‐41. [DOI] [PubMed] [Google Scholar]
- 164. Puras G, Martínez‐Navarrete G, Mashal M, et al. Protamine/DNA/Niosome ternary nonviral vectors for gene delivery to the retina: the role of protamine. Mol Pharm. 2015;12(10):3658‐3671. [DOI] [PubMed] [Google Scholar]
- 165. Siewert CD, Haas H, Cornet V, et al. Hybrid biopolymer and lipid nanoparticles with improved transfection efficacy for mRNA. Cells. 2020;9(9):2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Pattipeiluhu R, Arias‐Alpizar G, Basha G, et al. Anionic lipid nanoparticles preferentially deliver mRNA to the hepatic reticuloendothelial system. Adv Mater. 2022;34(16):e2201095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Pardi N, Tuyishime S, Muramatsu H, et al. Expression kinetics of nucleoside‐modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release. 2015;217:345‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Kranz LM, Diken M, Haas H, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534(7607):396‐401. [DOI] [PubMed] [Google Scholar]
- 169. Dolgin E. Better lipids to power next generation of mRNA vaccines. Science. 2022;376(6594):680‐681. [DOI] [PubMed] [Google Scholar]
- 170. Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov. 2021;20(11):817‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Teo SP. Review of COVID‐19 mRNA vaccines: bNT162b2 and mRNA‐1273. J Pharm Pract. 2021. 10.1177/08971900211009650 [DOI] [PubMed] [Google Scholar]
- 172. Lee P, Kim CU, Seo SH, et al. Current status of COVID‐19 vaccine development: focusing on antigen design and clinical trials on later stages. Immune Netw. 2021;21(1):e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zoltán K, Zain R. How to make enough vaccine for the world in one year. Public Citizen Report. 2021;26. https://heatinformatics.com/sites/default/files/images-videosFileContent/mRNA-vaccine-roadmap-May-26.pdf [Google Scholar]
- 174. Vogel AB, Kanevsky I, Che Y, et al. BNT162b vaccines protect rhesus macaques from SARS‐CoV‐2. Nature. 2021;592(7853):283‐289. [DOI] [PubMed] [Google Scholar]
- 175. Chivukula S, Plitnik T, Tibbitts T, et al. Development of multivalent mRNA vaccine candidates for seasonal or pandemic influenza. NPJ Vaccines. 2021;6(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wu CY, Cheng CW, Kung CC, et al. Glycosite‐deleted mRNA of SARS‐CoV‐2 spike protein as a broad‐spectrum vaccine. Proc Natl Acad Sci U S A. 2022;119(9):e2119995119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ahlén G, Frelin L, Nikouyan N, et al. The SARS‐CoV‐2 N protein is a good component in a vaccine. J Virol. 2020;94(18):e01279‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Kames J, Holcomb DD, Kimchi O, et al. Sequence analysis of SARS‐CoV‐2 genome reveals features important for vaccine design. Sci Rep. 2020;10(1):15643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Darby AC, Hiscox JA. Covid‐19: variants and vaccination. BMJ. 2021;372:n771. [DOI] [PubMed] [Google Scholar]
- 180. Joag V, Wijeyesinghe S, Stolley JM, et al. Cutting edge: mouse SARS‐CoV‐2 epitope reveals infection and vaccine‐elicited CD8 T cell responses. J Immunol. 2021;206(5):931‐935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ahmad S, Navid A, Farid R, et al. Design of a novel multi epitope‐based vaccine for pandemic coronavirus disease (COVID‐19) by vaccinomics and probable prevention strategy against avenging zoonotics. Eur J Pharm Sci. 2020;151:105387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Ong E, Wong MU, Huffman A, et al. COVID‐19 coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol. 2020;11:1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Khan S, Shaker B, Ahmad S, et al. Towards a novel peptide vaccine for Middle East respiratory syndrome coronavirus and its possible use against pandemic COVID‐19. J Mol Liq. 2021;324:114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Jeyanathan M, Afkhami S, Smaill F, et al. Immunological considerations for COVID‐19 vaccine strategies. Nat Rev Immunol. 2020;20(10):615‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Noh JY, Jeong HW, Kim JH, et al. T cell‐oriented strategies for controlling the COVID‐19 pandemic. Nat Rev Immunol. 2021;21(11):687‐688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Mateus J, Dan JM, Zhang Z, et al. Low‐dose mRNA‐1273 COVID‐19 vaccine generates durable memory enhanced by cross‐reactive T cells. Science. 2021;374(6566):eabj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Lederer K, Bettini E, Parvathaneni K, et al. Germinal center responses to SARS‐CoV‐2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022;185(6):1008‐1024.e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Röltgen K, Nielsen SCA, Silva O, et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS‐CoV‐2 infection and vaccination. Cell. 2022;185(6):1025‐1040.e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Levine‐Tiefenbrun M, Yelin I, Alapi H, et al. Viral loads of Delta‐variant SARS‐CoV‐2 breakthrough infections after vaccination and booster with BNT162b2. Nat Med. 2021;27(12):2108‐2110. [DOI] [PubMed] [Google Scholar]
- 190. Tartof SY, Slezak JM, Puzniak L, et al. Effectiveness of a third dose of BNT162b2 mRNA COVID‐19 vaccine in a large US health system: a retrospective cohort study. Lancet Reg Health Am. 2022;9:100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Evans JP, Zeng C, Carlin C, et al. Neutralizing antibody responses elicited by SARS‐CoV‐2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci Transl Med. 2022;14(637):eabn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Naaber P, Tserel L, Kangro K, et al. Dynamics of antibody response to BNT162b2 vaccine after six months: a longitudinal prospective study. Lancet Reg Health Eur. 2021;10:100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Falsey AR, j r Frenck RW, Walsh EE, et al. SARS‐CoV‐2 neutralization with BNT162b2 vaccine dose 3. N Engl J Med. 2021;385(17):1627‐1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Azzi L, Gasperina DD, Veronesi G, et al. Mucosal immune response in BNT162b2 COVID‐19 vaccine recipients. EBioMedicine. 2022;75:103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Lapuente D, Fuchs J, Willar J, et al. Protective mucosal immunity against SARS‐CoV‐2 after heterologous systemic prime‐mucosal boost immunization. Nat Commun. 2021;12(1):6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Chemaitelly H, Ayoub HH, AlMukdad S, et al. Duration of mRNA vaccine protection against SARS‐CoV‐2 Omicron BA.1 and BA.2 subvariants in Qatar. Nat Commun. 2022;13(1):3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Thompson MG, Burgess JL, Naleway AL, et al. Prevention and attenuation of Covid‐19 with the BNT162b2 and mRNA‐1273 vaccines. N Engl J Med. 2021;385(4):320‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA Covid‐19 vaccine in a nationwide mass vaccination setting. N Engl J Med. 2021;384(15):1412‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zacay G, Shasha D, Bareket R, et al. BNT162b2 vaccine effectiveness in preventing asymptomatic infection with SARS‐CoV‐2 virus: a nationwide historical cohort study. Open Forum Infect Dis. 2021;8(6):ofab262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Hall VJ, Foulkes S, Saei A, et al. COVID‐19 vaccine coverage in health‐care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): a prospective, multicentre, cohort study. Lancet. 2021;397(10286):1725‐1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Chodick G, Tene L, Rotem RS, et al. The effectiveness of the two‐dose BNT162b2 vaccine: analysis of real‐world data. Clin Infect Dis. 2022;74(3):472‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Bruxvoort KJ, Sy LS, Qian L, et al. Real‐world effectiveness of the mRNA‐1273 vaccine against COVID‐19: interim results from a prospective observational cohort study. Lancet Reg Health Am. 2022;6:100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Lauring AS, Tenforde MW, Chappell JD, et al. Clinical severity of, and effectiveness of mRNA vaccines against, covid‐19 from omicron, delta, and alpha SARS‐CoV‐2 variants in the United States: prospective observational study. Bmj. 2022;376:e069761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Garcia‐Beltran WF, Lam EC, St Denis K, et al. Multiple SARS‐CoV‐2 variants escape neutralization by vaccine‐induced humoral immunity. Cell. 2021;184(9):2372‐2383.e2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Lucas C, Vogels CBF, Yildirim I, et al. Impact of circulating SARS‐CoV‐2 variants on mRNA vaccine‐induced immunity. Nature. 2021;600(7889):523‐529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS‐CoV‐2. Nature. 2022;602(7898):676‐681. [DOI] [PubMed] [Google Scholar]
- 207. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature. 2021;592(7855):616‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Accorsi EK, Britton A, Fleming‐Dutra KE, et al. Association between 3 doses of mRNA COVID‐19 vaccine and symptomatic infection caused by the SARS‐CoV‐2 Omicron and delta variants. JAMA. 2022;327(7):639‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Garcia‐Beltran WF, Hoelzemer A, Denis KJS, et al. mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 Omicron variant. Cell. 2022;185(3):457‐466.e454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Abu‐Raddad LJ, Chemaitelly H, Ayoub HH, et al. Effect of mRNA vaccine boosters against SARS‐CoV‐2 Omicron infection in Qatar. N Engl J Med. 2022;386(19):1804‐1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Gruell H, Vanshylla K, Tober‐Lau P, et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS‐CoV‐2 Omicron variant. Nat Med. 2022;28(3):477‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Altmann DM, Boyton RJ. COVID‐19 vaccination: the road ahead. Science. 2022;375(6585):1127‐1132. [DOI] [PubMed] [Google Scholar]
- 213. Freyn AW, da Silva JR, Rosado VC,et al. A multi‐targeting, nucleoside‐modified mRNA influenza virus vaccine provides broad protection in mice. Mol Ther. 2020;28(7):1569‐1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Pardi N, Hogan MJ, Pelc RS, et al. Zika virus protection by a single low‐dose nucleoside‐modified mRNA vaccination. Nature. 2017;543(7644):248‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Alberer M, Gnad‐Vogt U, Hong HS, et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open‐label, non‐randomised, prospective, first‐in‐human phase 1 clinical trial. Lancet. 2017;390(10101):1511‐1520. [DOI] [PubMed] [Google Scholar]
- 216. Aldrich C, Leroux‐Roels I, Huang KB, et al. Proof‐of‐concept of a low‐dose unmodified mRNA‐based rabies vaccine formulated with lipid nanoparticles in human volunteers: a phase 1 trial. Vaccine. 2021;39(8):1310‐1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Wei J, Hui AM. The paradigm shift in treatment from Covid‐19 to oncology with mRNA vaccines. Cancer Treat Rev. 2022;107:102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. zur Hausen H. Viruses in human cancers. Science. 1991;254(5035):1167‐1173. [DOI] [PubMed] [Google Scholar]
- 219. Schiller JT, Lowy DR. Virus infection and human cancer: an overview. Recent Results Cancer Res. 2014;193:1‐10. [DOI] [PubMed] [Google Scholar]
- 220. McLaughlin‐Drubin ME, Munger K. Viruses associated with human cancer. Biochim Biophys Acta. 2008;1782(3):127‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Schiller JT, Lowy DR. An introduction to virus infections and human cancer. Recent Results Cancer Res. 2021;217:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Chen CJ, You SL, Hsu WL, et al. Epidemiology of virus infection and human cancer. Recent Results Cancer Res. 2021;217:13‐45. [DOI] [PubMed] [Google Scholar]
- 223. Fan YN, Li M, Luo YL, et al. Cationic lipid‐assisted nanoparticles for delivery of mRNA cancer vaccine. Biomater Sci. 2018;6(11):3009‐3018. [DOI] [PubMed] [Google Scholar]
- 224. Igyártó BZ, Jacobsen S, Ndeupen S. Future considerations for the mRNA‐lipid nanoparticle vaccine platform. Curr Opin Virol. 2021;48:65‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Hussain A, Yang H, Zhang M, et al. mRNA vaccines for COVID‐19 and diverse diseases. J Control Release. 2022;345:314‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Benteyn D, Heirman C, Bonehill A, et al. mRNA‐based dendritic cell vaccines. Expert Rev Vaccines. 2015;14(2):161‐176. [DOI] [PubMed] [Google Scholar]
- 227. Gu YZ, Zhao X, Song XR. Ex vivo pulsed dendritic cell vaccination against cancer. Acta Pharmacol Sin. 2020;41(7):959‐969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Hotz C, Wagenaar TR, Gieseke F, et al. Local delivery of mRNA‐encoded cytokines promotes antitumor immunity and tumor eradication across multiple preclinical tumor models. Sci Transl Med. 2021;13(610):eabc7804. [DOI] [PubMed] [Google Scholar]
- 229. Bauman J, Burris H, Clarke J, et al. 798 Safety, tolerability, and immunogenicity of mRNA‐4157 in combination with pembrolizumab in subjects with unresectable solid tumors (KEYNOTE‐603): an update. J ImmunoTher Cancer. 2020;8:A846‐A846. [Google Scholar]
- 230. Bechter O, Utikal J, Baurain J‐F, et al. 391 A first‐in‐human study of intratumoral SAR441000, an mRNA mixture encoding IL‐12sc, interferon alpha2b, GM‐CSF and IL‐15sushi as monotherapy and in combination with cemiplimab in advanced solid tumors. J ImmunoTher Cancer. 2020;8:A416‐A416. [Google Scholar]
- 231. Kroemer G, Zitvogel L, Galluzzi L. Victories and deceptions in tumor immunology: stimuvax(®). Oncoimmunology. 2013;2(1):e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Dizier B, Callegaro A, Debois M, et al. A Th1/IFNγ gene signature is prognostic in the adjuvant setting of resectable high‐risk melanoma but not in non‐small cell lung cancer. Clin Cancer Res. 2020;26(7):1725‐1735. [DOI] [PubMed] [Google Scholar]
- 233. Nguyen T, Urban J, Kalinski P. Therapeutic cancer vaccines and combination immunotherapies involving vaccination. Immunotargets Ther. 2014;3:135‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Melero I, Gaudernack G, Gerritsen W, et al. Therapeutic vaccines for cancer: an overview of clinical trials. Nat Rev Clin Oncol. 2014;11(9):509‐524. [DOI] [PubMed] [Google Scholar]
- 235. Rittig SM, Haentschel M, Weimer KJ, et al. Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol Ther. 2011;19(5):990‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Buonaguro L, Tagliamonte M. Selecting target antigens for cancer vaccine development. Vaccines (Basel). 2020;8(4):615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Hollingsworth RE, Jansen K. Turning the corner on therapeutic cancer vaccines. NPJ Vaccines. 2019;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Hodi FS. Well‐defined melanoma antigens as progression markers for melanoma: insights into differential expression and host response based on stage. Clin Cancer Res. 2006;12(3 Pt 1):673‐678. [DOI] [PubMed] [Google Scholar]
- 239. Sahin U, Oehm P, Derhovanessian E, et al. An RNA vaccine drives immunity in checkpoint‐inhibitor‐treated melanoma. Nature. 2020;585(7823):107‐112. [DOI] [PubMed] [Google Scholar]
- 240. Barbier AJ, Jiang AY, Zhang P, et al. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol. 2022;40(6):840‐854. [DOI] [PubMed] [Google Scholar]
- 241. Ricciuti B, Wang X, Alessi JV, et al. Association of high tumor mutation burden in non‐small cell lung cancers with increased immune infiltration and improved clinical outcomes of PD‐L1 blockade across PD‐L1 expression levels. JAMA Oncol. 2022. doi:10.1001/jamaoncol.2022.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Gautam S, Kachroo S, DeClue RW, et al. Clinical and utilization outcomes associated with tumor mutational burden in a real‐world pan‐tumor population. J Comp Eff Res. 2021;10(10):857‐868. [DOI] [PubMed] [Google Scholar]
- 243. Kang K, Xie F, Mao J, et al. Significance of tumor mutation burden in immune infiltration and prognosis in cutaneous melanoma. Front Oncol. 2020;10:573141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Davis EJ, Johnson DB, Sosman JA, et al. Melanoma: what do all the mutations mean?. Cancer. 2018;124(17):3490‐3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Naik PP. Current trends of immunotherapy in the treatment of cutaneous melanoma: a review. Dermatol Ther (Heidelb). 2021;11(5):1481‐1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Sabari J, Ramirez KA, Schwarzenberger P, et al. Abstract B209: phase 1/2 study of mRNA vaccine therapy+ durvalumab (durva)±tremelimumab (treme) in patients with metastatic non‐small cell lung cancer (NSCLC). Cancer Immunol Res. 2019;7(2):B209‐B209. [Google Scholar]
- 247. Morse MA, Gwin WR, Mitchell DA. Vaccine therapies for cancer: then and now. Target Oncol. 2021;16(2):121‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Mapara MY, Sykes M. Tolerance and cancer: mechanisms of tumor evasion and strategies for breaking tolerance. J Clin Oncol. 2004;22(6):1136‐1151. [DOI] [PubMed] [Google Scholar]
- 249. Esprit A, de Mey W, Bahadur Shahi R, et al. Neo‐antigen mRNA vaccines. Vaccines (Basel). 2020;8(4):776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Cheng R, Xu Z, Luo M, et al. Identification of alternative splicing‐derived cancer neoantigens for mRNA vaccine development. Brief Bioinform. 2022;23(2):bbab553. [DOI] [PubMed] [Google Scholar]
- 251. Blass E, Ott PA. Advances in the development of personalized neoantigen‐based therapeutic cancer vaccines. Nat Rev Clin Oncol. 2021;18(4):215‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Guo Y, Lei K, Tang L. Neoantigen vaccine delivery for personalized anticancer immunotherapy. Front Immunol. 2018;9:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Zhao X, Pan X, Wang Y, et al. Targeting neoantigens for cancer immunotherapy. Biomark Res. 2021;9(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Lang F, Schrörs B, Löwer M, et al. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat Rev Drug Discov. 2022;21(4):261‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Tran E, Ahmadzadeh M, Lu YC, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387‐1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Deniger DC, Pasetto A, Robbins PF, et al. T‐cell responses to TP53 “hotspot” mutations and unique neoantigens expressed by human ovarian cancers. Clin Cancer Res. 2018;24(22):5562‐5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Hecht JR, Shergill A, Goldstein MG, et al. Phase 2/3, randomized, open‐label study of an individualized neoantigen vaccine (self‐amplifying mRNA and adenoviral vectors) plus immune checkpoint blockade as maintenance for patients with newly diagnosed metastatic colorectal cancer (GRANITE). J Clin Oncol. 2020;40. https://ascopubs.org/doi/abs/10.1200/JCO.2022.40.16_suppl.TPS3635 [Google Scholar]
- 259. Hu Z, Leet DE, Allesøe RL, et al. Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nat Med. 2021;27(3):515‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Kreiter S, Vormehr M, van de Roemer N, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692‐696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Kalnin KV, Plitnik T, Kishko M, et al. Immunogenicity and efficacy of mRNA COVID‐19 vaccine MRT5500 in preclinical animal models. NPJ Vaccines. 2021;6(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Zhang R, Yuan F, Shu Y, et al. Personalized neoantigen‐pulsed dendritic cell vaccines show superior immunogenicity to neoantigen‐adjuvant vaccines in mouse tumor models. Cancer Immunol Immunother. 2020;69(1):135‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Linette GP, Carreno BM. Neoantigen vaccines pass the immunogenicity test. Trends Mol Med. 2017;23(10):869‐871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Ni Q, Zhang F, Liu Y, et al. A bi‐adjuvant nanovaccine that potentiates immunogenicity of neoantigen for combination immunotherapy of colorectal cancer. Sci Adv. 2020;6(12):eaaw6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Lancaster EM, Jablons D, Kratz JR. Applications of next‐generation sequencing in neoantigen prediction and cancer vaccine development. Genet Test Mol Biomarkers. 2020;24(2):59‐66. [DOI] [PubMed] [Google Scholar]
- 266. Zhang S, So A, Kaplan S, et al. Abstract A09: a bioinformatics workflow to identify neoantigens using next‐generation sequencing. Cancer Immunol Res. 2017;5(3):A09‐A09. [Google Scholar]
- 267. Keskin DB, Anandappa AJ, Sun J, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565(7738):234‐239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Hilf N, Kuttruff‐Coqui S, Frenzel K, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565(7738):240‐245. [DOI] [PubMed] [Google Scholar]
- 269. Burris HA, Patel MR, Cho DC, et al. A phase I multicenter study to assess the safety, tolerability, and immunogenicity of mRNA‐4157 alone in patients with resected solid tumors and in combination with pembrolizumab in patients with unresectable solid tumors. J Clin Oncol. 2019;37:2523‐2523. [Google Scholar]
- 270. Lopez JS, Camidge R, Iafolla M, et al. Abstract CT301: a phase Ib study to evaluate RO7198457, an individualized neoantigen specific immunoTherapy (iNeST), in combination with atezolizumab in patients with locally advanced or metastatic solid tumors. Cancer Res. 2020;80(16):CT301‐CT301. [Google Scholar]
- 271. Ott PA, Hu‐Lieskovan S, Chmielowski B, et al. A phase Ib trial of personalized neoantigen therapy plus anti‐PD‐1 in patients with advanced melanoma, non‐small cell lung cancer, or bladder cancer. Cell. 2020;183(2):347‐362.e324. [DOI] [PubMed] [Google Scholar]
- 272. Peng M, Mo Y, Wang Y, et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol Cancer. 2019;18(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Reynolds CR, Tran S, Jain M, et al. Neoantigen cancer vaccines: generation, optimization, and therapeutic targeting strategies. Vaccines (Basel). 2022;10(2):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Ingels J, De Cock L, Mayer RL, et al. Small‐scale manufacturing of neoantigen‐encoding messenger RNA for early‐phase clinical trials. Cytotherapy. 2022;24(2):213‐222. [DOI] [PubMed] [Google Scholar]
- 275. Heiser A, Coleman D, Dannull J, et al. Autologous dendritic cells transfected with prostate‐specific antigen RNA stimulate CTL responses against metastatic prostate tumors. J Clin Invest. 2002;109(3):409‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Cheever MA, Higano CS. PROVENGE (Sipuleucel‐T) in prostate cancer: the first FDA‐approved therapeutic cancer vaccine. Clin Cancer Res. 2011;17(11):3520‐3526. [DOI] [PubMed] [Google Scholar]
- 277. Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel‐T immunotherapy for castration‐resistant prostate cancer. N Engl J Med. 2010;363(5):411‐422. [DOI] [PubMed] [Google Scholar]
- 278. Sutherland SIM, Ju X, Horvath LG, et al. Moving on from sipuleucel‐T: new dendritic cell vaccine strategies for prostate cancer. Front Immunol. 2021;12:641307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Anassi E, Ndefo UA. Sipuleucel‐T (provenge) injection: the first immunotherapy agent (vaccine) for hormone‐refractory prostate cancer. P t. 2011;36(4):197‐202. [PMC free article] [PubMed] [Google Scholar]
- 280. Borch TH, Engell‐Noerregaard L, Zeeberg Iversen T, et al. mRNA‐transfected dendritic cell vaccine in combination with metronomic cyclophosphamide as treatment for patients with advanced malignant melanoma. Oncoimmunology. 2016;5(9):e1207842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Bonehill A, Heirman C, Tuyaerts S, et al. Messenger RNA‐electroporated dendritic cells presenting MAGE‐A3 simultaneously in HLA class I and class II molecules. J Immunol. 2004;172(11):6649‐6657. [DOI] [PubMed] [Google Scholar]
- 282. Bol KF, Figdor CG, Aarntzen EH, et al. Intranodal vaccination with mRNA‐optimized dendritic cells in metastatic melanoma patients. Oncoimmunology. 2015;4(8):e1019197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Bontkes HJ, Kramer D, Ruizendaal JJ, et al. Dendritic cells transfected with interleukin‐12 and tumor‐associated antigen messenger RNA induce high avidity cytotoxic T cells. Gene Ther. 2007;14(4):366‐375. [DOI] [PubMed] [Google Scholar]
- 284. Grünebach F, Kayser K, Weck MM, et al. Cotransfection of dendritic cells with RNA coding for HER‐2/neu and 4‐1BBL increases the induction of tumor antigen specific cytotoxic T lymphocytes. Cancer Gene Ther. 2005;12(9):749‐756. [DOI] [PubMed] [Google Scholar]
- 285. Van Lint S, Goyvaerts C, Maenhout S, et al. Preclinical evaluation of TriMix and antigen mRNA‐based antitumor therapy. Cancer Res. 2012;72(7):1661‐1671. [DOI] [PubMed] [Google Scholar]
- 286. Wilgenhof S, Van Nuffel A, Benteyn D, et al. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann Oncol. 2013;24(10):2686‐2693. [DOI] [PubMed] [Google Scholar]
- 287. Wilgenhof S, Corthals J, Heirman C, et al. Phase II study of autologous monocyte‐derived mRNA electroporated dendritic cells (TriMixDC‐MEL) plus ipilimumab in patients with pretreated advanced melanoma. J Clin Oncol. 2016;34(12):1330‐1338. [DOI] [PubMed] [Google Scholar]
- 288. Liu T, Liang Y, Huang L. Development and delivery systems of mRNA vaccines. Front Bioeng Biotechnol. 2021;9:718753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Pichon C, Perche FJTB. Design and delivery of messenger RNA‐based vaccines. The Biochemist. 2021;43(4):4‐7.34219990 [Google Scholar]
- 290. To KKW, Cho WCS. An overview of rational design of mRNA‐based therapeutics and vaccines. Expert Opin Drug Discov. 2021;16(11):1307‐1317. [DOI] [PubMed] [Google Scholar]
- 291. Cao Y, Gao GF. mRNA vaccines: a matter of delivery. EClinicalMedicine. 2021;32:100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Servick K, This mysterious $2 billion biotech is revealing the secrets behind its new drugs and vaccines. Accessed March 5, 2020. https://www.science.org/content/article/mysterious‐2‐billion‐biotech‐revealing‐secrets‐behind‐its‐new‐drugs‐and‐vaccines
- 293. Khurana A, Allawadhi P, Khurana I, et al. Role of nanotechnology behind the success of mRNA vaccines for COVID‐19. Nano Today. 2021;38:101142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Bochicchio S, Lamberti G, Barba AA. Polymer‐lipid pharmaceutical nanocarriers: innovations by new formulations and production technologies. Pharmaceutics. 2021;13(2):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Shepherd SJ, Issadore D, Mitchell MJ. Microfluidic formulation of nanoparticles for biomedical applications. Biomaterials. 2021;274:120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Edwards DK, Jasny E, Yoon H, et al. Adjuvant effects of a sequence‐engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J Transl Med. 2017;15(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Zhu W, Dong C, Wei L, et al. Promising adjuvants and platforms for influenza vaccine development. Pharmaceutics. 2021;13(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Pardi N, Hogan MJ, Naradikian MS, et al. Nucleoside‐modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med. 2018;215(6):1571‐1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Alameh M‐G, Weissman D. Nucleoside modifications of in vitro transcribed mRNA to reduce immunogenicity and improve translation of prophylactic and therapeutic antigens. RNA Therapeutics. 2022;141‐169. 10.1016/B978-0-12-821595-1.00014-2 [DOI] [Google Scholar]
- 300. McKay PF, Hu K, Blakney AK, et al. Self‐amplifying RNA SARS‐CoV‐2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11(1):3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Bansal D, Reimers MA, Knoche EM, et al. Immunotherapy and immunotherapy combinations in metastatic castration‐resistant prostate cancer. Cancers (Basel). 2021;13(2):334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Salvatori E, Lione L, Compagnone M, et al. Neoantigen cancer vaccine augments anti‐CTLA‐4 efficacy. NPJ Vaccines. 2022;7(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Bahl K, Senn JJ, Yuzhakov O, et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther. 2017;25(6):1316‐1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


