Abstract
Fusarium infections in humans (fusariosis) and in economically important plants involve species of several Fusarium species complexes. Species of the Fusarium solani species complex (FSSC) are the most frequent cause of human fusariosis. The FSSC comprises more than 60 closely related species that can be separated into three major clades by multi-locus sequence typing (MLST) using translation elongation factor 1-alpha (TEF1-α) and RNA polymerase II (RPB2) DNA sequences. The MLST nomenclature for clade 3 of the FSSC assigns numbers to species types (e.g., FSSC 2) and lowercase letters to identify unique haplotypes. The aim of this study was to analyse the genotypic and phenotypic characteristics of 15 environmental and 15 clinical FSSC isolates from Malaysia. MLST was used for the genotypic characterisation of FSSC isolates from various locations within Malaysia, which was complemented by their morphological characterisation on potato dextrose and carnation leaf agar. MLST identified eight different FSSC species: thirteen Fusarium keratoplasticum (i.e., FSSC 2), six Fusarium suttonianum (FSSC 20), five Fusarium falciforme (FSSC 3+4), two Fusarium cyanescens (FSSC 27), and one each of Fusarium petroliphilum (FSSC 1), Fusarium waltergamsii (FSSC 7), Fusarium sp. (FSSC 12), and Fusarium striatum (FSSC 21). Consistent with previous reports from Malaysia, most (11 of 15) clinical FSSC isolates were F. keratoplasticum and the majority (9 of 15) of environmental isolates were F. suttonianum (5) or F. falciforme (4) strains. The taxonomic relationships of the isolates were resolved phylogenetically. The eight Fusarium species also showed distinct morphological characteristics, but these were less clearly defined and reached across species boundaries. Although TEF1-α and RPB2 sequences were sufficient for the species identification of most FSSC isolates, a more precise MLST scheme needs to be established to reliably assign individual isolates of the species-rich FSSC to their geographically-, epidemiologically-, and host-associated sub-lineages.
Keywords: Fusarium solani, FSSC, MLST, TEF1-α, RPB2
1. Introduction
Fusarium, a genetically complex genus, belongs to the fungal order Hypocreales, in the class Sordariomycetes. The fusaria are globally distributed in environments such as soil, water, plants, and human habitats. Fusaria are significant plant pathogens causing severe vascular wilt and root rot disease in agriculturally important crops [1,2,3]. The Fusarium head blight of wheat [4] and Fusarium wilt of bananas [5] are amongst the most devastating diseases in plant hosts, causing heavy losses in global food production with an enormous impact on the communities that depend on these crops [6,7]. Fusarium wilt of banana, also known as Panama disease [8], has caused several devastating crop losses to the global banana industry [9,10].
Fusaria are also among the most frequent causes of invasive mould infections (IMIs), second only to aspergilli [11]. The increasing number of immunocompromised patients [12,13] heightens the concern for IMIs as they typically have high mortality rates (56%) [14], especially in neutropenic patients where the mortality rate reaches as high as 100% [15]. Fusaria comprise 20 species complexes [16]. The Fusarium solani species complex (FSSC) containing more than 60 different species accounts for ~60% of fusariosis cases worldwide [17,18], ranging from localised skin, nail, and eye infections to life-threatening disseminated IMIs [19].
Historically, species of the FSSC were simply referred to as Fusarium solani based on morphological features, host-specific pathogenicity, and sexual/asexual compatibility [20]. However, a Fusarium keratitis outbreak in the United States [21] changed that approach through the application of multi-locus sequence typing (MLST). FSSC isolates separated into three distinct clades based on the ribosomal RNA (rRNA) internal transcribed spacer (ITS) region, the D1 and D2 domains of the nuclear large-subunit (LSU) rRNA genes, translation elongation factor 1-alpha (TEF1-α), and RNA polymerase II (RPB2) sequences [22]. Clade 1 and clade 2 species of the FSSC are geographically restricted and exclusively associated with plants. They appear to be endemic to New Zealand and South America, respectively [23]. Members of FSSC clade 3 are more common in highly populated areas, and they seem to grow faster and produce more conidia than members of clades 1 or 2 [24]. The informal haplotype nomenclature of FSSC clade 3 species [21,22] was developed to facilitate sharing of more accurate epidemiological data by assigning individual isolates to a particular MLST. For example, FSSC clade 3 isolates with the MLST 1-b or 2-a are F. petroliphilum (FSSC 1) or F. keratoplasticum (FSSC 2) isolates, respectively. In 2018, Sandoval-Denis et al. proposed renaming species of the FSSC as Neocosmospora [25,26]. This proposal was, however, subsequently rebutted [27], and there is no final decision as to which nomenclature is more accurate or useful in a clinical setting [28].
Infections by species of the FSSC are problematic because most are innately resistant to mainstream azole antifungals, including voriconazole [29,30], which is the recommended treatment option for invasive fusariosis (IF) [31,32]. Amphotericin B seems to be the only antifungal to which most FSSC species are susceptible [29,30]. This hinders the efficacy of treating immunocompromised patients with IF [33,34,35,36]. The widespread use of fungicides in agriculture [37] has led to the rise and global spread of azole resistant Aspergillus fumigatus isolates [38] into hospital settings [39,40,41], some of which are beginning to exhibit resistance to multiple classes of antifungals [42]. A similar hypothesis was recently put forward for the emergence of Candida auris, a drug resistant human fungal pathogen of serious clinical concern. An investigation by Yadav et al. [43], provided strong evidence for the possible emergence and spread of multidrug resistant C. auris clinical isolates, usually found in marine habitats, through the application of fungicides on stored apples. A detailed review of the concerning trend for the selection of drug resistant fungal pathogens, including various Candida species and Cryptococcus neoformans, through the extensive use of agrochemicals in agriculture and the wood industry is provided by Bastos et al. [44].
The azole target, lanosterol-14α-demethylase, encoded by CYP51 genes [45], has three orthologues in Fusarium: CYP51A, CYP51B, and CYP51C [46]. We have recently reported a strong association between a CYP51A promoter deletion and voriconazole resistance in a collection of 25 Malaysian FSSC isolates, 20 of which were included in this study [46]. The 23 bp CYP51A promoter deletion was present in all voriconazole resistant FSSC isolates (six F. keratoplasticum and three F. suttonianum isolates) that could be sequenced. Unfortunately, the presence of the 23 bp CYP51A promoter deletion could not be confirmed for two additional voriconazole resistant clinical F. falciforme isolates (Ff541 and Ff0020), because neither DNA oligomer primer pair used could amplify this region for DNA sequencing. Interestingly, however, this region could be amplified from the remaining three voriconazole sensitive environmental F. falciforme isolates. As expected, none of these three isolates contained the 23 bp CYP51A promoter deletion [46]. These findings indicated a possible DNA exchange of the 23 bp CYP51A promoter deletion between the ancestors of the voriconazole resistant F. keratoplasticum and F. suttonianum and possibly also F. falciforme isolates via asexual recombination and positive selection by azole fungicide use in agriculture [46,47,48]. In the Fusarium fujikuroi species complex [49], however, antifungal resistance appears to be species specific [1]. Thus, a carefully designed MLST scheme for the identification of individual Fusarium isolates is essential to determine appropriate antifungal treatment therapy.
In Malaysia, most reports of Fusarium infections are related to plant infections of oil palm [50], pineapple [51], or paddy field soils [52], while another study reported that sea turtles were infected with Fusarium [53]. A study of 1449 environmental Fusarium isolates from the Malaysian highlands found that 66.1% belonged to the FSSC [54], highlighting its predominance in Malaysia. The first fusariosis case in Malaysia, a patient diagnosed with Fusarium keratitis, was reported in 1981 [55], and a 5-year retrospective review (2007–2011) of fungal keratitis at the Universiti Sains Malaysia Hospital found that almost half (46%; 19 of 41) of all fungal keratitis cases were caused by FSSC species [56].
The present study aimed to determine the phenotypic and genotypic relationships of FSSC isolates and their species distribution in the environment and in clinical samples. We hypothesized that genotypic identification through a molecular approach is more accurate and informative in a clinical context, as the nature of antifungal resistance in fusaria is species-specific and the taxonomic clade of the FSSC is diverse. The comparison between phenotypic and genotypic characteristics is an important first step to enable accurate and rapid identification of isolates of the FSSC so that effective antifungal treatment can be implemented for Fusarium infections of humans, animals, and plants. This study describes the species distribution of 30 FSSC isolates collected in Malaysia, and it provides a detailed phenotypic and morphological characterization of 10 clinical and 15 environmental isolates from that collection.
2. Materials and Methods
2.1. Fungal Isolates
The 15 clinical Fusarium isolates were obtained from the Hospital Canselor Tuanku Muhriz UKM and the Institute for Medical Research, Malaysia. They had originally been collected from nail, skin, corneal scrapings, and blood as a part of routine diagnostic procedures (Table 1). No identifying data from any patients were obtained or utilised in this study. A further 15 environmental isolates were from soil and plant debris located across six states in Peninsular Malaysia: Terengganu, Pahang, Kelantan, Perak, Kedah, and Selangor (Table 1). All isolates were presumptively identified as FSSC species based on their conidia morphology [57]. The antifungal susceptibilities of 25 of the 30 isolates to itraconazole/posaconazole, voriconazole, and amphotericin B were previously reported [46].
Table 1.
List of 15 clinical and 15 environmental FSSC isolates from Malaysia, their MLST type and their GenBank accession numbers for TEF1-α and RPB2.
| Isolate | Source 1 | Species | MLST Type 2 | GenBank Accession Number | |
|---|---|---|---|---|---|
| TEF1-α | RPB2 | ||||
| Clinical Isolates | |||||
| Fp667 | Eye | F. petroliphilum | 1-b | MN178239 | MN263125 |
| Fk620 | Skin | F. keratoplasticum | 2-a | MN178238 | MN263124 |
| Fk2781 | Nail | F. keratoplasticum | 2-a | MN178234 | MN263120 |
| Fk2309 | Nail | F. keratoplasticum | 2-f | MN178231 | MN263117 |
| Fk553 | Skin | F. keratoplasticum | 2-h | MN178237 | MN263123 |
| Fk2353 | Nail | F. keratoplasticum | 2-h | MN178232 | MN263118 |
| Fk994 | Nail | F. keratoplasticum | 2-h | MN178240 | MN263126 |
| Fk0168 | Blood | F. keratoplasticum | 2-h | MN178228 | MN263114 |
| Fk2622 | Nail | F. keratoplasticum | 2-h | MN178233 | MN263119 |
| Fk1049 * | Nail | F. keratoplasticum | 2-h | MN178241 | MN263127 |
| Fk1931 * | Nail | F. keratoplasticum | 2-k | MN178230 | MN263116 |
| Fk1930 * | Nail | F. keratoplasticum | 2-k | MN178229 | MN263115 |
| Ff0020 * | Eye | F. falciforme | 3+4-k | MN178227 | MN263113 |
| Fstr541 | Blood | F. striatum | 21 | MN178236 | MN263122 |
| Fs263 * | Eye | F. suttonianum | 20-c | MN178235 | MN263121 |
| Environmental isolates | |||||
| FkDI17 | Grass | F. keratoplasticum | 2-a | MN178221 | MN263107 |
| FkDir61 | Grass | F. keratoplasticum | 2-a | MN178225 | MN263111 |
| Ff4225 | Tobacco | F. falciforme | 3+4-k | MN178212 | MN263098 |
| Ff4290 | Straw compost | F. falciforme | 3+4-k | MN178215 | MN263101 |
| Ff4324 ** | Soil | F. falciforme | 3+4-k | MN178216 | MN263102 |
| Ff4325 | Honeydew | F. falciforme | 3+4-k | MN178217 | MN263103 |
| FwgDE4 ** | Soil | F. waltergamsii | 7-b | MN178218 | MN263104 |
| FspDE40 ** | Soil | Fusarium sp. | 12-a | MN178220 | MN263106 |
| Fs3769 | Coconut tree | F. suttonianum | 20-c | MN178207 | MN263093 |
| Fs3784 | Mangrove | F. suttonianum | 20-c | MN178209 | MN263094 |
| Fs3873 | Grass | F. suttonianum | 20-c | MN178208 | MN263095 |
| Fs3924 | Sugarcane | F. suttonianum | 20-c | MN178210 | MN263096 |
| Fs4279 | Dragon fruit | F. suttonianum | 20-c | MN178214 | MN263100 |
| FcDir16 ** | Soil | F. cyanescens | 27-a | MN178223 | MN263109 |
| FcDir23 ** | Soil | F. cyanescens | 27-a | MN178224 | MN263110 |
1 The body site or plant or soil environment from which the samples were collected. 2 The MLST was determined based on polyphasic identification using the Fusarium MLST database. Numbers were assigned to designate species and lowercase letters to identify unique haplotypes. * These isolates were excluded from the morphological investigations. ** These are additional isolates that were not included in the study by James et al. [46].
2.2. Morphological Examination
Macromorphological examinations were conducted on isolates grown on potato dextrose agar (PDA; Merck, Kenilworth, NJ, USA) plates incubated at 28 °C for fourteen days. The diameter and the colour of the colony surface and substrate mycelia (reverse side/bottom of agar plate) were recorded. For microscopic examination, each isolate was sub-cultured on approximately 1 cm2 blocks of agar containing a piece (0.5 cm2) of sterile carnation leaf agar (CLA). The agar blocks were placed on a sterile microscope slide, covered with a sterile cover slip, and incubated inside a petri dish for 12 h periods of light (day) and 12 h periods in the dark (night) by using a 103 V fluorescent light bulb (TL-D Standard Colours; Philips & Co., Eindhoven, The Netherlands) as a light source at room temperature (28 °C) for four to seven days until sufficient hyphal growth was observed on the cover slip. For microscopic observation, a drop of lactophenol cotton blue (Sigma-Aldrich, St. Louis, MO, USA) was applied to the fungal mycelia on the cover slip and the cover slip was placed face-down onto a microscope slide. The length and width of 30 randomly selected macroconidia were measured to determine the mean length and width for each isolate. Descriptions of the morphological characteristics were adopted from the Fusarium Laboratory Manual [57].
2.3. DNA Extraction and PCR Amplification
Fungal genomic DNA was extracted as previously described [46], and subsequently used for polymerase chain reaction (PCR) amplification of TEF1-α and RPB2 using previously described primers [22], listed in Table S1. The PCR mixture (20 μL) contained 1× GoTaq G2 Green Master Mix (Promega, Madison, WI, USA), 0.8 μM primers, and 10 ng template DNA. DNA amplification was performed with an initial denaturation of 1 min at 95 °C followed by 35 cycles of 30 s at 95 °C, 60 s at 55 °C, and 90 s at 72 °C and a final extension of 5 min at 72 °C. PCR products were purified using the QIAquick PCR Purification kit (QIAGEN Inc., Valencia, CA, USA) following the manufacturer’s instructions.
2.4. DNA Sequencing and Analysis of TEF1-α and RPB2 Sequences
DNA sequencing of PCR products was carried out by First BASE Laboratories Sdn Bhd (now known as Apical Scientific Sdn Bhd, Seri Kembangan, Malaysia). The TEF1-α and RPB2 sequences were compared with other fungal DNA sequences using the online NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi; 27 February 2022). For Fusarium species identification, the TEF1-α and RPB2 sequences were used in a BLAST search against the Fusarium MLST database (https://fusarium.mycobank.org/page/Fusarium_identification; 27 February 2022). The accession numbers for the TEF1-α and RPB2 sequences are listed in Table 1.
2.5. Phylogenetic Analyses
A representative dataset of TEF1-α and RPB2 sequences of FSSC clades 1, 2, and 3 species were extracted from the GenBank DNA repository (Table S2). The sequences were selected to best represent the global distribution of clinical and environmental isolates of the FSSC. The phylogenetic relationships between the concatenated TEF1-α and RPB2 sequences were investigated with two independent methods: Maximum Parsimony (MP) and Maximum Likelihood (ML). MP analysis was performed using MEGA 10.0.5 [58]. Trees were generated for 100 replicates of random stepwise addition of sequences and the Subtree-Pruning-Regrafting (SPR) algorithm [59], with all characters given equal weight. Branch support for the MP analysis was estimated by performing 1000 bootstrap replicates with a heuristic search of 10 random–addition replicates for each bootstrap replicate [60]. Phylogenetic trees were visualized using the program Treeview [61]. ML analysis was performed using RAxML-HPC2 version 8.2.12 on XSEDE [62] via the CIPRES Science Gateway Portal version 3.1 [63]. The best evolutionary model TIM2+I+G4 for the TEF1a-RPB2 dataset was calculated using ModelTest-NG on XSEDE [64].
2.6. Fusarium Growth Characteristics
Mycelial dry weights were used to determine the growth behaviour of selected isolates. Using optical densities to measure growth was not possible, because Fusarium cells grew as large, fluffy balls of mycelia after ~15 h incubation in liquid medium.
Inoculum suspensions of microconidia were prepared by adding 5 mL sterile saline to 5-day old PDA plate cultures, gently resuspending the cells by scraping with the end of a 1 mL pipette tip, and filtering the cell suspension through a two-layered cheesecloth. The cells were harvested in a 15 mL centrifuge tube by centrifugation for 5 min at 8000× g and resuspension of the cell pellet in the required volume of potato dextrose broth (PDB), containing 20% glycerol, to adjust the cell suspension to OD530 = 10 (~1 × 108 cfu/mL). The microconidia suspension was aliquoted in 1.5 mL microcentrifuge tubes and used immediately for inoculation or stored at −80 °C until required.
The growth characteristics of F. keratoplasticum Fk2309 and FkDir61, F. falciforme Ff4290 and F. striatum Fstr541 were determined for two media: PDB and RPMI-1640 (R6504; Sigma-Aldrich, St. Louis, MO, USA) supplemented with 2% glucose (i.e., RPMI-1640 supplemented with 18 g glucose). Ten microlitre cell suspensions (i.e., 106 cfu) of microconidia were used to inoculate flasks containing 50 mL medium, one for each time point of harvest. Inoculated media were incubated at 28 °C with shaking at 200 rpm. The cell suspensions were harvested by filtration through a glass fibre filter (No. 6; Schleicher & Schuell BioScience GmbH, Dassel, Germany) using a vacuum pump. The filters were dehydrated in a 30 °C incubator for 24 h and the cell dry weight was determined by subtracting the dry weight of the same filter measured before cell harvest.
3. Results
3.1. Molecular Identification
The 30 FSSC isolates from Malaysia were designated to species using their TEF1-α and RPB2 sequences and the polyphasic identification system of the Fusarium MLST database (Table 1). The 30 FSSC isolates comprised thirteen F. keratoplasticum (FSSC 2), six F. suttonianum (FSSC 20), five F. falciforme FSSC 3+4), two F. cyanescens (FSSC 27), and one isolate each of F. petroliphilum (FSSC 1), F. waltergamsii (FSSC 7), Fusarium sp. (FSSC 12) and F. striatum (FSSC 21). Most (11; 85%) F. keratoplasticum strains were clinical isolates as were the single isolates of F. petroliphilum and F. striatum while most (5; 83%) F. suttonianum strains and the single isolates of F. waltergamsii, Fusarium sp. of FSSC 12 and F. cyanescens were of environmental origin.
An ML phylogenetic tree for the concatenated TEF1-α and RPB2 sequences (2359 nucleotides) of 62 selected FSSC isolates from across the globe, including the 30 isolates from Malaysia, is presented in Figure 1. The majority (23 out of 30) of the Malaysian FSSC isolates (bold in Figure 1) belonged to three closely related FSSC species: F. keratoplasticum (FSSC 2), F. falciforme (FSSC 3+4) and F. suttonianum (FSSC 20). Isolates of the FSSC 2 or the FSSC 20 lineage could be clearly distinguished with the base-root branch of each showing 100% bootstrap support for both the MP and ML algorithms (Figure 1). However, the assignment of F. falciforme isolates to the FSSC 3+4 lineage was less clearly defined, with the base-root branch showing only 90% MP and 94% ML bootstrap support. One of the 30 Malaysian isolates (Fp667) was placed in the FSSC 1 lineage of F. petroliphilum. The F. striatum isolate Fstr541, a human blood isolate, formed a distinct branch (100% bootstrap support) with two closely related F. striatum sequences, one an insect-isolate from Columbia (NRRL52699) and the other an environmental isolate from Panama (NRRL22101; Figure 1). The sub-division of isolates within the FSSC 2 and FSSC 3+4 lineages (e.g., F. keratoplasticum FSSC 2-a, -f, -h, -k) mostly followed their haplotype assignment [22], although with less convincing bootstrap support for the individual sub-groups (Figure 1).
Figure 1.
Maximum likelihood (ML) phylogram of the concatenated TEF1-α and RPB2 sequences for the 30 Malaysian FSSC isolates and 37 publicly available sequences of globally distributed FSSC isolates. DNA sequences of two close relatives (F. cicatricum and F. staphyleae) were used as the outgroup. Isolates characterised in this study are highlighted in bold font. The source of individual isolates (E = environmental, H = hospital, P = plant) is shown in the column to the right of the strain identifier. The voriconazole resistant (MIC > 32 mg/L) isolates with the 23 bp CYP51A promoter deletion and the voriconazole susceptible (MIC ≤ 12 mg/L) isolates are indicated with R and S, respectively. The voriconazole susceptibilities of isolates marked with a question mark (?) were not determined. Technical issues prevented the confirmation of the 23 bp CYP51A promoter deletion in the voriconazole resistant F. falciforme (Ff0020) and F. striatum (Fstr541) isolates (marked with asterisks; *). The roots, with 100% bootstrap support, of the three major FSSC clades (clades 1, 2 and 3) are in bold. Numbers at internodes represent the percentage maximum parsimony (MP) and maximum likelihood (ML) bootstrap support (MP-BS/ML-BS) of 1000 replicates; a single value means that both values were identical. The scale bar indicates the number of nucleotide substitutions per position.
3.2. Morphological Characteristics
To investigate whether morphological characteristics correspond with the molecular species identification, we examined the macroscopic and microscopic characteristics of 25 of the 30 FSSC isolates (Table 1). The species of individual FSSC isolates could not be identified with certainty based on their colony morphology alone (Figure 2A,B). The macroconidia morphologies (Figure 3) provided more distinctive species-specific characteristics, although these too were imprecise and crossed species boundaries.
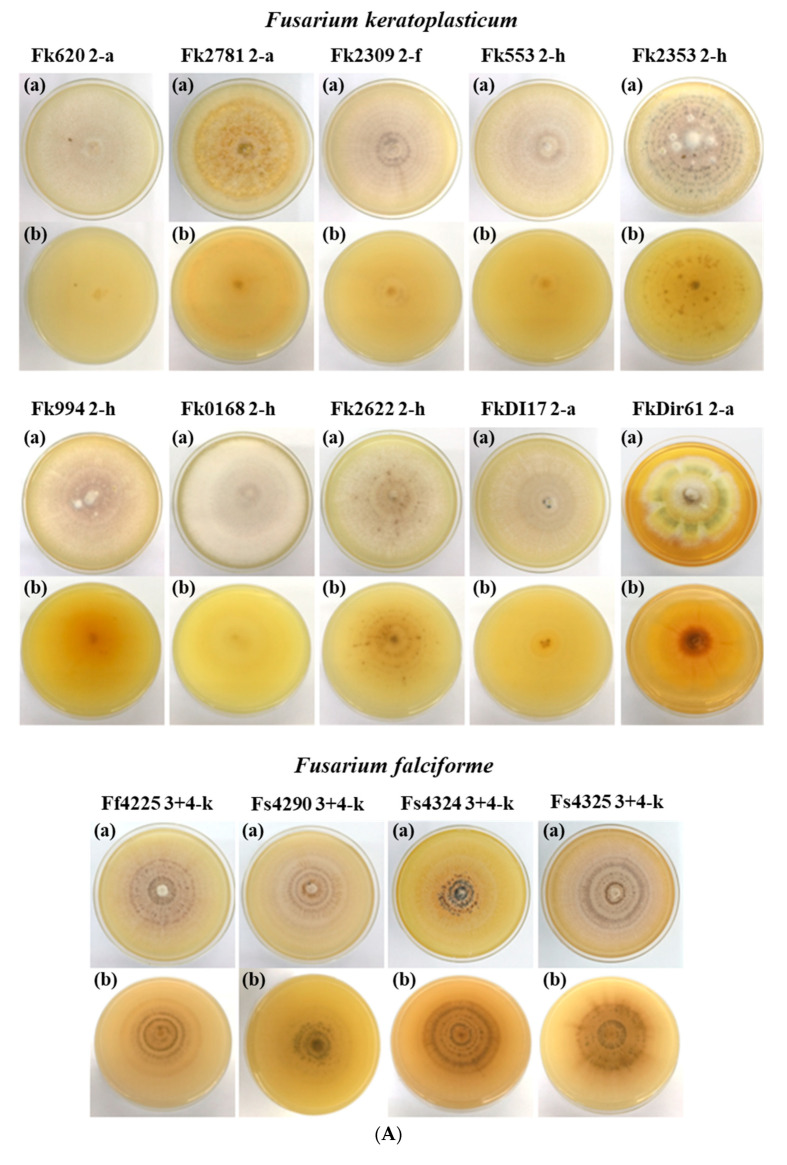
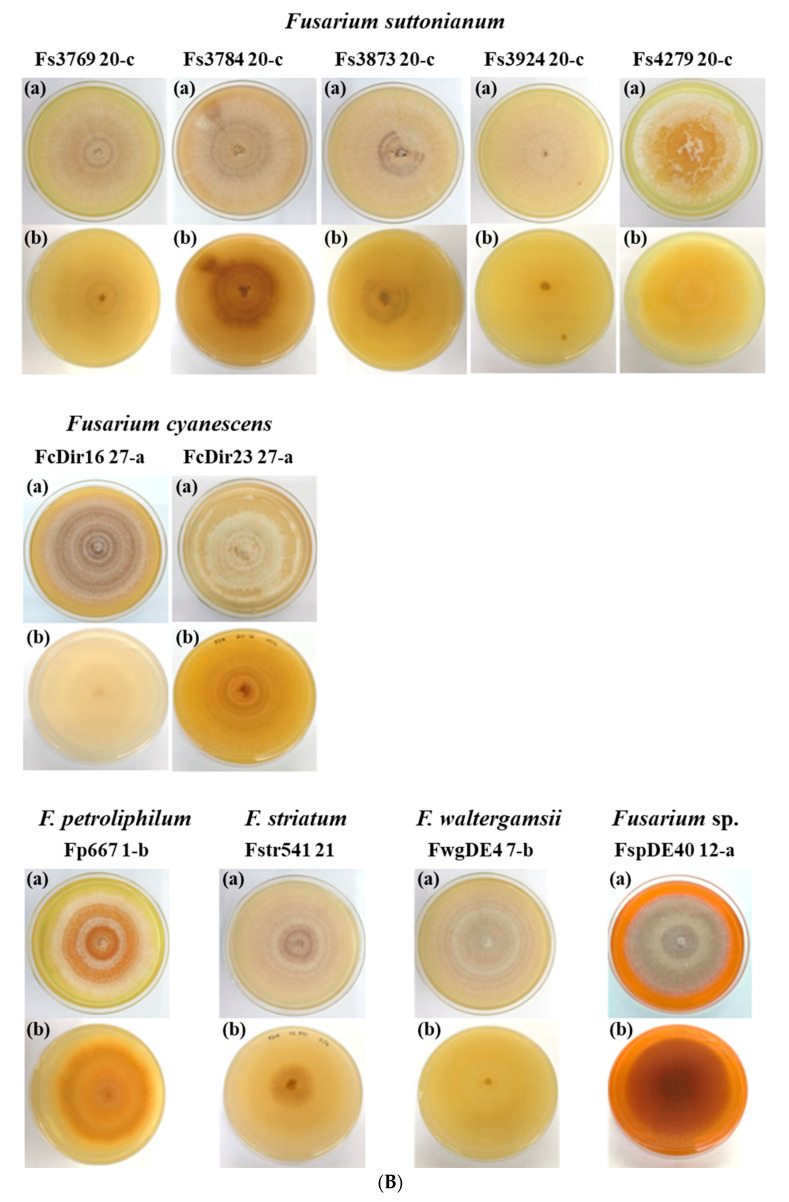
Figure 2.
(A). Colony morphologies of F. keratoplasticum (ten) and F. falciforme (four) isolates grown on PDA at 28 °C for two weeks with alternating 12 h light and 12 h dark periods. View from the top (a) and underneath (b). (B). Colony morphologies of F. suttonianum (five), F. cyanescens. (two), and one each of F. petroliphilum, F. striatum, F. waltergamsii and Fusarium sp. isolates grown on PDA at 28 °C for two weeks with alternating 12 h light and 12 h dark periods. View from the top (a) and underneath (b).
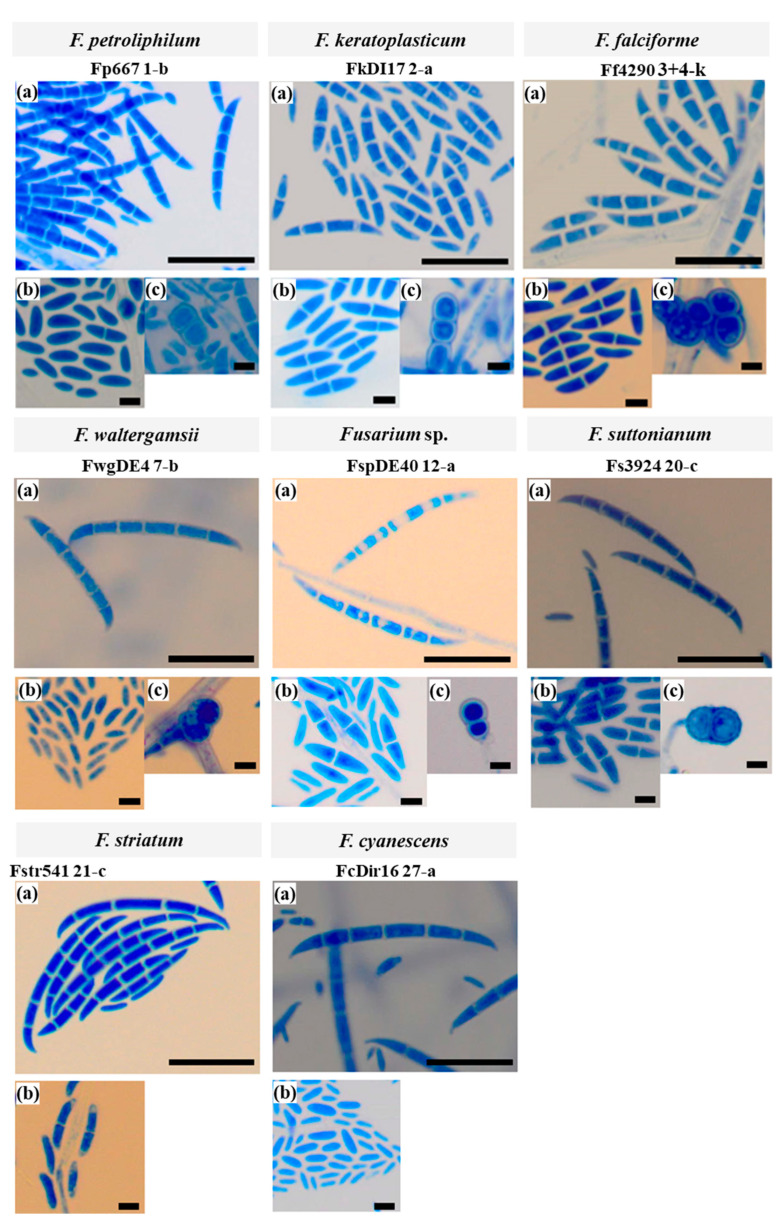
Figure 3.
Microscopic images at 40× magnification of eight FSSC isolates representing the eight species identified among the collection of the 30 Malaysian FSSC isolates. Cells were grown on CLA at 28 °C for four to seven days with alternating 12 h light and 12 h dark periods. Cells were stained with lactophenol cotton blue. Macroconidia (a; scale bar: 100 µm), microconidia (b; scale bar: 20 µm), and chlamydospores (c; scale bar: 20 µm).
3.2.1. Colony Morphology
The growth rate of colonies grown on PDA for nine days at 28 °C with 12 h light (day) and 12 h (night) dark periods ranged from 6.1 to 9.4 mm (diameter)/day (Table 2). Early hyphae were hyaline and sparse, radiating out from the centre of the agar plug containing the mycelial inoculum. The aerial mycelia of all young colonies (3–5 days) were white, but their colour and appearance changed with age resulting in rings of white interspersed by coloured patches ranging from green to yellow, orange, brown and dark brown (see images labelled (a) in Figure 2A,B). The colour of the colony after two weeks of growth, viewed from underneath, ranged from shades of yellow to dark brown (see images labelled (b) in Figure 2A,B). The mean colony diameters on day-9 incubation for the ten F. keratoplasticum, four F. falciforme and five F. suttonianum isolates were 63 ± 8.39, 75 ± 3.40, and 81 ± 4.41 mm, respectively (Table 2).
Table 2.
Morphological features of FSSC isolates.
| Isolate 1 | Mycelial Growth on PDA at 28 °C (Day-9) |
Length and Width of Macroconidia (µm) 2,3,4 |
|||||
|---|---|---|---|---|---|---|---|
| Top View |
Viewed from Underneath | Colony Diameter (mm) |
2 and 3 Septa | 4 and 5 Septa | |||
| Length | Width | Length | Width | ||||
| CLINICAL ISOLATES (10) | |||||||
| Fp667 | white, brown | brown, yellow | 55 | 27.10 ± 3.65 | 2.79 ± 0.41 | 30.61 ± 2.68 | 2.99 ± 0.39 |
| Fk620 | White | white | 70 | none | none | none | none |
| Fk2781 | white brownish | yellow | 66 | 23.68 ± 2.95 | 3.05 ± 0.52 | none | none |
| Fk2309 | white with purple stripes | white | 55 | 21.37 ± 2.56 | 3.04 ± 0.47 | none | none |
| Fk553 | White | yellow | 66 | none | none | none | none |
| Fk2353 | white with purplish dark spots | white | 60 | 20.85 ± 1.93 | 3.02 ± 0.46 | none | none |
| Fk994 | white, brown | orange | 63 | 21.71 ± 3.22 | 2.91 ± 0.49 | none | none |
| Fk0168 | White | yellow | 68 | none | none | none | none |
| Fk2622 | White | white | 58 | none | none | none | none |
| Fstr541 | dark brown, white | dark brown, yellow | 65 | 26.63 ± 3.47 | 2.44 ± 0.47 | 31.49 ± 9.90 | 2.81 ± 0.49 |
| ENVIRONMENTAL ISOLATES (15) | |||||||
| FkDI17 | White | yellow | 78 | 24.56 ± 1.92 | 2.95 ± 0.50 | none | none |
| FkDir61 | white, dark brown, yellow, green | dark brown, orange, yellow | 46 | none | none | none | none |
| Ff4225 | white, dark brown | dark brown, orange, yellow | 78 | 21.50 ± 1.64 | 3.37 ± 0.35 | none | none |
| Ff4290 | White | Yellow | 76 | 25.51 ± 3.31 | 3.53 ± 0.53 | none | none |
| Ff4324 | white, purple | white, purple | 78 | 25.82 ± 2.00 | 3.27 ± 0.51 | none | none |
| Ff4325 | white, brown | Brown | 70 | 30.09 ± 2.71 | 3.07 ± 0.39 | none | none |
| FwgDE4 | white, brown | brown, yellow | 75 | 30.68 ± 3.51 | 2.95 ± 0.52 | 40.56 ± 2.79 | 2.92 ± 0.45 |
| FspDE40 | white, dark brown | dark brown, orange | 49 | 40.71 ± 2.15 | 2.98 ± 0.49 | 47.65 ± 4.27 | 2.94 ± 0.43 |
| Fs3769 | White | yellow | 80 | 24.83 ± 3.04 | 3.17 ± 0.42 | 41.02 ± 2.91 | 3.29 ± 0.41 |
| Fs3784 | dark brown, white | dark brown, yellow | 85 | none | none | none | none |
| Fs3873 | White | white | 80 | 33.18 ± 2.14 | 3.30 ± 0.54 | 41.32 ± 2.43 | 3.07 ± 0.52 |
| Fs3924 | White | yellow | 85 | 25.29 ± 5.02 | 3.31 ± 0.39 | none | none |
| Fs4279 | White | yellow | 73 | none | none | none | none |
| FcDir16 | white, dark brown | dark brown, orange, yellow | 63 | 41.56 ± 2.83 | 2.86 ± 0.41 | 50.76 ± 3.86 | 2.74 ± 0.39 |
| FcDir23 | White | white | 63 | 29.85 ± 4.89 | 3.02 ± 0.51 | 36.71 ± 1.37 | 2.95 ± 0.36 |
1 Species abbreviation: F. petroliphilum (Fp), F. keratoplasticum (Fk), F. striatum (Fstr), F. falciforme (Ff), F. waltergamsii (Fwg), Fusarium sp. (Fsp), F. suttonianum (Fs), F. cyanescence (Fc). 2 Size of macroconidia grown on CLA for four to seven days at 28 °C. 3 Mean values of 30 random conidia ± standard deviation. 4 none means not present.
3.2.2. Microconidia and Chlamydospores
Microconidia were present in all 25 FSSC isolates examined, with either single oval shaped cells or two cells separated by a septum and reniform (kidney-shaped). Representative examples for isolates of each of the eight FSSC species are presented in the images labelled (b) in Figure 3. Chlamydospores, images labelled (c) in Figure 3, were only observed in 14 of the 25 isolates: two F. falciforme (Ff4290, Ff4324), three F. suttonianum (Fs3769, Fs3784, Fs3924), seven F. keratoplasticum isolates (Fk553, Fk994, Fk2309, Fk2353, Fk2622, FkDI17, FkDir61), one each of F. waltergamsii (FwgDE4) and Fusarium sp. (FspDE40). When present, they were all globose and terminal rather than intercalary, and existed singly, paired, or in chains. F. petroliphilum, F. keratoplasticum, and F. falciforme had smooth-walled chlamydospores while F. suttonianum had rough-walled chlamydospores (Figure 3). Chlamydospores were absent in Fstr541 and FcDir16 when grown on CLA medium.
3.2.3. Macroconidia
As expected, the macroconidia morphology was the most distinctive feature for the identification of Fusarium species [57]. All four F. falciforme isolates, but only about half of the F. keratoplasticum and F. suttonianum isolates, had macroconidia with at least 2 septa (Table 2). None of the ten F. keratoplasticum or the four F. falciforme isolates, and only two F. suttonianum isolates, had macroconidia with 4 or 5 septa (Table 2). Yet, all isolates of the remaining five species, F. petroliphilum, F. striatum, F. waltergamsii, Fusarium sp., and F. cyanescens had macroconidia with both 2 or 3 and 4 or 5 septa (Table 2). The mean lengths and widths of the macroconidia for isolates are presented in Table 2. The mean lengths ranged from 21–42 µm for 2 or 3 septa, and 31–51 µm for 4 or 5 septa (Table 2). Although there was a clear difference in the average length of the macroconidia with 2 or 3 septa between the five F. keratoplasticum (22.43 ± 1.60 µm), the four F. falciforme (25.70 ± 3.51 µm), and the three F. suttonianum isolates (27.77 µm ± 4.69; Table 3), inter-species differences were not statistically significant; there was too much of an overlap between individual isolates to use macroconidia length to assign isolates to a particular species (Table 2).
Table 3.
Average length and width of Fusarium species macroconidia with 2 or 3 septa. Values are presented as the means ± standard deviation.
| Species | Macroconidia 1 | |
|---|---|---|
| Length (µm) | Width (µm) | |
| F. keratoplasticum (n = 5) 2 | 22.43 ± 1.60 | 2.99 ± 0.06 |
| F. falciforme (n = 4) | 25.70 ± 3.51 | 3.32 ± 0.19 |
| F. suttonianum (n = 3) 3 | 27.77 ± 4.69 | 3.26 ± 0.06 |
Representative images of macroconidia for the eight species identified in this study, F. petroliphilum, F. keratoplasticum, F. falciforme, F. waltergamsii, Fusarium sp., F. suttonianum, F. cyanescens and F. striatum, are presented in Figure 3 (labelled (a)). The shape of the macroconidia of all eight species was straight or slightly curved. F. petroliphilum macroconidia were much more abundant than for any of the other FSSC species investigated. F. petroliphilum macroconidia were elongate and slightly curved in shape and most contained 4 to 5 septa. The triangular-shaped apical and basal cells were blunt and barely notched, respectively. F. keratoplasticum macroconidia were straight and cylindrical to slightly curved with up to 3 septa for 50% (5/10) of the isolates. Similar to F. petroliphilum, the triangular shaped apical cells were blunt, and the basal cells were barely notched.
The macroconidia of the F. keratoplasticum isolates were also noticeably shorter than for most other isolates (Table 2 and Table 3). All four F. falciforme isolates only had macroconidia with 2–3 septa. They were slightly curved and larger than the F. keratoplasticum macroconidia, but they possessed similar triangular-shaped blunt apical and barely notched basal cells. F. waltergamsii macroconidia had distinctly hooked apical cells and notched to foot-like basal cells. Fusarium sp. of the FSSC 12 had macroconidia with slightly straighter dorsal, straight apical and barely notched basal cells. F. suttonianum macroconidia were somewhat straight on both dorsal and ventral lines, with moderate curvature that was more prominent in the apical and basal cells with 4–5 septa. The F. striatum macroconidia with 2 or 3 septa were slightly thinner (2.44 ± 0.47 µm; Table 2), straight, and almost needle-like whereas the widths for most other isolates ranged from 2.79 µm for Fp667 to 3.53 µm for Ff4290, most being ~3 µm in width (Table 2). The F. striatum macroconidia had hooked apical and barely notched, foot-shaped basal cells (Figure 3). F. cyanescens macroconidia were slightly curved, with hooked apical cells and barely notched basal cells.
Seven (Fk620, Fk553, Fk0168, Fk2622, FkDir61, Fs3784, Fs4279) of the 25 isolates grown on CLA did not produce any macroconidia (i.e., 5 of 10 F. keratoplasticum, equally distributed among clinical and environmental isolates, and 2 of 5 F. suttonianum isolates; Table 2). The remaining 5 F. keratoplasticum isolates (Fk2781, Fk2309, Fk2353, Fk994, FkDI17) only produced macroconidia with up to 3 septa, as did all 4 F. falciforme isolates (Ff4225, Ff4290, Ff4324, Ff4325). Only 8 of the 25 isolates (Fp667, Fstr541, FwgDE4, FspDE40, Fs3769, Fs3783, FcDir16, FcDir23) produced macroconidia with 4 or 5 septa. They included all isolates of F. petroliphilum (1), F. striatum (1), F. waltergamsii (1), Fusarium sp. (1) and F. cyanescence (2) and 2 of the 5 F. suttonianum isolates (Table 2 and Figure 3).
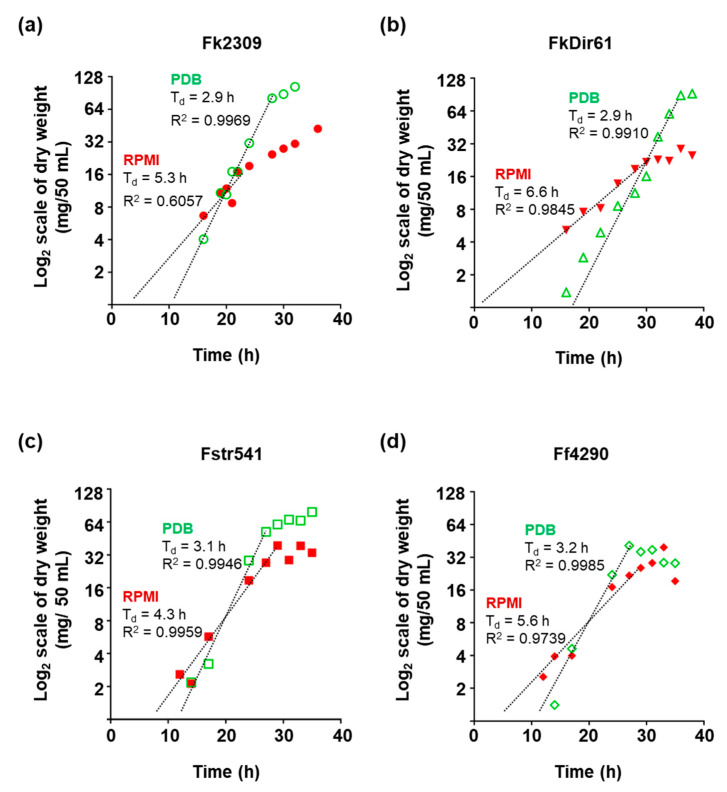
3.3. Growth of F. keratoplasticum, F. striatum and F. falciforme in PDB and RPMI
Mycelial dry weight measurements were used to create growth curves for F. keratoplasticum Fk2309 and FkDir61, F. striatum Fstr541, and F. falciforme Ff4290. The generation time (Td) for all four strains in PDB was ~ 3 h while their generation time in RPMI ranged from 4.3 h to 6.6 h with FkDir61 being the slowest growing strain (Figure 4). An extrapolation of the exponential growth lines towards the initial inoculum size of <0.5 mg/50 mL provided an estimate for the lag phase of each strain (Figure 4). All four strains took extended periods of time (they each had a ~10–15 h lag phase) to adapt to growth in PDB liquid medium, even though the microconidia inoculum was prepared from cells grown on PDA solid medium. Although all four Fusarium isolates grew much slower (~1.5–2 times slower) in RPMI, the estimated lag phases were surprisingly short (~7 h) for Fstr541 and Ff4290 and almost non-existent for Fk2309 and FkDir61 (<1–3 h) (Figure 4). All four isolates reached much higher (~3 times) cell densities (~100 mg/50 mL) in stationary phase in PDB than in RPMI (~32 mg/50 mL), except for Ff4290 which reached similarly low cell densities of ~32 mg/50 mL in both media (Figure 4). Based on these results, mid-logarithmic phase is reached after 21 h incubation in PDB.
Figure 4.
Growth curves of two F. keratoplasticum strains, Fk2309 (a) and FkDir61 (b), and of F. striatum and F. falciforme isolates Fstr541 (c) and Ff4290 (d) grown in PDB (green circles) or RPMI (red circles) medium, respectively. Dotted lines are the trendlines for the exponential growth phase that were used to calculate the generation time (Td). The results presented were from a single experiment.
4. Discussion
Invasive fusariosis is a severe disease of the immunocompromised, mostly those with hematologic malignancies [36]. Disseminated fusariosis is usually confirmed by a positive blood culture and has a poor prognosis with a high mortality rate [11]. Only two (Fk0168 and Fstr541) of the 15 clinical isolates were from positive blood cultures. The patients’ underlying conditions were, unfortunately, not recorded. The remaining 13 clinical FSSC isolates were from superficial infection sites; eight were from nail, two from skin, and three from eye infections. Thus, the majority (87%; 13 of 15) of the fusariosis cases were superficial nail (8 F. keratoplasticum isolates), eye (3; one isolate each of F. petroliphilum, F. falciforme and F. suttonianum), and skin infections (2 F. keratoplasticum isolates) similar to previous reports [11]. The species distribution of the 15 clinical FSSC isolates agreed with previous reports that found F. keratoplasticum (FSSC 2) to be one of the FSSC species most frequently isolated in the clinic [21,65].
Three of the eight FSSC species contained both environmental and clinical isolates: F. keratoplasticum, F. falciforme, and F. suttonianum (Table 1). F. keratoplasticum (FSSC 2) was the most prevalent clinical isolate (73%; 11 of 15), all but one (blood isolate Fk0168; Table 1) were skin or nail isolates, in good agreement with previous reports [21,65,66,67,68,69,70]. F. keratoplasticum is also an important veterinary pathogen, causing infections in equine and marine vertebrates as well as in invertebrates [71]. In contrast, the majority of the F. falciforme (80%; 4 of 5) and F. suttonianum (83%; 5 of 6) isolates were of environmental origin. Ff0020 and Fs263 were the only two isolates of human origin (eye; Table 1).
The other five FSSC species were isolated at a much lower frequency. They were either clinical (F. petroliphilum and F. striatum, one each) or environmental isolates (F. waltergamsii and Fusarium sp., one each, and two F. cyanescens isolates; Table 1). The F. suttonianum isolates accounted for 33% (5 of 15) of all environmental isolates. Others have also recovered F. suttonianum isolates from skin, nail, corneal ulcer, and blood samples [25]. The F. falciforme isolates accounted for ~27% (4 of 15) of the environmental isolates, and, like F. suttonianum, only one was of clinical origin (eye). Interestingly, F. falciforme has been reported as the predominant species isolated in South India accounting for 83% [72] and 93% [73] of all FSSC isolates. The only F. petroliphilum (FSSC 1) isolate was of clinical origin (eye). F. petroliphilum isolates have been previously obtained from air samples which caused airborne transmission of IF [74] and even fatal disseminated fusariosis [75].
F. suttonianum appears innately resistant to most azole antifungals. Based on the azole susceptibility data that we previously reported [46], the six F. suttonianum isolates in this study were resistant to itraconazole, posaconazole, and also voriconazole, and all carried the 23 bp CYP51A promoter deletion that was tightly associated with voriconazole resistance. F. suttonianum resistance to other triazoles including the agricultural fungicides tebuconazole and propiconazole has also been reported [66]. The voriconazole resistant F. keratoplasticum isolates Fk2309, Fk2781, and FkDI17 also carried the 23 bp CYP51A promoter deletion (Figure 1 and [46]) which suggested cross-species DNA exchange between their common ancestors. The fact that the voriconazole resistant F. suttonianum and F. keratoplasticum isolates were of both environmental and clinical origin (Figure 1) supports the hypothesis that voriconazole resistance was selected for by fungicide use in agriculture. Whether the two additional voriconazole resistant isolates, Ff0020 and Fstr541 (marked with an asterisk in Figure 1), also contain the 23 bp CYP51A deletion remains to be investigated. It is quite likely that the rather distantly related isolate Fstr541 (Figure 1) and possibly also the only voriconazole resistant F. falciforme isolate Ff0020, both of clinical origin, have different voriconazole resistance conferring mutations. One possibility may be gain of function mutations in a transcription factor that causes the overexpression of orthologs of the recently discovered multidrug efflux pump, F. keratoplasticum Abc1 [76].
The identity of 25 of the 30 FSSC isolates was initially assessed with traditional identification methods by carefully analysing the macroscopic and microscopic features of cell cultures grown on CLA medium [77]. The shape and size of the microconidia and clamydospores was rather uniform and provided no distinctive power for accurate species identification. However, the shape and size of the macroconidia provided more distinctive features. The presence of larger macroconidia with 3–5 septa, the shape of their spores and their apical and basal cells, the number of septa and the size of the macroconidia are key characteristics for the identification of FSSC species [77]. The macroconidia of most FSSC isolates (72%; 18 of 25) had 2 or 3 septa and only eight of them also had macroconidia with 4 or 5 septa (Table 2). A previous study of Malaysian FSSC isolates [78] reported similar results for four F. keratoplasticum and four F. falciforme isolates, although there were notable differences. All F. keratoplasticum and F. falciforme isolates had macroconidia with 3 or 4 septa and, as in our study, none had 5 septa. However, their average macroconidia size (length × width) [78] was ~50% greater (33.75 × 4.88 µm and 41.75 × 5.10 µm, respectively) than the average F. keratoplasticum and F. falciforme macroconidia sizes determined in the present investigation (Table 2 and Table 3). These differences are likely due to the different growth conditions used by the different laboratories. Cheri et al. (2015), for instance, incubated the CLA plate cultures at room temperature (25 ± 2 °C) rather than at 28 °C. Another study [79] investigated the morphology of F. keratoplasticum and F. petroliphilum isolates grown at 22 °C with alternating 12 h light/dark cycles using a 120 V UV light bulb as a light source instead of a 103 V cool white fluorescent light bulb. And a third study [25] of five FSSC species (FSSC 6, 7, 9, 20 and 43) used synthetic-nutrient-agar [80] with and without carnation leave pieces incubated at ‘room temperature’, a term that could vary significantly between different laboratories. Contrary to these studies [25,79,80], about half of the presently investigated F. keratoplasticum (5 of 10) and F. suttonianum (2 of 5) isolates had no apparent macroconidia at all. This could be due to subtle variations in the culturing conditions or a reflection of their different geographical origins.
The average colony diameters of isolates of the three major FSSC species F. keratoplasticum, F. falciforme and F. suttonianum grown on PDA for nine days at 28 °C were measured. However, as with any of the other morphological observations, there were strain variations that made it impossible to assign individual isolates to a particular FSSC species based on these measurements. There was also no other morphological characteristic that could distinguish between isolates of the various FSSC MLST sub-types (e.g., F. keratoplasticum FSSC 2-a, 2-f, 2-h; Figure 2A). One of the limitations of this study was the small number of FSSC isolates investigated which naturally precludes an accurate and representative description of the morphology of individual species.
We also investigated the growth characteristics of Fusarium isolates in PDB and RPMI media frequently used to cultivate fungi (PDB) or determine the drug susceptibilities (RPMI) of clinical and environmental fungal isolates. For fair comparisons, RPMI was adjusted to 2% glucose. The Fusarium isolates chosen included two clinical isolates from nail (Fk2309) and blood (Fstr541) and two environmental isolates (FkDir61, Ff4290). The two F. keratoplasticum isolates represented a voriconazole resistant (Fk2309) isolate with the 23 bp CYP51A promoter deletion and a voriconazole sensitive isolate with the wild type CYP51A promoter. All four strains grew equally fast in PDB with a ~3 h generation time. Their growth was ~1.5- to 2-times slower in RPMI (4.3–6.6 h). Interestingly, however, all strains appeared to have a shorter lag phase (~5–8 h) in RPMI, especially the two F. keratoplasticum isolates Fk2309 (~5 h) and FkDir61 (~7 h; Figure 4); the lag times for all four strains were longer (~10–15 h) when grown in PDB medium. A major difference between the two media is the pH. While the pH of PDB (pH 5.2) is designed for optimum fungal growth, RPMI (pH 7.2) mimics the growth conditions the fungus would encounter in humans. Whether the reduced lag times in RPMI or the inability to produce macroconidia with more than 3 septa by any of the F. keratoplasticum and F. falciforme isolates have anything to do with their increased likelihood to infect human tissue remains to be investigated.
The TEF1-α and RPB2 sequences alone were not enough to correctly identify one of the 30 FSSC isolates, Fstr541. This isolate was initially identified as F. falciforme Ff541 (MLST 3+4 oo) [46]. Only the phylogenetic relationship of the 30 FSSC isolates with 35 closely related published FSSC sequences could identify this isolate as F. striatum. The concatenated TEF1-α and RPB2 sequences were also not able to clearly separate isolates of different sub-lineages (e.g., 2-a, -d, -f, -h, -k or 3+4-k, -ii, -tt, -oo) for the two largest and rather diverse FSSC species, F. keratoplasticum and F. falciforme (Figure 1). F. falciforme isolates accounted for approximately one-third (63) of 191 unique haplotypes (i.e., FSSC 3+4-a to -kkk) of clade 3 of the FSSC in a previous investigation [22]. Previous studies used additional sequences for FSSC species identification; i.e., the entire internal transcribed spacer region (ITS) plus the D1+D2 fragments of the large rRNA subunit (LSU) with [81] or without [22] the calcium-binding messenger protein gene, calmodulin (CAM). LSU and ITS, however, were the least informative sequences to distinguish between closely related FSSC species [16,22]. This is why we did not use them in the present study. However, our study clearly demonstrates the need for additional housekeeping gene sequences such as RPB1 and TUB2 to distinguish individual F. keratoplasticum and F. falciforme isolates more clearly from each other. The Candida albicans MLST scheme, for instance, uses seven housekeeping genes (AAT1a, ACC1, ADP1, MPIb, SYA1, VPS13, and ZWF1b) [82]. The MLST schemes of several other pathogenic Candida species (C. krusei, C. tropicalis, C. glabrata, and C. dubliniensis) use at least six gene sequences, eight sequences are used for C. dubliniensis [83], and the A. fumigatus MLST scheme uses seven gene sequences (ANXC4, BGT1, CAT1, LIP, MAT1-2, SODB, and ZRF2) [84] to accurately assign isolates to the various geographically-, epidemiologically-, or specific host-associated sub-lineages.
5. Conclusions
F. keratoplasticum isolates were by far the most frequent clinical (11 of 15) isolates. F. suttonianum (5 of 15) and F. falciforme (4 of 15) were the most frequent environmental isolates. The remaining five FSSC species were only isolated once (F. petroliphilum, F. striatum, F. waltergamsii, Fusarium sp.) or twice (F. cyanescens). Interestingly, none of the F. keratoplasticum or F. falciforme isolates investigated (10 and 4, respectively) produced macroconidia with 4 or 5 septa, and only about half of the F. keratoplasticum and F. suttonianum isolates produced any macroconidia at all. Yet, all isolates of the other five FSSC species produced macroconidia with 4 or 5 septa. None of the morphologic or microscopic observations were sufficient for the species level identification of any of the FSSC isolates. Far more accurate results were obtained with phylogenetic analysis using the TEF1-α and RPB2 sequences. However, the inclusion of additional gene sequences is recommended for a more accurate resolution of their phylogenetic relationships, especially for the two most diverse species F. keratoplasticum and F. falciforme, which are also the two main FSSC species causing life-threatening IF.
Acknowledgments
We would like to thank Universiti Kebangsaan Malaysia Medical Centre and the Institute for Medical Research Malaysia for providing the isolates used in this study.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof8080845/s1, Table S1: Primers used for PCR amplification of Fusarium TEF1-α and RPB2 genes. Table S2: Source, species and MLST of 37 Fusarium isolates selected for the phylogenetic analysis of TEF1-α and RPB2 sequences presented in Figure 1.
Author Contributions
Conceptualization, methodology, validation and data curation J.E.J. and J.S. (Jacinta Santhanam); software, J.S. (Jariya Sakayaroj) and S.S.; resources, M.F.A.R. and L.Z.; writing—original draft preparation, J.E.J.; supervision and writing—review and editing, J.S. (Jacinta Santhanam), E.L. and R.D.C.; visualization, J.E.J., N.M.R. and M.A.B.; project administration and funding acquisition, J.S. (Jacinta Santhanam); All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data (DNA sequences) can be obtained freely from GenBank using the accession numbers provided or by contacting the first or corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Ministry of Education Malaysia under Fundamental Research Grant Scheme (FRGS/1/2018/SKK11/UKM/02/1). J.E.J. was the recipient of a Sir Thomas Kay Sidey Postgraduate Visiting Fellowship from the University of Otago Faculty of Dentistry.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coleman J.J. The Fusarium solani species complex: Ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 2016;17:146–158. doi: 10.1111/mpp.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazan K., Gardiner D.M. Transcriptomics of cereal—Fusarium graminearum interactions: What we have learned so far. Mol. Plant Pathol. 2018;19:764–778. doi: 10.1111/mpp.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urbaniak C., Massa G., Hummerick M., Khodadad C., Schuerger A., Venkateswaran K. Draft genome sequences of two Fusarium oxysporum isolates cultured from infected Zinnia hybrida plants grown on the International Space Station. Genome Announc. 2018;6:e00326-18. doi: 10.1128/genomeA.00326-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dweba C., Figlan S., Shimelis H., Motaung T., Sydenham S., Mwadzingeni L., Tsilo T. Fusarium head blight of wheat: Pathogenesis and control strategies. Crop Prot. 2017;91:114–122. doi: 10.1016/j.cropro.2016.10.002. [DOI] [Google Scholar]
- 5.Maryani N., Lombard L., Poerba Y., Subandiyah S., Crous P., Kema G. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 2019;92:155–194. doi: 10.1016/j.simyco.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stover R. Fusarium wilt of banana: Some history and current status of the disease; Proceedings of the First International Conference on Fusarial Wilt of Banana; Miami, FL, USA. 27–30 August 1989; pp. 1–7. [Google Scholar]
- 7.Windels C.E. Economic and social impacts of Fusarium head blight: Changing farms and rural communities in the Northern Great Plains. Phytopathology. 2000;90:17–21. doi: 10.1094/PHYTO.2000.90.1.17. [DOI] [PubMed] [Google Scholar]
- 8.Ordonez N., Seidl M.F., Waalwijk C., Drenth A., Kilian A., Thomma B.P., Ploetz R.C., Kema G.H. Worse comes to worst: Bananas and Panama disease-when plant and pathogen clones meet. PLoS Pathog. 2015;11:e1005197. doi: 10.1371/journal.ppat.1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook D.C., Taylor A.S., Meldrum R.A., Drenth A. Potential economic impact of Panama disease (tropical race 4) on the Australian banana industry. J. Plant Dis. Prot. 2015;122:229–237. doi: 10.1007/BF03356557. [DOI] [Google Scholar]
- 10.Ploetz R.C. Panama disease: An old nemesis rears its ugly head: Part 1. the beginnings of the banana export trades. Plant Health Prog. 2005;6:18. doi: 10.1094/PHP-2005-1221-01-RV. [DOI] [Google Scholar]
- 11.Nucci M., Anaissie E. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 2007;20:695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rotjanapan P., Chen Y.C., Chakrabarti A., Li R.Y., Rudramurthy S.M., Yu J., Kung H.C., Watcharananan S., Tan A.L., Saffari S.E., et al. Epidemiology and clinical characteristics of invasive mould infections: A multicenter, retrospective analysis in five Asian countries. Med. Mycol. 2018;56:186–196. doi: 10.1093/mmy/myx029. [DOI] [PubMed] [Google Scholar]
- 13.Lass-Flörl C., Cuenca-Estrella M. Changes in the epidemiological landscape of invasive mould infections and disease. J. Antimicrob. Chemother. 2017;72:i5–i11. doi: 10.1093/jac/dkx028. [DOI] [PubMed] [Google Scholar]
- 14.Horn D.L., Freifeld A.G., Schuster M.G., Azie N.E., Franks B., Kauffman C.A. Treatment and outcomes of invasive fusariosis: Review of 65 cases from the PATH Alliance((R)) registry. Mycoses. 2014;57:652–658. doi: 10.1111/myc.12212. [DOI] [PubMed] [Google Scholar]
- 15.Nucci M., Anaissie E. Cutaneous infection by Fusarium species in healthy and immunocompromised hosts: Implications for diagnosis and management. Clin. Infect. Dis. 2002;35:909–920. doi: 10.1086/342328. [DOI] [PubMed] [Google Scholar]
- 16.O’Donnell K., Rooney A.P., Proctor R.H., Brown D.W., McCormick S.P., Ward T.J., Frandsen R.J., Lysøe E., Rehner S.A., Aoki T., et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013;52:20–31. doi: 10.1016/j.fgb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 17.O’Donnell K., Sutton D.A., Rinaldi M.G., Sarver B.A., Balajee S.A., Schroers H.J., Summerbell R.C., Robert V.A., Crous P.W., Zhang N., et al. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010;48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroers H.J., Samuels G.J., Zhang N., Short D.P., Juba J., Geiser D.M. Epitypification of Fusisporium (Fusarium) solani and its assignment to a common phylogenetic species in the Fusarium solani species complex. Mycologia. 2016;108:806–819. doi: 10.3852/15-255. [DOI] [PubMed] [Google Scholar]
- 19.Al-Hatmi A.M.S., Bonifaz A., Ranque S., Sybren de Hoog G., Verweij P.E., Meis J.F. Current antifungal treatment of fusariosis. Int. J. Antimicrob. Agents. 2018;51:326–332. doi: 10.1016/j.ijantimicag.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Matuo T., Snyder W.C. Use of morphology and mating populations in the identification of formae speciales in Fusarium solani. J. Phytopathol. 1973;63:562–565. doi: 10.1094/Phyto-63-562. [DOI] [Google Scholar]
- 21.Chang D.C., Grant G.B., O’Donnell K., Wannemuehler K.A., Noble-Wang J., Rao C.Y., Jacobson L.M., Crowell C.S., Sneed R.S., Lewis F.M., et al. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. J. Am. Med. Assoc. 2006;296:953–963. doi: 10.1001/jama.296.8.953. [DOI] [PubMed] [Google Scholar]
- 22.O’Donnell K., Sutton D.A., Fothergill A., McCarthy D., Rinaldi M.G., Brandt M.E., Zhang N., Geiser D.M. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 2008;46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donnell K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia. 2000;92:919–938. doi: 10.1080/00275514.2000.12061237. [DOI] [Google Scholar]
- 24.Aoki T., O’Donnell K., Homma Y., Lattanzi A.R. Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex-F. virguliforme in North America and F. tucumaniae in South America. Mycologia. 2003;95:660–684. doi: 10.1080/15572536.2004.11833070. [DOI] [PubMed] [Google Scholar]
- 25.Sandoval-Denis M., Crous P. Removing chaos from confusion: Assigning names to common human and animal pathogens in Neocosmospora. Persoonia. 2018;41:109–129. doi: 10.3767/persoonia.2018.41.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoval-Denis M., Lombard L., Crous P.W. Back to the roots: A reappraisal of Neocosmospora. Persoonia. 2019;43:90–185. doi: 10.3767/persoonia.2019.43.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donnell K., Al-Hatmi A.M.S., Aoki T., Brankovics B., Cano-Lira J.F., Coleman J.J., de Hoog G.S., Di Pietro A., Frandsen R.J.N., Geiser D.M., et al. No to Neocosmospora: Phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the genus Fusarium. mSphere. 2020;5:e00810-20. doi: 10.1128/mSphere.00810-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crous P.W., Lombard L., Sandoval-Denis M., Seifert K.A., Schroers H.J., Chaverri P., Gene J., Guarro J., Hirooka Y., Bensch K., et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021;98:100116. doi: 10.1016/j.simyco.2021.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Espinel-Ingroff A., Colombo A.L., Cordoba S., Dufresne P.J., Fuller J., Ghannoum M., Gonzalez G.M., Guarro J., Kidd S.E., Meis J.F., et al. International evaluation of MIC distributions and epidemiological cutoff value (ECV) definitions for Fusarium species identified by molecular methods for the CLSI broth microdilution method. Antimicrob. Agents Chemother. 2016;60:1079–1084. doi: 10.1128/AAC.02456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaize M., Normand A.C., Imbert S., Al-Hatmi A.M.S., Chryssanthou E., Cassaing S., Schuttler C., Hasseine L., Mahinc C., Costa D., et al. Antifungal susceptibility of 182 Fusarium species isolates from 20 European centers: Comparison between EUCAST and gradient concentration strip methods. Antimicrob. Agents Chemother. 2021;65:e0149521. doi: 10.1128/AAC.01495-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stempel J.M., Hammond S.P., Sutton D.A., Weiser L.M., Marty F.M. Invasive fusariosis in the voriconazole era: Single-center 13-year experience. Open Forum Infect. Dis. 2015;2:ofv099. doi: 10.1093/ofid/ofv099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lortholary O., Obenga G., Biswas P., Caillot D., Chachaty E., Bienvenu A.L., Cornet M., Greene J., Herbrecht R., Lacroix C., et al. International retrospective analysis of 73 cases of invasive fusariosis treated with voriconazole. Antimicrob. Agents Chemother. 2010;54:4446–4450. doi: 10.1128/AAC.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada K., Endo T., Hashimoto D., Saga T., Ara T., Ogasawara R., Yasumoto A., Ibata M., Takahata M., Shigematsu A., et al. Disseminated fusariosis emerged from prolonged local genital infection after cord blood transplantation. J. Infect. Chemother. 2018;24:660–663. doi: 10.1016/j.jiac.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Silva G.M., Silveira A.R., Betania C.A., Macedo D.P., Neves R.P. Disseminated fusariosis secondary to neuroblastoma with fatal outcome. Mycopathologia. 2013;176:233–236. doi: 10.1007/s11046-013-9674-8. [DOI] [PubMed] [Google Scholar]
- 35.Esnakula A.K., Summers I., Naab T.J. Fatal disseminated Fusarium infection in a human immunodeficiency virus positive patient. Case Rep. Infect. Dis. 2013;2013:379320. doi: 10.1155/2013/379320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nucci M., Anaissie E.J., Queiroz-Telles F., Martins C.A., Trabasso P., Solza C., Mangini C., Simoes B.P., Colombo A.L., Vaz J. Outcome predictors of 84 patients with hematologic malignancies and Fusarium infection. Cancer. 2003;98:315–319. doi: 10.1002/cncr.11510. [DOI] [PubMed] [Google Scholar]
- 37.Zubrod J.P., Bundschuh M., Arts G., Bruhl C.A., Imfeld G., Knabel A., Payraudeau S., Rasmussen J.J., Rohr J., Scharmuller A., et al. Fungicides: An Overlooked Pesticide Class? Environ. Sci. Technol. 2019;53:3347–3365. doi: 10.1021/acs.est.8b04392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meis J.F., Chowdhary A., Rhodes J.L., Fisher M.C., Verweij P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R Soc. Lond. B Biol. Sci. 2016;371:20150460. doi: 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chowdhary A., Kathuria S., Xu J., Meis J.F. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013;9:e1003633. doi: 10.1371/annotation/4ffcf1da-b180-4149-834c-9c723c5dbf9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao D., Wang F., Yu S., Dong S., Wu R., Cui N., Ren J., Xu T., Wang S., Wang M., et al. Prevalence of azole-resistant Aspergillus fumigatus is highly associated with azole fungicide residues in the fields. Environ. Sci. Technol. 2021;55:3041–3049. doi: 10.1021/acs.est.0c03958. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., van den Heuvel J., Debets A.J.M., Verweij P.E., Melchers W.J.G., Zwaan B.J., Schoustra S.E. Evolution of cross-resistance to medical triazoles in Aspergillus fumigatus through selection pressure of environmental fungicides. Proc. Biol. Sci. 2017;284:20170635. doi: 10.1098/rspb.2017.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang S.E., Sumabat L.G., Melie T., Mangum B., Momany M., Brewer M.T. Evidence for the agricultural origin of resistance to multiple antimicrobials in Aspergillus fumigatus, a fungal pathogen of humans. G3. 2022;12:jkab427. doi: 10.1093/g3journal/jkab427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yadav A., Jain K., Wang Y., Pawar K., Kaur H., Sharma K.K., Tripathy V., Singh A., Xu J., Chowdhary A. Candida auris on apples: Diversity and clinical significance. mBio. 2022;13:e0051822. doi: 10.1128/mbio.00518-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bastos R.W., Rossato L., Goldman G.H., Santos D.A. Fungicide effects on human fungal pathogens: Cross-resistance to medical drugs and beyond. PLoS Pathog. 2021;17:e1010073. doi: 10.1371/journal.ppat.1010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellado E., Diaz-Guerra T.M., Cuenca-Estrella M., Rodriguez-Tudela J.L. Identification of two different 14-alpha sterol demethylase-related genes (CYP51A and CYP51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 2001;39:2431–2438. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James J.E., Lamping E., Santhanam J., Milne T.J., Abd Razak M.F., Zakaria L., Cannon R.D. A 23 bp cyp51A promoter deletion associated with voriconazole resistance in clinical and environmental isolates of Neocosmospora keratoplastica. Front. Microbiol. 2020;11:272. doi: 10.3389/fmicb.2020.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becher R., Hettwer U., Karlovsky P., Deising H.B., Wirsel S.G. Adaptation of Fusarium graminearum to tebuconazole yielded descendants diverging for levels of fitness, fungicide resistance, virulence, and mycotoxin production. Phytopathology. 2010;100:444–453. doi: 10.1094/PHYTO-100-5-0444. [DOI] [PubMed] [Google Scholar]
- 48.Chen J., Wei J., Fu L., Wang S., Liu J., Guo Q., Jiang J., Tian Y., Che Z., Chen G., et al. Tebuconazole resistance of Fusarium graminearum field populations from wheat in Henan Province. J. Phytopathol. 2021;169:525–532. doi: 10.1111/jph.13021. [DOI] [Google Scholar]
- 49.Al-Hatmi A.M., van Diepeningen A.D., Curfs-Breuker I., de Hoog G.S., Meis J.F. Specific antifungal susceptibility profiles of opportunists in the Fusarium fujikuroi complex. J. Antimicrob. Chemother. 2015;70:1068–1071. doi: 10.1093/jac/dku505. [DOI] [PubMed] [Google Scholar]
- 50.Hafizi R., Salleh B., Latiffah Z. Morphological and molecular characterization of Fusarium solani and F. oxysporum associated with crown disease of oil palm. Braz. J. Microbiol. 2013;44:959–968. doi: 10.1590/S1517-83822013000300047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ibrahim N.F., Mohd M.H., Nor N.M.I.M., Zakaria L. Fusarium fujikuroi causing fusariosis of pineapple in peninsular Malaysia. Australas. Plant Dis. Notes. 2016;11:21. doi: 10.1007/s13314-016-0206-5. [DOI] [Google Scholar]
- 52.Lim G. Fusarium in paddy soils of West Malaysia. Plant Soil. 1972;36:47–51. doi: 10.1007/BF01373455. [DOI] [Google Scholar]
- 53.Sidique S.N.M., Azuddin N.F., Joseph J. First report of Fusarium species at nesting sites of endangered sea turtles in Terengganu and Melaka, Malaysia. Malays. Appl. Biol. 2017;46:195–205. [Google Scholar]
- 54.Manshor N., Rosli H., Ismail N.A., Salleh B., Zakaria L. Diversity of Fusarium species from highland areas in Malaysia. Trop. Life Sci. Res. 2012;23:1–15. [PMC free article] [PubMed] [Google Scholar]
- 55.Singh H., Jamal F., Marahakim M., Song C. Fusarium solani keratitis. First report from Malaysia. Int. Med. J. Malays. 1981;36:89–91. [PubMed] [Google Scholar]
- 56.Mohd-Tahir F., Norhayati A., Siti-Raihan I., Ibrahim M. A 5-year retrospective review of fungal keratitis at Hospital Universiti Sains Malaysia. Interdiscip. Perspect. Infect. Dis. 2012;2012:851653. doi: 10.1155/2012/851563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leslie J., Summerell B. The Fusarium Laboratory Manual. 1st ed. Blackwell Publishing; Ames, IA, USA: 2006. pp. 113–117. [Google Scholar]
- 58.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York, NY, USA: 2000. [Google Scholar]
- 60.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 61.Page R.D. Tree View: An application to display phylogenetic trees on personal computers. J. Bioinform. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 62.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. J. Bioinform. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 64.Darriba D., Posada D., Kozlov A.M., Stamatakis A., Morel B., Flouri T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2019;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Donnell K., Humber R.A., Geiser D.M., Kang S., Park B., Robert V.A., Crous P.W., Johnston P.R., Aoki T., Rooney A.P., et al. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia. 2012;104:427–445. doi: 10.3852/11-179. [DOI] [PubMed] [Google Scholar]
- 66.Herkert P.F., Al-Hatmi A.M.S., de Oliveira Salvador G.L., Muro M.D., Pinheiro R.L., Nucci M., Queiroz-Telles F., de Hoog G.S., Meis J.F. Molecular characterization and antifungal susceptibility of clinical Fusarium species from Brazil. Front. Microbiol. 2019;10:737. doi: 10.3389/fmicb.2019.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chiewchanvit S., Chongkae S., Mahanupab P., Nosanchuk J.D., Pornsuwan S., Vanittanakom N., Youngchim S. Melanization of Fusarium keratoplasticum (F. solani species complex) during disseminated fusariosis in a patient with acute leukemia. Mycopathologia. 2017;182:879–885. doi: 10.1007/s11046-017-0156-2. [DOI] [PubMed] [Google Scholar]
- 68.Tupaki-Sreepurna A., Al-Hatmi A.M., Kindo A.J., Sundaram M., de Hoog G.S. Multidrug-resistant Fusarium in keratitis: A clinico-mycological study of keratitis infections in Chennai, India. Mycoses. 2017;60:230–233. doi: 10.1111/myc.12578. [DOI] [PubMed] [Google Scholar]
- 69.Guevara-Suarez M., Cano-Lira J.F., de Garcia M.C., Sopo L., De Bedout C., Cano L.E., Garcia A.M., Motta A., Amezquita A., Cardenas M., et al. Genotyping of Fusarium isolates from onychomycoses in Colombia: Detection of two new species within the Fusarium solani species complex and in vitro antifungal susceptibility testing. Mycopathologia. 2016;181:165–174. doi: 10.1007/s11046-016-9983-9. [DOI] [PubMed] [Google Scholar]
- 70.Al-Hatmi A.M., Bonifaz A., Tirado-Sanchez A., Meis J.F., de Hoog G.S., Ahmed S.A. Fusarium species causing eumycetoma: Report of two cases and comprehensive review of the literature. Mycoses. 2017;60:204–212. doi: 10.1111/myc.12590. [DOI] [PubMed] [Google Scholar]
- 71.O’Donnell K., Sutton D.A., Wiederhold N., Robert V.A., Crous P.W., Geiser D.M. Veterinary fusarioses within the United States. J. Clin. Microbiol. 2016;54:2813–2819. doi: 10.1128/JCM.01607-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Homa M., Shobana C.S., Singh Y.R., Manikandan P., Selvam K.P., Kredics L., Narendran V., Vagvolgyi C., Galgoczy L. Fusarium keratitis in South India: Causative agents, their antifungal susceptibilities and a rapid identification method for the Fusarium solani species complex. Mycoses. 2013;56:501–511. doi: 10.1111/myc.12062. [DOI] [PubMed] [Google Scholar]
- 73.Homa M., Galgóczy L., Manikandan P., Narendran V., Sinka R., Csernetics Á., Vágvölgyi C., Kredics L., Papp T. South Indian isolates of the Fusarium solani species complex from clinical and environmental samples: Identification, antifungal susceptibilities, and virulence. Front. Microbiol. 2018;9:1052. doi: 10.3389/fmicb.2018.01052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moretti M.L., Busso-Lopes A.F., Tararam C.A., Moraes R., Muraosa Y., Mikami Y., Gonoi T., Taguchi H., Lyra L., Reichert-Lima F., et al. Airborne transmission of invasive fusariosis in patients with hematologic malignancies. PLoS ONE. 2018;13:e0196426. doi: 10.1371/journal.pone.0196426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ersal T., Al-Hatmi A.S., Cilo B.D., Curfs-Breuker I., Meis J.F., Ozkalemkas F., Ener B., van Diepeningen A.D. Fatal disseminated infection with Fusarium petroliphilum. Mycopathologia. 2015;179:119–124. doi: 10.1007/s11046-014-9813-x. [DOI] [PubMed] [Google Scholar]
- 76.James J.E., Lamping E., Santhanam J., Cannon R.D. PDR transporter ABC1 is involved in the innate azole resistance of the human fungal pathogen Fusarium keratoplasticum. Front. Microbiol. 2021;12:673206. doi: 10.3389/fmicb.2021.673206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Summerell B.A., Salleh B., Leslie J.F. A utilitarian approach to Fusarium identification. Plant Dis. 2003;87:117–128. doi: 10.1094/PDIS.2003.87.2.117. [DOI] [PubMed] [Google Scholar]
- 78.Chehri K., Salleh B., Zakaria L. Morphological and phylogenetic analysis of Fusarium solani species complex in Malaysia. Microb. Ecol. 2015;69:457–471. doi: 10.1007/s00248-014-0494-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Short D.P., O’Donnell K., Thrane U., Nielsen K.F., Zhang N., Juba J.H., Geiser D.M. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet. Biol. 2013;53:59–70. doi: 10.1016/j.fgb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 80.Nash S.M., Snyder W.C. Quantitative estimations by plate counts of propagules of the bean root rot Fusarium in field soils. Phytopathology. 1962;52:567–572. [Google Scholar]
- 81.O’Donnell K., Sutton D.A., Rinaldi M.G., Gueidan C., Crous P.W., Geiser D.M. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J. Clin. Microbiol. 2009;47:3851–3861. doi: 10.1128/JCM.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bougnoux M.E., Tavanti A., Bouchier C., Gow N.A.R., Magnier A., Davidson A.D., Maiden M.C.J., Enfert C., Odds F.C. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 2003;41:5265–5266. doi: 10.1128/JCM.41.11.5265-5266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Odds F.C., Jacobsen M.D. Multilocus sequence typing of pathogenic Candida species. Eukaryot. Cell. 2008;7:1075–1084. doi: 10.1128/EC.00062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bain J.M., Tavanti A., Davidson A.D., Jacobsen M.D., Shaw D., Gow N.A.R., Odds F.C. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J. Clin. Microbiol. 2007;45:1469–1477. doi: 10.1128/JCM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data (DNA sequences) can be obtained freely from GenBank using the accession numbers provided or by contacting the first or corresponding author.