Abstract
Expression of the Escherichia coli leuV operon, which contains three tRNA1Leu genes, is regulated by several mechanisms including growth-rate-dependent control (GRDC) and stringent control (SC). Structural variants of the leuV promoter which differentially affect these regulatory responses have been identified, suggesting that promoter targets for GRDC and SC may be different and that GRDC of the leuV promoter occurs in the absence of guanosine 3′,5′-bisdiphosphate. To determine the mechanisms of the leuV promoter regulation, we have examined the stability of promoter open complexes and the effects of nucleotide triphosphate (NTP) concentration on the efficiency of the leuV promoter and its structural variants in vitro and in vivo. The leuV promoter open complexes were an order of magnitude more stable to heparin challenge than those of rrnBp1. The major initiating nucleotide GTP as well as other NTPs increased the stability of the leuV promoter open complexes. When the cellular level of purine triphosphates was increased at slower growth rates by pyrimidine limitation, a 10% reduction in leuV promoter activity was seen. It therefore appears that transcription initiation from the leuV promoter is less sensitive to changes in intracellular NTP concentration than that from rrnBp1. Comparative analysis of regulation of the leuV promoter with and without upstream activating sequences (UAS) demonstrated that the binding site for factor of inversion stimulation (FIS) located in UAS is essential for maximal GRDC. Moreover, the presence of UAS overcame the effects of leuV promoter mutations, which abolished GRDC of the leuV core promoter. However, although the presence of putative FIS binding site was essential for optimal GRDC, both mutant and wild-type leuV promoters containing UAS showed improved GRDC in a fis mutant background, suggesting that FIS protein is an important but not unique participant in the regulation of the leuV promoter.
The tRNA multigene family of Escherichia coli consists of 84 structural genes, which produce 46 tRNA species (9). The production of tRNA is closely regulated such that under various physiological conditions the amount of tRNA produced is optimal for protein synthesis (16). It has become apparent that the use of synonymous codons for a given amino acid is not random but strongly biased so that the codon chosen is precisely correlated with the relative abundance of the respective tRNA species among the isoacceptors. This is particularly true for highly expressed genes, while, in weakly expressed genes, synonymous codons recognized by rare tRNA species are used with appreciable frequency. It is therefore important and interesting to understand the factors which ensure that the cellular complement of anticodons is optimal for all physiological conditions. Earlier estimates showed that as the growth rate (expressed as doublings per hour) increases from 0.6 to 2.5, tRNA concentration increases from 6.3 × 104 to 7 × 105 molecules per cell (10), which has been referred to as growth-rate-dependent control (GRDC). A recent study has demonstrated that the cellular concentration of numerous tRNA species is under GRDC and that factor for inversion stimulation (FIS) is required for the regulation of several tRNA species (35).
When protein synthesis is inhibited in E. coli by amino acid starvation or with analogues, tRNA synthesis, like rRNA synthesis, is strongly curtailed and the cellular level of guanosine 3′,5′-bisdiphosphate (ppGpp) is dramatically increased. This response has been referred to as the stringent response. Analyses of the synthesis of individual tRNA species suggest that most if not all species are subject to stringent control (SC) (7, 26, 43). It has been proposed that both GRDC and SC are modulated by the intracellular level of ppGpp, but this remains controversial since it has been previously reported that rRNA promoters appear to display growth-rate-dependent activity in cells which cannot synthesize ppGpp (19). It seems clear from a number of studies that ppGpp is absolutely required for the stringent response (12, 13, 19, 22–24).
Added complexities of the control of rRNA genes have emerged from studies that demonstrated that these highly expressed genes show some form of feedback inhibition, perhaps involving ribosomes or factors which interact with them (20, 21). Other data which were interpreted to be in conflict with this idea have been presented (3). A study by Vogel et al. reported that when a pyrBI strain was exposed to partial pyrimidine starvation, levels of ppGpp correlated directly with growth rate and the rates of rRNA synthesis (44). In addition, it was proposed that postinitiation effects of ppGpp may be an important factor in SC and/or GRDC (45). A number of studies have shown clearly that ppGpp affects RNA polymerase elongation rates in vivo and in vitro (28, 29, 46).
Yet another level of control for rrn promoters has recently been proposed (18). It was shown that at least two E. coli rrn promoters (rrnBp1 and rrnDp1) require relatively high levels of initiating nucleotide triphosphates (NTPs) for optimal activity and that unstable open complexes of promoters are stabilized by high concentrations of initiating NTPs. It was reported that promoter activity increased as a function of NTP concentration in vivo and in vitro. In view of these results, it was proposed that the GRDC of rrnp1 promoters as well as the homeostatic regulation of ribosomal synthesis might be explained by the response of these promoters to intracellular NTP concentrations (18).
The leuV operon is located at 98 min of the E. coli genome from which three tRNA1Leu genes are transcribed. In our previous studies, we have shown that the leuV promoter (leuVp) shares several features with rrnp1 promoters. First, the leuV promoter contains upstream activating sequences (UAS) and an upstream element (UP) (7). In addition, FIS appears to bind the UAS and is required for optimal promoter activity (39). We have also shown that the core promoter lacking UP and UAS sequences displays some GRDC but that sequences which flank the core promoter are required for optimal GRDC as well (15).
In this study, we have attempted to determine the extent to which mechanisms for GRDC are employed by tRNA genes. In addition, we wished to determine the extent to which the various mechanisms for GRDC might be shared with the well-studied rrnp1 promoters. In particular, we have examined the putative role of NTP concentration and UAS in regulation of the leuV promoter. We report here that a tRNA promoter displays important differences from rrnBp1 in this regard and that multiple mechanisms are clearly required for the expression of this tRNA gene.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli DJ480 (MG1655 lacZ) was used as a host for measurements of GRDC and was a gift from Ding Jun Jin (National Cancer Institute, National Institutes of Health [NIH]). Strain RLG1072, which harbors a λ lysogen with polylinker cloning sites upstream of the trp-lacZ fusion, and fis::kan767 strain RLG1379 were a gift from Richard L. Gourse (University of Wisconsin at Madison). Three lysogenic strains containing leuV promoter-lacZ fusions with deletions or mutations in the upstream FIS binding site (containing residues −107 to +11, for example, and/or amino acid changes shown in parentheses) were also obtained from the Gourse laboratory. They are RLG4043 [leuV(−107 +11)-lacZ], RLG4045 [leuV(−107 +11, T-72G, T-71G)-lacZ], and RLG4044 [leuV(−47 +11)-lacZ] (39). All three are lysogens prepared with the MG1655-derived strain, RLG4006 (lacΔ145thi-39::Tn10). Another MG1655 derivative, CF1693 (relA251 spoT207) producing no detectable ppGpp was a gift from M. Cashel (NIH, Bethesda, Md.) (47). The pyrimidine auxotroph strain, CLT246 (car-403::Tn10ΔlacIZ) (38) was utilized to provoke changes in NTP levels in vivo. This strain was constructed by transduction of car-403::Tn10 into strain VH1000, a lacIZ pyrE+ derivative of strain MG1655. VH1000 was a gift of J. Hernandez.
M9 minimal medium (30) for growth of MG1655 derivatives was routinely supplemented with uracil (50 mg/liter) and thiamine (10 mg/liter). Media used for GRDC experiments were as follows: medium 1, M9 plus 0.2% glycerol; medium 2, M9 plus 0.2% glucose; medium 3, M9 plus 0.2% glucose and 0.2% Casamino Acids; and medium 4, M9 plus 0.4% glucose and 0.5% Casamino Acids. Luria broth was used for routine cloning procedures and as medium 5 for GRDC experiments.
Ultrapure NTPs were purchased from Pharmacia Biotech, 32P-labeled nucleotides were obtained from NEN DuPont. Supercoiled plasmid DNA was isolated by double ultracentrifugation in CsCl. Enzymes for molecular cloning were purchased from New England Biolabs. E. coli RNA polymerase was purchased from Amersham. Avian myeloblastosis virus reverse transcriptase was purchased from Promega.
Preparation of mutant promoters.
The parental plasmid in this study was pLC76 (8), which was originally derived from pKK232-8 (11). It contains the leuV(−50 +11) promoter located just upstream of the chloramphenicol acetyltransferase (cat) gene flanked by recognition sites for unique restriction endonucleases ClaI and HindIII. Plasmid pLTC76 was derived by the insertion of the trp attenuator termination of transcription sequence into a unique HindIII site of plasmid pLC76. This site was located between the leuV promoter and the reporter cat gene. The inserted trp attenuator sequence contained a unique NsiI site which was used for further cloning procedures. PCR fragments containing relevant portions of the leuV operon were directionally cloned into pLTC76 digested with restriction endonucleases ClaI and NsiI. This provided a termination signal for in vitro transcription initiated at the leuV promoter. Transcripts ranging from 86 to 194 nucleotides long were produced from these constructions during in vitro transcription. Single-base substitution G to C at position +7 of the leuV operon created a recognition site for the unique restriction endonuclease KpnI. We have observed no effect of this substitution on transcription in vitro and on regulation of the leuV operon expression in vivo. Mutations in the promoter region were generated by oligonucleotide-directed mutagenesis using PCR. Previously described sites for restriction endonucleases ClaI and KpnI were used to clone mutant versions of the leuV promoter. The same sites were used to clone the tac promoter sequence which initiates the transcription of the leuV operon with GTP. The sequence of this promoter flanked by recognition sites for endonucleases ClaI and KpnI was −39GAGCTGTTGACAATTAATCATCGGCTCGTATAATGTGTGGG+1. pRLG1617 was a gift from Richard L. Gourse (University of Wisconsin at Madison).
Single-copy λ lysogens containing leuV promoter-lacZ fusions were constructed as previously described (32). Appropriate leuV promoter derivatives prepared in this laboratory contained either residues −50 to +11 for core promoter or residues −107 to +11 for promoter with UAS, with the exception of the leuV promoter used for measurements of GRDC in the ppGpp− strain, which contained residues −107 to +55 (15). Promoter fragments were prepared by PCR, digested with restriction enzymes EcoRI and HindIII, ligated to the left and right arms of λ, packaged in vitro, and used to lysogenize appropriate hosts as described previously (7, 8). All mutations and λ lysogens were confirmed by DNA sequence analyses. All strains were checked for double lysogens as previously outlined (37).
β-Galactosidase assays for GRDC.
Measurements were performed on promoter-lacZ fusions as described previously (31) with minor modifications. Lysogen cultures were grown at 30°C in defined media listed above. β-Galactosidase specific activity was determined in extracts prepared by sonication of cells collected at mid-log phase by centrifugation and resuspended in appropriate buffer. Values were expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside (ONGP) cleaved per minute per milligram of protein. Protein concentrations were measured by using the Bio-Rad protein assay reagent according to provided protocols with immunoglobulin G as protein standard.
Primer extension assay for SC.
Single-copy λ lysogens containing the appropriate leuV promoter-lacZ fusions were used for SC measurements. Cultures of test strains were grown overnight at 30°C in Luria broth or medium 2 (see above). After dilution to an A600 of 0.05, cells were grown to an A600 of approximately 0.6 and each test culture was divided into two aliquots. The stringent response was provoked in the first aliquot of each culture by the addition of serine hydroxamate (Sigma) to a final concentration of 1 mg/ml. The other half received an equal volume of distilled water. Thirty minutes after the addition of serine hydroxamate or water, 300 μl of mid-log reference culture was added to each test sample. Total RNA was then isolated as previously described (41). The above reference culture was strain MG1655 harboring the plasmid pLTC76 which contains the leuV(−50 +11) promoter inserted into the polylinker site upstream of the trp attenuator. Unique transcripts derived from this reference construct serve to correct for variations in RNA isolation and efficiency of primer extension reactions. Each RNA sample was split and probed with test or reference primers. Primer extension of RNA derived from the reference strain was performed by using the unique sequence primer 5′GCTTATCGATACCGTCGACCTCGAGGGG3′. This probe primes only transcripts initiated at the plasmid-borne leuV promoter and produces reverse transcripts 44 nucleotides long. Since only a few residues of the tRNA gene sequences were present, tRNA processing pathways were not operative and therefore not a factor in quantitations.
In addition to the reference primer, all samples were also probed as described previously (2) using the test primer 5′CTACCAATTCCGCCACCTTCGCATACCATC3′. This sequence is located within the β-galactosidase gene and yields reverse transcripts 68 nucleotides long.
After electrophoresis in 8% denaturing polyacrylamide gels (7 M urea included), bands were visualized by autoradiography and quantified by radioanalytical imaging using ImageQuant PhosphorImager software (Molecular Dynamics, Sunnyvale, Calif.).
Determination of promoter activity in response to in vivo pyrimidine limitation.
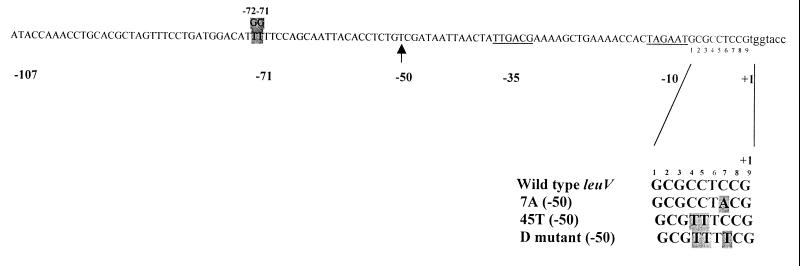
To address the question of leuV promoter responses to in vivo purine levels, the pyrimidine auxotroph CLT246 was lysogenized with one of two phages containing leuV promoter-lacZ fusions. The constructions included the leuV(−107 +55) promoter and the D mutant, which contains T substitutions at positions 4, 5, and 7 in the discriminator region (Fig. 1). In the pyrimidine auxotroph strain, CLT246, the first enzyme in the pyrimidine biosynthesis pathway is insertionally inactivated (38). Cellular pyrimidine concentrations and growth rate were modulated by growing cells in C medium (1) supplemented with 1 mM arginine, 0.4% glucose, 0.25 mM UMP, and MgSO4 ranging in concentrations from 0.2 to 0.8 mM. β-Galactosidase activities were measured as described above.
FIG. 1.
leuV promoter sequence and structural variants. −10 and −35 regions are underlined, and the numbering of bases in the discriminator starts from the −10 region. Note the G substitutions at positions −72 and −71. Promoter variants in the discriminator region are shown in bold type, and all contain sequences from positions −50 to +11 of the leuV promoter (14) fused directly to trp-lacZ fusion sequences in the λ phage employed for lysogen formation.
In vitro transcription.
Reaction mixtures (10 μl) contained the following: 50 mM Tris-HCl (pH 7.9); 5 mM MgCl2; 0.1 mM dithiothreitol; 0.1 mg of bovine serum albumin per ml; 200 μM (each) ATP, CTP, or GTP; 20 μM UTP including [α-32P]UTP; supercoiled DNA; RNA polymerase; and various concentrations of KCl or potassium glutamate. When other α-32P-labeled NTPs were used, the amount of unlabeled nucleotide was 10-fold lower and the concentration of unlabeled UTP was increased up to 200 μM. For multiple rounds of transcription, 0.5 nM DNA template and defined KCl concentrations were used. Reactions were initiated by the addition of 5 nM RNA polymerase and allowed to proceed for 10 min. Single rounds of transcription were done in the presence of 70 mM KCl. In all experiments, 5 nM RNA polymerase was preincubated with 3 nM DNA template for 10 min. Transcription was initiated by addition of a mixture of NTPs added 10 s after heparin addition to a final concentration of 100 μg/ml. Transcription was performed at 23°C for 10 min and stopped by the addition of formamide loading buffer. Transcripts were analyzed on 6% sequencing gels, followed by autoradiography, and quantified with ImageQuant PhosphorImager (Molecular Dynamics). To determine the stability of promoter open complexes, RNA polymerase was preincubated with DNA template in the presence of different effectors if indicated for 10 min. Heparin (100 μg/ml) was added at time zero. Transcription was initiated by the addition of NTPs at various times after the addition of heparin, and reactions were allowed to proceed 10 more min.
RESULTS
Promoter mutations that affect in vivo regulation.
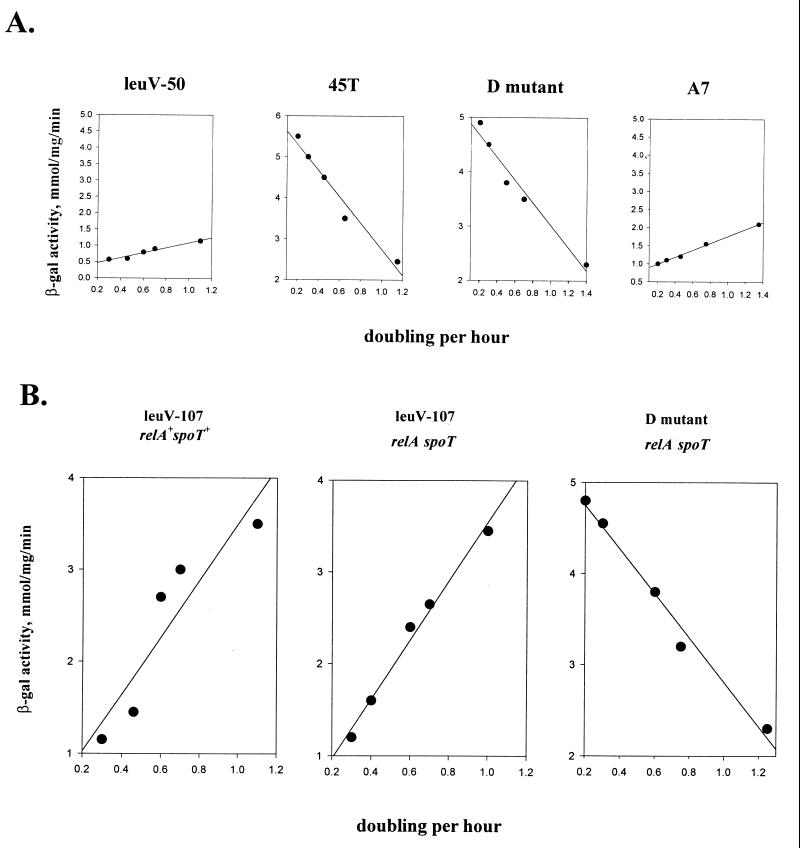
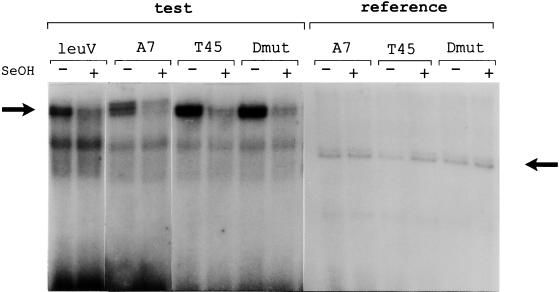
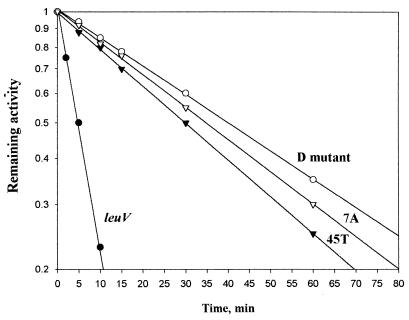
First, we wanted to determine whether the promoter sequences required for GRDC and SC are the same in leuVp and whether these regulatory responses employ the same mechanisms for controlling tRNA gene transcription. Initially, we addressed this question by analyzing regulation of the leuV promoter and its structural variants in vivo. Our hypothesis was that if both responses require the same promoter sequences, then all promoter mutations should affect both responses equally. As can be seen in Fig. 2A, sequence variations in the discriminator region can have large effects on GRDC. Two mutant promoters, D and T45 (Fig. 1), with substitutions lying in a middle portion of the discriminator region displayed disturbed GRDC (Fig. 2A) yet exhibited normal SC (Fig. 3). In contrast, mutant A7 (Fig. 2A) displayed normal GRDC, but its SC was affected. In this case, a 70% reduction in promoter activity was observed after induction of amino acid starvation compared to 90% reduction for the wild-type promoter (Fig. 3). These results clearly indicate that mutations in the promoter can differentially affect GRDC and SC.
FIG. 2.
(A) GRDC of the leuV(−50 +10) promoter and its mutants. β-Galactosidase (β-gal) activities of promoter–trp-lacZ fusions are plotted against growth rate expressed as doublings per hour. Each datum point represents the average of at least three independent experiments. The slopes determined for the different promoters using Sigma Plot linear regression analyses were as follows: leuV, 0.75; T45 mutant, −3; D mutant, −2; and A7 mutant, 0.73. (B) GRDC of leuV(−107 +55) promoter and D mutant (residues −50 to +11) in relA spoT mutant strain and control strain MG1655.
FIG. 3.
SC of the leuV(−50 +10) promoter and mutant derivatives. The strains from which RNA was extracted and whether the cultures were treated (+) or not treated (−) with serine hydroxymate (SeOH) to induce the stringent response are shown above the lanes. Transcripts were detected with the test primer which is complementary to β-galactosidase mRNA sequences or with the reference primer complementary to unique plasmid transcripts (see Materials and Methods). These mixtures were electrophoresed in separate wells to avoid overlapping with nonspecific bands. All samples were quantitated by using an ImageQuant PhosphorImager. Actual values (in pixels × 1,000 per microgram of total RNA) of promoter activity were as follows: leuV, 20.8 ± 4.2 before induction and 3 ± 0.7 after induction; 45T, 48.5 ± 5.7 before induction and 7.5 ± 0.65 after induction; D mutant (Dmut), 49 ± 6.8 before induction and 8 ± 0.9 after induction; and A7 mutant, 37.5 ± 5.2 before induction and 13 ± 2.3 after induction. Values reported are the averages of five different independent experiments.
The putative role of ppGpp in GRDC and SC of ribosomal promoter rrnBp1 has been recently examined (19). It was shown that in the relA spoT mutant, rrnBp1 was relaxed for the stringent response, but its GRDC was unimpaired. Thus, it was proposed that SC required ppGpp but that GRDC did not. To determine whether this was also the case for the leuV promoter, appropriate promoter reporter fusions were inserted into control and relA spoT strains and then GRDC and SC were assessed. As expected, it was found that the stringent response of the leuV promoter in the relA spoT mutant was drastically affected, revealing further accumulation of message after induction of amino acid starvation (data not shown). Next we tested GRDC of the complete leuV promoter in the relA spoT mutant and in the control strain. As can be seen, GRDC was not affected in the absence of ppGpp (Fig. 2B). This strongly suggested that similar to the rrnBp1 promoter, GRDC of leuVp occurs in the absence of ppGpp. The D mutant, which exhibited a reverse response to growth rate in the wild-type strain was also tested in the relA spoT mutant background. In this case, the negative slope of activity versus growth rate was essentially the same as that of the control strain (Fig. 2A). This indicates that the promoter activity of D mutant is unaffected at all growth rates tested in the absence of ppGpp. Moreover, it suggests that other promoters showing negative growth regulation such as lac may, like the D mutant, be regulated in the absence of ppGpp. Overall, these studies support the hypothesis that GRDC and SC involve different mechanisms, which is consistent with results previously reported for the rrnBp1 promoter (27).
Stability and initiating nucleotide dependence of promoter open complexes.
Recent studies have shown that rrnBp1 promoter open complexes are extremely unstable in vitro and are stabilized by high concentrations of the initiating nucleotide ATP. It was shown that the apparent Ks for ATP in vitro is more than 1,500 μM in the presence of 200 mM KCl and that in vivo promoter activity is proportional to ATP concentration. It was also shown that a rrnBp1 mutant, which displayed more-stable open complex of promoters lost GRDC. Based on these results, a model referred to as NTP sensing was proposed, which suggested that the in vivo level of purine triphosphates determines the steady-state level of rrnp1 open complexes and therefore directly controls the rate of transcription initiation (18).
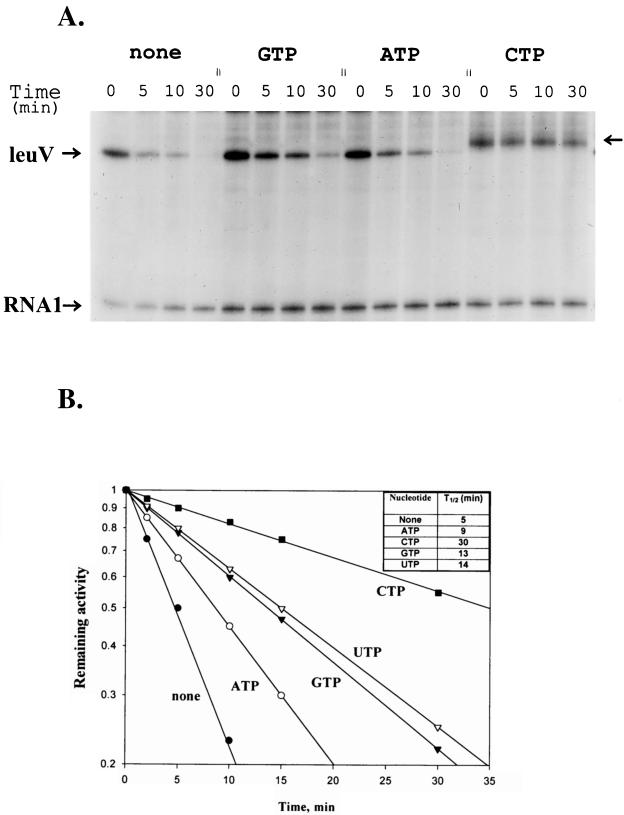
In view of the apparent structural resemblance between rrnBp1 and leuV promoters, and similar responses to SC and GRDC, we have examined the stability of the leuV promoter open complex to heparin challenge in the presence of different nucleotides. We wished to determine whether the leuV promoter open complexes could be stabilized by the presence of initiating nucleotide. If this were so, it would be possible that the nucleotide sensing mechanism proposed for the ribosomal promoter was operative for leuVp as well. Figure 4 shows that the leuV promoter forms relatively unstable promoter open complexes (half-life [t1/2], 5 min), but it is substantially more stable than those of rrnBp1. The t1/2 of rrnBp1 was about 30 s under the same assay conditions. Preincubation with the initiating nucleotide, GTP, as well as both ATP and UTP increased the stability of the leuV promoter open complex from two- to threefold (Fig. 4B). Interestingly, CTP caused the formation of the most stable complex which is based primarily on reiterative transcription of leuVp (36a).
FIG. 4.
Stability of leuV promoter open complexes preincubated in the absence or presence of nucleotides. (A) Typical experiment showing separation of transcripts derived from a single round of transcription at various times after heparin addition at time zero. The supercoiled plasmid template employed contained sequences from the leuV operon from positions −50 to +136 inserted into pLTC76 (see Materials and Methods). Transcripts terminated at the trp attenuator derived from this construction were 194 nucleotides long. Experiments were performed in the absence (none) or presence of 1 mM (each) GTP, ATP, and CTP in the preincubation mixture. (B) Half-lives of leuV promoter open complexes formed in the absence or presence of 1 mM of each nucleotide in the preincubation mixture. Band counts derived from transcription of the above plasmid template were plotted semilogarithmically as a function of time. Data presented are the averages of at least three independent experiments.
In order to estimate the level of NTPs required for optimal activity of the leuV promoter, we performed multiple rounds of transcription of the leuV promoter and its mutants in vitro as a function of NTP concentrations. In addition, it was important to determine whether Ks values for initiating nucleotide are relevant to the changes in GRDC for mutant promoters.
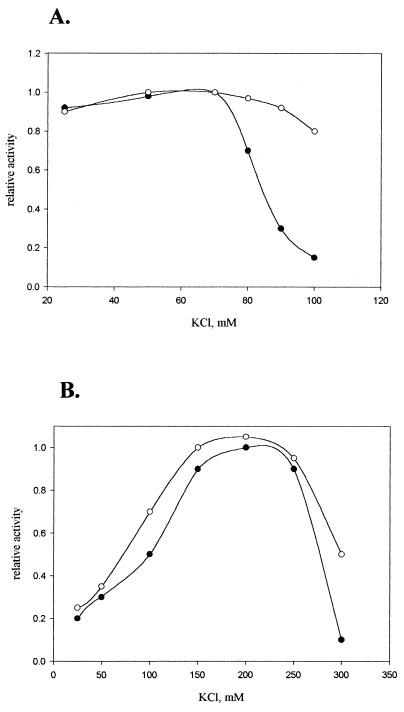
First, we determined optimal salt concentrations for transcription of the wild-type leuV core promoter. This was particularly important, given the strong effect salt had upon Ks values reported for rrnBp1 (18). We found a significant difference in KCl optima between single and multiple rounds of transcription (Fig. 5). In a single round of transcription, the optimal KCl concentration was about 70 mM, with a substantial drop in promoter efficiency at higher salt concentrations. The D mutant was efficiently transcribed over a wider range of KCl concentrations up to 200 mM (Fig. 5A). In multiple rounds of transcription (Fig. 5B), both the wild type and D mutant showed similar KCl dependencies with broad optima between 150 to 250 mM KCl. Results obtained with the A7 and T45 mutant promoters were essentially the same as that for the D mutant (data not shown).
FIG. 5.
KCl dependence of the leuV promoter transcription in vitro. (A) Single round of transcription. Supercoiled plasmid templates containing either the wild-type leuV promoter (filled circles) or the D mutant promoter (open circles) were preincubated with RNA polymerase at different concentrations of KCl. Reactions were initiated by the addition of all NTPs and heparin as indicated in Materials and Methods. Templates employed for transcription contained leuV operon sequences from positions −50 to +67 inserted into pLTC76 (see Materials and Methods), producing 132-nucleotide transcripts. (B) Multiple rounds of transcription. Transcription was initiated by the addition of RNA polymerase to supercoiled plasmid templates described above containing either the wild type (filled circles) or the D mutant (open circles). The activities of these templates at increasing concentrations of KCl are indicated.
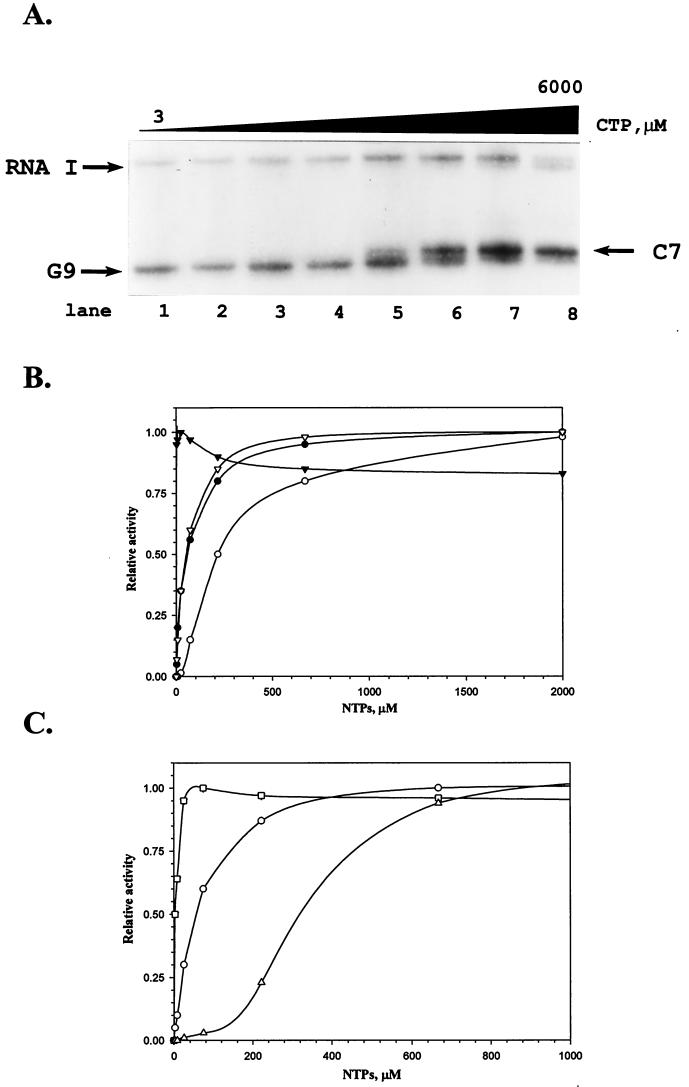
To visualize transcripts which differed in length by 1 nucleotide, we used a supercoiled plasmid, which produced 86-base transcript (see Materials and Methods). Multiple initiation sites emanating primarily from positions C7 and G9 were observed. At low GTP concentrations, initiation occurred primarily from position C7, while at low CTP concentrations, transcription was predominately initiated at position G9 (Fig. 6A). It was important to quantitate transcripts initiated only at G9 or C7, independent of an alternative initiating nucleotide. Results of in vitro transcription as a function of nucleotide concentration in the presence of 200 mM KCl are presented in Fig. 6B. The apparent Ks values for GTP determined in the presence of both 50 and 200 mM of KCl were about 50 μM. Essentially the same Ks value was shown for UTP; however, the apparent Ks for CTP at 240 μM was significantly higher. The maximal efficiency of RNA synthesis with various ATP concentrations was observed at a concentration of ATP as low as 2.7 μM. The fact that polymerase utilizes both CTP and GTP as initiation nucleotides is quite different from ribosomal promoters, which have a single initiation site. For example, rrnBp1 is strongly dependent on high levels of ATP, whereas rrnDp1 is dependent, but less so, on levels of GTP for maximal activity in multiple rounds of transcription at high salt concentrations (18). This might reflect the levels of these nucleotides in vivo. Given these differences, we have compared the apparent Ks values for initiating nucleotides for leuVp with two other promoters, including a derivative of the tac promoter, which initiates the transcription with GTP, and rrnBp1, which initiates with ATP. Here it can be seen that the Ks values varied over almost 2 orders of magnitude (Fig. 6C), and apparent Ks values for initiating nucleotide were 5, 50, and 330 μM for tacp, leuVp, and rrnBp1, respectively. In addition, we found that the apparent Ks for GTP for all mutant versions of the leuV promoter was approximately 80 μM, which was slightly higher than that for leuVp (data not shown).
FIG. 6.
Multiple rounds of transcription in vitro of the leuV(−50 +11) promoter as a function of NTP concentration in the presence of 200 mM KCl. (A) Autoradiogram of 5% denaturing polyacrylamide gel representing the separation of the transcription products in the presence of a fixed concentration of GTP (200 μM) and increasing concentrations of CTP. The concentrations of CTP are 3, 8, 24, 72, 216, 670, 2,000, and 6,000 μM (lanes 1 to 8, respectively). The template employed contained leuV promoter sequences from positions −50 to +31, which produced 86-nucleotide transcripts. (B) Nucleotide dependence of the leuV promoter in multiple rounds of transcription. Increasing concentrations of ATP (filled triangles), CTP (open circles), GTP (filled circles) and UTP (open triangles) are shown. In the case of ATP and UTP titrations, two bands corresponding to initiation at C7 and G9 were seen at all concentrations. In this case, values plotted are the sum of these two transcripts. In the case of CTP, only counts corresponding to initiation at C7 were quantitated and plotted. Similarly, in the case of GTP titrations, only counts in G9 were quantitated and plotted. (C) Initiating nucleotide dependence of promoters in multiple rounds of transcription. Transcriptional activities as a function of initiating nucleotide concentrations are presented for leuVp (circles), tacp (squares), and rrnBp1 (triangles). Multiple rounds of transcription were performed in the presence of 200 mM KCl as described in Materials and Methods. In the case of leuV and tac promoters, various concentrations of GTP were employed. For rrnBp1, increasing ATP concentrations were used.
Taken together, the data from these experiments show that there are distinct differences between the leuV and rrnBp1 promoters with respect to their responses to the concentration of the initiating nucleotide and the effect of salt on promoter efficiency. These results impinge on the important question of the role of initiating nucleotide concentration on in vivo promoter activity.
Role of the promoter open complex stability on GRDC.
Recently it was proposed that the stability of promoter open complexes is directly related to the regulation of stringently controlled promoters (49). That is, these promoters might invariably display relatively unstable promoter open complexes. Another study has suggested that this property may be important for GRDC of rRNA gene expression as well (18). It has been further speculated that the intrinsic instability of the complexes of these promoters correlates with the GC richness of the discriminator region located between positions −10 and +1 of the promoter (33) and that the sequence of the discriminator region might determine the energy barrier for promoter open complex formation (36). In view of these reports, we wished to determine whether the leuV promoter shares these properties with other promoters studied and, in particular, whether promoter open complex instability is always a property of promoter subject to GRDC and SC. Our initial approach was to examine promoter open complex stability of leuVp structural variants which differ in GRDC and SC.
As can be seen, all three mutants displayed higher stability of promoter open complexes than that of the wild type (Fig. 7). As discussed above, two derivatives, T45 (t1/2, 28 min) and D (t1/2, 40 min), displayed significantly altered GRDC (Fig. 2A). Importantly, GRDC of the third variant studied, A7 (t1/2, 35 min) was indistinguishable from that of the wild-type promoter (Fig. 2A), but its SC was measurably affected (Fig. 3). None of the other mutations, which abolished GRDC affected the stringent response promoter. All mutations described here decreased GC richness of the discriminator, which might affect kinetics of promoter open complex formation. It is therefore not surprising that all the substitutions increased the half-life of the promoter open complex, what was accompanied by reduced KCl sensitivity of promoters in a single round of transcription (Fig. 5A and unpublished results).
FIG. 7.
Stability of open complexes formed with mutants of leuV promoter. Band counts derived from transcription of the supercoiled plasmid templates were plotted semilogarithmically as a function of time. Templates employed for these transcriptions contained leuV operon sequences from positions −50 to +67 (see Materials and Methods), producing 132-nucleotide transcripts. Data presented are the averages of at least three independent experiments. Transcription was performed with the wild-type leuV promoter D mutant, T45, and 7A.
Effect of NTP concentration in vivo.
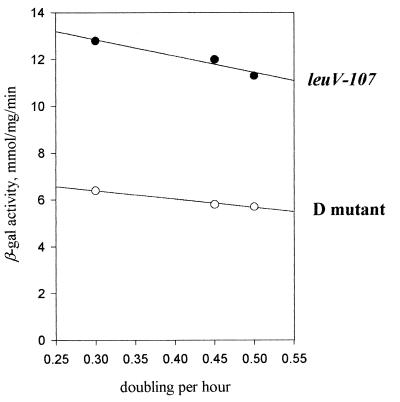
Overall, the experiments above indicate that the leuV promoter is less responsive at least in vitro to NTP concentrations than rrnBp1. Moreover, it appears that the apparent Ks value for the major initiating nucleotide, GTP, is relatively low compared to estimated in vivo purine levels at any growth rate (34). If this is the case, we might predict that the leuV promoter would be less responsive to changes in the intracellular level of NTPs. Therefore, the following experiments were done. We employed the approach recently used by others for the study of rrnBp1 (18). Restriction of exogenous pyrimidines causes a marked reduction of the intracellular concentration of UTP and CTP and of the growth rate for the strain CLT246, which exhibits defective pyrimidine biosynthesis (38). The lowered concentration of pyrimidine triphosphates limits RNA synthesis, and as a result, increases the concentration of intracellular ATP and GTP. With this approach, it was previously reported that the efficiency of the rrnBp1 promoter was decreased almost 50% when the intracellular ATP level was reduced around twofold (18). For the leuV promoter, we detected an approximately 10% reduction in promoter activity under the same growth conditions (Fig. 8), in contrast to an increase of approximately 40% in promoter activity, which occurs in a control strain not starved for pyrimidines at the growth rates tested. These results are not inconsistent with the model that the leuV promoter is sensitive to NTP concentration in vivo and that this mechanism is involved in GRDC of leuVp. It is clear, however, that leuVp is less sensitive to this control mechanism compared to recent results obtained for rrnBp1.
FIG. 8.
GRDC of the leuV(−107 +55) promoter and D mutant (residues −50 to +11) in CLT246 (car::Tn10) strain. The complete leuV promoter and the D mutant, containing T substitutions at positions 4, 5, and 7, are shown. β-gal, β-galactosidase.
Role of UAS in GRDC.
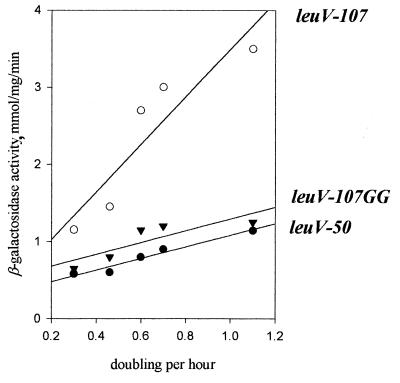
We previously observed that flanking sequences affected GRDC of the leuV promoter. Deuster et al. showed that promoter constructions containing sequences downstream of the promoter displayed greater GRDC than one without these sequences (15). Later, Bauer et al. showed that the core leuV promoter is compromised in GRDC (8). We have examined this question here in a relA+ strain using comparable constructions. These experiments were particularly interesting because it had been shown that the rrnBp1 core promoter is necessary and sufficient for GRDC (6). As can be seen in Fig. 9, optimal GRDC was strongly dependent on upstream sequences. Note that not all GRDC is eliminated when UAS are deleted. The substitution of two T’s for two G’s at positions −71 and −72 completely abolished the effect of UAS. We have recently shown that these sequences contain a functional FIS site (39).
FIG. 9.
Effects of upstream sequences on leuV GRDC. β-Galactosidase activities of the leuV(−107 +11) promoter, the leuV(−50 +11) promoter, and the leuV(−107 +11) promoter containing substitutions at positions −71 and −72 of the fis site are shown.
Interaction of UAS and core promoter elements.
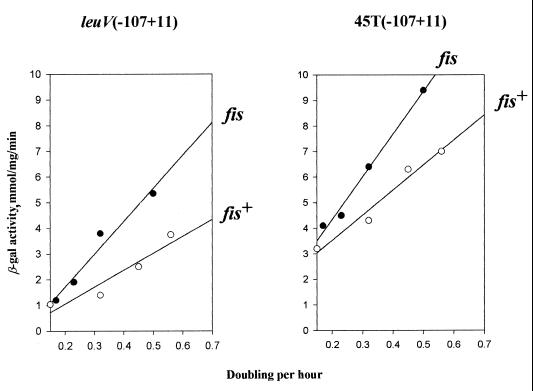
Above we have shown that the wild-type leuV promoter requires upstream sequences not only for maximal activity but also for a full response to changes in growth rate. Moreover, a functional Fis binding site must be present in UAS to mediate these activities. We have shown as well that particular promoter mutations disrupt GRDC of leuVp when introduced into the core promoter. What then will be observed when the fis gene is disrupted, and what effects do UAS have on mutations in the core promoter? The first question has been initially addressed in other communications (35, 39). It was shown that although purified FIS protein stimulates leuVp transcription about threefold in vitro, the activity of the leuV promoter was greater in fis mutant background (39). Nevertheless, elevated levels of tRNA1Leu compared to 16S rRNA were reported in fis+ cells (35). Therefore, FIS is clearly involved in regulation of leuVp in vivo and in vitro. However, it has not been determined yet whether inactivation of the fis gene is reflected somehow on GRDC of the leuV promoter. To determine directly whether FIS contributes to GRDC of the leuV promoter, we measured β-galactosidase activities derived from chromosomal single copy of wild-type and T45 mutant leuVp-lacZ fusions as a function of growth rate in fis and fis+ strains (Fig. 10). First, Fig. 10 illustrates that the presence of UAS not only optimized GRDC for wild-type leuVp but restored GRDC for the T45 mutant, which displayed completely disrupted GRDC as the core promoter. Second, it can be seen that promoter activities were increased rather than decreased in a fis mutant background and that GRDC of both promoters was increased at least by 15% in fis mutant cells compared to wild-type cells. Similar results were obtained with the D mutant (data not shown), and the absence of functional FIS protein did not affect the regulation of the leuV core promoter (data not shown). Overall, these experiments show that UAS can abolish the effects of core promoter mutation and that these sequences are the dominant determinant for GRDC of the leuV promoter.
FIG. 10.
GRDC of the leuV and T45 mutant promoters with upstream sequences in fis+ and fis strains. β-Galactosidase (β-gal) activities of promoter-lacZ fusions are plotted against growth rate expressed as doublings per hour. Each datum point represents the average of at least three independent experiments.
DISCUSSION
We have shown here that the targets for SC and GRDC in the leuV promoter are not identical, since mutations which differentially affect these regulatory responses have been identified. We have also demonstrated that GRDC but not SC of the leuV promoter occurs in the absence of ppGpp. Similar findings have been reported for the rrnBp1 promoter (19, 27). However, a body of evidence has been put forth suggesting that ppGpp is involved in both SC and GRDC (25, 48). This conclusion is based in part on the observation that ppGpp levels varied inversely with the expression of stable RNA genes at various growth rates (42). In addition, work with mutants of RNA polymerase (4) and studies focused on tyrT operon regulation (43) further support a role for ppGpp in both regulatory mechanisms. Based on our results and that of others, we conclude that GRDC and SC for leuVp and rrnBp1 must proceed at least in part via different mechanisms.
It is not surprising that similar regulatory strategies are used for the expression of stable RNAs to ensure close regulation of protein synthesis under various growth conditions. However, recent in vivo studies strongly suggested that tRNA gene expression is heterogeneous (17, 40). It was originally shown that some tRNA species exhibited positive GRDC, whereas the level of others did not increase, or even decreased, as growth rates were increased (16, 17). Now it is generally accepted that the intracellular concentration of all tRNA species is increased between growth rates of 0.4 and 2.5 doublings per h (14). The rate of increase is specific for each tRNA and can vary as a function of input signals involved in regulation. For example, one level of control of tRNA biosynthesis involves FIS. When synthesis of tRNA species was compared in fis+ and fis strains, the expression of some tRNAs as a function of growth rate was strongly dependent upon the presence of FIS, whereas others showed no requirement for FIS or even appeared to be more abundant in the fis mutant at all growth rates tested (35). It is therefore perplexing why leuVp displays even better growth rate regulation in the absence of an active fis gene. This suggests that even further complexities exist for controlling the level of tRNA promoters in vivo. Recently, it has been proposed that in the case of a rRNA promoter, FIS and another DNA binding protein H-NS interact with DNA at overlapping sites, thereby working in concert to orchestrate promoter activity as a function of growth. It is conceivable that other factors such as supercoiling of DNA or other DNA binding proteins may be involved as well in these interactions. Therefore, the interpretation of data derived for strains lacking any of these factors becomes difficult. For example, in the absence of FIS, other factors may operate independently and FIS might then serve a negative role in the presence of particular combinations of factors or conditions. It has been reported that FIS may act via contacts with the alpha subunit of RNA polymerase, which in turn may alter the actual site and kinetics of promoter-polymerase interactions. Moreover, it has been shown that the concentration of FIS increases with increasing growth rate. The simplest model in this case would be that FIS simply increases in concentration and subsequently stimulates promoter activity at higher concentrations. Whatever the case, it is clear from these experiments that a simple model whereby FIS alone modulates promoter activity as growth changes seems very unlikely.
We have previously shown that at least one tRNA core promoter (leuXp) does not exhibit GRDC (40). This may not be surprising, since this promoter contained no UAS, suggesting that GRDC of leuXp might be solely mediated by UAS. It is important to note that the argT, metT, and leuV core promoters displayed moderate GRDC and consequently must have the appropriate sequences for UAS-independent GRDC. It will be interesting to determine whether tRNA promoters differ in the relative contributions of core and flanking promoter sequences for GRDC. We should point out, however, that other postinitiation regulatory mechanisms might determine final levels of tRNA isoacceptors. For example, transcription elongation rates, tRNA processing, modification, and turnover all affect cellular levels of tRNA at a given growth rate.
As we have outlined above, we also examined the differential effects of mutations in the leuV core promoter on SC and GRDC. Selected discriminator mutations in the core promoter (D and T45 mutants) clearly disturbed GRDC, such that promoter activity varied inversely with growth rate (Fig. 2A). Neither of these mutants displayed measurable changes in SC. Conversely, the A7 mutation was modestly affected in SC but still displayed normal GRDC. It is striking from our experiments and those of others that many mutations can disrupt GRDC in both rrnBp1 and leuV promoters (7, 27, 36b) and with the exception of the A7 mutation reported here, no single base substitution in the discriminator motif of leuVp and rrnBp1 affecting SC has been identified so far. However, multiple mutations in the discriminator region of rrnBp1 rrnBp2 and tyrTp promoters have been reported to significantly affect both SC and GRDC (27, 43, 48). It is possible, therefore, that the leuV promoter may utilize different sequence requirements and regulatory strategies than these systems. Our experiments suggest that whatever mechanisms are employed for GRDC or SC, targets in the leuV promoter responsible for regulation at steady-state growth and during amino acid starvation are not necessarily the same. We wish to emphasize that all these promoter variants lack UAS sequences, and effects seen must therefore be UAS independent. We believe that more-extensive mutagenesis of the leuV promoter may be required to fully describe targets of the various forms of control.
The recent report of Gaal et al. has provided evidence for the role of intracellular levels of initiating nucleotides in GRDC for ribosomal promoters (18). These researchers showed that polymerase-rrnBp1 complexes are intrinsically unstable, as judged by heparin challenge experiments, and that the initiating nucleotide ATP stabilizes these complexes. Based on the observations that both ribosome production and intracellular purine levels are increased as a function of growth rate (10, 34), the researchers proposed a model for the control of ribosome synthesis by “NTP sensing.” This model states that the high concentration of initiating nucleotide stabilizes rrnp1 open complexes in vivo and may account for the increased activities of the promoters as a function of growth and increased intracellular purine concentrations.
We have examined this issue for the leuV promoter, and it appears that this mechanism could be operative for the leuV promoter as well. However, its relative contribution to GRDC is much less than that for rrnBp1. We showed that the leuV promoter is less sensitive to salt than rrnBp1 in single and multiple rounds of transcription. These results reflect the effect of salt concentration on promoter opening. Extreme salt sensitivity of rrnBp1 may have been in part responsible for the high Ks value of ATP reported for rrnBp1. In this case, ATP presumably operated as a ligand, increasing the half-life of promoter open complexes and thereby increasing the number of subsequent productive elongation events. The binding of ATP stimulated the initiation of rrnBp1 transcription in vitro, facilitating the formation of promoter open complexes under conditions where the KCl concentration was still optimal for polymerase functioning but was high enough for efficient polymerase-rrnBp1 open complex formation. In the case of leuVp, which forms more-stable open promoter complexes than rrnBp1, all NTPs increased the stability of promoter open complex in vitro (Fig. 4). Therefore, we conclude that the concentration of all nucleotides could play a role in increasing leuVp open complex stability and promoter efficiency in vivo. This is consistent with the finding of multiple initiating sites for the leuV promoter in vivo and in vitro, which could make it responsive to the level of more than one nucleotide. Our results are not inconsistent with the idea of a special role of initiating NTPs in transcription, but our experiments suggest that the level of GTP alone may not account for all changes in promoter activity seen as a function of growth.
In accordance with our results, GRDC of the leuV promoter was less affected in β′ Δ215-220 mutant of RNA polymerase than were rrnBp1 and rrnDp1 (5). It was found that the β′ Δ215-220 mutant polymerase forms less-stable complexes with both rrnBp1 and λPR, and therefore requires higher concentrations of initiating nucleotides than for wild-type polymerase. This mutant has been shown to reduce the increase in promoter activities compared to wild-type strain for rrnBp1 and rrnDp1 from 5.8 to 1.8 and from 4.4 to 2, respectively, between the growth rates of 0.7 and 1.4. The leuV promoter displayed a twofold increase in promoter activity in the wild type and a 1.4-fold increase was observed in the β′ Δ215-220 mutant under the same growth conditions. In agreement with our conclusions, it was suggested that the leuV promoter was less responsive to the NTP sensing (5).
All described mutants of leuVp formed more-stable open complexes of promoters (Fig. 7), but only two (T45 and D mutants) had disturbed GRDC. We interpret this to mean that the stability of promoter open complexes alone does not determine the GRDC of the leuV promoter. How then can we account for GRDC of the leuV promoter if NTP levels and stability of promoter open complexes are only partially involved in regulation if at all?
We propose the following summary for how the leuV promoter might be regulated. First, it is clear that ppGpp is required for the SC of the leuV operon and functions via interaction directly with RNA polymerase. Second, ppGpp is not essential for GRDC as we have shown here and may not be involved in changes in promoter activity under the conditions tested. Intracellular concentrations of NTP may not be a major factor for GRDC of promoter activity. Finally, UAS sequences are required for optimal GRDC. It will be interesting to determine the nature of other factors involved in regulation of leuVp. It will be important to understand in detail the interactions between FIS-mediated control and core promoter mechanisms such as the effects of NTP concentrations. It is conceivable that these two mechanisms are interactive. For, example, could binding of factors to UAS affect either the kinetics of promoter open complex formation or modulate the parameters of NTP sensing? Overall, it is clear that no single model which explains GRDC can be proposed for all stable RNA genes.
Given these points, it becomes important to know how other tRNA promoters may have evolved to respond to differences in growth rate. It has been reported that tRNA genes appear to be heterogeneous with respect to responses to growth rate and requirements for FIS (35). Taken together, the work presented here and elsewhere suggests that tRNA genes may in fact have evolved a variety of strategies for GRDC. This idea is supported by the observation that primary sequences in tRNA promoters show considerable heterogeneity. Finally, it will be interesting to determine whether similar diversity of tRNA gene expression is seen with regard to mechanisms of the stringent response.
ACKNOWLEDGMENTS
This work was supported in part by grants GM50747 from the National Institute of Health to W.M.H. and GM29466 to C.L.T.
We thank Wilma Ross, Rick Gourse, Michael Bartlett, and Cathy Squires for very important and useful discussions, and we thank Tamas Gaal and Wilma Ross for lysogens.
REFERENCES
- 1.Alper M D, Ames B N. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J Bacteriol. 1978;133:149–157. doi: 10.1128/jb.133.1.149-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1987. pp. 4.8.1.–4.8.4.. [Google Scholar]
- 3.Baracchini E, Bremer H. Control of rRNA synthesis in Escherichia coli at increased rrn gene dosage. Role of guanosine tetraphosphate and ribosome feedback. J Biol Chem. 1991;266:11753–11760. [PubMed] [Google Scholar]
- 4.Baracchini E, Glass R, Bremer H. Studies in vivo on Escherichia coli RNA polymerase mutants altered in the stringent response. Mol Gen Genet. 1988;213:379–387. doi: 10.1007/BF00339606. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett M S, Gaal T, Ross W, Gourse R L. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett M S, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer B F, Elford R M, Holmes W M. Mutagenesis and functional analysis of the Escherichia coli tRNA(1Leu) promoter. Mol Microbiol. 1993;7:265–273. doi: 10.1111/j.1365-2958.1993.tb01117.x. [DOI] [PubMed] [Google Scholar]
- 8.Bauer B F, Kar E G, Elford R M, Holmes W M. Sequence determinants for promoter strength in the leuV operon of Escherichia coli. Gene. 1988;63:123–134. doi: 10.1016/0378-1119(88)90551-3. [DOI] [PubMed] [Google Scholar]
- 9.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. . [Comment.] [DOI] [PubMed] [Google Scholar]
- 10.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 11.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 12.Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 13.Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 14.Dong H, Nilsson L, Kurland C G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 15.Duester G, Elford R M, Holmes W M. Fusion of the Escherichia coli tRNALeu1 promoter to the galK gene: analysis of sequences necessary for growth-rate-dependent regulation. Cell. 1982;30:855–864. doi: 10.1016/0092-8674(82)90290-2. [DOI] [PubMed] [Google Scholar]
- 16.Emilsson V, Kurland C G. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J. 1990;9:4359–4366. doi: 10.1002/j.1460-2075.1990.tb07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emilsson V, Naslund A K, Kurland C G. Growth-rate-dependent accumulation of twelve tRNA species in Escherichia coli. J Mol Biol. 1993;230:483–491. doi: 10.1006/jmbi.1993.1165. [DOI] [PubMed] [Google Scholar]
- 18.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 19.Gaal T, Gourse R L. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gourse R L, de Boer H A, Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 21.Gourse R L, Takebe Y, Sharrock R A, Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc Natl Acad Sci USA. 1985;82:1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamming J, Ab G, Gruber M. E. coli RNA polymerase-rRNA promoter interaction and the effect of ppGpp. Nucleic Acids Res. 1980;8:3947–3963. doi: 10.1093/nar/8.17.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haseltine W A, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haseltine W A, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature. 1972;238:381–384. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez V J, Bremer H. Guanosine tetraphosphate (ppGpp) dependence of the growth rate control of rrnB P1 promoter activity in Escherichia coli. J Biol Chem. 1990;265:11605–11614. [PubMed] [Google Scholar]
- 26.Ikemura T, Dahlberg J E. Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J Biol Chem. 1973;248:5033–5041. [PubMed] [Google Scholar]
- 27.Josaitis C A, Gaal T, Gourse R L. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingston R E, Nierman W C, Chamberlin M J. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem. 1981;256:2787–2797. [PubMed] [Google Scholar]
- 29.Krohn M, Wagner R. Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. J Biol Chem. 1996;271:23884–23894. doi: 10.1074/jbc.271.39.23884. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 32.Miura A, Krueger J H, Itoh S, de Boer H A, Nomura M. Growth-rate-dependent regulation of ribosome synthesis in E. coli: expression of the lacZ and galK genes fused to ribosomal promoters. Cell. 1981;25:773–782. doi: 10.1016/0092-8674(81)90185-9. [DOI] [PubMed] [Google Scholar]
- 33.Mizushima-Sugano J, Kaziro Y. Regulation of the expression of the tufB operon: DNA sequences directly involved in the stringent control. EMBO J. 1985;4:1053–1058. doi: 10.1002/j.1460-2075.1985.tb03738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuhard J, Nygaard P. Purines and pyrimidines. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 445–473. [Google Scholar]
- 35.Nilsson L, Emilsson V. Factor for inversion stimulation-dependent growth rate regulation of individual tRNA species in Escherichia coli. J Biol Chem. 1994;269:9460–9465. [PubMed] [Google Scholar]
- 36.Ohlsen K L, Gralla J D. Interrelated effects of DNA supercoiling, ppGpp, and low salt on melting within the Escherichia coli ribosomal RNA rrnB P1 promoter. Mol Microbiol. 1992;6:2243–2251. doi: 10.1111/j.1365-2958.1992.tb01400.x. [DOI] [PubMed] [Google Scholar]
- 36a.Pokholok, D. K., and W. M. Holmes. Unpublished results.
- 36b.Pokholok, D. K., et al. Unpublished results.
- 37.Powell B S, Rivas M P, Court D L, Nakamura Y, Turnbough C L., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi F, Turnbough C L., Jr Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J Mol Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. [DOI] [PubMed] [Google Scholar]
- 39.Ross W, Salomon J, Holmes W M, Gourse R L. Activation of Escherichia coli leuV transcription by FIS. J Bacteriol. 1999;181:3864–3868. doi: 10.1128/jb.181.12.3864-3868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowley K B, Elford R M, Roberts I, Holmes W M. In vivo regulatory responses of four Escherichia coli operons which encode leucyl-tRNAs. J Bacteriol. 1993;175:1309–1315. doi: 10.1128/jb.175.5.1309-1315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarmientos P, Sylvester J E, Contente S, Cashel M. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell. 1983;32:1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- 42.Sarubbi E, Rudd K E, Cashel M. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- 43.Travers A A, Lamond A I, Weeks J R. Alteration of the growth-rate-dependent regulation of Escherichia coli tyrT expression by promoter mutations. J Mol Biol. 1986;189:251–255. doi: 10.1016/0022-2836(86)90397-9. [DOI] [PubMed] [Google Scholar]
- 44.Vogel U, Pedersen S, Jensen K F. An unusual correlation between ppGpp pool size and rate of ribosome synthesis during partial pyrimidine starvation of Escherichia coli. J Bacteriol. 1991;173:1168–1174. doi: 10.1128/jb.173.3.1168-1174.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel U, Jensen K F. Effects of guanosine 3′,5′-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J Biol Chem. 1994;269:16236–16241. [PubMed] [Google Scholar]
- 46.Vogel U, Jensen K F. Effects of the antiterminator BoxA on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J Biol Chem. 1995;270:18335–18340. doi: 10.1074/jbc.270.31.18335. [DOI] [PubMed] [Google Scholar]
- 47.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 48.Zacharias M, Goringer H U, Wagner R. Influence of the GCGC discriminator motif introduced into the ribosomal RNA P2- and tac promoter on growth rate control and stringent sensitivity. EMBO J. 1989;8:3357–3363. doi: 10.1002/j.1460-2075.1989.tb08498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y N, Jin D J. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]