Abstract
Starved cultures of Escherichia coli undergo successive rounds of population takeovers by mutants of increasing fitness. These mutants express the growth advantage in stationary phase (GASP) phenotype. Previous work identified the rpoS819 allele as a GASP mutation allowing cells to take over stationary-phase cultures after growth in rich media (M. M. Zambrano, D. A. Siegele, M. A. Almirón, A. Tormo, and R. Kolter, Science 259:1757–1760, 1993). Here we have identified three new GASP loci from an aged rpoS819 strain: sgaA, sgaB, and sgaC. Each locus is capable of conferring GASP on the rpoS819 parent, and they can provide successively higher fitnesses for the bacteria in the starved cultures. All four GASP mutations isolated thus far allow for faster growth on both individual and mixtures of amino acids. Each mutation confers a growth advantage on a different subset of amino acids, and these mutations act in concert to increase the overall catabolic capacity of the cell. We present a model whereby this enhanced ability to catabolize amino acids is responsible for the fitness gain during carbon starvation, as it may allow GASP mutants to outcompete the parental cells when growing on the amino acids released by dying cells.
In the natural environment, chemoorganotrophs such as Escherichia coli obtain both their carbon and energy from organic matter released by other cells. The mechanisms of organic nutrient release are variable, ranging from regulated extrusion of metabolic end products to release as a result of death and lysis of donor cells (3, 21–23, 34). However, the actual bioavailability of carbon in nature is low due to intense competition (22, 23). As a result, natural microbial populations spend the majority of their lives under starvation stress, interspersed with sporadic and short-lived periods of growth as nutrients become available.
Our laboratory uses carbon-starved cultures of E. coli as an experimental model to understand the processes of survival and evolution in natural microbial populations. E. coli can survive extended periods of starvation. In aerated rich medium (Luria-Bertani [LB] broth), E. coli ceases growth due to carbon limitation (37). During the first several days of starvation, the population loses 90 to 99% of the viable counts (40). However, the viable counts nearly level off after these first few days, and populations can survive for several years in this spent LB medium aerated at 37°C without further addition of carbon (7, 8). As the cultures consume exogenous carbon during exponential growth, the biomass is the most likely source of carbon during extended survival, which becomes available when the cells die.
While the overall population of stationary-phase E. coli cultures may be considered starved in that there is no net increase in biomass, there are subpopulations that are clearly not starved, as they are able to grow as a subculture and take over the population (8, 38–40). These subpopulations consist of mutants with enhanced fitness during starvation. The ability to grow during starvation has been termed the growth advantage in stationary phase (GASP) phenotype (38). Studies on cultures starved for extended periods demonstrate that the GASP phenomenon is continuous: multiple rounds of population takeovers occur throughout the starvation period (7, 40). Interestingly, as the cultures age, they increase in diversity, as several genetically distinct subpopulations coexist (7).
The first mutation conferring the GASP phenotype after growth in rich media was identified as an allele of rpoS (40), a gene whose product, ςS, is responsible for the regulation of many genes during starvation stress (12). Transduction of the GASP allele of rpoS (rpoS819) into an otherwise wild-type strain was sufficient to confer the GASP phenotype (40). The rpoS819 allele is a 46-bp duplication at the 3′ end of the gene, which results in a replacement of the last four residues in ςS with 39 new amino acids. Expression of two ςS-dependent genes, katE (25) and bolA (4, 15), are both reduced in the rpoS819 strain (40), indicating a reduction of function in this allele. The physiological basis for the fitness gain of the rpoS819 mutation is not yet known.
The purpose of our investigation was to understand how GASP mutations alter cell physiology to provide fitness gains in stationary phase. To this end, we sought to identify and characterize new GASP mutations. ZK1141, an isolate from an aged culture of the rpoS819 strain, was capable of outcompeting its rpoS819 parent, indicating that additional GASP mutations accumulated in this strain (38). In this study, we have demonstrated that the ZK1141 strain has acquired three new GASP mutations, each of which can confer the GASP phenotype on the rpoS819 parent. Each of these newly identified GASP alleles, as well as the rpoS819 allele, increased starvation survival fitness in an additive manner. Each of these four GASP alleles also conferred growth advantages on amino acids as the sole sources of carbon and energy. Similar to the competitive fitnesses, these growth phenotypes were additive.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this study are listed in Table 1.
TABLE 1.
E. coli strains used in this study
| Strain | Genotype or phenotype | Reference or source |
|---|---|---|
| GASP strain | ||
| ZK126 | W3110 tna2 ΔlacU169; G0 | 6 |
| ZK819 | ZK126 rpoS819 rpsL Smr; GI | 40 |
| ZK820 | ZK126 rpoS819 gyrA Nalr; GI | 40 |
| ZK1141 | ZK819 sgaA sgaB sgaC; GII | 38 |
| ZK2552 | ZK819 ilvGMEDA+ (spontaneous suppressor) Valr | This study |
| ZK2553 | ZK819 bgl+ (spontaneous suppressor) Bgl+ | This study |
| ZK2554 | ZK1141 ilvGMEDA+ (same allele as in ZK2552) Valr | This study |
| ZK2555 | ZK1141 bgl+ (spontaneous suppressor) Bgl+ | This study |
| ZK2557 | ZK2553 rpoS+ | This study |
| ZK2559 | ZK2553 sgaB | This study |
| ZK2561 | ZK2553 sgaA | This study |
| ZK2563 | ZK2553 sgaC | This study |
| ZK2564 | ZK2553 sgaA sgaB sgaC | This study |
| ZK2618 | ZK1141 trpB::Tn10Tcr | This study |
| Other strains | ||
| CAG12173 | MG1655 cysC95::Tn10Tcr | 31 |
| CAG18501 | MG1655 rbsD::Tn10Tcr | 31 |
| CAG18528 | MG1655 zbj-3110::Tn10Kanr | 31 |
| KER176 | rpsL lipA150::Tn1000dKanr | 35 |
| NU1107 | VJS433 serC::mini-MudI194 Kanr | 14 |
| ZK173 | MB2 (Hfr Δgal trpB::Tn10 proC::Tn5 lacU169) | Lab stock |
| ZK1000 | ZK126 ΔrpoS::Kanr | 4 |
Media and growth conditions.
All experiments were performed at 37°C, except where noted. The media used in this study have been previously described (19). M63 minimal medium was supplemented with 1 μg of thiamine per ml and 1 mM MgSO4. All amino acids used in this study were of the l configuration. Where appropriate, LB plates were supplemented with streptomycin (25 μg/ml), nalidixic acid (20 μg/ml), tetracycline (15 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (30 μg/ml). All chemicals were from Sigma. Optical density (OD) was monitored with a Spectronic 20D+ Spectrophotometer (Milton Roy).
Genetic techniques.
Phage P1vir transduction and Hfr conjugation using the Singer et al. (31) strain collections were performed as described elsewhere (19). Insertional mutagenesis with mini-Tn10 transposons from the vectors λNK1323 (Tcr), λNK1316 (Kanr), and λNK1324 (Cmr) was performed as described previously (13).
Construction of GASP strains.
Because incorporation of new genetic markers can alter the fitness of bacteria, we constructed our strains such that the final strains differed from the parental strains only by the allele(s) of the GASP loci. This was achieved by first bringing an auxotrophy mutation or the streptomycin-sensitive (Sms) allele of rpsL that mapped near the GASP loci (see Results) into the recipient by P1vir transduction. We could then cotransduce the GASP alleles with P1vir grown on ZK1141 or ZK126 into these strains by selecting for prototrophy or Smr, and then testing among those transductants for the cotransduction of the GASP allele, by assaying directly for the GASP phenotype or another physiological phenotype where appropriate (see below).
The rpoS+ strains were constructed by using the cysC95::Tn10Tcr mutation from CAG12173 and the ΔrpoS::Kanr mutation from ZK1000 (4). Strains carrying the sgaA allele of ZK1141 were constructed with the lipA150::Tn1000dKanr marker from strain KER176 (35). Mutants with the sgaA GASP allele were identified by their larger colony sizes when grown on M63 glutamate (0.5%) plates (see Results). Strains carrying the sgaB allele of ZK1141 were constructed with the serC::mini-MudI194 allele from strain NU1107 (14). Mutants with the sgaB GASP allele were identified by their increased sensitivity to serine, determined by a filter disc technique and confirmed by assaying for mucoidy at 30°C (see Results). Strains carrying the sgaC GASP allele were constructed with the rpsL+ (Sms) allele of ZK126. Mutants with the sgaC GASP allele were identified by scoring for the sgaC GASP phenotype (see below).
Stationary-phase competitions.
Competition experiments were adapted from those of Zambrano et al. (40). For competitions in LB, initial cultures were inoculated from frozen glycerol stocks into 3 ml of LB and grown overnight. These were then subcultured 1:100 into fresh LB and incubated for 24 h before being mixed for the competitions. The two populations were monitored by serial dilution in M63 medium and plating on minimal salicin and minimal glucose-plus-valine plates. We verified that the majority of the population remained prototrophic (and hence detectable on the selection media) by comparing the counts on the selection media with those on LB. In no case did we observe a difference in total viable counts on minimal and rich media. For competitions in M63-serine (0.5%)-isoleucine (0.03%)-valine (0.03%)-leucine (0.03%)-NaCl (0.5%), colonies were inoculated from an LB plate into the defined medium, and the cultures were incubated until they reached stationary phase (1 to 4 days). Cultures of strain ZK2618 (trpB::Tn10) were also supplemented with 0.004% tryptophan to facilitate growth. The ZK1141 and ZK2618 cultures were washed in M63 medium before inoculating as a 1:10,000 minority into the ZK820 cultures. The two populations were monitored by serial dilution in M63 medium and plating on LB-streptomycin and LB-nalidixic acid.
Molecular techniques.
The DNA flanking the mini-Tn10Cmr transposons was determined by using an arbitrary PCR-based protocol (5), with the modifications described by Pratt and Kolter (28). PCR products were subjected to sequence analysis by the Micro Core Facility, Department of Microbiology and Molecular Genetics, Harvard Medical School, and the sequences were compared with the GenBank DNA database by using the BLAST program (1).
RESULTS
Isolation and mapping of three new GASP loci from an aged rpoS819 strain.
Our approach to investigate the physiology of the GASP phenomenon was to identify and characterize several mutations able to confer the GASP phenotype. That many rounds of GASP takeover occur in starved cultures of E. coli (7, 40) implies that the survivors have acquired multiple GASP mutations. As we were interested in exploring potential genetic interactions of accumulated GASP mutations, our strategy was to isolate and investigate the GASP mutations of a single mutant survivor (ZK1141) from a culture that had undergone several rounds of population takeover.
We have devised a nomenclature that describes the relationship of the GASP mutants within a single lineage (Table 2). Gn (short for GASPn) denotes an isolate from an aged culture of strain Gn − 1 that is capable of outcompeting the Gn − 1 strain in a stationary-phase competition. In this study we analyzed the GII strain ZK1141 (rpoS819 sgaA sgaB sgaC), which is descended from the GI strain ZK819 (rpoS819), which is descended from the G0 wild-type strain ZK126 (all Gn designations in this work refer to these strains).
TABLE 2.
GASP nomenclature
| GASP mutant designation | Strain used in this study | Strain history | GASP mutations |
|---|---|---|---|
| G0 | ZK126 | Initial strain | None (wild type) |
| GI | ZK819 | G0 strain aged in LB | rpoS819 |
| GII | ZK1141 | GI strain aged in LB | rpoS819 sgaA sgaB sgaC |
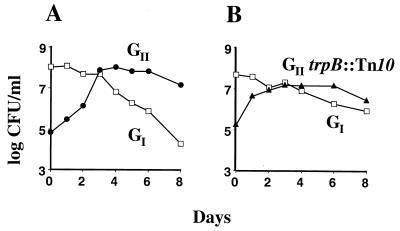
ZK1141 was isolated as a Smr survivor of a mixed culture; a 10-day-old culture of ZK819 and a 1-day-old culture of the Sms nalidixic acid-resistant (Nalr) version of ZK819 (ZK820) were grown together in fresh LB and then incubated for a week under starvation conditions (37). ZK1141 has the GII phenotype: it is capable of completely taking over a 1-day-old population of GI when inoculated as a 1-day-old minority (38).
The mutation responsible for the GI GASP phenotype of ZK819 has been identified as an allele of rpoS called rpoS819 (40). The GII phenotype of ZK1141 was initially thought to be due to a single mutation, which was termed sga, for stationary-phase growth advantage (38). However, genetic analysis of ZK1141 (see below) has demonstrated that there are three GASP mutations that each contribute to the GII GASP phenotype.
We identified the mutations responsible for the GII phenotype of ZK1141 with a genetic selection technique adapted from Zambrano et al. (40): when introduced into the GI strain, the GII GASP alleles conferred a GASP phenotype versus the GI parent. We made a pool of approximately 1,000 GII mutants with randomly inserted mini-Tn10Tcr or Kanr transposons in the chromosome. We then selected for linkage of the mini-Tn10 to the GASP alleles by making a P1vir lysate of the pool and infecting GI with this lysate to obtain a new pool of about 500 Tcr or Kanr transductants. These pools were then inoculated as a minority into 1-day-old ZK820 (the Nalr Sms GI strain) cultures, and the cultures were allowed to further starve to select from the pool those GI transductants carrying GII alleles: these transductants could grow in the starved culture, whereas the GI transductants not carrying the GII GASP alleles could not. The GI transductants carrying the GII GASP alleles were isolated from the culture several days after the pool was inoculated by titering the culture onto LB-streptomycin Sm plates. We then confirmed that the mutants isolated on the LB-streptomycin titer plate had the GII GASP alleles by moving their mini-Tn10 alleles into a fresh GI background by P1vir transduction and testing among those transductants for cotransduction of the GII GASP phenotype by competition versus the Nalr GI. In this manner, we identified three distinct mutations harbored by GII that conferred a GASP phenotype to GI. These mutations were designated sgaA, sgaB, and sgaC.
Mapping the GII GASP loci.
During our investigations of strain ZK1141, we discovered that it has two phenotypes that its ZK819 parent lacks: mucoid growth on glucose at 30°C (but not at 37°C) and an enhanced sensitivity to the amino acid serine. Serine is a competitive inhibitor of homoserine dehydrogenase I, and high levels of intracellular serine result in isoleucine starvation (9, 10). We scored relative serine sensitivities by streaking the strains on an M63-glucose plate toward a filter disc soaked with 10% serine placed in the center of the plate and determining the relative sizes of the growth inhibition zones. Instrumental in the mapping of the sgaB locus was the discovery that the sgaB GASP allele is responsible for both the mucoidy and serine sensitivity phenotypes of ZK1141. The sgaB mutation was mapped to min 20 by using the Hfr and P1 mapping sets (31); the sgaB mutation was 95% linked to the zbj-3110::Tn10Kanr marker of CAG18528, located at min 19.8 (2, 31).
The sgaA and sgaC mutations were each mapped by determining the location of random mini-Tn10Cmr insertions linked to the initial mini-Tn10Tcr or mini-Tn10Kanr insertions used to isolate the GASP alleles (see above). These mini-Tn10Cmr markers were mapped by the arbitrarily primed PCR technique. The sgaA-linked mini-Tn10Cmr was found to be in ybdN at min 13.7 and was 60% linked to the mini-Tn10Tcr, which in turn was 10% linked to sgaA. The sgaC-linked mini-Tn10Cmr was found to be in gspA at min 74.4, which was 66% linked to the mini-Tn10Kanr, which in turn was 50% linked to sgaA.
The GASP phenotypes of three new GASP loci: sgaA, sgaB, and sgaC.
Having isolated and mapped three GASP alleles of strain GII, we wanted to test whether each of these alleles alone could confer a selective advantage over the GI parent during stationary phase. We assayed for the GASP phenotype by mixing 24-h-old cultures of the two strains in question and monitoring changes in each population by viable count assay. The two populations were distinguished because they carry different neutral markers. Marker neutrality was confirmed empirically by switching the markers between the strains and performing all mixes reciprocally. Viable counts of each population are determined by titering the culture on the two relevant selection plates. Previous reports suggested that the rpsL allele conferring Smr and the gyrA allele conferring Nalr are neutral in stationary-phase competitions in E. coli (7, 40). We confirmed that the Smr and Nalr markers are neutral during extended starvation. However, during the first 4 days of competition of a 1:1 mix (see below), viable counts were consistently 2- to 10-fold higher for the Smr strain than for the Nalr strain, although the Smr counts eventually dropped to equal those of the Nalr population. Since the first 4 days of the competitions in this study were critical, we differentially marked the ZK819 (Smr) strain with two new markers that remain neutral throughout the competition: valine-resistant growth on glucose (Valr) and the ability to grow on β-glucosides (Bgl+). The Valr and Bgl+ markers were isolated as spontaneous mutations conferring the ability to grow on M63-glucose-valine or M63-salicin plates, respectively. Unless otherwise noted, this pair of markers was used for all competitions described below.
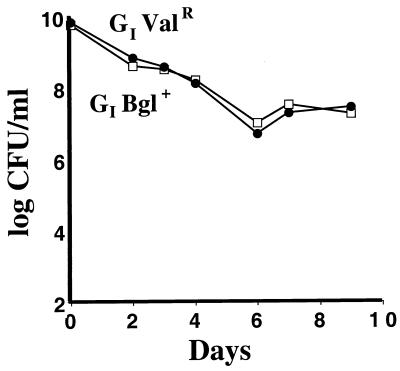
The Valr mutation mapped to the ilvGMEDA operon. Since E. coli K-12 has a frameshift in ilvG, which prevents expression of the one Valr isozyme of acetohydroxy acid synthase of E. coli, encoded by ilvGM (16), the Valr mutation is most likely a suppressor of this frameshift. The Bgl+ mutants arose at high frequency in our ZK819 and ZK1141 strains (about 1 in 107 plated cells), which prevented transduction of the same allele into either background. We therefore selected for Bgl+ mutants in both ZK819 and ZK1141 backgrounds. Both Bgl+ mutations mapped to the bgl operon and are likely to be insertions or point mutations in the bglR regulatory locus (30). The neutrality of the Valr and Bgl+ mutations throughout the starvation period was demonstrated by competitions of the Valr and Bgl+ derivatives of the ZK819 and ZK1141 strains. Neither marker conferred a competitive advantage or disadvantage, as determined by 1:1 competitions (Fig. 1). Furthermore, neither strain grew as a 1:1,000 minority versus the other strain (data not shown). Finally, neither of the two mutations altered the fitness of the strains versus the Vals Bgl− parent (data not shown). Each mix was performed at least four times.
FIG. 1.
The Valr and Bgl+ markers are selectively neutral in stationary phase. One-day-old cultures of the Valr (ZK2552) (●) and Bgl+ (ZK2553) (□) derivatives of the GI mutant (ZK819) were mixed 1:1 and cocultured. Viable counts were assayed on M63-glucose-valine or M63-salicin plates. Neither strain was outcompeted in four competitions.
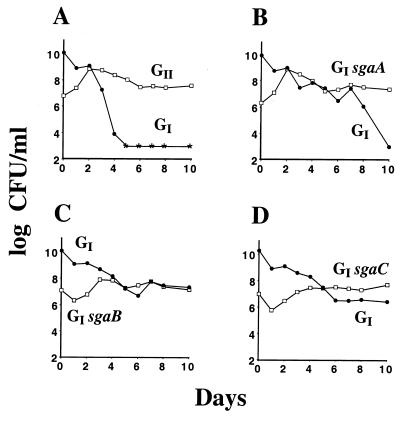
The fact that we can isolate mutants with the GASP phenotype from starved bacterial cultures suggests that these GASP mutants, starting from a single mutant cell, are able to increase in number during the starvation period to establish themselves as the majority population. One assay for the GASP phenotype is thus the ability of the mutant, when placed as a minority population in a culture of the parent, to grow relative to the parent and eventually establish itself as the majority population. To demonstrate this aspect of GASP for each of the GII GASP alleles, we constructed the GI sgaA (the GI strain that has the sgaA allele of GII), GI sgaB, and GI sgaC strains with the Valr or Bgl+ selectable marker and competed them as a 1,000-fold minority with the GI parent that had the other selectable marker. Figure 2 demonstrates that each of the three GASP alleles confers the ability to grow when inoculated as a minority and take over the population. Like the GII mutant (Fig. 2A), the GI sgaA mutant grew on the first day of the competition (Fig. 2B), while the GI sgaB and GI sgaC mutants experienced a 1-day lag period before growth (Fig. 2C and D). However, none of the GI mutants with a single GII GASP allele was able to eliminate the majority population as rapidly or as completely as the GII mutant, suggesting that their fitness advantages are additive, a possibility addressed below.
FIG. 2.
The GII alleles of ZK1141 confer the GASP phenotype. Into a 1-day-old culture of the GI mutant (ZK2552) (●) was inoculated as a 1,000-fold minority of a 1-day-old culture of the GII strain (ZK2555) (A), the GI sgaA strain (ZK2561) (B), the GI sgaB strain (ZK2559) (C), or the GI sgaC strain (ZK2563) (□) (D). Asterisks indicate that viable counts fell below detectable levels (<103 CFU/ml). The patterns of GASP takeovers were identical in six replicate mixtures, including ones where the selectable markers were switched between competing strains.
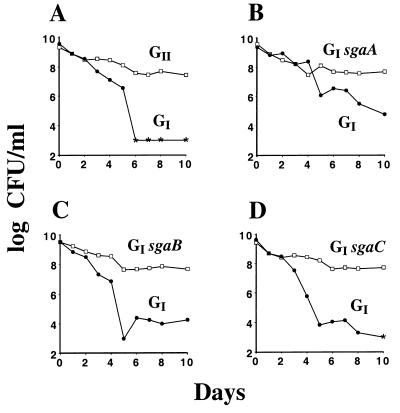
Another component of the GASP phenotype is the ability of the GASP mutant to directly outcompete the parent, resulting in the death of the parent. This ability is assayed by mixing the two cultures in a 1:1 ratio and looking for the decline of one of the two populations during the starvation period. Figure 3 demonstrates that all three of the GI mutants with a GII GASP allele are capable of outcompeting the GI parent. Interestingly, during the first 2 days of the 1:1,000 and 1:1 mixes for both GII and GI sgaA mutants (Fig. 2A and B and 3A and B), the GASP mutant could grow as a minority population but did not yet outcompete the GI parent when mixed in equal numbers. This finding implies that while there may be utilizable carbon for the strains available during the first 2 days, the competition for those nutrients does not become lethal for the parental strain until the environmental conditions change as a result of continued starvation.
FIG. 3.
The GII alleles confer a competitive advantage to GI cells. A 1-day-old culture of the GI mutant (ZK2552) (●) was mixed 1:1 with a 1-day-old culture of the GII strain (ZK2555) (A), the GI sgaA strain (ZK2561) (B), the GI sgaB strain (ZK2559) (C), or the GI sgaC strain (ZK2563) (□) (D). Asterisks indicate that viable counts fell below detectable levels (<103 CFU/ml). The patterns of GASP takeovers were identical in six replicate mixtures, including ones where the selectable markers were switched between the competing strains.
The three GII GASP mutations are necessary and sufficient for the GII GASP phenotype.
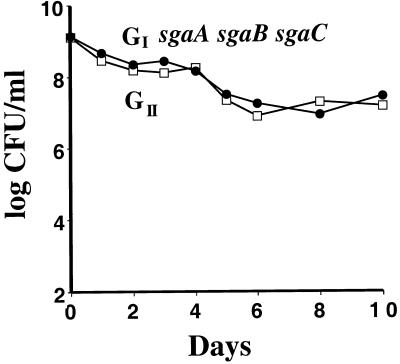
Our selection method identified three new distinct GASP loci on the chromosome of GII: sgaA, sgaB, and sgaC. To determine whether there are additional GASP mutations in GII, we examined whether the three GASP mutations identified so far were both necessary and sufficient for the GII GASP phenotype. Our first approach was to compete the GII mutant with a constructed GI sgaA sgaB sgaC mutant. Neither strain had a competitive advantage when competed in a 1:1 mix (Fig. 4). Furthermore, the GII mutant was unable to grow when inoculated as a 1:1,000 minority into a culture of the GI sgaA sgaB sgaC (data not shown). These results demonstrate that the GII strain lacks any additional GASP mutations that would confer a competitive advantage over the constructed strain. Additionally, the GI sgaA sgaB sgaC strain grew immediately and completely displaced the GI strain as the majority when inoculated as a 1:1,000 minority into the GI culture (data not shown); this GASP phenotype was indistinguishable from that of the GII strain (Fig. 2A). The results from these three different competition experiments thus demonstrate that the sgaA, sgaB, and sgaC mutations are sufficient for the GII GASP phenotype.
FIG. 4.
The sgaA, sgaB, and sgaC alleles are sufficient for the GII GASP phenotype of ZK1141. A 1-day-old culture of the reconstructed GII strain GI sgaA sgaB sgaC (ZK2564) (●) was mixed 1:1 with a 1-day-old culture of the GII strain (ZK2554) (□). Neither strain was outcompeted in eight competitions.
We next asked if all three GII GASP alleles were necessary for the GII GASP phenotype. We competed 1:1 the GII strain versus one of three constructed strains harboring two of the three GII GASP mutations: GI sgaA, sgaB, GI sgaA sgaC, or GI sgaB sgaC. In every case, the GII strain outcompeted the constructed strain (data not shown). Hence, all three reconstructed strains, each lacking one of the three GII alleles, were less fit than GII, indicating that each of the three GII alleles is necessary for the full fitness gain of the GII strain. That each is necessary for the GII GASP phenotype indicates that they act additively to confer higher and higher fitnesses in stationary phase.
GASP mutants obtain nutrients from dying cells in a chemically defined medium.
Because growth in LB ceases due to carbon limitation (37), it has been assumed that the GASP mutants obtain the nutrients required for growth from the dying majority population. To demonstrate directly that the dying cells release nutrients which the GASP mutants can utilize during growth, we identified a chemically defined medium in which the GII GASP strain could grow and outcompete the GI strain. Figure 5A shows the GASP phenotype of the GII strain versus GI after both cultures were grown to stationary phase in M63 salts medium supplemented with serine (0.5%), isoleucine, valine, and leucine (0.03% each), and NaCl (0.5%). As in LB, the GII strain grew rapidly as a 1:10,000 minority to take over the population. Like the prototroph, a tryptophan auxotrophic derivative of the GII strain (trpB::Tn10) was able to grow and take over the GI minority (Fig. 5B). This finding indicates that the GII strain can scavenge enough tryptophan to meet its growth requirement during the population takeover. As no exogenous tryptophan was supplied by the medium, the only remaining source of tryptophan is the cells of the dying majority population. This is the first evidence that the GASP mutants can scavenge nutrients released by the dying cells during carbon starvation.
FIG. 5.
GASP in a chemically defined medium. Stationary-phase cultures grown in M63-serine (0.5%)-isoleucine (0.03%)-valine (0.03%)-leucine (0.03%)-NaCl (0.5%) (plus tryptophan [0.004%] for the GII trpB::Tn10 strain [ZK2618]) were mixed 1:10,000 to assay the GASP phenotype. Both the GII strain (ZK1141; ●) (A) and the tryptophan auxotrophic GII trpB::Tn10 strain (ZK2618; ▴) (B) express the GASP phenotype as a minority versus the GI strain (ZK820; □). The patterns of GASP takeovers were identical in four competitions.
The three GII GASP alleles and the rpoS819 allele confer faster growth on amino acids.
We reasoned that the primary selective force acting on carbon-starved cells is the ability to utilize the carbon released by the dying majority population. Therefore, we asked whether the GASP mutations confer faster growth on carbon sources that may resemble the nutrients released by dying cells. The three GII GASP mutations were tested in the GI background, as it was from this background that GII was selected. We also tested the growth phenotypes of the GI GASP mutation, rpoS819, by comparing its growth rate with that of the GI strain carrying the rpoS+ allele (this is essentially a G0 strain, as the rpoS819 mutation is sufficient to confer the GI GASP phenotype of ZK819 [38]). We first assayed growth on LB, a rich medium containing many of the building blocks, vitamins, and energy sources necessary for growth. The growth rates on LB were indistinguishable between the different mutant strains (data not shown).
The composition of the medium during prolonged carbon starvation has not been characterized. However, we speculated that amino acids are the most abundant nutrients released by the dying cells, given that amino acids account for most of the dry weight of E. coli (26). Hence, we determined the relative growth rates of the GASP mutants on mixtures of amino acids, either as a combination of monomers and short peptides (tryptone) or only as monomers (Casamino Acids). While none of the individual GASP alleles had much of an effect, the GII strain harboring all three GII GASP mutations grew significantly faster than GI on the monomer and peptide mixture (Table 3). In contrast, all four GASP mutations conferred significantly faster growth on the mixture of the amino acid monomers. Interestingly, in every case, the strains with more GASP mutations grew faster, indicating that the GASP mutations act additively to confer faster growth on mixtures of amino acids.
TABLE 3.
Relative growth rates on amino acid mixtures
| Carbon and energy source (1.0%) | Relative growth rate (±SD, n = 3)a
|
|||||
|---|---|---|---|---|---|---|
| G0 (ZK2557) | GI (ZK2553) | GIsgaA (ZK2561) | GIsgaB (ZK2559) | GIsgaC (ZK2563) | GII (ZK2555) | |
| Tryptone | 1.00 (±0.01) | 0.98 (±0.01) | 1.00 (±0.03) | 1.03 (±0.01) | 1.03 (±0.02) | 1.14 (±0.03) |
| Casamino Acids | 1.00 (±0.02) | 1.16 (±0.01) | 1.34 (±0.01) | 1.31 (±0.05) | 1.21 (±0.02) | 1.43 (±0.02) |
A relative growth rate of 1.00 indicates a generation time (in hours) of 0.59 on tryptone or 1.28 on Casamino Acids.
Since all four GASP mutations confer faster growth on mixtures of amino acids, we attempted to identify individual amino acids that the GASP mutants could catabolize more rapidly. We assayed growth in liquid media containing M63 salts and the amino acid in question at 0.5% (wt/vol). Because of the potential problem of amino acids such as serine and cysteine inhibiting isoleucine biosynthesis and thus preventing growth on single amino acids (9, 10), we supplemented all of the growth media with isoleucine (0.03%). We assayed for the ability of the GASP mutants to grow on each of the 20 amino acids singly as the sole source of carbon and energy. At least one of the six strains tested could grow on alanine, asparagine, aspartate, glutamate, glutamine, proline, serine, or threonine; none grew on the other 11 amino acids. While E. coli K-12 can grow on tryptophan (32), our strains were not expected to grow, as they lack tryptophanase activity due to the tna2 mutation.
All four GASP alleles conferred growth advantages on several amino acids, manifested as a higher growth rate or, in some cases, a new ability to grow on the particular amino acid (Table 4). The rpoS819 GASP allele conferred upon the cell the new ability to utilize asparagine and glutamine as sole sources of carbon and energy. However, the rpoS819 mutants grew to an OD at 600 nm of only about 0.3 on glutamine. It is therefore uncertain whether the cells grew incompletely on glutamine or grew on an impurity in the glutamine supply instead. The rpoS819 allele alone conferred faster growth on glutamate, serine, threonine, and alanine. The sgaA allele conferred the new ability to grow on aspartate and conferred faster growth on asparagine and glutamate. The sgaB and sgaC alleles both conferred faster growth on alanine, threonine, and serine. Comparison of the growth rates for the G0, GI, and GII strains indicates that both the repertoire of amino acids and the growth rates on the amino acids increase as more GASP alleles are added and demonstrates that these GASP alleles act additively to increase the overall capacity to catabolize amino acids.
TABLE 4.
Relative growth rates on single amino acids as carbon sources
| Amino acid | Relative growth rate (±SD, n = 2)a
|
|||||
|---|---|---|---|---|---|---|
| G0 (ZK2557) | GI (ZK2553) | GIsgaA (ZK2561) | GIsgaB (ZK2559) | GIsgaC (ZK2563) | GII (ZK2555) | |
| Alanine | 1.00 (±0.01) | 1.04 (±0.02) | 0.88 (±0.00) | 1.25 (±0.01) | 1.11 (±0.02) | 1.21 (±0.01) |
| Asparagine | 0 | 1.00 (±0.03) | 1.90 (±0.04) | 0.69 (±0.03) | 0.72 (±0.04) | 1.60 (±0.03) |
| Aspartate | 0 | 0 | 1.00 (±0.01) | 0 | 0 | 0.53 (±0.00) |
| Glutamate | 1.00 (±0.03) | 2.93 (±0.01) | 9.14 (±0.00) | 1.92 (±0.07) | 2.17 (±0.03) | 9.29 (±0.00) |
| Glutamineb | 0 | 1.00 (±0.02) | 0.87 (±0.04) | 0.75 (±0.09) | 0.76 (±0.03) | 0.82 (±0.00) |
| Serine | 1.00 (±0.05) | 1.63 (±0.36) | 0.86 (±0.06) | 9.81 (±0.66) | 2.30 (±0.14) | 9.19 (±0.28) |
| Threonine | 1.00 (±0.02) | 2.17 (±0.11) | 1.16 (±0.02) | 3.66 (±0.00) | 3.92 (±0.11) | 5.01 (±0.04) |
| Prolinec | 1.00 (±0.1) | 0.72 (±0.11) | 2.1 (±0.1) | 0.45 (±0.05) | 1.6 (±0.1) | 10.5 (±0.7) |
For all amino acids except proline (see footnote c), growth rates in liquid M63 plus 0.5% amino acid (plus 0.03% isoleucine) medium were determined. A relative growth rate of 1.00 indicates a generation time (in hours) of 4.50 on alanine, 14.0 on asparagine, 11.2 on aspartate, 58.5 on glutamate, 12.0 on glutamine, 61.2 on serine, or 304 on threonine; a relative growth rate of zero indicates no growth.
Cultures stopped growing at an OD of 600 nm of 0.2 to 0.3. This was not due to alkalinization of the medium, as the pH remained below 7.5, nor was it due to end product inhibition, as the cultures grew readily when spiked with 0.2% glucose.
Overnight LB cultures were diluted into M63 medium and plated on M63 proline (0.5%) plus isoleucine (0.03%). Values indicate relative colony areas (±SD, n = 20) after 4 days of incubation.
We were not able to obtain relative growth rates on proline in liquid cultures, because the cultures were consistently taken over by faster-growing mutants. However, we were able to obtain estimates of relative growth rates by comparing the sizes (surface area) of colonies grown on M63-proline (plus isoleucine) plates (Table 4). The sgaA and sgaC alleles conferred faster growth on proline, and their effects on growth were strikingly additive, as the GII colonies were significantly larger than those of the GI mutants with either single allele alone.
Interestingly, the four GASP alleles conferred slower growth on several of the amino acids (Table 4). The rpoS819 mutation conferred slower growth on proline; the sgaA mutation conferred slower growth on alanine, glutamine, and serine; the sgaB mutation conferred slower growth on asparagine, glutamate, glutamine, and proline; and the sgaC mutation conferred slower growth on asparagine, glutamate, and glutamine. This indicates that while the individual GASP alleles confer fitness gains on some amino acids, each also confers a fitness loss on other amino acids. We discuss the implications of these observations below.
DISCUSSION
Previous work identified an allele of rpoS, rpoS819, as a mutation that can confer the GASP phenotype on E. coli (40). Genetic analysis of an isolate (GII) from a starved culture of the rpoS819 GASP strain (GI) has revealed three new GASP mutations: sgaA, sgaB, and sgaC. All four GASP mutations of this isolate map to different regions of the chromosome, suggesting that there are many loci that when mutated can confer fitness advantages in stationary phase.
As each of the three new GASP mutations acquired by the GII strain can confer a GASP phenotype on the GI parental strain, it is most likely that they were acquired as a result of three successive GASP takeover events. GII was isolated from a culture starved for a total of about 2.5 weeks, which suggests that in our experimental system population takeovers during starvation can happen faster than once per week. The fact that the three GII GASP mutations are necessary (and sufficient) for the GII GASP phenotype demonstrates directly that the acquisition of multiple GASP mutations can provide successively higher fitnesses for starved bacteria. The continual accumulation of GASP mutations by cells within the surviving population can account for the multiple rounds of population takeovers observed in starved cultures (7, 40). Our results thus support the growing body of evidence that starved populations are highly dynamic and undergo frequent population takeovers as a result of fitness differences among the competing subpopulations (7, 8, 40).
A major purpose of our investigation was to understand how GASP mutations alter cell physiology to provide fitness gains in stationary phase. Previous studies have demonstrated that fitness gains during conditions of limited substrate availability in chemostats or selections for growth on novel substrates were manifested as increases in catabolic potential for those substrates (11, 17, 20, 24, 33, 36). In our system, we have observed that dying cells in stationary-phase cultures release nutrients (e.g., tryptophan) that can be utilized by the growing GASP mutants, and we hypothesized that the GASP mutants selected are those that outcompete their parents for these limited substrates. We observed a direct correlation between relative GASP fitness and relative ability to catabolize amino acids, which are likely to be the most abundant nutrients released by dying cells (26). The GI GASP mutation rpoS819 and the three GII GASP mutations sgaA, sgaB, and sgaC all confer higher growth rates on an amino acid mixture (Casamino Acids). To our knowledge, this is the first report that mutations in rpoS can alter amino acid catabolism during exponential growth, extending previous observations that wild-type ςS affects the physiology of both arrested and growing cells (12, 27).
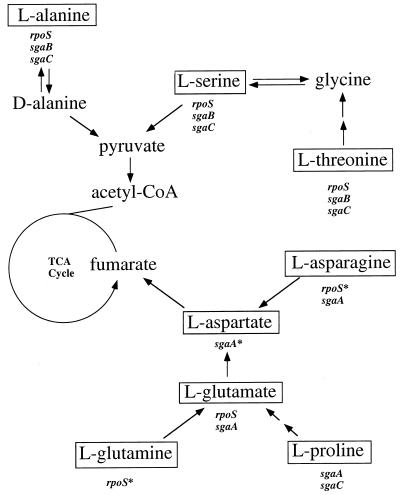
All four of the GASP mutations examined in this study also increase the ability of the cell to utilize certain amino acids singly as the sole source of carbon and energy. These changes were manifested as either a higher growth rate or a new ability to grow on the particular amino acid. The GASP mutations are pleiotropic in this respect, as each affects growth on several amino acids. The subsets of amino acids on which they confer a growth advantage are overlapping but distinct. In general, these subsets contain amino acids that enter catabolism along the same major degradative pathways (reviewed in reference 18) (Fig. 6). The sgaA allele confers a growth advantage on amino acids that enter the central metabolic pathway through aspartate and fumarate, while the sgaB and sgaC alleles confer growth advantages on amino acids that enter this pathway through pyruvate (the effect of sgaC on proline metabolism is the one exception). The rpoS819 mutation, on the other hand, affected both of these degradative pathways. It is tempting to speculate that the GASP mutations, especially rpoS819, alter the regulation of the enzymes of these degradative pathways. We are currently investigating the mechanistic bases of these physiological changes.
FIG. 6.
The primary catabolic pathways for the amino acids on which the GASP mutants have a growth advantage (reviewed by McFall and Newman [18]). The arrows indicate enzymatic steps between metabolites. Boxed amino acids denote amino acids that can serve as sole sources of carbon and energy for at least one of the six strains tested. Listed under the boxed amino acids are the loci whose GASP alleles confer a growth advantage on the amino acid. An asterisk indicates the GASP allele that confers the novel ability to grow on the amino acid.
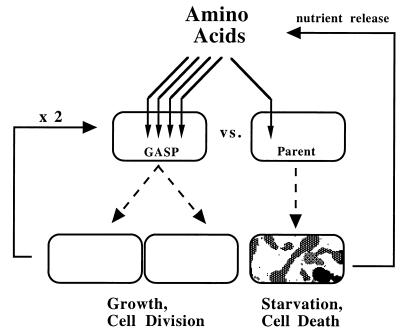
Based on our findings, we propose a model for a physiological basis of GASP (Fig. 7). We hypothesize that the primary selective force acting on carbon-starved cells is the ability to scavenge carbon sources released by the dying cells for the purposes of cell maintenance and growth. Cells unable to obtain a sufficient supply of carbon and energy can no longer maintain activities essential for viability, and they die. The fact that in every case studied the GASP fitness correlates directly with the capacity to catabolize amino acids leads us to propose that the most significant nutrients released are catabolizable amino acids. At the onset of starvation, the population is composed almost entirely of cells of the parental genotype that compete with equal fitness for carbon sources. Hence, the death during the first few days of starvation is stochastic, as all parental cells have an equal chance of scavenging the nutrients and surviving. However, the rare mutants within the population expressing the GASP phenotype outcompete their parents for the carbon resources because of their enhanced catabolic capabilities. These advantages provide the GASP cells with enough resources not only to survive but to grow and divide during starvation conditions. Once the GASP mutants grow to a significant cell density, they effectively decrease the amount of carbon available to the parental cells. These parental cells die as a result and release their nutrients into the medium. This model involves two positive feedback loops which can account for both the rapid growth of the GASP mutant and the rapid death of the parent. Supporting our model is the finding that if the GII strain ZK1141 lacks the respiratory enzyme NADH dehydrogenase I, essential for the utilization of several amino acids catabolized by GII (29), it loses the GASP phenotype versus the GI parental strain, ZK819 (38).
FIG. 7.
A model for the physiological basis of the GASP phenotype.
Interestingly, we observed that while GI mutants with a single GII allele grow faster than GI with certain amino acids as sole carbon sources, they grow slower with others. This result may seem inconsistent with our model for the physiological basis of GASP. However, growth on mixed amino acids may more accurately reflect the growth of the GASP mutants during starvation, since all amino acids are likely to be released at similar rates from the dying cells. We have demonstrated that all four GASP alleles confer an overall advantage when growing on mixed amino acids (Casamino Acids). Hence, our results suggest that when the GASP mutants are competing with their parents for the complex mixtures of nutrients released by the dying cells, their faster growth on some amino acids outweighs their slower growth on others.
ACKNOWLEDGMENTS
We thank J. E. Cronan, Jr., M. E. Winkler, and A. Wright for providing strains. We thank S. E. Finkel, G. A. O’Toole, and L. A. Pratt for critical reading of the manuscript and members of the Kolter lab for helpful comments.
This work was supported by grants from the National Science Foundation (MCB9728936) and the National Institutes of Health (GM55199).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Berlyn M K, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 3.Blackburn N, Fenchel T, Mitchell J. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science. 1998;282:2254–2256. doi: 10.1126/science.282.5397.2254. [DOI] [PubMed] [Google Scholar]
- 4.Bohannon D E, Connell N, Keener J, Tormo A, Espinosa-Urgel M, Zambrano M M, Kolter R. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and the role of ς70. J Bacteriol. 1991;173:4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caetano-Annoles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–92. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 6.Connell N, Han Z, Moreno F, Kolter R. An E. coli promoter induced by the cessation of growth. Mol Microbiol. 1987;1:195–201. doi: 10.1111/j.1365-2958.1987.tb00512.x. [DOI] [PubMed] [Google Scholar]
- 7.Finkel S E, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkel S E, Zinser E, Gupta S, Kolter R. Life and death in stationary phase. Mol Microbiol. 1997;103:3–16. [Google Scholar]
- 9.Hama H, Kayahara T, Tsuda M, Tsuchiya T. Inhibition of homoserine dehydrogenase I by l-serine in Escherichia coli. J Biochem. 1991;109:604–608. doi: 10.1093/oxfordjournals.jbchem.a123427. [DOI] [PubMed] [Google Scholar]
- 10.Hama H, Sumita Y, Kakutani Y, Tsuda M, Tsuchiya T. Target of serine inhibition in Escherichia coli. Biochem Biophys Res Commun. 1990;168:1211–1216. doi: 10.1016/0006-291x(90)91157-n. [DOI] [PubMed] [Google Scholar]
- 11.Helling R B, Vargas C N, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 13.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 14.Lam H M, Winkler M E. Metabolic relationships between pyridoxine (vitamin B6) and serine biosynthesis in Escherichia coli. J Bacteriol. 1990;172:6518–6528. doi: 10.1128/jb.172.11.6518-6528.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of Escherichia coli cells is controlled by the novel sigma factor, ςS (rpoS) J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawther R P, Calhoun D H, Adams C W, Hauser C A, Gray J, Hatfield G W. Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci USA. 1981;78:922–925. doi: 10.1073/pnas.78.2.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenski R E, Mongold J A, Sniegowski P D, Travisano M, Vasi F, Gerrish P J, Schmidt T M. Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another? Antonie Leeuwenhoek. 1998;73:35–47. doi: 10.1023/a:1000675521611. [DOI] [PubMed] [Google Scholar]
- 18.McFall E, Newman E B. Amino acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 358–379. [Google Scholar]
- 19.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 20.Miller R D, Dykhuizen D E, Green L, Hartl D L. Specific deletion occurring in the directed evolution of 6-phosphogluconate dehydrogenase in Escherichia coli. Genetics. 1984;108:765–772. doi: 10.1093/genetics/108.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriarty D J W, Bell R T. Bacterial growth and starvation in aquatic environments. In: Kjelleberg S J, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 25–53. [Google Scholar]
- 22.Morita R Y. Bioavailability of energy and its relationship to growth and starvation survival in nature. Can J Microbiol. 1988;34:436–441. [Google Scholar]
- 23.Morita R Y. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 24.Mortlock R P. Metabolic acquisitions through laboratory selection. Annu Rev Microbiol. 1982;36:259–284. doi: 10.1146/annurev.mi.36.100182.001355. [DOI] [PubMed] [Google Scholar]
- 25.Mulvey M R, Sorby P A, Triggs-Raine B L, Loewen P C. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene. 1988;73:337–345. doi: 10.1016/0378-1119(88)90498-2. [DOI] [PubMed] [Google Scholar]
- 26.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 13–28. [Google Scholar]
- 27.Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 29.Prüß B M, Nelms J M, Park C, Wolfe A J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds A E, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon of E. coli. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- 31.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snell E E. Tryptophanase: structure, catalytic activities and mechanisms of action. Adv Enzymol. 1975;42:287–333. doi: 10.1002/9780470122877.ch6. [DOI] [PubMed] [Google Scholar]
- 33.Sonti R V, Roth J R. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor B L. How do bacteria find the optimal concentration of oxygen? Trends Biochem Sci. 1983;8:438–441. [Google Scholar]
- 35.Vanden Boom T J, Reed K E, Cronan J E., Jr Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J Bacteriol. 1991;173:6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weikert C, Sauer U, Bailey J E. Use of a glycerol-limited, long-term chemostat for isolation of Escherichia coli mutants with improved physiological properties. Microbiology. 1997;143:1567–1574. doi: 10.1099/00221287-143-5-1567. [DOI] [PubMed] [Google Scholar]
- 37.Zambrano M M. Ph.D. thesis. Cambridge, Mass: Harvard University; 1993. [Google Scholar]
- 38.Zambrano M M, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase-I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zambrano M M, Kolter R. GASPing for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 40.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]