Abstract
Bipolaris species are known to be important plant pathogens that commonly cause leaf spot, root rot, and seedling blight in a wide range of hosts worldwide. In 2017, complex symptomatic cases of maydis leaf blight (caused by Bipolaris maydis) and maize leaf spot (caused by Curvularia lunata) have become increasingly significant in the main maize-growing regions of India. A total of 186 samples of maydis leaf blight and 129 maize leaf spot samples were collected, in 2017, from 20 sampling sites in the main maize-growing regions of India to explore the diversity and identity of this pathogenic causal agent. A total of 77 Bipolaris maydis isolates and 74 Curvularia lunata isolates were screened based on morphological and molecular characterization and phylogenetic analysis based on ribosomal markers—nuclear ribosomal DNA (rDNA) internal transcribed spacer (ITS) region, 28S nuclear ribosomal large subunit rRNA gene (LSU), D1/D2 domain of large-subunit (LSU) ribosomal DNA (rDNA), and protein-coding gene-glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Due to a dearth of molecular data from ex-type cultures, the use of few gene regions for species resolution, and overlapping morphological features, species recognition in Bipolaris has proven difficult. The present study used the multi-gene phylogenetic approach for proper identification and diversity of geographically distributed B. maydis and C. lunata isolates in Indian settings and provides useful insight into and explanation of its quantitative findings.
Keywords: Bipolaris maydis, Curvularia lunata, ITS, GAPDH, LSU, maize, multi-gene phylogeny
1. Introduction
Maize (Zea mays L.) is one of the most diverse crops with great adaptability under a range of broad agro-climatic zones. It is the third-most commonly cultivated cereal after wheat and rice, is known as the ‘Queen of the Cereals’ globally because of its significant potential for genetic yield among cereals, and is grown in approximately 150 m ha across about 160 countries with wide soil properties, climate, biodiversity, and management practices [1]. Maize is cultivated on 9.2 million hectares in India, with an average yield of 2965 kg/ha and a total production of 27.8 million tons [2,3]. The major producing states of maize in India are, by zone, East: Bihar, Orissa; North: Uttar Pradesh, Punjab, Haryana, Himachal Pradesh, Jammu, Kashmir; West: Rajasthan, Gujarat, Maharashtra; South: Andhra Pradesh; Central: Madhya Pradesh [4,5].
On a global basis, maize is affected by 112 diseases, but in Indian agroclimatic conditions, there are 35 main diseases that attack maize [6]. Worldwide yield losses in maize up to 9% have been estimated due to fungal, bacterial, and nematode diseases [7]. The severe maize diseases are spread across a broad disease spectrum and include downy mildew, rust, leaf blight, and stalk rot [8]. These diseases also include maydis leaf blight (MLB), one of the most devastating diseases that has emerged as an economically significant problem [9].
Worldwide, Curvularia and Bipolaris species attack cereal and grasses, causing leaf spot diseases on rice, maize, sorghum, wheat, and barley, resulting in severe grain yield losses [10,11,12,13]. An important genus of plant pathogens is Bipolaris (an anamorph of the ascomycetes genus Cochliobolus), with more than 100 species. Bipolaris species mostly attacks members of the poaecae family and causes root rot, leaf spot or blight, seedling blight, and ear rot and blight [14]. Due to being seed-borne in nature, Bipolaris spp. causes significant damage to field crops, mostly as post-harvest damage that ultimately impairs market value [15,16]. Bipolaris spp. has caused severe epidemics, such as the Great Bengal famine, Northern leaf spot blight, and Southern corn leaf blight, which ultimately caused significant economic losses in recent decades [17].
Worldwide, in maize producing regions, the maydis leaf blight, also known as Southern corn leaf blight (SCLB), is a severe maize crop disease caused by Bipolaris maydis, an anamorph of Cochliobolus heterostrophus. It is considered as a graminicolous species of Helminthosporium [11,17,18]. In USA and UK, Bipolaris maydis caused severe epidemics in 1970. The decline in the development of maize is possibly due to maydis leaf blight in India.
Maydis leaf blight (MLB or SCLB) generally occurs in moderately humid and warm conditions with temperatures of 20–32 °C and causes potential damage and yield losses in major maize-growing regions of India. During the Kharif season, the disease spreads rapidly throughout India and is most prevalent in plains, highlands, and peninsular regions [19]. Sharma and Rai (2000) reported that maydis leaf blight caused by B. maydis qualifies as a major disease of maize is capable of inflicting significant losses in productivity, to the extent of 41 percent [20].
Bipolaris and Curvularia species mainly have been characterized based on cultural and morphological characteristics in the past few decades. Dothideomycetes fungi, the most diverse class of fungi, include Curvularia and Bipolaris species [21]. All members of Drechslera and Exserohilum once grouped to the graminicolous Helminthosporium species [12]. Species of Bipolaris were previously known as Helminthosporium. The genus Helminthosporium is divided into four genera based on taxonomic refinements: Drechslera, Bipolaris, Exserohilum, and Curvularia. They are morphologically similar, and particularly, these four genera are called helminthosporioid fungi [12,18]. Some of these related genera have caused maize leaf spot and mixed infections, caused by the genera Curvularia and Exserohilum, C. intermedia, C. lunata, C. eragrostidis, C. pallescens, C. clavata, C. spicifera, C. papendorfii and C. inaequalis [22,23], while leaves, sheaths, and maize bracts were reportedly infected by E. rostratum and Exserohilum turcicum, which have also been reported to cause extreme leaf spot in maize [24,25].
Under field conditions, the symptoms of maydis leaf blight caused by those species were prevalent and complex. It is difficult to identify these species reliably based on symptoms alone. Despite their significance, species identification remains problematic in Bipolaris and Curvularia since numerous species have morphological traits that overlap, making it difficult to differentiate between these two genera under different conditions [12,26]. It might be difficult to distinguish between the genera and species of Bipolaris and Curvularia. Due to substantial intraspecies and interspecies variability and the difficulty of generating sexual phases under laboratory circumstances, asexual morphological traits are insufficient to correctly identify species [26,27]. However, combined study can differentiate these two genera based on sexual, conidia morphology, and molecular studies.
Curvularia has conidia with overly enlarged intermediate cells, which contribute to the curvature, whereas Bipolaris has a continuous curvature throughout the conidium duration. In addition, conidia are usually longer in Bipolaris than those in Curvularia [28]. Based on the morphology of ascospores, Curvularia forms loosely packed ascospores in the ascus, while Bipolaris produces supercoiled ascospores [29].
Representatives of the Consortium for the Barcode of Life’s Fungal Working Group (FWG) evaluated different markers, three nuclear ribosomal DNA regions (ITS, LSU, SSU), and one protein-coding gene (RPB1), and proposed the ITS region as the key fungal barcode marker [30]. Species identification [31], species level phylogenies [32], and DNA barcoding studies [33] have all been conducted in the region. ITS data in the International Nucleotide Sequence Database (INSD: GenBank, EMBL, and DDBJ) demonstrated that this region is not equally variable in all fungi taxa, although most studies have designated it as the standard barcode marker for fungi.
These taxa have minimal or no barcode gaps in the ITS sequence, according to Stielow et al., 2015 [34]. The ITS region alone may not be sufficient for some species identification. Mycologists, particularly those working with ascomycetous fungi, frequently explore the feasibility of a two-marker barcoding system for fungi [7]). LSU [35,36] and protein-coding genes can be employed with ITS [36]. Using slowly evolving protein-coding genes, in particular, can significantly improve the phylogenetic resolution of deep divergences [37].
Bipolaris species are now typically identified based on morphological traits; however, other species exhibit numerous similarities, and conidial characteristics are frequently varied depending on isolates and culture circumstances. Due to ambiguities in morphological characteristics, DNA sequences of multiple loci are preferred to molecular identification and define Bipolaris and Curvularia species [28,38]. Currently, phylogenetic analysis studies are based on protein-coding gene-glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Ribosomal markers of rDNA internal transcribed spacer (ITS) and a large subunit of nuclear ribosomal DNA (LSU) have been used to evaluate the phylogenetic relationships among the Bipolaris genus isolates and with their sister genera [11,28,38]. The ITS, LSU regions, and GAPDH gene phylogeny provided a better resolution for taxa at the terminal clades, which has also been reported for other Dothideomycetes [39,40,41]. Based on these loci, phylogenetic studies have allowed Cochliobolus (sexual morph) species to be reallocated to either Bipolaris or Curvularia [29].
RAxML (Randomized Axelerated Maximum Likelihood) is a tool for inferring complex phylogenetic trees using maximum likelihood in a sequential and parallel way. It can also be used to perform post-analysis on phylogenetic tree sets, alignment analyses, and short-read evolutionary placement. RAxML is among the most widely used and effective software methods for creating a maximum likelihood (ML) tree. This would be the most accurate method for evaluation, but it is also the most resource-intensive. The model developed by RAxML is believed to be a good evolution model. Under this model, there was no major change in the likelihood of the inferred tree fitting the genuine tree [42].
It is difficult to distinguish between species solely based on their morphological characteristics because many of their morphological features overlap. Bhunjan et al., 2020 estimated 132 species epithets in Index Fungorum under Bipolaris, of which 15 had been moved to Curvularia, and only 43 are approved as Bipolaris species [43].
Based on the above studies, the combined study of morphological and molecular data gave a precise and correct method for identifying Bipolaris spp. Thus, the objectives of this study were as follows: (i) identification of Bipolaris maydis as the cause of maydis leaf blight, and Curvularia lunata as the cause of leaf spot of maize based on morphological analysis, (ii) precise identification and phylogenetic analysis based on ITS, GAPDH, LSU, and D1 and D2 regions of LSU to know the diversity of Bipolaris maydis and Curvularia lunata isolates in different geographical locations of major maize-growing regions in India.
2. Materials and Methods
2.1. Collection of Maydis-Leaf-Blight-Symptomatic Samples from Major Maize-Growing Regions of India
Sampling
During 2017, leaf samples showing characteristic symptoms of maydis leaf blight and leaf spot were collected from Bihar, Uttar Pradesh, Punjab, and Uttarakhand, covering five agro-climatic zones as follows: Western Himalayan Region, Uttarakhand; Lower Gangetic Plains region, Eastern Bihar; Middle Gangetic Plains region, parts of Uttar Pradesh and Bihar; Upper Gangetic Plains region, central and western parts of Uttar Pradesh and the Hardwar and Udham Nagar districts of Uttarakhand; Trans Gangetic Plains region, Punjab, under maize-cropping regions in India (Table 1). Around 186 symptomatic samples of maydis leaf blight were collected based on the characteristic disease symptoms of diamond-shaped lesions that vary in size, with 3–22 mm length and 2–6 mm width, that become rectangular- or spindle-shaped lesions with chlorotic halos at later stages. In the case of leaf spot of maize, 129 maize-leaf-spot-symptomatictic samples in the early stages of maize leaves that had yellow necrotic spots, which eventually expand to circular, spindle-shaped, oval, or strip lesions, were collected. Twenty sampling districts were covered during sampling, and their district locations are depicted in Supplementary Figure S1.
Table 1.
Sampling sites of maydis leaf blight and maize leaf spot samples from major maize-growing regions of India.
| S. No. | Sampling Sites | District Coordinates (Latitude and Longitude) |
Maydis-Leaf-Blight-Symptomatic Samples (186) | Maize Leaf Spot Samples (129) |
|---|---|---|---|---|
| 1. | Jaunpur, Uttar Pradesh | 25.62 N; 82.98 E | 09 | 07 |
| 2. | Sant Kabir Nagar, Uttar Pradesh | 25.59 N; 82.98 E | 06 | 04 |
| 3. | Faizabad, Uttar Pradesh | 26.73 N; 82.15 E | 04 | 03 |
| 4. | Maharajganj, Uttar Pradesh | 27.22 N; 83.82 E | 09 | 05 |
| 5. | MauNathBhanjan, Uttar Pradesh | 25.87 N; 83.47 E | 15 | 04 |
| 6. | Ballia, Uttar Pradesh | 25.77 N; 84.16 E | 13 | 07 |
| 7. | Azamgarh, Uttar Pradesh | 25.73 N; 82.99 E | 09 | 05 |
| 8. | Udham Singh Nagar, Uttarakhand | 28.99 N; 79.51 E | 13 | 05 |
| 9. | Almora, Uttarakhand | 29.62 N; 79.67 E | 07 | 08 |
| 10. | Haldwani, Uttarakhand | 29.16 N; 79.51 E | 06 | 03 |
| 11. | Samastipur, Bihar | 25.87 N; 85.81 E | 08 | 11 |
| 12. | Khagaria, Bihar | 25.51 N; 86.50 E | 12 | 07 |
| 13. | Begusarai, Bihar | 25.38 N; 86.18 E | 18 | 12 |
| 14. | Rohtas, Bihar | 24.63 N; 83.92 E | 06 | 06 |
| 15. | Baruni, Bihar | 25.45 N; 86.00 E | 09 | 07 |
| 16. | Bhagalpur, Bihar | 25.25 N; 86.95 E | 06 | 04 |
| 17. | Katihar, Bihar | 25.39 N; 87.63 E | 08 | 02 |
| 18. | Ludhiana, Punjab | 30.90 N; 75.79 E | 09 | 11 |
| 19. | Jalandhar, Punjab | 30.99 N; 75.74 E | 11 | 06 |
| 20. | Allowal, Punjab | 31.15 N; 75.67 E | 08 | 12 |
2.2. Isolation and Morphological Characterization of Pathogens
The symptomatic tissue was cut from the margins of lesions and surface-sterilized with 1% sodium hypochlorite for 1 min and then rinsed with distilled water three times. The surface-sterilized leaf tissues (5–6 mm2) with lesions were placed on Petri plates containing potato dextrose medium (HiMedia, Mumbai, India) amended with chloramphenicol (0.05 g/L) (HiMedia, Mumbai, India) and incubated at 28 ± 2 °C for three days [44]. The agar disc (7 mm) containing the mycelia from the 3-day-old colonies was placed on the center of a Petri plate with fresh PDA and then incubated at 28 ± 2 °C. The pure cultures were obtained through single-spore isolation [29] and maintained on agar slants with regular subculturing.
Observations based on cultural characteristics of the different isolates were recorded over a 15-day incubation period of PDA containing sporulating cultures. Three replications of each isolate were maintained. Microscopic features of isolates, i.e., conidia shape, size, number of septa, color were taken as parameters by using a Olympus BX41 Epi and Trans Fluorescence Microscope (Olympus, Tokyo, Japan) for differentiation of genera [45]. While measuring the morphological features of different genera, thirty observations were randomly documented. The isolates were used to study the morphological characteristics described earlier [10,12,45,46].
Conidia and conidiophores were mounted in distilled water and viewed in an Olympus BX41 Phase Contrast and Dark Field Microscope (Olympus, Tokyo, Japan). Measurements of conidial width were recorded from the widest part of each conidium. Olympus BX41TF Epi and Trans Fluorescence Microscope (Olympus, Hamburg, Germany) measured their lengths and widths based on previous studies [10,12]. On average, 30 measurements of conidial length and width of each isolate was recorded. Mean ± standard deviation for length and width ranges of each conidium was recorded.
Bipolaris maydis isolates were cultured on PDA (Himedia, Mumbai, India) for 15 days at 28 °C. Actively growing fungal cultures of agar blocks were cut and fixed overnight at 0.1 M sodium phosphate buffer (pH 7.3) containing 2% glutaraldehyde. In phosphate buffer, each fungal mat was thoroughly washed three times for 15 min and samples were dehydrated for 15 min by using a series of graded ethanol (10, 20, 30, 40, 50, 60, 70, 80, 90, and 100%).The dehydrated and fixed samples of Bipolaris maydis isolates were further dried for 5 min with CO2 and then immediately fixed on aluminum stubs and sputter-coated with carbon in a Polaron E-500 sputter coater (Polaron Equipment, Watford, England) and viewed under a visible scanning electron microscope (S3400N-Hitachi, Tokyo, Japan) immediately.
Koch’s postulates test [47] for pathogenicity of the isolates of Bipolaris maydis and Curvularia lunata were carried out in the glass house. In glass house, five maize seeds of Kanchan variety were sown in 45 cm plastic pots containing sterilized soil and were kept in glass house at 26 °C with 18 h photoperiod. One plant per pot was maintained. Bipolaris maydis and Curvularia lunata conidial suspension was prepared by scraping all conidia from the culture plate after 10 days of incubation followed by suspension in deionized water containing 0.02% tween 20 (Himedia, Mumbai, India). The final pathogen concentration was adjusted to 1 × 106 conidia L−1, as suggested in earlier reports [48]. The final spore suspension concentration (106 spores/mL) was inoculated on the leaves of 25-day-old maize plants using a hand atomizer until leaf run-off. Uninoculated plants were sprayed with sterile distilled water in the same way. Each treatment had three replicates, and the maize plants were kept in a glass house at 26 ± 2 °C with 12 h of darkness and 12 h of light, with the treated plants covered with polythene bags to maintain a relative humidity of more than 90% as per the method described by Manzar et al. [49]. After the infection appeared on the leaves, the fungus was re-isolated from the infected leaves and the identity was reconfirmed, to prove Koch’s postulates. The pathogenicity test was carried out thrice (Supplementary Figure S5a,b). The cultures of the isolate were deposited to the IDA-approved National Agriculturally Important Microbial Culture Collection (NAIMCC) and the accession number was obtained.
2.3. Molecular Characterization
2.3.1. DNA Extraction, PCR, and Sequencing
Via genomic DNA extraction, the isolates were cultured on PDA medium for 15 days at 28 °C for DNA isolation. Mycelial scrapings (50–60 mg) were obtained from the fully grown cultures in the Petri plate. Harvested mycelium was transferred into an autoclaved mortar and then powdered in a pestle mortar with liquid nitrogen. To extract genomic DNA, Nucleopore GDNA Fungus Kit- NP-7006D (Genetix Biotech Asia Pvt. Ltd., New Delhi, India) was used, as described in a previous report [29]. Quantification and assessment of the quality of genomic DNA were checked using a spectrophotometer (Shimadzu, Tokyo, Japan) by recording the absorbance at 245 nm.
2.3.2. Amplification and Purification of PCR Products
The isolates were amplified using the PCR primers and conditions listed in Manamgoda et al. [29], for ITS (ITS1 and ITS4), GAPDH (gpd1 and gpd2), LSU (LSU(F) and LSU®), and D1 and D2 region of LSU (NL1 and NL4), i.e., ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), LSU(F) (5′-ACCCGCTGAACTTAAGC-3′); LSU(R) (5′-TCCTGAGGGAAACTTCG-3′); gpd1 (5′-ATACACTGCCACCCAGAAGG-3′) and gpd2 (5′-TCGATGCGAACAGTCAAGTC-3′); and NL1 (5′-GCATATCAATAAGCGGAGGAAAA-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) [29,38,50,51,52]. PCR (Peqlab-95-06002-PEQSTAR-96 gradient thermocycler, VWR International GmbH, Darmstadt, Germany) was performed in a 25 μL volume containing 1X Taq Buffer with KCl (2.5 μL), 200 µM dNTP (0.5 μL), 1.5 mM MgCl2 (1.5 μL), 10 pM forward primer (1 μL), 10 pM reverse primer (1 μL), 1U/reaction Taq polymerase (0.2 μL) and 1X nuclease-free water (17.3 μL) (Genetix Biotech Asia Pvt. Ltd., New Delhi, India), and 100 ng genomic DNA (1 μL). PCR conditions listed in Manamgoda et al. (2012) for ITS, GAPDH, LSU, and D1 and D2 region of LSU gene. Amplified products (10 μL) were separated from 1.5% agarose gel containing ethidium bromide (0.5 μg/mL) by gel electrophoresis performed at 2 V cm−1 of the gel length until the bands were resolved and visualized, documented, and photographed in the gel-documentation unit (Bio-Rad, Philadelphia, PA, USA). Gel extraction kit (Sure extract spin PCR clean/Gel extraction kit, Genetix Biotech Asia Pvt. Ltd., New Delhi, India) was used to purify ITS, LSU, GAPDH, D1 and D2 regions of LSU of the amplified PCR products. All amplicons were bidirectional-sequenced through Sanger sequencing method (Eurofins Pvt. Ltd. Bangalore, India) in order to get maximum length.
2.3.3. Sequence Alignment and Phylogenetic Analyses
Newly generated ITS, GAPDH, and LSU sequences were analyzed separately with all available ex-type available sequences downloaded from GenBank and DDBJ to determine preliminary identifications of all the isolates. Raw sequences were assembled with Sequencher v. 4.9 for Windows (Gene Codes Corp., Ann Arbor, MI, USA). The assembled consensus sequences were initially aligned with MAFFT v. 7 using default settings (http://mafft.cbrc.jp/alignment/server/; accessed on 26 June 2021) and adjusted manually where necessary [53]; poorly aligned regions of the alignments and hyper-variable regions where alignment was ambiguous were removed using Trimal [54] and exported in a standard PHYLIP file. To fully resolve closely related species, all isolates were subjected to a multi-gene combined analysis. Phylogenetic reconstructions of concatenated and individual gene trees were performed using maximum likelihood (ML) criteria. Maximum likelihood trees were generated using RAxML 8.1.15 [55] under a GTRGAMMA model. Bipartitioned information was also input into RAxML software [56] for maximum likelihood (ML) analysis. A rapid bootstrap (BS) analysis was conducted with 1000 replications, the software produced a best-scoring ML tree with BS probabilities, and the results were obtained in ‘TRE’ files. The combined multi-marker dataset (ITS, GAPDH and LSU) was bipartitioned by gene region and was analyzed using RAxML as described above. Phylogenetic trees were viewed in MEGA v. 11 [57]. All sequences generated were deposited in GenBank (Table 2).
Table 2.
Details of the isolates used in this study, including the hosts, locations, and GenBank accession numbers of the generated sequences of the ribosomal marker (ITS, LSU, and D1 and D2 region of LSU) and protein-coding gene GAPDH.
| Isolates Code |
Host | Location | Identified Organism |
ITS1 and ITS4 Accession Number |
GAPDH Accession Number |
LSU Accession Number |
D1 and D2 Region of LSU Accession Number |
NAIMCC Accession Number |
Reference |
|---|---|---|---|---|---|---|---|---|---|
| E43 | Zea mays | Hariharpur, Sant Kabir Nagar, Uttar Pradesh, India | Curvularia lunata | MH104636 | LC543643 | MT416023 | LC546663 | NAIMCC-F-03913 | This study |
| E36 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MH104637 | LC543644 | MT416024 | LC546664 | Yet to be submitted | This study |
| E50 | Zea mays | Pantnagar, Udham Singh Nagar, Uttarakhand, India |
Bipolaris maydis | MH145411 | LC543968 | MT416015 | LC546657 | NAIMCC-F-03997 | This study |
| E7 | Zea mays | Nagla, Udham Singh Nagar, Uttarakhand, India | Bipolaris maydis | MH145412 | LC508971 | MT416016 | LC546658 | NAIMCC-F-03919 | This study |
| E10 | Zea mays | HardasChak, Khagaria, Bihar, India | Bipolaris maydis | MH145413 | LC543969 | MT416017 | LC546659 | NAIMCC-F-03990 | This study |
| E15 | Zea mays | Godargama, Begusarai, Bihar, India |
Bipolaris maydis | MH145414 | LC538220 | MT416018 | LC546660 | NAIMCC-F-03991 | This study |
| E13 | Zea mays | Khargajepur, MauNath Bhanjan Uttar Pradesh, India |
Curvularia lunata | MH183189 | LC524133 | MT416004 | LC546646 | NAIMCC-F-03917 | This study |
| E27 | Zea mays | KhiriyaMau Nath BhanjanUttar Pradesh, India | Bipolaris maydis | MH183190 | LC544122 | MT416005 | LC546647 | NAIMCC-F-03916 | This study |
| E25 | Zea mays | Kasuet, MaunathBhanjanUttar Pradesh, India | Bipolaris maydis | MH183191 | LC529408 | MT416006 | LC546648 | NAIMCC-F-03988 | This study |
| E37 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MH183192 | LC544123 | MT416007 | LC546649 | NAIMCC-F-03915 | This study |
| E41 | Zea mays | Azamgarh, Uttar Pradesh, India | Curvularia lunata | MH183193 | LC529749 | MT416008 | LC546650 | NAIMCC-F-03914 | This study |
| E16 | Zea mays | Sari Tole SanicharaAsthan, Samastipur, Bihar, India | Curvularia lunata | MH183194 | LC538355 | MT416009 | LC546651 | Yet to be submitted | This study |
| E39 | Zea mays | Mau Nath BhanjanUttar Pradesh, India | Curvularia lunata | MH183195 | LC544124 | MT416010 | LC546652 | Yet to be submitted | This study |
| E12 | Zea mays | Singhaw, FatehpurUttarPradesh, India | Curvularia lunata | MH183196 | LC508972 | MT416011 | LC546653 | NAIMCC-F-03918 | This study |
| E19 | Zea mays | Saidpur Ama, Begusarai, Bihar | Bipolaris maydis | MH183197 | LC541581 | MT416012 | LC546654 | NAIMCC-F-03992 | This study |
| E34 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MH183198 | LC545392 | MT416013 | LC546655 | Yet to be submitted | This study |
| E3 | Zea mays | Godargama, Begusarai, Bihar, India | Bipolaris maydis | MT799977 | LC538353 | MT799989 | LC546665 | NAIMCC-F-03989 | This study |
| E14 | Zea mays | Lakho, Begusarai, Bihar, India | Curvularia lunata | MT799978 | LC538354 | MT799990 | LC546666 | NAIMCC-F-04001 | This study |
| E24 | Zea mays | Jigni, Rohtas, Bihar, India | Bipolaris maydis | MT524321 | LC552295 | MT516300 | MT533838 | NAIMCC-F-03993 | This study |
| E30 | Zea mays | BhindKund, Ballia, Uttar Pradesh, India | Bipolaris maydis | MT524322 | LC552288 | MT516301 | MT533839 | NAIMCC-F-03998 | This study |
| E35 | Zea mays | Ludhiana, Punjab, India | Curvularia lunata | MT524323 | LC552292 | MT516302 | MT533840 | Yet to be submitted | This study |
| E28 | Zea mays | Bhar, MauNathBhanjan, Uttar Pradesh, India | Curvularia lunata | MT524324 | LC552287 | MT516303 | MT533841 | Yet to be submitted | This study |
| E33 | Zea mays | Khojpur, Jalandhar, Punjab, India | Curvularia lunata | MT524325 | LC552290 | MT516304 | MT533842 | NAIMCC-F-03986 | This study |
| E6 | Zea mays | Talwandi Bharo, Jalandhar, Punjab, India | Curvularia lunata | MT524326 | LC552294 | MT516305 | MT533843 | Yet to be submitted | This study |
| E38 | Zea mays | Jalandhar, Punjab, India | Bipolaris maydis | MT524327 | LC552293 | MT516306 | MT533844 | NAIMCC-F-03996 | This study |
| E32 | Zea mays | Allowal, Punjab, India | Curvularia lunata | MT524328 | LC552289 | MT516307 | MT533845 | Yet to be submitted | This study |
| E31 | Zea mays | BarharaChargaha, Maharajganj, Uttar Pradesh, India | Curvularia lunata | MT524329 | LC552683 | MT516308 | MT533846 | NAIMCC-F-04002 | This study |
| E26 | Zea mays | Mustafabad, FaizabadUttar Pradesh, India | Bipolaris maydis | MT524331 | LC552286 | MT516310 | MT533848 | NAIMCC-F-03994 | This study |
| AR 5183 | Zea mays | Japan | Bipolaris maydis | KM230390 | KM034848 | NA | NA | NA | [11] |
| CBS 136.29 | Zea mays | Japan | Bipolaris maydis | KJ909769 | KM034845 | NA | NA | NA | [11] |
| AR 5182 | Zea mays | Japan | Bipolaris maydis | KM230388 | KM034844 | NA | NA | NA | [11] |
| CBS 137271/C5 | Zea mays | USA | Bipolaris maydis | AF071325 | KM034846 | NA | NA | NA | [11] |
| CBS137271/C5 | Zea mays | USA | Bipolaris maydis | AF071325 | KM034846 | KM243280 | NA | NA | [11] |
| Zea mays | NA | ||||||||
| CBS157.34 | Indonesia | Bipolaris maydis | JX256430 | JX276442 | - | NA | NA | [42] | |
| CPC 28832 | Triticum aestivum | Thailand | Bipolaris sorokiniana | MF490812 | MF490834 | - | NA | NA | [59] |
| M 1122/C4 | Zea mays | USA | Bipolaris maydis | KM230389 | KM034847 | - | NA | NA | [11] |
| CBS 108941 | NA | = | Dreschlera erythrospila | AY004782 | AY004813 | - | NA | NA | [60] |
| MFLUCC 10-0706 | Oryza sativa, | Thailand | Curvularia lunata | JX256431 | JX276443 | JX256398 | NA | NA | [11] |
| CBS730.96 | Human lung biopsy | USA | Curvularia lunata | JX256429 | JX276441 | JX256396 | NA | NA | [11] |
| MFLUCC 10-0695 | Panicum sp. | Thailand | Curvularia lunata | JX256432 | JX276444 | JX256399 | NA | NA | [11] |
| CBS 136.29 | Zea mays | Japan | Bipolaris maydis | KJ909769 | KM034845 | - | NA | NA | [11] |
| CBS 307.64 | Zea mays | USA | Bipolaris maydis | HF934925 | HG779085 | HF934875 | NA | NA | [61] |
| CBS 130.26 | Zea mays | - | Bipolaris maydis | HF934923 | HG779084 | HF934873 | NA | NA | [61] |
| CBS573.73 | Zea mays | USA | Bipolaris maydis | HF934924 | NA | MH872501 | HF934881 | NA | [61] |
| CBS 307.84 | Avena sativa | Sweden | Pyrenophora avenicola | MK539972 | MK540180 | MK540042 | NA | NA | [62] |
| CBS330.53 | = | Japan | Rhizopus oryzae | MH857229 | NA | MH868766 | NA | NA | [63] |
| ICMP 6128 | Cynodon dactylon | New Zealand | Bipolaris cynodontis | JX256412 | JX276427 | JX256380 | NA | NA | [42] |
| BRIP 12898 | Melinis munitiflora | Australia | Bipolaris melinidis | JN601035 | JN600972 | JX256411 | NA | NA | [42] |
| CBS 280.91 | Microlaenae stipoidis | Australia | Bipolaris microlaenae | JN601032 | JN600974 | JN600995 | NA | NA | [42] |
| MFLUCC 10-0694 | Oryza sativa | Thailand | Bipolaris oryzae | JX256413 | JX276428 | JX256381 | NA | NA | [42] |
| BRIP 12790 | Cynodon dactylon | Australia | Bipolaris peregianensis | JN601034 | JN600977 | JN601000 | NA | NA | [42] |
| MFLUCC 10-0705 | Panicum sp. | Thailand | Curvularia alcornii | JX256421 | JX276434 | JX256388 | NA | NA | [42] |
| MFLUCC 10-0687 | Oryza sativa | Thailand | Curvularia asianensis | JX256422 | JX276435 | JX256389 | NA | NA | [42] |
| CBS 193.62 | Air | Pakistan | Curvularia ellisii | JN192375 | JN600963 | JN600985 | NA | NA | [42] |
| ICMP 6160 | Gladiolus sp. | New Zealand | Curvularia gladioli | JX256426 | JX276438 | JX256393 | NA | NA | [42] |
| BRIP 23186a | - | Australia | Curvularia graminicola | JN192376 | JN600964 | JN600986 | NA | NA | [42] |
| BRIP 15933 | Chloris gayana | Australia | Curvularia hawaiiensis | JN601028 | JN600965 | JN600987 | NA | NA | [42] |
| CBS 284.91 | Heteropogon contortus | Australia | Curvularia heteropogonis | JN192379 | JN600969 | JN600990 | NA | NA | [42] |
| ICMP 6172 | Ischaemum indicum | Solomon Islands | Curvularia ischaemi | JX256428 | JX276440 | JX256395 | NA | NA | [42] |
| BRIP 15882a | Eragrostis interrupta | Australia | Curvularia ovariicola | JN601031 | JN600971 | JN600992 | NA | NA | [42] |
| BRIP 13165a | Sporobolus fertilis | Australia | Curvularia ravenelii | JN192386 | JN600978 | JN601001 | NA | NA | [42] |
| CBS 274.52a | Soil | Spain | Curvularia spicifera | JN192387 | JN600979 | JX256400 | NA | NA | [42] |
| BRIP 12375 | Unknown | Australia | Curvularia tripogonis | JN192388 | JN600980 | JN601002 | NA | NA | [42] |
| CBS 146.63 | Unknown | India | Curvularia tuberculata | MH858243 | LT715830 | MH869845 | NA | NA | [42] |
| CBS 192.29 | Japan | Curvularia coicis | MH855040 | HG779130 | MH866505 | NA | NA | [63] | |
| BRIP 14845 | Coffea arabica | Kenya | B. coffeana | KJ415525 | KJ415421 | KJ415478 | NA | NA | [64] |
| BRIP 12530 | Dactyloctenium radulan | Australia | B. clavate | KJ415524 | KJ415422 | KJ415477 | NA | NA | [64] |
| CBS274.91 | Eleusine indica | USA | Bipolaris eleusines | KJ909768 | KM034820 | KM243289 | NA | NA | [11] |
| BRIP 14838 | Croton sp. | - | Bipolaris crotonis | KJ415526 | KJ415420 | KJ415479 | NA | NA | [64] |
| CBS 109894 | C. dactylon | Hungary | Bipolaris cynodontis | KJ909767 | KM034838 | KM243288 | NA | NA | [11] |
| CBS241.92 | Hevea sp. | Nigeria | B.heveae | KJ909763 | KM034843 | KM243294 | NA | NA | [11] |
| BRIP 14840 | Gossypium sp. | Kenya | B. gossypina | KJ415528 | KJ415418 | KJ415481 | NA | NA | [11] |
| BRIP 15613 | Microlaena stipoides | Australia | B. microlaenae | JN601032 | JN600974 | JN600995 | NA | NA | [18] |
| CBS 199.29 | P. miliaceum | Japan | B. panici-miliace | KJ909773 | KM042896 | KM243281 | NA | NA | [18] |
| BRIP 12790 | C. dactylon | Australia | B. peregianensis | JN601034 | JN600977 | JN601000 | NA | NA | [18] |
| BRIP 14839 | E. coracana | Zambia | B. pluriseptata | KJ41553 | KJ415414 | KJ41548 | NA | NA | [64] |
| IMI 228224 | Salvinia auriculata | Brazil | B. salviniae | KJ922390 | KM034829 | KM243283 | NA | NA | [11] |
| CBS 120.24 | Italy | B. sorokiniana | KJ909776 | KM034821 | KM243278 | NA | NA | [11] | |
| CBS 624.68 | Dichanthium annulatum | USA | C. robusta | KJ909783 | KM083613 | KM243297 | NA | NA | [11] |
| CBS 349.90 | S. creber | Australia | C. riley | KJ909766 | KM083612 | KM243267 | NA | NA | [11] |
| CBS 239.48 | - | USA | Curvularia portulacae | MH856324 | LT715903 | MH867878 | NA | NA | [63] |
| CBS 350.90a | Perotis rara | Australia | Curvularia perotidis | JN192385 | HG779138 | JN600999 | NA | NA | [18] |
| CBS 156.35 | Air | Jawa | C. pallescens | KJ922380 | KM083606 | KM243269 | NA | NA | [11] |
| CBS 160.58 | Eleusine Indica | USA | C. nodulosa | JN601033 | JN600975 | JN600997 | NA | NA | [11] |
| CBS 656.74 | Desert soil | Egypt | C. subpapendorfii | KJ909777 | KM061791 | KM243266 | NA | NA | [11] |
| CBS 327.64 | Avena sativa | USA | B. victoriae | KJ909778 | KM034811 | KM243271 | NA | NA | [11] |
| BRIP 17186 | Heliconia psittacorum | Australia | Bipolaris heliconiae | KJ415530 | KJ415417 | KJ415483 | NA | NA | [64] |
| BRIP 15900 | Sorghum bicolor | Australia | Curvularia sorghina | KJ415558 | KJ415388 | KJ415512 | NA | NA | [64] |
| BRIP 14845 | Coffea arabica | Kenya | Bipolaris coffeana | KJ415525 | KJ415421 | KJ415478 | NA | NA | [64] |
| E29 | Zea mays | India | Curvularia geniculata | MT524330 | LC552684 | MT533847 | NA | NA | [49] |
CBS: CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; MFLUCC: Mae Fah Luang University Culture Collection, Center of Excellence in Fungal Research, Thailand; BRIP: Biosecurity Queensland Plant Pathology Herbarium, Brisbane, Queensland, Australia. NA: not available.
2.3.4. Statistical Analysis
Statistical analysis of length and width of each isolate were analyzed by using a one-way analysis of variance [58] and Duncan’s multiple range test (DMRT) using Statistical Product and Service Solution (SPSS) version 16.0 software Developed by SPSS Inc., now IBM SPSS Armonk, NY, USA. On average, 30 measurements of conidial length and width of each isolate were recorded. Mean ± standard deviation for length and width ranges of each conidium was recorded.
3. Results
3.1. Morphological Identification of Bipolaris Isolates (Maydis Leaf Blight)
Twenty sampling district sites were chosen from major maize-growing regions of India, from which a total of 186 leaf blight symptomatic samples were collected. Seventy-seven isolates of Bipolaris were identified from the total collected maydis-leaf-blight-symptomatic samples (Table 1). The 77 isolates were divided into five groups as per cultural and morphological characteristics (Table 3). Group A consisted of 20 isolates, representing 25.97%, Group B consisted of 9 isolates, representing 11.68%, Group C consisted of 25 isolates accounting for 32.46%, Group D had 11 isolates accounting for 14.28%, and Group E consisted of 12 isolates which accounted for 15.58%. Table 3, give a brief overview of the morphological data for these Bipolaris isolates in groups A–E. Characteristics of the colony: variable colony morphology on PDA was found after 15 days for each group. Group A isolates had gray colonies with spots, raised mycelia was observed with irregular margin; in Group B isolates, colony color was gray with smooth raised mycelia and irregular margins. The isolates in Group C produced gray colonies with appressed mycelia and irregular margin. The colonies from Group D were blackish gray with white spot, smooth appressed mycelia, and regular margin, whereas Group E had whitish gray colonies, rough raised mycelia, and irregular margin. The conidia length-width ratio, color and shape, and conidial morphology was grouped into five groups. Group A had a larger length–width ratio than Group C and Group D. All the isolates of the five groups produced slightly curved, light-brown to brown, fusiform conidia (Supplementary Figure S2).
Table 3.
Morphological characteristics of Bipolaris isolates.
| Group | Colony Characteristics | Conidia | ||
|---|---|---|---|---|
| Average Length (µm) | Average Width (µm) | Shape | ||
| A (n = 20) | Grey with white spots, rough raised mycelia, irregular margin | 80.44 ± 0.71 a (64–89) |
14.64 ± 0.28 ab (13–16) |
Fusiform, slightly curved, dark brown, 4–9 distoseptate |
| B (n = 9) | Grey, smooth raised mycelia, irregular margin | 65.38 ± 0.64 e (53–71) |
12.43 ± 0.59 d (11–13) |
Fusiform, slightly curved, light brown, 4 distoseptate |
| C (n = 25) | Grey, smooth appressed mycelia, irregular margin | 74.05 ± 0.14 c (63–97) |
15.14 ± 0.35 a (14–16) |
Brown, 6–8 distoseptate |
| D (n = 11) | Blackish grey with white spot, smooth appressed mycelia, regular margin | 76.68 ± 0.54 b (68–84) |
13.44 ± 0.44 c (12–14) |
Brown, 4–5 distoseptate |
| E (n = 12) | Whitish grey, rough raised mycelia, irregular margin | 71.29 ± 0.80 d (51–83) |
13.89 ± 0.38 bc (12–15) |
Light brown, 5–9 distoseptate |
The values in the column represent the average of 30 measurements, followed by the standard deviation. Duncan’s multiple range tests (DMRT) show that values with different alphabetical (a–e) superscripts within a column are significantly different (p ≤ 0.05).
SEM observations: Bipolaris conidia had smooth walls, were fusiform or slightly curved, and placed on a geniculate conidiophore. Bipolaris spp. conidial attachment to conidiophores, conidial hilum, and conidia were all seen (Supplementary Figure S3).
3.2. Morphological Identification of Curvularia Isolates (Maize Leaf Spot)
A total of 129 maize-leaf-spot-symptomatic samples were collected from 20 sampling districts from five major maize-growing regions and 74 Curvularia isolates were identified based on their morphological characteristics (Table 4). Seventy-four isolates were divided into five groups based on their cultural and morphological characteristics. Group F consisted of 15 isolates, representing 20.27%, Group G included 8 isolates, accounting for 10.81%, Group H consisted of 21 isolates, accounting for 28.37%, Group I had 14 isolates, accounting for 18.91%, and Group J consisted of 16 isolates, which accounted for 21.62%. A brief description of the cultural and morphological characteristics of Curvularia isolates as described in F–J groups can be found in Table 4. Different colony morphology was found on PDA for each distinct group after 7 days of incubation. Group F isolates had black colonies and smooth velvety mycelia with regular margin; Group G isolates had colonies that were gray in color with smooth floccose mycelia and irregular margins. The isolates in Group H produced gray colonies with smooth appressed mycelia and irregular margin. The colonies from Group I were blackish gray with smooth appressed mycelia and regular margin. The colonies produced by Group J isolates were whitish gray with smooth velvety mycelia and regular margin. Conidial morphology: conidial types were observed among the five groups based on the following conidia shapes, color and length–width ratio: The length–width ratio of Group I was larger than those of Group G and Group J. All Group isolates produced 3–5 distoseptate conidia with light-brown to brown color (Supplementary Figure S4).
Table 4.
Morphological characteristics of Curvularia isolates.
| Group | Colony Characteristics | Conidia | ||
|---|---|---|---|---|
| Average Length | Average Width | Shape | ||
| F (n = 15) |
Black, smooth velvety mycelia, regular margin | 20.86 ± 0.85 bc (19–22) |
10.23 ± 0.35 b (8–11) |
Light brown, 3–4 distoseptate |
| G (n = 8) |
Grey, smooth floccose mycelia, irregular margin | 27.29 ± 0.79 a (25–29) |
12.71 ± 0.72 a (12–13) |
Light yellow, 3–4 distoseptate |
| H (n = 21) |
Grey, smooth appressed mycelia, irregular margin | 29.15 ± 0.27 a (25–35) |
11.96 ± 0.78 a (10–12) |
Brown, 4–5 distoseptate |
| I (n = 14) |
Blackish grey, smooth appressed mycelia, regular margin | 19.69 ± 0.46 c (16–24) |
9.58 ± 0.44 b (9–10) |
Light brown, 3–4 distoseptate |
| J (n = 16) |
Whitish grey, smooth velvety mycelia, regular margin | 22.82 ± 0.24 b (22–23) |
8.77 ± 0.67 b (8–9) |
Light brown, 3–4 distoseptate |
The values in the column represent the average of 30 measurements, followed by the standard deviation. Duncan’s multiple range tests (DMRT) show that values with different alphabetical (a–c) superscripts within a column are significantly different (p ≤ 0.05).
3.3. Pathogenicity of the Isolate
Koch’s postulates experiments were performed for the isolates and were positive for causing MLB disease and leaf spot of maize. The maydis leaf blight symptoms appeared as small yellow necrotic spots that later become spindle- or elliptical-shaped lesions and were observed in the inoculated plants, Zea mays c.v. Kanchan (Supplementary Figure S5a). In maize leaf spot, the symptoms appeared as yellow necrotic spots in the early stages of maize leaves, which eventually expanded to circular, oval, or strip lesions formed after 10 days of inoculation, identical to those seen in the field for leaf spot of maize. (Supplementary Figure S5b).
3.4. Phylogenetic Study of Bipolaris Isolates (Causal Organism of Maydis Leaf Blight)
PCR-amplified product of Bipolaris spp. and Curvularia spp. using primer pairs ITS1/ITS4, GAPDH1/GAPDH2, D1/D2 LSU yielded specific amplified products of approximately 650 bp (ITS), 472 bp (gapdh), and 917 bp (LSU). Maximum likelihood analyses were performed both individually and in combination based on the collected sequenced data to produce phylograms. Forty-three ex-type closely related species of Bipolaris and Curvularia were used as reference sequences. The isolates were characterized at molecular level using thee three above-mentioned gene loci; upon NCBI BLAST search, they showed 99–100% similarity with concern pathogen and were identified as Bipolaris maydis and Curvularia lunata. These isolates were submitted to GenBank and IDA (International Depositary Authority)-approved culture collection center NAIMCC after approval of cultures authenticity from the authority. Accession no. of culture collection is mentioned and described in Table 2.
3.4.1. Phylogeny Based on the Ribosomal Marker ITS Regions of Bipolaris maydis
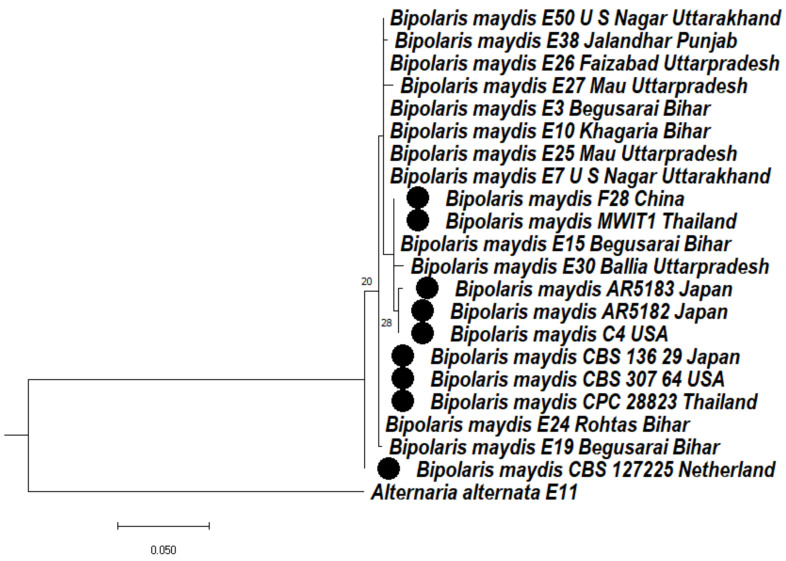
Based on the sequences of the ITS regions of Bipolaris isolates, a phylogenetic tree was made to ascertain the relationship among the isolates of Bipolaris maydis. In the phylogenetic tree, two clades were formed. In clade 1, two subclades were formed; subclade 1A formed two subclades, 1Aa and 1Ab. In subclade 1Aa, Bipolaris maydis isolates E50, E38, E26, E27, E3, E10, E25, E7 were clustered together, whereas F28, MWT1, E15, E30 were clustered together, AR5183, AR5182, C4 were clustered together, and CBS136.29, CBS-307.64, CPC-28823, E24 were clustered together. Subclade 1Ab consists of E19. Clade 1B consists of CBS127225. Alternaria alternata (E11) was used as an outgroup in clade 2 (Figure 1).
Figure 1.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS). Sequenced data of the Bipolaris maydis isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
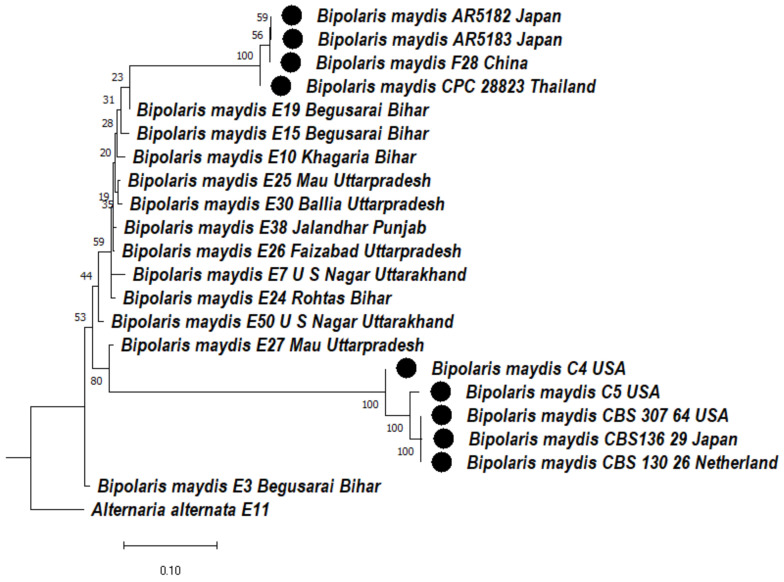
3.4.2. Phylogenetic Analysis of ITS + GAPDH Gene of Bipolaris maydis
A phylogenetic tree was constructed based on the aligned sequences of ITS + GAPDH region to ascertain the phylogenetic relationship between the Bipolaris maydis isolates. The cladogram was divided into two clades. In clade 1, there are two subclades: in subclade 1Aa, the Bipolaris maydis isolates AR5182, AR5183, F28 were clustered together with a bootstrap value of 100% and CPC28823, E19, E15, E10, E25, E30, E38, E26, E7, E24 formed a sister clade with them, whereas subclade 1Ab consists of E50 isolate, which formed a distinct clade. Subclade 1Ba consists of E27 isolate with a bootstrap value of 80%, subclade 1Bb consists of CBS307.64, CBS136.29, CBS130.26, which were clustered together with a bootstrap value of 100%, whereas C5, C4 formed a sister clade with them, with a bootstrap value of 100%. Subclade 2 consists of E3 isolate. Alternaria alternata E11 was used as an outgroup in clade 2 (Figure 2).
Figure 2.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Sequenced data of the Bipolaris maydis isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
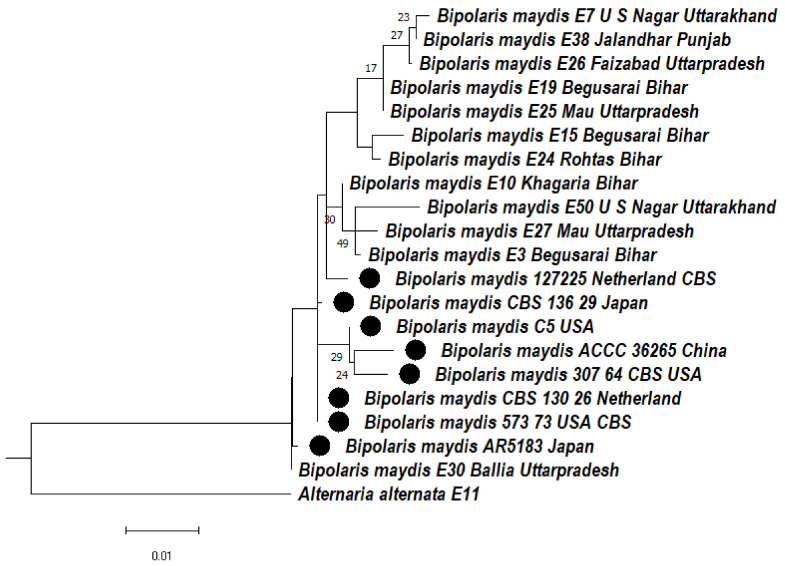
3.4.3. Phylogeny Based on the ITS + LSU Gene of Bipolaris maydis
The phylogenetic tree was constructed based on the aligned sequences of ITS + LSU region to ascertain the evolutionary relationship among the Bipolaris maydis isolates. The cladogram was divided into two clades. Clade 1 divided into two subclades (1 and 2). In subclade 1, two subclades were formed, 1A and 1B. In subclade 1Aaa, Bipolaris maydis isolates E7, E38, E26 were clustered together, whereas E19, E25 formed a sister clade with them, and E15, E24 were clustered together. In subclade 1Abb, E50, E27, E3 were clustered together, whereas E10 formed a distinct clade with them. Subclade 1Ab consists of 127, 225 isolates. Subclade 1B consists of CBS 136.29, whereas in subclade 1C, C5, ACCC36265, 307.64 were clustered together. In subclade 1D, Bipolaris maydis isolates CBS130.26, CBS 573.73, AR 5183 formed a distinct clade. Subclade 2 consists of E30 isolate. In clade 2, Alternaria alternata (E11) was used as an outgroup that formed a separate clade (Figure 3).
Figure 3.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS) and large Subunit gene regions (LSU). Sequenced data of the Bipolaris maydis isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
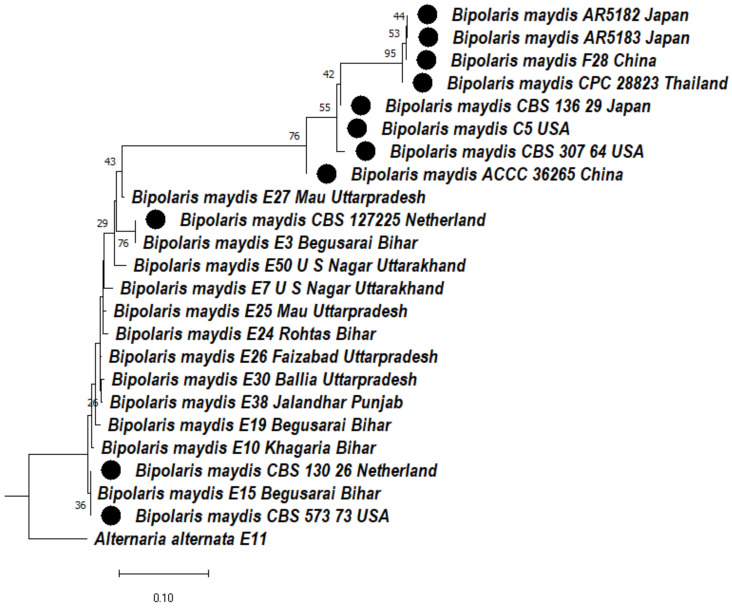
3.4.4. Phylogeny Based on the ITS + LSU + GAPDH Gene of Bipolaris maydis
A cladogram was constructed based on the concatenated aligned multilocus sequences of Bipolaris maydis isolates to ascertain the phylogenetic relationship among the Bipolaris maydis isolates. The cladogram was divided into two clades (Figure 4). In clade 1, two subclades formed. Subclade 1A consists of subclade 1Aa and 1Ab; in subclade 1Aa, AR5182, AR518312, F28, CPC28823 were clustered together with a bootstrap value of 95% and CBS136.29, CBS307.64, C5 were clustered together, whereas in subclade 1Ab, ACCC36265, E27 formed a sister clade with them with a bootstrap value of 76%. In subclade 1B, Bipolaris maydis isolates CBS127225, E3 were clustered together with a bootstrap value of 76%, whereas E50, E7, E25, E24, E26, E30, E38, E19, E10 formed a sister clade with them. In subclade 1C, CBS130.26, E15, CBS573.73 were clustered together. In clade 2, E11 Alternaria alternata was used as an outgroup that formed a separate clade (Figure 4).
Figure 4.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS) and large Subunit gene regions (LSU) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Sequenced data of the Bipolaris maydis isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
3.5. Phylogenetic Study of Curvularia lunata Isolates (Causal Organism of Maize Leaf Spot)
3.5.1. Phylogeny Based on the ITS of Curvularia lunata
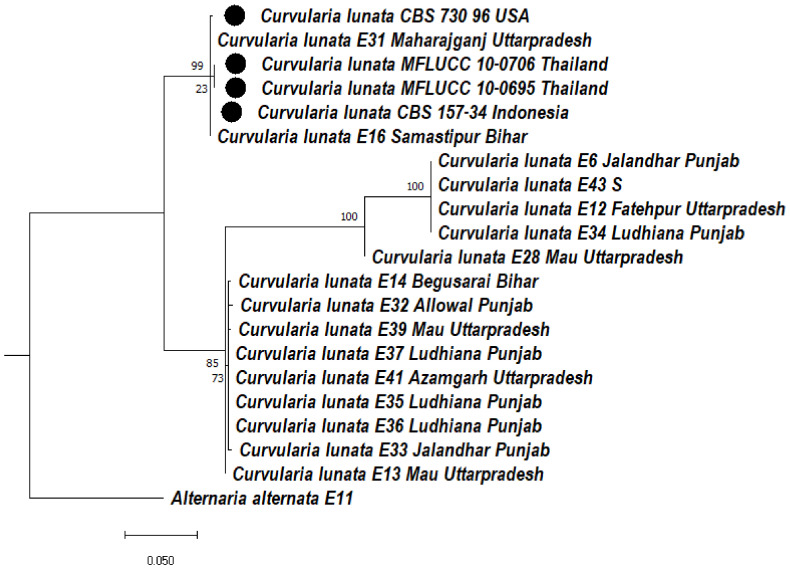
Based on the ITS region, the Curvularia lunata (maize leaf spot) isolates formed two clades. In clade 1, two subclades, 1A and 1B, were formed. In subclade 1A, two subclades, 1A and 1B, were formed. In subclades 1A, Curvularia lunata isolates CBS730.96, E31, CBS 157-34, E16 were clustered together, whereas MFLUCC 10-0706, MFLUCC 10-0695 were clustered together with a bootstrap value of 99%. In subclade 1Ba, E6, E43, E12, E34 were clustered with each other and formed a distinct clade with a bootstrap value of 100%, whereas E28 formed a distinct clade with a bootstrap value 100%. In subclade 1Bb, E14, E32, E39, E37, E41, E35, E36, E33 isolates formed a distinct clade with a bootstrap value 85%, whereas E13 formed a distinct clade with a bootstrap value of 73%. In clade 2, E11 Alternaria alternata was used as an outgroup that formed a separate clade (Figure 5).
Figure 5.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS), and large subunit gene regions (LSU) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Sequenced data of the Curvularia lunata isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
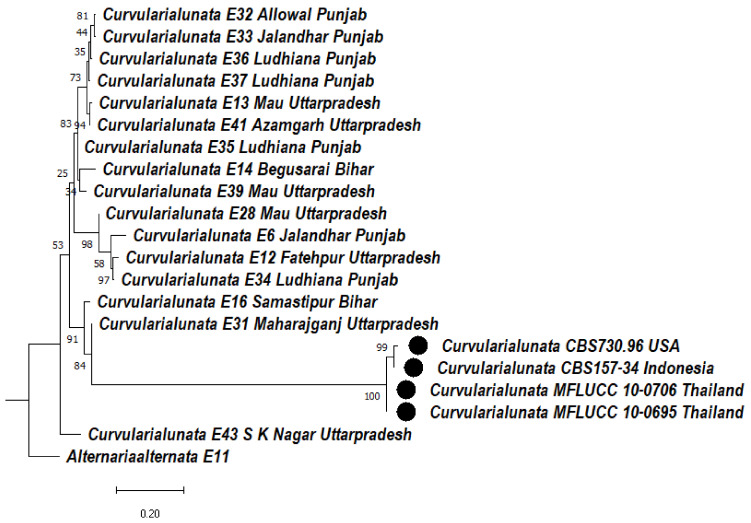
3.5.2. Phylogeny Based on the ITS + GAPDH of Curvularia lunata
Based on the ITS + GAPDH region, the Curvularia lunata isolates formed two clades formed. In clade 1, there are two subclades, 1 and 2. In subclade 1A, E32, E33 isolates were clustered together with a bootstrap value of 81%, whereas E36, E37 formed a sister clade with them with a bootstrap value of 73%; E13, E41 were clustered together with a bootstrap value of 94% and formed a sister clade with them. In subclade 1B, E35, E14, E39, were clustered together. In subclade 1C, E28, E6, E12, E34 were clustered together with a bootstrap value of 98%. Subclade 1Da consists of E16 with a bootstrap value of 91% and subclade 1Db consists of E31 with a bootstrap value of 84%, whereas in subclade 1Dc, CBS 730.96, CBS 157-34 were clustered together with a bootstrap value of 99% and MFLUCC 10-0706, MFLUCC 10-0695 were clustered together with a bootstrap value of 100%. Subclade 2 consists of Curvularia lunata isolate E43. In clade 2, Alternaria alternata (E11) was used as an outgroup that formed a separate clade (Figure 6).
Figure 6.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Sequenced data of the Curvularia lunata isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
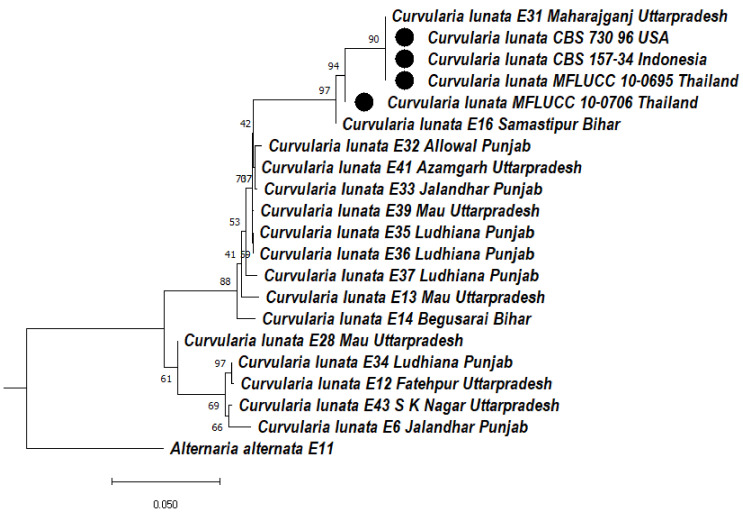
3.5.3. Phylogeny Based on the ITS + LSU of Curvularia lunata
Based on the ITS + LSU region, two clades were formed; in clade 1, two subclades formed, subclade 1 and subclade 2. In subclade 1Aa, Curvularia lunata isolates E31, CBS 730.96, CBS 157-34, MFLUCC 10-0695 were clustered together with a bootstrap value of 90%, MFLUCC 10-0706 formed a sister clade with a bootstrap value of 94%, and E16 formed a sister clade with a bootstrap value of 97%. In subclade 1Ab, E32, E41, E33 were clustered together with a bootstrap value of 76% and E39, E35, E36 formed a sister clade with them, whereas E37, E13, E14 formed a distinct clade with a bootstrap value of 88%. Subclade 1Ca consists of E28 with a bootstrap value of 61%, whereas in subclade 1Cb, E34, E12 were clustered together with a bootstrap value of 97% and E43, E6 were clustered together with a bootstrap value of 69%. In clade 2, Alternaria alternata (E11) was used as an outgroup to form a distinct clade (Figure 7).
Figure 7.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS) and large subunit gene regions (LSU). Sequenced data of the Curvularia lunata isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicates analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
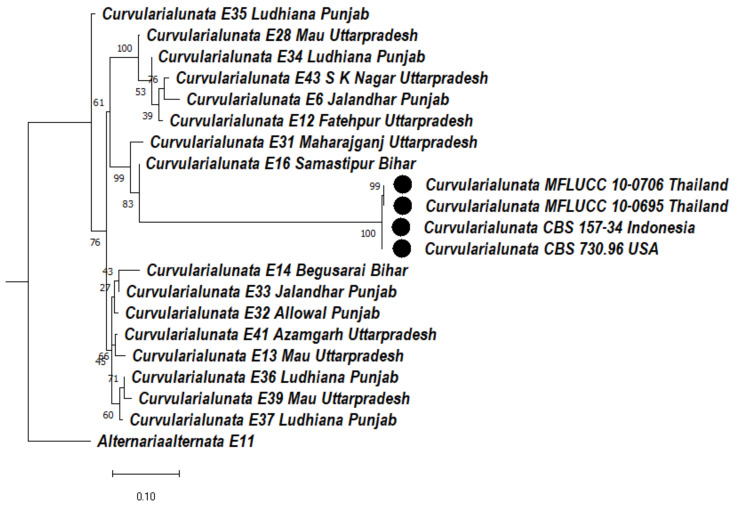
3.5.4. Phylogeny Based on the Multigene of Curvularia lunata
A cladogram was constructed based on the concatenated sequences of ITS, LSU, and GAPDH gene regions. Two clades were formed, clade 1 and clade 2. In clade 1 there are two subclades, subclade 1 and subclade 2. In subclade 1, Curvularia lunata isolate E35 formed a distinct clade. In subclade 2Aa, E28 formed a distinct clade with a bootstrap value of 100%. In subclade 2Ab, E34 formed a distinct clade, E43, E6 were clustered together with a bootstrap value of 76%, and E12 formed a sister clade with them. In subclade 2Aca, two clades formed. In subclade 2Ac, E31 formed a clade with a bootstrap value of 99%; in subclade 2Acb, MFLUCC 10-706, MFLUCC 10-0695 were clustered together with a bootstrap value of 99% and CBS 157-34, CBS 730.96 were clustered with each other and formed a distinct clade with a bootstrap value of 96%, whereas E16 formed a sister clade with them with a bootstrap value of 83%. In subclade 2Ad, E14, E33 were clustered together and E32 formed a sister clade with them, whereas E41, E13 were clustered together with a bootstrap value of 66%, and E36, E39 were clustered together with a bootstrap value of 71% and E37 formed a sister clade with them. In clade 2, Alternaria alternata (E11) was used as an outgroup formed a distinct clade (Figure 8).
Figure 8.
Dendrogram constructed from maximum likelihood method based on combined internal transcribed spacer (ITS), large subunit gene regions (LSU), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Sequenced data of the Curvularia lunata isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. The symbol ● denotes as ‘ex-type’ closely related species sequences obtained from GenBank. Alternaria alternata used as an outgroup.
3.6. Phylogenetic Studies between Bipolaris and Curvularia Complex
Based on the combined sequences of ITS gene regions of Bipolaris maydis and Curvularia lunata, a cladogram was constructed based on the multilocus sequences to know the phylogenetic relationship among the 26 isolates of Bipolaris maydis and 21 isolates of Curvularia lunata. Rhizopus oryzae was used as an outgroup, and 43 ex-type closely related Curvularia and Bipolaris species were used as a reference sequence.
3.6.1. Phylogeny Based on the Combined Sequences of ITS + GAPDH of Bipolaris maydis and Curvularia lunata
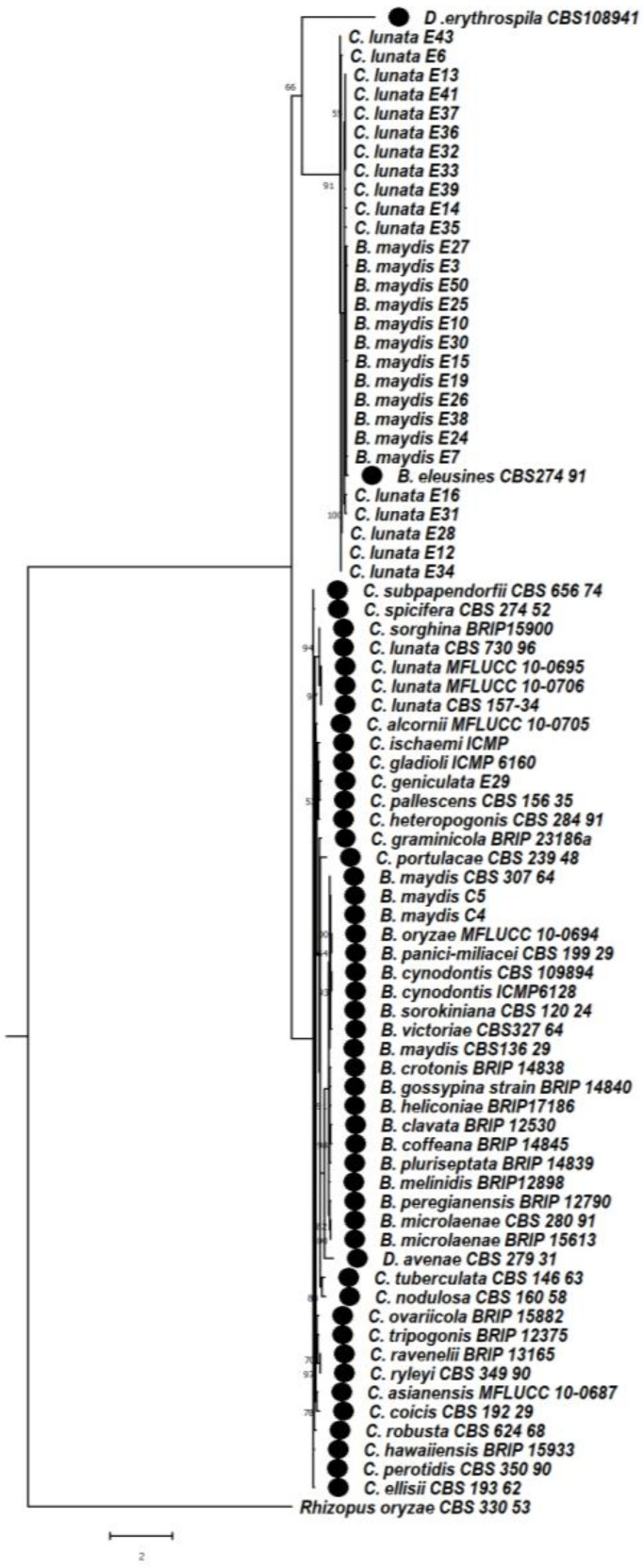
A multilocus phylogenetic analysis of the combined sequences of the ITS and GAPDH gene regions was performed among isolates of Bipolaris maydis and Curvularia lunata. Drechslera species were used as a related genera and Rhizopus oryzae was used as an outgroup to determine the phylogenetic relationship among the isolates of Bipolaris maydis and Curvularia lunata. On the basis of the phylogenetic tree, two separate clades formed. In clade 1, two subclades formed, subclade 1 and subclade 2. Subclade 1A consists of Drechslera erythospila isolate with a bootstrap value of 66%, subclade 1Ba consist of distinct genus Curvularia lunata isolates E43, E6, E13, E41, E37, E36, E32, E33, E39, E14, E35, which were clustered together with a bootstrap value of 91%, whereas subclade 1Bb formed with a genus of Bipolaris maydis isolates E27, E3, E50, E25, E10, E30, E15, E19, E26, E38, E24, E7, with Bipolaris eleusines CBS274.91 clustered together. In subclade 1Bc, Curvularia lunata isolates E16, E31, E28, E12, E34 were clustered together with a bootstrap value of 100%.
Subclade 2A consists of Curvularia subpapendorfii (CBS 656.74), Curvularia spicifera (CBS 274.52), Curvularia sorghina (BRIP15900), Curvularia lunata (CBS 730.96) Curvularia lunata ex-type species MFLUCC 10-0695, MFLUCC 10-0706, CBS 157-34, MFLUCC 10-0706, CBS157-34, Curvularia ischaemi (ICMP 6172), Curvularia gladioli (ICMP 6160), Curvularia geniculata (E29), Curvularia pallescens (CBS 156.35), Curvularia heteropogonis (CBS 284.91), Curvularia graminicola (BRIP 23186a), Curvularia portulacae (CBS 239.48). In subclade 2B, Bipolaris maydis isolates CBS 307.64, C5, C4, Bipolaris oryzae (MFLUCC 10-0694), Bipolaris panici-miliacei (CBS 199.29), Bipolaris cynodontis (CBS 109894), Bipolaris cynodontis (ICMP 6128), Bipolaris sorokiniana (CBS 120.24), Bipolaris victoriae (CBS 327.64), Bipolaris maydis (CBS 136.29), Bipolaris crotonis (BRIP 14838), Bipolaris gossypina (BRIP 14840), Bipolaris heliconiae (BRIP 17186), Bipolaris clavata_(BRIP 12530), Bipolaris coffeana (BRIP 14845), Bipolaris pluriseptata (BRIP 14839), Bipolaris melinidis (BRIP12898), Bipolaris peregianensis (BRIP 12790), Bipolaris microlaenae (CBS 280.91), Bipolaris microlaenae (BRIP 15613) were clustered together, whereas Drechslera avenae (CBS279.31) formed a sister clade with them with a bootstrap value of 97%. Subclade 2C consists of Curvularia tuberculata (CBS 146.63) and Curvularia nodulosa (CBS 160.58), which were clustered together with a bootstrap value of 88%; Curvularia ovariicola_(BRIP 15882), Curvularia tripogonis (BRIP 12375), Curvularia ravenelii (BRIP 13165), Curvularia ryleyi (CBS 349.90), Curvularia asianensis (MFLUCC 10-0687), Curvularia coicis (CBS 192.29), Curvularia robusta (CBS 624.68), Curvularia hawaiiensis (BRIP 15933), Curvularia perotidis (CBS 350.90), Curvularia ellisii (CBS 193.62) were clustered together.
In clade2, Rhizopus oryzae (CBS 330.53) is considered as an outgroup which is entirely different from Helminthosporoid genera (Figure 9).
Figure 9.
Maximum likelihood phylogeny based on combined internal transcribed spacer (ITS) and (GAPDH). Sequenced data of the Bipolaris maydis and Curvularia lunata isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicates analysis. Drechslera avenae CBS 279.31, Drechslera erythospila (sexual stage-Pyrenophora erythospila) CBS 108,941 were used as other genera from Helminthosporoid group. The symbol ● denotes as ‘ex-type’ 43 closely related species sequences obtained from GenBank. Rhizopus oryzae used as an outgroup.
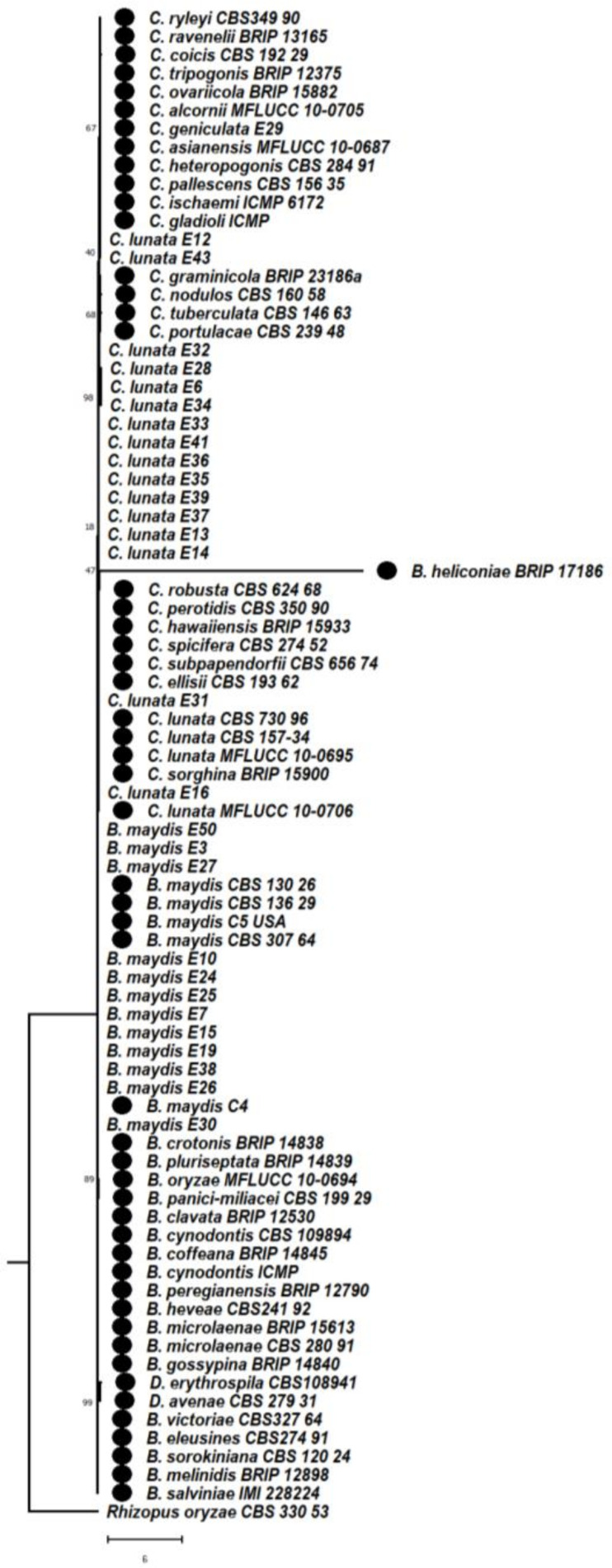
3.6.2. Phylogeny Based on the Combined Sequences of ITS + LSU of Bipolaris maydis and Curvularia lunata
A multilocus phylogenetic analysis of the combined sequences of the ITS and LSU gene regions was performed on the basis of the phylogenetic tree, and two clades were formed. Clade 1A consists of Curvularia ryleyi (CBS 349.90), Curvularia ravenelii (BRIP 13165), Curvularia coicis (CBS192.29), Curvularia tripogonis (BRIP12375), Curvularia ovaricola (BRIP15882), Curvularia alcornii (MFLUCC 10-0705), Curvularia geniculata (E29), Curvularia asianensis (MFLUCC 10-0687), Curvularia pallescens CBS 156.35, Curvularia heteropogonis (CBS 284.91), Curvularia ischaemi (ICMP 6172), Curvularia gladioli (ICMP 6160), Curvularia lunata isolates E12, E43, Curvularia graminicola_(BRIP 23186a), Curvularia nodulosa (CBS 160.58), Curvularia tuberculata (CBS 146.63), Curvularia portulacae (CBS 239.48), Curvularia lunata isolates E32, E28, E6, E34, E33, E41, E36, E35, E39, E37, E13, E14, Bipolaris heliconiae (BRIP 17186), Curvularia robusta (CBS 624.68), Curvularia perotidis (CBS 350.90), Curvularia hawaiiensis (BRIP 15933), Curvularia spicifera (CBS 274.52), Curvularia ellisii (CBS 193.62), Curvularia lunata isolates:E31, CBS 730.96, CBS 157.34, MFLUCC 10-0695, MFLUCC 10-0706, E16, Curvularia subpapendorfii (CBS 656.74), Curvularia sorghina (BRIP 15900), Bipolaris maydis isolates E50, E3, E27, CBS 136.29, CBS 130.26, C5, CBS307.64, E10, E24, E25, E7, E15, E19, E38, E26, C4, E30, Bipolaris crotonis (BRIP 14838), Bipolaris pluriseptata (BRIP 14839), Bipolaris oryzae (MFLUCC 10-0694), Bipolaris panici-miliacei (CBS 199.29), Bipolaris clavata_(BRIP 12530), Bipolaris cynodontis (ICMP 6128), Bipolaris coffeana (BRIP 14845), Bipolaris cynodontis (CBS 109894), Bipolaris peregianensis (BRIP 12790), Bipolaris heveae (CBS241.9), Bipolaris microlaenae (CBS 280.91), Bipolaris microlaenae (BRIP 15613), Bipolaris gossypina (BRIP 14840), Drechslera avenae (CBS 279.31), Drechslera erythospila (sexual stage-Pyrenophora erythospila CBS108941), Bipolaris victoriae_(CBS327.64), Bipolaris melinidis (BRIP 12898), Bipolaris sorokiniana (CBS 120.24), Bipolaris salviniae (IMI 228224), which were clustered with each other. In clade 2, Rhizopus oryzae (CBS 330.53) is considered as an outgroup which is entirely different from Helminthosporoid genera (Figure 10).
Figure 10.
Maximum likelihood phylogeny based on combined internal transcribed spacer (ITS) and large subunit gene regions (LSU). Sequenced data of the Bipolaris maydis, and Curvularia lunata isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. Drechslera avenae CBS 279.31, Drechslera erythospila (sexual stage-Pyrenophora erythospila) CBS 108,941 were used as other genera from Helminthosporoid group. The symbol ● denotes as ‘ex-type’ 43 closely related species sequences obtained from GenBank. Rhizopus oryzae used as an outgroup. Rhizopus oryzae used as an outgroup.
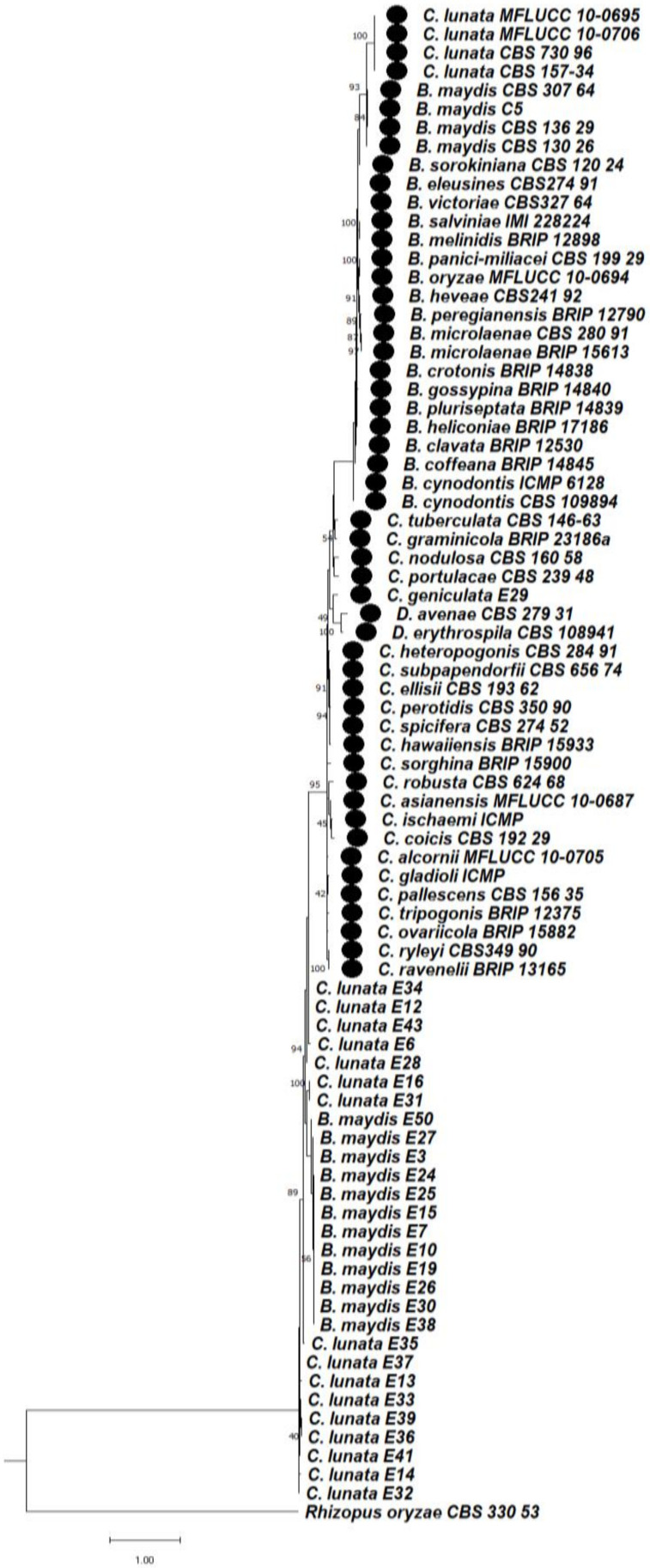
3.6.3. Phylogeny Based on the Combined Regions Multigene for Bipolaris and Curvularia Complex
A multilocus phylogenetic analysis of the concatenated sequences of ITS + GAPDH + LSU region was performed among the Bipolaris maydis and Curvularia lunata isolates, where Drechslera avenae and Rhizopus oryzae were used as an outgroup. On the basis of the phylogenetic tree, two separate clades formed. In clade 1, three subclades were formed; in clade 2, Rhizopus oryzae (CBS 330.53) was considered as an outgroup which is completely different from Helminthosporoid genera.
In subclade 1Aa, Curvularialunata (MFLUCC10-0695), Curvularialunata (MFLUCC10-0706), Curvularia lunata isolates (CBS730.96), (CBS157.34) were clustered together. In subclade1Ab, Bipolaris maydis isolates CBS 307.64, CBS 136.29, C5, CBS 157.34, CBS 130.26, Bipolaris sorokiniana (CBS 120.24), Bipolaris eleusines (CBS274.91), Bipolaris pluriseptata (BRIP 14839), Bipolaris clavata (BRIP 12530), Bipolaris coffeana (BRIP 14845), Bipolaris panici-miliacei (CBS 199.29), Bipolaris oryzae (MFLUCC 10-0694), Bipolaris heveae (CBS 241.92), Bipolaris heliconiae (BRIP 17186), Bipolaris gossypina (BRIP 14840), Bipolaris crotonis (BRIP 14838), Bipolaris microlaenae (CBS 280.91), Bipolaris microlaenae (BRIP 15613), Bipolaris peregianensis (BRIP 12790), Bipolaris melinidis (BRIP 12898), Bipolaris salviniae (IMI 228224), Bipolaris eleusines (CBS274.91), Bipolaris pluriseptata (BRIP 14839), Bipolaris clavata (BRIP 12530), Bipolaris coffeana (BRIP 14845), Bipolaris cynodontis (ICMP 6128), Bipolaris cynodontis (CBS 109894) were clustered together. In subclade 1Ac, Curvularia tuberculata (CBS 146-63), Curvularia graminicola (BRIP 23186a), Curvularia nodulosa (CBS 160.58), Curvularia portulacae (CBS 239.48), Curvularia geniculata (E29), Drechslera avenae (CBS 279.31), Drechslera erythospila (sexual stage-Pyrenophora erythospila) (CBS 108941), Curvularia heteropogonis (CBS289.91), Curvularia ovaricola (BRIP15882), Curvularia tripogonis (BRIP 12,375), Curvularia sorghina (BRIP15900), Curvularia coicis (CBS 192.29), Curvularia ischaemi (ICMP 6172), Curvularia asianensis (MFLUCC 10-0687), Curvularia alcornii (MFLUCC 10-0705) Curvularia gladioli (ICMP), Curvularia pallescens (CBS 156.35), Curvularia ryleyi (CBS 349.90), Curvularia ravenelli (BRIP 13165) were clustered together.
In subclade 2Aa, Curvularia lunata isolates E34, E12, E43, E6, E28, E16, E31 were clustered together. In subclade 2Ab, Bipolaris maydis isolates E50, E27, E3, E24, E25, E15, E7, E10, E19, E26, E30, E38 were clustered together. In subclade 2Ac, Curvularia lunata isolates E35, E37, E13, E33, E39, E36, E41, E14, E32 were clustered together.
In clade 2, Rhizopus oryzae (CBS 330.53) is considered as an outgroup that is entirely different from Helminthosporoid genera (Figure 11).
Figure 11.
Maximum likelihood phylogeny based on combined internal transcribed spacer (ITS), large subunit gene regions (LSU), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Sequenced data of the Bipolaris maydis and Curvularia lunata isolates were inferred with Raxml based on GTR + Gamma model. Numerical value presented in the node indicates bootstrap value with a 1000 non-parametric bootstrap replicate analysis. Drechslera avenae CBS 279.31, Drechslera erythospila (sexual stage-Pyrenophora erythospila) CBS 108,941 were used as other genera from Helminthosporoid group. The symbol ● denotes as ‘ex-type’ 43 closely related species sequences obtained from GenBank. Rhizopus oryzae used as an outgroup.
4. Discussion
Bipolaris and Curvularia species that are anamorphs of Cochliobolus species are important pathogens of the Poaceae family. Leaf spots, blight, root rot, and crown rots are diseases caused by these pathogens [11,12,38]. Payak and Sharma (1981) reported 61 diseases in maize [65]. Among the diseases, maydis leaf blight causes yield losses of up to 41 percent [20]. Considerable phenotypic, pathogenic, and genetic variations occur among the pathogens under the genus Helminthosporium [66,67,68]. Bipolaris maydis, previously called Helminthosporium maydis, also shows enormous variability. The studies on diversity analysis based on morphological variation and phylogenetic basis in B. maydis-Zea mays pathosystem have not been carried out very extensively. Keeping this fact in mind, the present study was undertaken, and the results are here discussed. The present work focused on elucidating the biodiversity of the Bipolaris maydis, causing maydis leaf blight, and Curvularia lunata, causing maize leaf spot. To this end, a vast collection of Bipolaris maydis isolates obtained mainly from maize from the major maize-growing regions of Bihar, Punjab, Uttar Pradesh, and Uttarakhand were characterized based on morphological and molecular characteristics.
Seventy-seven maydis leaf blight isolates were morphologically characterized and formed into five groups based on their morphology. The data showed that different isolate lengths and widths of conidia varied between 65.38–80.44 μm and 12.43–14.64 μm, respectively (Table 1). In Bipolaris maydis isolates, Group A had the largest size of conidia, having a mean length of 80.44 μm and width of 14.64 μm, whereas in Group B, the size of conidia was the smallest (65.38 × 12.43 μm) and showed a distinct variation among the isolates based on conidia length and width. Similar observations were made by Sun et al., 2020; Pal et al., 2015; Leonard and Suggs, 1974 [14,69,70]. Comparison of the conidium sizes of the isolates of Bipolaris maydis and Curvularia lunata examined in this study was consistent with results from previous studies [12,71,72]. The number of septa of various isolates ranged between four and nine, and the average number of septa was highest for Group E (5–9); in Group D, the number of septa was smallest (4–5). The isolates showed two textural classes, namely rough and smooth, and colony pattern was either appressed or raised type. Group A and Group E isolates showed rough texture with raised colony pattern, whereas Group C showed smooth texture with an appressed colony. Group A, B, C, and E grown on PDA showed regular margins, while Group D exhibited irregular margins. Similar variations in number of septa among the isolates of Bipolaris maydis were found in the same agroclimatic zones [14,70,73].
In Curvularia lunata isolates, Group H had the largest size of conidia, having a mean length of 29.15 μm and width of 11.96 μm, whereas in Group I, the size of conidia was smallest (19.69 × 9.58 μm) and showed a distinct variation among the isolates based on conidia length and width. The distoseptate conidia were smooth-walled and curved at subterminal cell, three-septate, four-celled, and brown. The Group H, and Group I produced grey, smooth mycelia, whereas Group F produced black smooth velvety mycelia. These morphological characters are similar to the characters of Curvularia lunata-caused leaf spot of maize described earlier [74,75] and standard descriptions from Ellis (1971) [10].
Our study mainly highlights the correct way of identifying of Bipolaris maydis and Curvularia lunata isolates, as both show complex symptoms in field conditions that can cause confusion in their identification. In this study, we screened out indigenous isolates based on morphological characteristics and ribosomal markers ITS and LSU, while protein-coding GAPDH genes were used for molecular identification and phylogenetic analysis. A phylogenetic tree was constructed based on the concatenated aligned multigene sequences of ribosomal markers ITS and LSU and protein-coding GAPDH gene regions of Bipolaris maydis isolates and found that maydis leaf blight isolates of Bihar, Uttar Pradesh, and Punjab isolates were clustered together, whereas Uttatrakhand isolates were clustered only with Bihar and Uttarpradesh isolates. In other words, Upper Gangetic Plain region and Middle Gangetic Plain region isolates of Bipolaris maydis showed similarity with each other. Whiles Bipolaris maydis isolates constructed phylogeny only on the basis ITS sequences, the Bihar, Uttarpradesh, Punjab, and Uttarakhand isolates clustered together, and the aligned multigene sequences of ribosomal marker ITS and protein-coding GAPDH gene regions revealed that the isolates of Bipolaris maydis from other countries such as Japan, China, USA, and the Netherlands did not cluster with the Indian isolates, but when phylogeny was constructed based only on ITS sequences, Bipolaris maydis isolates from Japan, China, USA, and the Netherlands clustered with the Indian isolates. This indicates that nuclear ribosomal DNA (rDNA) internal transcribed spacer (ITS) sequences alone could not differentiate the isolates based on their geographical locations, but that combined ITS, LSU, and GAPDH phylogeny was able to differentiate among the isolates based on said locations. A cladogram was constructed based on the concatenated sequences of ribosomal markers ITS and LSU and protein-coding GAPDH gene regions of Curvularia lunata isolates. The combined analysis of ITS, LSU, and GAPDH phylogeny of Curvularia lunata isolates revealed that most of the isolates of Punjab and Uttarakhand were clustered together, whereas Bihar isolates only clustered with Uttarpradesh isolates. Similarly, the Curvularia lunata isolates of Thailand, Indonesia, and USA were clustered together but did not cluster with Indian-origin isolates. However, when phylogeny was made on the basis of ITS sequences or combined sequences of ITS and LSU the Curvularia lunata isolates of Thailand, Indonesia, and USA were clustered with Bihar and Uttarpradesh isolates. Similar observations were found as reported earlier [11,14,29].
The only morphological distinction between Curvularia and the helminthosporioid genus Bipolaris is the conidial curvature and length. Both genera have species with intermediate morphology, requiring sequencing data to distinguish them adequately. The overlapping morphological features of several Curvularia taxa make species identification difficult [20,66]. Only morphological evidence with the subjective determination of phenotypic characteristics such as spore size has been used to identify Bipolaris and Curvularia spp., while some were identified entirely on the basis of host relationship. Several duplicate sequences of Bipolaris spp. with different names were submitted in GenBank. When searching for generic placement, ITS BLAST search should be used because it is the most efficient. Notably, Xue et al. (2016) included eight species of Bipolaris in their phylogenetic analysis, which led to incorrect identification of two of the three strains. Therefore, sequencing data are critical for reliable species identification with ITS, GAPDH, and LSU gene regions [28], although the ITS and GAPDH alone can still resolve the majority of taxa in Curvularia [26]. Although the ITS regions alone can resolve the majority of taxa in Curvularia [26], sequences of GAPDH and LSU regions are needed to clarify if it is a different species. In our study, for accurate identification of Curvularia lunata and Bipolaris maydis isolates, phylogenetic analyses using multi-genes (ITS or LSU) or protein-coding genes such as GAPDH were performed; the multigene analysis highlights the similarity and relationships between different isolates from different geographical locations [26,38,43].
5. Conclusions
It is difficult to differentiate sister genera (Bipoplaris and Curvularia) solely based on morphological examination, because many of their morphological features overlap. Therefore, other identification methods, such as molecular identification, are necessary for identification. Here, we used GAPDH, a protein-coding gene, and other ribosomal markers like ITS or LSU for phylogenetic analysis in Bipolaris and Curvularia spp. to ensure precise identification. We used precise taxon sampling for accurate results by adding 43 ex-type closely related Bipolaris and Curvularia spp. sequences from GenBank that derived from type material to identify the delineate species in Bipolaris and Curvularia. Based on the phylogenetic tree, multigene analysis (ITS + LSU + GAPDH) yielded accurate identification of fungal isolates such as B. maydis and C. lunata. While the multigene analysis highlights the similarity and relationships between the different isolates from different geographical locations, the information is comparatively degenerate in single-gene-based phylogeny. In this study, an unclear bifurcation of Drechslera genera was also observed in the case of (ITS + GAPDH) phylogeny, as Drechslera erythrospila CBS108941 were clustered with Curvularia lunata, but this case was resolved after using a multigene phylogenetic approach (ITS + LSU + GAPDH). Upon so doing, the Drechslera genera formed a separate cluster, which gives a clue that single- or oligogene-based molecular identification may be incorrect sometimes for correct identification. Nonetheless, multiple-gene-based concatenated phylogenies are much accurate and useful for identification in cases such as the sister genera Bipolaris-Curvularia complex, and they also can be utilized to differentiate isolates of fungi from different geographical locations. This research contributes to understanding Bipolaris maydis and Curvularia lunata maize disease complexity and offers an approach for correct diagnosis using concatenated gene sequence analysis. We found that B. maydis and C. lunata were the dominant species infecting maize in all geographical locations surveyed of major maize-growing regions. Future studies should focus on expanding genetic information on Bipolaris and Curvularia spp., and proper identification should continue to rely on molecular data, not on morphological parameters. The habitat and the host range of many Bipolaris species are incompletely understood. Extensive sampling and accumulation of molecular data will improve understanding of the host range and ecological significance of Bipolaris and Curvularia spp.
Acknowledgments
The authors extend their gratitude to the Indian Council of Agricultural Research, New Delhi, ICAR National Bureau of Agriculturally Important Microorganisms and Maunath Bhanjan, U.P. India, for providing financial assistance during the study. The authors wish to express their sincere thanks to Kumar, M and Hillol Chakdar for providing timely PCR facilities and also a special thanks to Alok Upadhyay, Sr. Technician, NAIMCC and Pusparaj, Contractual, Plant Pathology Lab for the maintenance of fungal cultures during the time period of this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8080802/s1, Figure S1: Sampling district location shown in Map of India.; Figure S2: Representative Bipolaris isolates showing the morphological description from Group A to E; Figure S3: Scanning electron microscope showing conidiophore, conidia and hilum of Bipolaris isolates; Figure S4: Representative isolates of Curvularia spp. showing the morphological descriptions from Group F- Group J; Figure S5: (5a-a) Pot experiment for pathogenicity proving of maize leaf spot (b) maize leaf spot. symptomatic leaves (c) Fully grown Curvularia lunata culture plate (d) Microscopic image of conidia of Curvularia lunata at 40X; (5b-a) Pot experiment for pathogenicity proving of maydis leaf blight (b) maydis leaf blight symptomatic leaves (c) Fully grown Bipolaris maydis culture plate (d) Microscopic image of conidia of Bipolaris maydis at 40X.
Author Contributions
Conceptualization, writing—original draft preparation, N.M.; Writing—review and editing, A.S.K. and N.M.; writing—original draft preparation, investigation, A.S.K., N.M.; Editing, A.M. and M.V.S.R.; supervision, funding acquisition, H.V.S., A.K.S. (Alok Kumar Srivastava), A.K.S. (Anil Kumar Saxena), M.R. and P.K.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by ICAR-NBAIM, Maunath Bhanjan, Uttar Pradesh. Project ID-IXX13356.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Parihar C., Jat S., Singh A., Kumar R.S., Hooda K., GK C., Singh D. Maize production technologies in India. DMR Tech. Bull. Dir. Maize Res. Pusa Campus New Delhi. 2011;30:1–7. [Google Scholar]
- 2.Ngoune Tandzi L., Mutengwa C.S. Estimation of maize (Zea mays L.) yield per harvest area: Appropriate methods. Agronomy. 2020;10:29. doi: 10.3390/agronomy10010029. [DOI] [Google Scholar]
- 3.Faostat F. Food and Agriculture Organization of the United Nations-Statistic Division. 2019. [(accessed on 23 May 2021)]. Available online: https://www.fao.org/faostat/en/#data.
- 4.Otieno H.M. Assessment Of Key Kharif Season Maize Production Constraints in East Champaran District, India. Int. J. Sci. Technol. Res. 2019;8:920–924. [Google Scholar]
- 5.Venkata Rao P., Subbaiah G., Veeraraghavaiah R. Productivity and nutrient uptake of rice fallow maize (Zea mays L.) as influenced by plant density and fertilizer N under no-till conditions. Int. J. Curr. Microbiol. Appl. Sci. 2016;5:826–836. [Google Scholar]
- 6.Revilla P., Anibas C., Tracy W. Sweet Corn Research around the World 2015–2020. Agronomy. 2021;11:534. doi: 10.3390/agronomy11030534. [DOI] [Google Scholar]
- 7.Singh M., Mehra R., Malik V.K. Evaluation of Maize Genotypes against Maydis Leaf Blight Caused by Bipolaris maydis (Nisikado and Miyake) Shoemaker under Artificial Epiphytotic Conditions. Int. J. Curr. Microbiol. App. Sci. 2018;7:1006–1013. doi: 10.20546/ijcmas.2018.705.125. [DOI] [Google Scholar]
- 8.Kumar R., Hooda K., Olakh D., Kaur H., Malik V., Kumar S. Reaction of QPM inbred lines against Maydish Leaf Blight (MLB) and Charcol Rot. Electron. J. Plant Breed. 2013;4:1280–1283. [Google Scholar]
- 9.Malik V.K., Singh M., Hooda K.S., Yadav N.K., Chauhan P.K. Efficacy of newer molecules, bioagents and botanicals against maydis leaf blight and banded leaf and sheath blight of maize. Plant Pathol. J. 2018;34:121. doi: 10.5423/PPJ.OA.11.2017.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis M. Dematiaceous Hyphomycetes, Commonwealth Mycol. Inst. Kew Surrey Lond. Engl. 1971;1:1–608. [Google Scholar]
- 11.Manamgoda D., Rossman A.Y., Castlebury L., Crous P.W., Madrid H., Chukeatirote E., Hyde K.D. The genus bipolaris. Stud. Mycol. 2014;79:221–288. doi: 10.1016/j.simyco.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivanesan A. Graminicolous Species of Bipolaris, Curvularia, Drechslera, Exserohilum and Their Teleomorphs. CAB International; Wallingford, UK: 1987. [Google Scholar]
- 13.Tan Y.P., Crous P.W., Shivas R.G. Eight novel Bipolaris species identified from John L. Alcorn’s collections at the Queensland Plant Pathology Herbarium (BRIP) Mycol. Prog. 2016;15:1203–1214. doi: 10.1007/s11557-016-1240-6. [DOI] [Google Scholar]
- 14.Sun X., Qi X., Wang W., Liu X., Zhao H., Wu C., Chang X., Zhang M., Chen H., Gong G. Etiology and Symptoms of Maize Leaf Spot Caused by Bipolaris spp. in Sichuan, China. Pathogens. 2020;9:229. doi: 10.3390/pathogens9030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh B., Chand D., Maurya A., Sharma R., Agarwal P. Pathogenic fungi intercepted in introduced transgenics during 2006–2013. Plant Pathol. Quar. 2014;4:99–107. doi: 10.5943/ppq/4/2/4. [DOI] [Google Scholar]
- 16.Yago J.I., Roh J.-H., Bae S.-D., Yoon Y.-N., Kim H.-J., Nam M.-H. The effect of seed-borne mycoflora from sorghum and foxtail millet seeds on germination and disease transmission. Mycobiology. 2011;39:206–218. doi: 10.5941/MYCO.2011.39.3.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengyella L., Yekwa E.L., Nawaz K., Iftikhar S., Tambo E., Alisoltani A., Feto N.A., Roy P. Global invasive Cochliobolus species: Cohort of destroyers with implications in food losses and insecurity in the twenty-first century. Arch. Microbiol. 2018;200:119–135. doi: 10.1007/s00203-017-1426-6. [DOI] [PubMed] [Google Scholar]
- 18.Manamgoda D.S., Cai L., Bahkali A.H., Chukeatirote E., Hyde K.D. Cochliobolus: An overview and current status of species. Fungal Divers. 2011;51:3–42. doi: 10.1007/s13225-011-0139-4. [DOI] [Google Scholar]
- 19.Gogoi R., Singh S., Singh P.K., Kulanthaivel S., Rai S. Genetic variability in the isolates of Bipolaris maydis causing maydis leaf blight of maize. Afr. J. Agric. Res. 2014;9:1906–1913. [Google Scholar]
- 20.Sharma R., Rai S. Proceedings of International Conference on Integrated Plant Disease Management for Sustainable Agriculture. Indian Phytopathological Society; Solan, India: 2000. Assessment and quantitative analysis of losses due to Drechslera maydis in maize; pp. 246–248. [Google Scholar]
- 21.Kirk P., Cannon P., Minter D., Stalpers J. Ainsworth & Bisby’s Dictionary of the Fungi. 10th ed. CAB International; Wallingford, UK: 2008. [Google Scholar]
- 22.Gan L., Dai Y., Yang X., Du Y., Ruan H., Shi N., Chen F. First report of leaf spot caused by Cochliobolus eragrostidis on corn (Zea mays L.) in Fujian Province, China. Plant. Dis. 2018;102:439. doi: 10.1094/PDIS-03-17-0372-PDN. [DOI] [Google Scholar]
- 23.Li G., Liu K., Xiao S., Lu Y., Xue C., Wang G. First report of leaf spot of maize (Zea mays) caused by Bipolaris spicifera in China. Plant Dis. 2016;100:855. doi: 10.1094/PDIS-07-15-0750-PDN. [DOI] [Google Scholar]
- 24.Tsai J., Tsai W., Chen J. Pathogenicity of Exserohilum rostratum on corn and weeds in the corn fields. Plant Pathol. Bull. 2001;10:181–186. [Google Scholar]
- 25.Weems J.D., Bradley C.A. Exserohilum turcicum race population distribution in the north central United States. Plant Dis. 2018;102:292–299. doi: 10.1094/PDIS-01-17-0128-RE. [DOI] [PubMed] [Google Scholar]
- 26.Manamgoda D.S., Rossman A.Y., Castlebury L.A., Chukeatirote E., Hyde K.D. A taxonomic and phylogenetic re-appraisal of the genus Curvularia (Pleosporaceae): Human and plant pathogens. Phytotaxa. 2015;212:175–198. doi: 10.11646/phytotaxa.212.3.1. [DOI] [Google Scholar]
- 27.Arabi M., Jawhar M. Molecular and pathogenic variation identified among isolates of Cochliobolus sativus. Aust. Plant Pathol. 2007;36:17–21. doi: 10.1071/AP06081. [DOI] [Google Scholar]
- 28.Marin-Felix Y., Groenewald J., Cai L., Chen Q., Marincowitz S., Barnes I., Bensch K., Braun U., Camporesi E., Damm U. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017;86:99–216. doi: 10.1016/j.simyco.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manamgoda D., Cai L., McKenzie E., Chukeatirote E., Hyde K. Two new Curvularia species from northern Thailand. Sydowia. 2012;64:255–266. [Google Scholar]
- 30.Tekpinar A., Kalmer A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwig. 2019;109:187–224. doi: 10.1127/nova_hedwigia/2019/0528. [DOI] [Google Scholar]
- 31.Smith M.E., Henkel T.W., Uehling J.K., Fremier A.K., Clarke H.D., Vilgalys R. The Ectomycorrhizal Fungal Community in a Neotropical Forest Dominated by the Endemic Dipterocarp Pakaraimaea dipterocarpacea. PLoS ONE. 2013;8:e55160. doi: 10.1371/journal.pone.0055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrower E., Ammirati J.F., Cappuccino A.A., Ceska O., Kranabetter J.M., Kroeger P., Lim S., Taylor T., Berbee M.L. Cortinarius species diversity in British Columbia and molecular phylogenetic comparison with European specimen sequences. Botany. 2011;89:799–810. doi: 10.1139/b11-065. [DOI] [Google Scholar]
- 33.Schoch C.L., Robbertse B., Robert V., Vu D., Cardinali G., Irinyi L., Meyer W., Nilsson R.H., Hughes K., Miller A.N., et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database. 2014;2014:21. doi: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stielow J.B., Lévesque C.A., Seifert K.A., Meyer W., Irinyi L., Smits D., Renfurm R., Verkley G.J.M., Groenewald M., Chaduli D., et al. One fungus, which genes? Development and Assessment of Universal Primers for Potential Secondary Fungal DNA Barcodes. Persoonia. 2015;35:242–263. doi: 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter T.M., Shokralla S., Baird D., Golding G.B., Hajibabaei M. Ribosomal DNA and Plastid Markers Used to Sample Fungal and Plant Communities from Wetland Soils Reveals Complementary Biotas. PLoS ONE. 2016;11:e0142759. doi: 10.1371/journal.pone.0142759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raja H.A., El-Elimat T., Oberlies N.H. Minutisphaerales (Dothideomycetes, Ascomycota): A new order of freshwater ascomycetes including a new family, Minutisphaeraceae, and two new species from North Carolina, USA. Mycologia. 2015;107:845–862. doi: 10.3852/15-013. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among Ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 38.Berbee M., Pirseyedi M., Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. doi: 10.1080/00275514.1999.12061106. [DOI] [Google Scholar]
- 39.Boonmee S., Zhang Y., Chomnunti P., Chukeatirote E., Tsui C.K., Bahkali A.H., Hyde K.D. Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers. 2011;51:63–102. doi: 10.1007/s13225-011-0147-4. [DOI] [Google Scholar]
- 40.Hunter G.C., Wingfield B.D., Crous P.W., Wingfield M.J. A multi-gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Stud. Mycol. 2006;55:147–161. doi: 10.3114/sim.55.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J.-K., Phookamsak R., Jones E.G., Zhang Y., Ko-Ko T.W., Hu H.-L., Boonmee S., Doilom M., Chukeatirote E., Bahkali A.H. Astrosphaeriella is polyphyletic, with species in Fissuroma gen. nov., and Neoastrosphaeriella gen. nov. Fungal Divers. 2011;51:135–154. doi: 10.1007/s13225-011-0142-9. [DOI] [Google Scholar]
- 42.Manamgoda D.S., Cai L., McKenzie E.H., Crous P.W., Madrid H., Chukeatirote E., Shivas R.G., Tan Y.P., Hyde K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012;56:131–144. doi: 10.1007/s13225-012-0189-2. [DOI] [Google Scholar]
- 43.Bhunjun C.S., Dong Y., Jayawardena R.S., Jeewon R., Phukhamsakda C., Bundhun D., Hyde K.D., Sheng J. A polyphasic approach to delineate species in Bipolaris. Fungal Divers. 2020;102:225–256. doi: 10.1007/s13225-020-00446-6. [DOI] [Google Scholar]
- 44.Dai Y., Yang X., Gan L., Chen F., Ruan H., Du Y., Shi N., Gao Z. First report of southern leaf blight caused by Cochliobolus heterostrophus on corn (Zea mays) in Fujian Province, China. Plant. Dis. 2016;100:1781. doi: 10.1094/PDIS-12-15-1490-PDN. [DOI] [Google Scholar]
- 45.Alcorn J. The taxonomy of “Helminthosporium” species. Ann. Rev. Phytopathol. 1988;26:37–56. doi: 10.1146/annurev.py.26.090188.000345. [DOI] [Google Scholar]
- 46.Subramanian C., Jain B. A revision of some graminicolous Helminthosporia. Curr. Sci. 1966;35:352–355. [Google Scholar]
- 47.Mullen J.M. Plant Disease Diagnosis. In: Trigiano R.N., Windham M.T., Windham A.S., editors. Plant Pathology Concepts and Laboratory Exercises. 2nd ed. CRC Press; Boca Raton, FL, USA: 2008. [Google Scholar]
- 48.Hooda K.S., Bagaria P.K., Khokhar M., Kaur H., Rakshit S. Mass Screening Techniques for Resistance to Maize Diseases. ICAR-Indian Institute of Maize Research, PAU Campus; Ludhiana, India: 2018. [Google Scholar]
- 49.Manzar N., Kashyap A.S., Sharma P.K., Saxena A.K. First report of leaf spot of maize caused by Curvularia geniculata in India. Plant Dis. 2021;105:4155. doi: 10.1094/PDIS-03-21-0637-PDN. [DOI] [PubMed] [Google Scholar]
- 50.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 51.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’donnell K. The fungal holomorph: Mitotic, meiotic and pleomorphic speciation in fungal systematics. Fusarium Its Near Relat. 1993;1:225–233. [Google Scholar]
- 53.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jayawardena R.S., Hyde K.D., Jeewon R., Ghobad-Nejhad M., Wanasinghe D.N., Liu N., Phillips A.J., Oliveira-Filho J.R.C., da Silva G.A., Gibertoni T.B. One stop shop II: Taxonomic update with molecular phylogeny for important phytopathogenic genera: 26–50 (2019) Fungal Divers. 2019;94:41–129. doi: 10.1007/s13225-019-00418-5. [DOI] [Google Scholar]
- 55.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raza M., Zhang Z.-F., Hyde K.D., Diao Y.-Z., Cai L. Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers. 2019;99:1–104. doi: 10.1007/s13225-019-00434-5. [DOI] [Google Scholar]
- 57.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trillas M.I., Casanova E., Cotxarrera L., Ordov’as J., Borrero C., Avil’es M. Composts from agricultural waste and the Trichoderma asperellum strain T-34 suppress Rhizoctonia solani in cucumber seedlings. Biol. Control. 2006;39:32–38. doi: 10.1016/j.biocontrol.2006.05.007. [DOI] [Google Scholar]
- 59.Marin-Felix Y., Senwanna C., Cheewangkoon R., Crous P.W. New species and records of Bipolaris and Curvularia from Thailand. Mycosphere. 2017;8:1555–1573. doi: 10.5943/mycosphere/8/9/11. [DOI] [Google Scholar]
- 60.Zhang G., Berbee M.L. TITLE Pyrenophora phylogenetics inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. J. Mycol. 2001;93:1048–1063. doi: 10.1080/00275514.2001.12063240. [DOI] [Google Scholar]
- 61.Madrid H., Da Cunha K., Gené J., Dijksterhuis J., Cano J., Sutton D., Guarro J., Crous P. Novel Curvularia species from clinical specimens. Pers. Mol. Phyl. Evol. Fungi. 2014;33:48. doi: 10.3767/003158514X683538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marin-Felix Y., Hernández-Restrepo M., Iturrieta-González I., García D., Gené J., Groenewald J., Cai L., Chen Q., Quaedvlieg W., Schumacher R. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019;94:1–124. doi: 10.1016/j.simyco.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vu D., Groenewald M., De Vries M., Gehrmann T., Stielow B., Eberhardt U., Al-Hatmi A., Groenewald J., Cardinali G., Houbraken J. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019;92:135–154. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan Y.P., Madrid H., Crous P.W., Shivas R.G. Johnalcornia gen. et. comb. nov., and nine new combinations in Curvularia based on molecular phylogenetic analysis. Aust. Plant. Pathol. 2014;43:589–603. doi: 10.1007/s13313-014-0315-6. [DOI] [Google Scholar]
- 65.Payak M., Sharma R. Maize diseases and approaches to their management in India. Int. J. Pest. Manag. 1985;31:302–310. doi: 10.1080/09670878509371006. [DOI] [Google Scholar]
- 66.Drechsleb C. A species of Hebninthosporium distinct from Helminthosporium sacohari, causing brown stripe of Sugar Cane. Phytopathology. 1928;18:135–136. [Google Scholar]
- 67.Luttrell E. A taxonomic revision of Helminthosporium sativum and related species. Am. J. Bot. 1955;42:57–68. doi: 10.1002/j.1537-2197.1955.tb11094.x. [DOI] [Google Scholar]
- 68.Potter P.E., Maynard J.B., Pryor W.A. Sedimentology of Shale: Study Guide and Reference Source. Springer Science & Business Media; Berlin/Heidelberg, Germany: 2012. [Google Scholar]
- 69.Leonard K., Suggs E.G. Setosphaeria prolata, the ascigerous state of Exserohilum prolatum. Mycologia. 1974;66:281–297. doi: 10.1080/00275514.1974.12019603. [DOI] [Google Scholar]
- 70.Pal I., Singh V., Gogoi R., Hooda K., Bedi N. Characterization of Bipolaris maydis isolates of different maize cropping zones of India. Ind. Phytopathol. 2015;68:63–66. [Google Scholar]
- 71.Morejon K.R., Moraes M.H.D., Bach E.E. Identification of Bipolaris bicolor and Bipolaris sorokiniana on wheat seeds (Triticum aestivum L.) in Brazil. Braz. J. Microbiol. 2006;37:247–250. doi: 10.1590/S1517-83822006000300010. [DOI] [Google Scholar]
- 72.Chun S.-C., Loo H.-M., Lee S.-H., Jung I.-M. A seedborne fungus Bipolaris spicifera detected from imported grass seeds. Plant Pathol. J. 2003;19:133–137. doi: 10.5423/PPJ.2003.19.3.133. [DOI] [Google Scholar]
- 73.Lakshmi P. Differential interactions between Helminthosporium maydis Nisikado & Miyake incitant of maydis leaf blight and cultivars of Zea mays LM Sc.(Ag.) Thesis. IARI. 1984;1:26–78. [Google Scholar]
- 74.Harlapur S., Kulkarni M., Yeshoda H., Srikant K. Variability in Exserohilum turcicum (Pass.) Leonard and Suggs., causal agent of turcicum leaf blight of maize. Karn. J. Agric. Sci. 2007;20:665–666. [Google Scholar]
- 75.Addangadi K.C., Harlapur S.I. Ph.D. Thesis. University of Agricultural Sciences Dharwad; Dharwad, India: 2015. Studies on Curvularia Leaf Spot of Maize. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.