Abstract
Antimicrobial resistance is an ancient natural phenomenon increasingly pressured by anthropogenic activities. Escherichia coli has been used as markers of environmental contamination and human-related activity. Seabirds may be bioindicators of clinically relevant bacterial pathogens and their antimicrobial resistance genes, including extended-spectrum-beta-lactamase (ESBL) and/or plasmid-encoded AmpC (pAmpC), in anthropized and remote areas. We evaluated cloacal swabs of 20 wild magnificent frigatebirds (Fregata magnificens) of the Alcatrazes Archipelago, the biggest breeding colony of magnificent frigatebirds in the southern Atlantic and a natural protected area with no history of human occupation, located in the anthropized southeastern Brazilian coast. We characterized a highly virulent multidrug-resistant ST648 (O153:H9) pandemic clone, harboring blaCTX–M–2, blaCMY–2, qnrB, tetB, sul1, sul2, aadA1, aac(3)-VIa and mdfA, and virulence genes characteristic of avian pathogenic (APEC) (hlyF, iroN, iss, iutA, and ompT) and other extraintestinal E. coli (ExPEC) (chuA, kpsMII, and papC). To our knowledge, this is the first report of ST648 E. coli co-producing ESBL and pAmpC in wild birds inhabiting insular environments. We suggest this potentially zoonotic and pathogenic lineage was likely acquired through indirect anthropogenic contamination of the marine environment, ingestion of contaminated seafood, or by intra and/or interspecific contact. Our findings reinforce the role of wild birds as anthropization sentinels in insular environments and the importance of wildlife surveillance studies on pathogens of critical priority classified by the World Health Organization.
Keywords: pAmpC, ESBL, antimicrobial resistance, island, wildlife, One Health
Introduction
Antimicrobial resistance result from a naturally occurring ancient phenomenon that has been severely affected by anthropogenic activities such as use, misuse and overuse of antimicrobials in human and veterinary medicine, aquaculture and agriculture, and release of pharmaceutical manufacturing, domestic and agricultural waste into the environment (Wright, 2007; West et al., 2010; Yang et al., 2013; Michael et al., 2014). Worryingly, the issue of antimicrobial resistance leads to great healthcare, social and economical burdens worldwide, thus considered a quintessential One Health issue (Michael et al., 2014; Ewbank et al., 2021). Escherichia coli (order Enterobacterales) has been broadly suggested and used as a marker of environmental contamination and anthropogenic activity (Bonnedahl et al., 2009; Tenaillon et al., 2010). Extended-spectrum-ß-lactamase (ESBL)- and plasmid-encoded AmpC (pAmpC)-producing E. coli are classified as critical priority pathogens within the One health interface by the World Health Organization (WHO) (Tacconelli et al., 2018; Mughini-Gras et al., 2019).
Seabirds have been used as environmental bioindicators of ESBL/pAmpC-positive E. coli in remote locations due to their potential as sentinels of natural and anthropogenic-related changes to the marine ecosystem health (Hernandez et al., 2010; Hernández and González-Acuña, 2016; Ewbank et al., 2022). Given that clinically-relevant antimicrobial resistance genes are considered environmental pollutants and markers of environmental anthropization (Pruden et al., 2006; Jobbins and Alexander, 2015), most ESBL/pAmpC-producing E. coli studies have focused in synanthropic seabird species inhabiting anthropized environments (e.g., urban areas and dumpsites) (Bonnedahl et al., 2009; Atterby et al., 2016; Ahlstrom et al., 2018). Yet, insular biomes not inhabited by humans represent an informative setting in the study of the One Health chain of antimicrobial resistance by providing valuable insight into: (i) the occurrence, diversity, and dissemination of antimicrobial resistance genes (ARGs) and antimicrobial-resistant bacteria (ARB), such as ESBL/pAmpC-producing E. coli; (ii) the indirect anthropogenic effects over the environment (e.g., marine pollution); and (iii) the potential influence of biological and ecological characteristics of their local avian fauna (e.g., migration, use of coastal areas) (Hernandez et al., 2010; Ewbank et al., 2021, 2022).
Herein we analyzed cloacal swabs of 20 wild magnificent frigatebirds (Fregata magnificens; family Fregatidae) from an uninhabited archipelago located in southeastern Brazil, using microbiological techniques and whole genome sequencing (WGS) to investigate the occurrence, and phenotypic and genotypic characteristics of ESBL- and pAmpC-producing E. coli classified by WHO as critical priority pathogens (Tacconelli et al., 2018), and further identify and characterize their bacterial lineages, serotypes, resistome, plasmidome and virulome.
Materials and methods
Study area
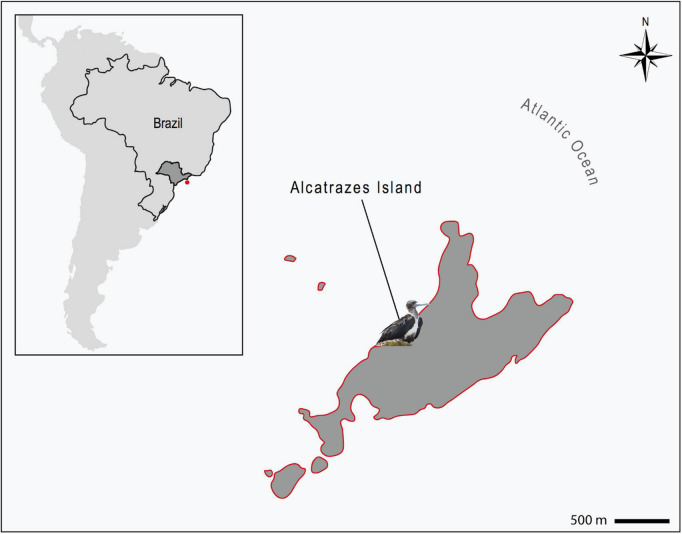
The Alcatrazes Island is the principal, among the five islands and four islets forming the Alcatrazes Archipelago (24° 05′ 44.69′′ S 45° 41′ 52.92′′ W), located at 36 km off the coast of São Sebastião, in São Paulo state, southeastern Brazil (Figure 1). The archipelago, including the Alcatrazes Island, have no records of onshore human occupation or tourist visitation, the latter limited to the offshore territory. In 1979, the Brazilian Navy started using the northeastern face of Alcatrazes Island as target for artillery practice. Later on, in 1987, the Tupinambás Ecological Station (Esec Tupinambás) was created, partially including the archipelago, and restricting visitation even more. In 2013, the Brazilian Navy moved its training grounds to a smaller island of Alcatrazes. Finally, in 2016, the archipelago and adjacent marine area (approximately 273 km2) were declared a conservation area - the Alcatrazes Archipelago Wildlife Refuge (Refuìgio de Vida Silvestre do Arquipeìlago de Alcatrazes - Refuìgio de Alcatrazes), focused specifically on the conservation of its local wildlife and flora, administered by the Chico Mendes Institute for Biodiversity Conservation (ICMBio), Brazilian Ministry of Environment (Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio), 2017). This study was performed in full compliance with the Biodiversity Information and Authorization System (SISBIO 59150-4), Brazilian Ministry of Environment and the Ethical Committee in Animal Research of the School of Veterinary Medicine and Animal Sciences, University of São Paulo (Process number 1753110716).
FIGURE 1.
Location of the Alcatrazes Archipelago (red dot), territory of São Paulo state (dark gray), southeastern Brazil (inserted map). Magnificent frigatebird (Fregata magnificens) sampled in Alcatrazes island, part of the Alcatrazes Archipelago (lined in red). Scale: 500 m.
Sampling and bacterial identification
Twenty magnificent frigatebirds (17 adults and 3 juveniles) were sampled in the main island (Alcatrazes Island), in January 2020. The evaluated birds comprised nine males, eight females and three individuals of undetermined sex. All birds were captured with a butterfly net, manually restrained and immediately released after sample collection. The cloacal swabs were maintained in Amies transport medium containing charcoal and maintained at room temperature until processed (within 7 days). In order to select ESBL- and pAmpC-producing E. coli strains, cloacal samples were streaked onto ceftriaxone (CRO, 2 mg/L)-supplemented MacConkey agar plates) and incubated overnight at 35 ± 2°C. Bacterial isolates were identified by Matrix Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS, Bruker Daltonik, Germany).
Antimicrobial susceptibility testing
Antimicrobial susceptibility was evaluated by the disc diffusion method using the following human and veterinary antimicrobials (Clinical and Laboratory Standards Institute [CLSI], 2018, 2019): amoxicillin/clavulanate, ceftriaxone, cefotaxime, ceftiofur, ceftazidime, cefepime, cefoxitin, imipenem, meropenem, ertapenem, enrofloxacin, ciprofloxacin, gentamicin, amikacin, chloramphenicol, trimethoprim-sulfamethoxazole, and tetracycline. The double-disc synergy test (DDST) was used for ESBL screening (EUCAST, 2017).
Whole genome sequence analysis
The genomic DNA of the ESBL/pAmpC-positive E. coli strain was extracted using a PureLinkTM Quick Gel Extraction Kit (Life Technologies, Carlsbad, CA, United States) and a genomic paired-end library (75 × 2 bp), prepared using a Nextera XT DNA Library Preparation Kit (Illumina Inc., Cambridge, United Kingdom), according to the manufacturer’s instructions. The whole genome was sequenced on the NextSeq platform (Illumina). De novo genome assembly was performed with CLC Genomics Workbench 12.0.3. The draft genome sequence was automatically annotated using the NCBI Prokaryotic Genome Annotation Pipeline v.3.2. The MLST 2.0, PlasmidFinder 2.0, ResFinder 4.1, VirulenceFinder 2.0 and SerotypeFinder 2.0 databases available at the Centre for Genomic Epidemiology1 were used to identify, respectively, the multilocus sequence type (MLST), plasmid replicons, resistome, virulome and serotype. A prediction filter of ≥98 and 100% were set for sequence identity and coverage thresholds, respectively. Additionally, phylogroup analysis was performed using the ClermonTyping database2. The nucleotide sequence data reported is available in the DDBJ/EMBL/GenBank databases under accession number NZ_JAGYFD010000000. The E. coli AA18 strain genomic information is available on the OneBR platform under ID number ONE1193.
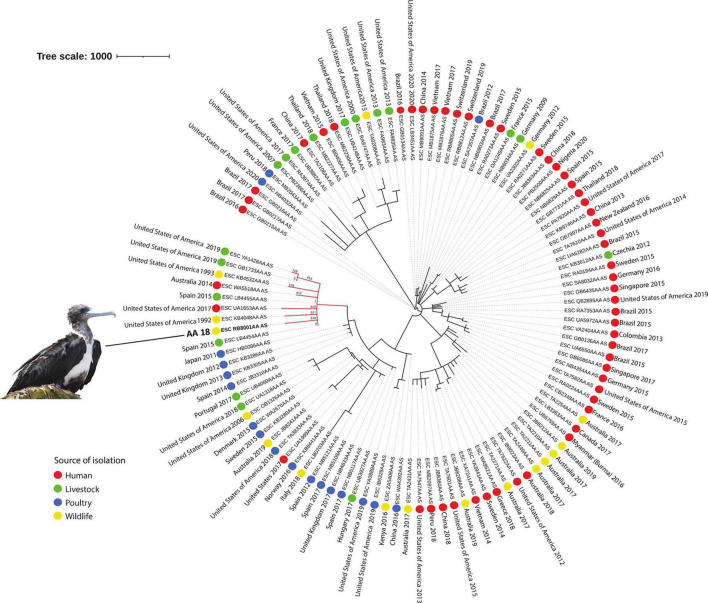
The MSTree V2 tool from Enterobase4 was used to generate a minimum spanning tree based on the wgMLST scheme and 25,002 loci considering our E. coli isolate and an international collection of 107 E. coli strains belonging to ST648, selected according to source of isolation (colored circles), and country and year of isolation (Figure 2). iTOL v.65 was used to edit and visualize the phylogenetic tree. An interactive version of the tree is available at https://itol.embl.de/tree/1791137681100671617229508.
FIGURE 2.
Phylogeny of CTX-M-2 and CMY-2-producing ST648 E. coli isolated from a magnificent frigatebird (Fregata magnificens) and global E. coli ST648. Each genome is shown in accordance to the source of isolation (colored circles), and country and year of isolation. In the red branch are the genomes that were phylogenetically closer to sample AA18. Tree scale: 1000.
Results
Overall, we found an ESBL/pAmpC-producing E. coli prevalence of 5% (1/20) in the evaluated individuals. Phenotypically, the E. coli isolate (designated AA18 strain) presented a multidrug resistant (MDR) profile to amoxicillin/clavulanic acid, ceftiofur, cefoxitin, cefepime, aztreonam, trimethoprim-sulfamethoxazole, gentamicin, and tetracycline; remaining susceptible to carbapenems ertapenem, imipenem and meropenem (Clinical and Laboratory Standards Institute [CLSI], 2018, 2019). Regarding to genomic data, trimmed paired-end reads were assembled into 137 contigs, with 425,81 x coverage, and a G + C content of 49% (Andrews, 2010). Briefly, strain AA18 presented a genome size calculated as 5.4 million base pairs (bp), with 5,145 protein-coding sequences, 87 pseudogenes, 83 tRNAs, 3 rRNAs and 10 non-coding RNAs genomic analysis revealed that the isolate harbored genes blaCTX–M–2, blaCMY–2, qnrB, tetB, sul1, sul2, aadA1, aac(3)-VIa and mdfA in its resistome (Table 1).
TABLE 1.
Genomic and epidemiological data of E. coli strain AA18.
| Characteristics | E. coli strain AA18 |
| Source | Cloacal swab |
| Genome size (Mbp) | 5,4 |
| No. of CDSa | 5,145 |
| G + C content (%) | 57,25 |
| tRNA (n) | 83 |
| rRNA (n) | 3 |
| Non-coding RNA (n) | 10 |
| Pseudogenes | 87 |
| CRISPR | 2 |
| MLST (ST)b | 648 |
| Serotype | O153:H9 |
| Resistome | |
| β-lactams | blaCXT–M–2, blaCMY–2 |
| Aminoglycosides | aa(3)-VIa, aadA1 |
| Fluoroquinolones | qnrB |
| Sulphonamides | sul1, sul2 |
| Tetracyclines | tetB |
| Macrolides | mdfA |
| Heavy metal | bhsA, cusF, cutA, dsbAB, fetAB, fieF, glpF, mntPR, modE, nfsA, phnE, pitA, rcnR, robA, sitBCD, tehB, sodAB, ychH, yieF, yodD, zinT, znuA, zur |
| Biocides | cba, chuA, cma, cvaC, eilA, etsC, gad, hlyF, hra, ireA, iroN, iss, iucC, iutA, kpsE, kpsMII, lpfA, mchF, ompT, papC, sitA, terC, traT, tsh, yfcV |
| Virulome | acrE, cpxA, mdtEF, tehB, sugE, ydeOP |
| Plasmidome | Col, IncFIB, IncFII |
| OneBR ID | ONE119 |
| GenBank accession number | NZ_JAGYFD010000000 |
aCDSs, coding sequences. bMLST, multilocus sequence type; ST, sequence type.
Multilocus sequence typing (MLST) and serotype analyses revealed that the isolate corresponded to ST648 and belonged to the O153:H9 group, respectively. Our isolate presented several relevant virulence genes characteristic of avian pathogenic (APEC) and other extraintestinal pathogenic E. coli (ExPEC), such as chuA (outer membrane hemin receptor), kpsMII (group 2 capsule synthesis), fimC (fimbriae type I), sitA (iron transport protein), and traT (transfer protein). Additionally, WGS analysis also identified genes encoding resistance to disinfectants (i.e., acridines, chlorhexidine, crystal violet, ethidium bromide, quaternary ammonium compounds, sodium dodecyl sulphate), heavy metals (i.e., lead, arsenic, copper, silver, antimony, zinc, tellurium, tungsten, cobalt, nickel, manganese, cadmium, mercury, iron, molybdenum, chromium, and vanadium), acid or basidic environment (i.e., H2O2, HCl and NaOH), and pesticide (glyphosate). The resistome, plasmidome and virulome are listed in Table 1.
Upon phylogenetic analysis, strain AA18 clustered with genomes from E. coli strains recovered from humans (Australia), livestock (Spain and United States), poultry (United States of America), and European herring gulls (Larus argentatus; United States of America), with a high variability ranging from 2 to 628 SNPs (Figure 2).
Discussion
Herein, we found an overall prevalence of 5% (1/20) of ESBL/AmpC-positive E. coli isolates in magnificent frigatebirds of Alcatrazes Archipelago, southeastern Brazilian coast: a highly virulent MDR avian pathogenic E. coli (APEC) isolate of the pandemic high-risk ST648 clone (serotype O153:H9) harboring genes blaCTX–M–2 and blaCMY–2. To the authors’ knowledge, this is the first report of the ST648 clone and pAmp-EC in wild birds inhabiting insular environments.
The CTX-M-2 and CMY-2 enzymes are, respectively, the most prevalent CTX-M ESBL in South America and pAmpC beta-lactamase worldwide (Jacoby, 2009; Rocha et al., 2015), reported in a variety of epidemiological settings in Brazil (Rocha et al., 2015; Cunha et al., 2017; Melo et al., 2018; de Carvalho et al., 2020; Fernandes et al., 2020b). Among wild birds, blaCTX–M–2 and blaCMY–2 genes have been described in bacterial pathogens colonizing gulls, corvids and Eurasian magpie (Pica pica) in Europe (Loncaric et al., 2013; Stedt et al., 2015; Alcalá et al., 2016; Jamborova et al., 2017; Athanasakopoulou et al., 2021), and in gulls and bald eagles (Haliaeetus leucocephalus) from the Americas (Poirel et al., 2012; Báez et al., 2015; Atterby et al., 2016; Liakopoulos et al., 2016; Ahlstrom et al., 2018). In Brazil, blaCTX–M–2-positive bacteria have been detected in wild birds of prey and parrots, and in Magellanic penguin (Spheniscus magellanicus); whereas blaCMY–2-positive bacteria have been described in birds of prey (Sellera et al., 2017; Batalha de Jesus et al., 2019; de Carvalho et al., 2020).
The international clone ST648 is predominantly MDR and virulent, and one of the most commonly reported international sequence types (STs) in the human–animal–environmental interface worldwide, suggesting great host adaptation (Hu et al., 2013; Fernandes et al., 2018; de Carvalho et al., 2020). Of note, ST648 has been detected in wild birds from almost all continents, including Europe (Guenther et al., 2010; Schaufler et al., 2019), the Americas (Poirel et al., 2012; Báez et al., 2015), Asia (Hasan et al., 2012; Yang et al., 2016), and Oceania (Mukerji et al., 2019). In South America, this clone has been described in wild birds of prey in Brazil (Batalha de Jesus et al., 2019; de Carvalho et al., 2020) and gulls in Chile (Báez et al., 2015).
Strain AA18 carried several virulence genes of concern characteristic of highly pathogenic avian pathogenic E. coli (APEC) isolates: cvaC (colicin V), fimC (fimbriae type I), hlyF (hemolysin F), iroN (salmochelin), iss (increased serum survival), iucC (aerobactin production) iutA (ferric aerobactin receptor), ompT (outer membrane protein), sitA (iron transport protein), tsh (temperature-sensitive hemagglutinin) and traT (transfer protein) (Ewers et al., 2007; Sarowska et al., 2019). Additionally, we also found virulence genes characteristic of the other ExPEC: chuA (outer membrane hemin receptor), kpsMII, papC (outer membrane usher protein), and yfcV (major subunit of a putative chaperone-usher fimbria) (Grimwood et al., 2000; Kim, 2002; Sarowska et al., 2019). APEC strains may cause colibacillosis – multiple systemic and localized avian infection that may lead to high mortality and decreased production, capable of imposing severe economic losses to the poultry industry worldwide (Kemmett et al., 2014). Of note, some of the virulence factors found in our isolate were previously reported in ExPEC sampled from magnificent frigatebirds from the Alcatrazes Archipelago: cvaC, fimH, hlyF, iroN, iss, iutA, ompT and papC (Saviolli et al., 2016). Although APEC and other ExPEC strains are phylogenetically close, sharing some of the same virulence genes, APEC may carry other genes not common in other ExPEC isolates, such as those present in the colicin V (ColV) plasmid (Rodriguez-Siek et al., 2005; Bélanger et al., 2011). These characteristics suggest that APEC strains are potentially zoonotic, and could be a reservoir and source of virulence genes for other ExPEC strains (Ewers et al., 2007; Bélanger et al., 2011). In humans, APEC infections could take place through consumption of undercooked food from animal origin (especially retail poultry products), and direct contact with birds and their feces (Dziva and Stevens, 2008). Yet, despite the hypothetical zoonotic and pathogenic potential of our isolate (Ewers et al., 2014; Maluta et al., 2014; Sarowska et al., 2019), our findings must be carefully interpreted in light of the low prevalence of ESBL/AmpC-positive E. coli found herein (5%; 1/20) and the apparently healthy condition (with no signs of disease) presented by frigatebirds in Alcatrazes (also described by Saviolli et al., 2016). Furthermore, our strain also harbored genes encoding resistance to heavy metals, QACs and pesticides (Table 1), which may promote the development of AMR and co-selection of ARGs (Zou et al., 2014; Ramakrishnan et al., 2019; Mazhar et al., 2021).
Anthropization has been suggested as a driving factor in the epidemiology of ARGs in wildlife (Ahlstrom et al., 2018; Sacristaìn et al., 2020; Ewbank et al., 2021). Although occasionally visited or exploited for commercial guano harvesting until the mid-20th century, to this date, there are no reports of human occupation or settlements in the archipelago (Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio), 2017). Nevertheless, Alcatrazes is located in the highly anthropized southeastern Brazilian coast, subjected to intense tourism activities, fishing, and oil exploitation, that also harbors the largest port complex (Santos Port), and oil and derivatives terminal in Latin America (Almirante Barroso Maritime Terminal - TEBAR). Of note, recent studies assessing antimicrobial resistance pollution in the marine ecosystem of the southeastern Brazilian coast showed that the local resistome is indeed under severe anthropogenic pressure (Fernandes et al., 2017, 2020a,b; Sellera et al., 2018a,b).
The Alcatrazes Archipelago is the largest insular bird breeding site of the southeastern Brazilian coast and the biggest breeding colony of magnificent frigatebirds in the southern Atlantic (Alcatrazes Island) (Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio), 2017). Magnificent frigatebirds are non-synanthropic, non-migratory, and highly colonial seabird species that prefer insular over coastal environments, and also known for their particular feeding techniques (e.g., kleptoparasitism and fisheries interaction) (Saviolli et al., 2016; BirdLife International, 2021). Such characteristics infer that the studied Alcatrazes individuals most likely sustain very limited to no direct contact with humans, but that due to their philopatric (site fidelity) behavior, and limited roosting and nesting area of the island, are continuously interacting with the other frigatebird specimens and bird species using the area (especially with brown boobies (Sula leucogaster) and black vultures (Coragyps atratus), A.C. Ewbank, personal observation). Consequently, such close contact and active exchange of body fluids may be a possible route of infection by ESBL/pAmpC-positive E. coli, as seen in other avian pathogens (de Thoisy et al., 2009; Niemeyer et al., 2017).
Thus, in light of the above, we suggest that our isolate was likely acquired through one or more of the following: (i) indirect colonization by a bacterium released from human sources into the local marine environment (e.g., sewage) (Fernandes et al., 2017, 2018, 2020a,b); (ii) ingestion of contaminated seafood (Brahmi et al., 2015; Sellera et al., 2018a,b); and (iii) direct intra and/or interspecies contact (Ewbank et al., 2022).
Interestingly, according to the phylogenetic results, our isolate was not closely related to the selected ST648 isolates from other geographical regions or ecological sources included in the analysis. This indicates that, even though the origin of our isolate was likely related to human sources, this phylogenetic cluster seems to be restricted to the specific coastal/insular geographical area of Alcatrazes, southeastern Brazil. Nevertheless, additional studies in the region are necessary in order to confirm this hypothesis.
Previous studies have discussed the hypothetical potential of wild birds as reservoirs and disseminators of ARGs and ARB to insular biomes (Hernandez et al., 2010; Ewbank et al., 2021, 2022). Nevertheless, in spite of experimental studies assessing the shedding, contamination and potential transmission of ARGs and ARB by wild birds (Sandegren et al., 2018; Franklin et al., 2020), their potential role as dispersers under real-world conditions is still unknown. Our findings demonstrate that even in the absence of regular human presence, insular resistomes are indirectly pressured by anthropogenic activities, suggesting that contamination of the marine ecosystem and inter and/or intraspecific bird interactions should also be considered in the study of antimicrobial resistance in these biomes.
Herein we reported the genomic background of a critical priority E. coli strain belonging to the pandemic high-risk clone ST648 E.coli with a hypothetical zoonotic and avian pathogenic potential colonizing a wild magnificent frigatebird of an insular biome. Our findings reinforce, within a One Health perspective, the importance of surveillance studies of WHO critical priority pathogens in wildlife and the role of wild birds as anthropization sentinels in insular environments. Future studies evaluating the occurrence and diversity of ESBL/pAmpC-positive E. coli in magnificent frigatebirds on the Alcatrazes Archipelago should rely on continuous temporal sampling to assess a larger number of specimens, evaluate interacting species (i.e., brown boobies and black vultures), and environmental samples (i.e., sea water and soil), including local marine life (i.e., fish), in order to monitor these populations through a One Health approach and further elucidate the epidemiology of ESBL/pAmpC-positive E. coli in this insular environment.
Data availability statement
The data presented in this study are deposited in the DDBJ/EMBL/GenBank repository, accession number NZ_JAGYFD010000000. The sequence has been released and is available at the repository: https://www.ncbi.nlm.nih.gov/nuccore/NZ_JAGYFD000000000.1.
Ethics statement
This study was performed in full compliance with the Biodiversity Information and Authorization System (SISBIO 59150-4), Brazilian Ministry of Environment and the Ethical Committee in Animal Research of the School of Veterinary Medicine and Animal Sciences, University of São Paulo (Process no. 1753110716).
Author contributions
AE: conceptualization, methodology, investigation, writing original draft, supervision, project administration, and funding acquisition. DF-C: methodology, investigation, writing original draft, and supervision. CS: conceptualization, methodology, investigation, funding acquisition, and writing original draft. FE: methodology, formal analysis, and writing original draft. BF and BC: methodology, and formal analysis. SG: conceptualization, investigation, resources, and funding acquisition. RZ and MG: investigation and resources. JC-D and NL: conceptualization, methodology, resources, writing–review and editing, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank the staff of the Refúgio de Vida Silvestre do Arquipélago de Alcatrazes – Institute Chico Mendes for Biodiversity Conservation (ICMBio), José F. Delgado-Blas, Marcos Paulo Vieira Cunha and Terezinha Knöbl for the logistical and scientific support. We also thank Chris H. Gardiner, for contributing in the review of this manuscript.
Footnotes
Funding
This study was supported by the Bill & Melinda Gates Foundation (Grand Challenges Explorations Brazil OPP1193112). Under the grant conditions of the Foundation, a CC BY or equivalent license is applied to the Author Accepted Manuscript version arising from this submission. Additionally, this study was financed by the Coordination for the Improvement of Higher Education Personnel (CAPES), National Council for Scientific and Technological Development (CNPq) (AMR 443819/2018-1, 433128/2018-6), and the São Paulo State Research Foundation (FAPESP) (2020/08224-9). AE and FE receive doctoral fellowships from FAPESP (process no. 2018/20956-0 and 2019/15578-4, respectively), BC from CAPES (88882.333054/2019-01), and RZ from CNPq (process no. 165364/2018-1). BF was a recipient of a post-doctoral fellowship from CAPES (88887.358057/2019-00). CS was the recipient of a Juan the la Cierva Incorporation Fellowship IJC2020-046019-I. JC-D was the recipient of a professorship from CNPq (304999-18) and NL was a research fellow of CNPq (314336/2021-4)].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Ahlstrom C. A., Bonnedahl J., Woksepp H., Hernandez J., Olsen B., Ramey A. M. (2018). Acquisition and dissemination of cephalosporin-resistant E. coli in migratory birds sampled at an Alaska landfill as inferred through genomic analysis. Sci. Rep. 8:7361. 10.1038/s41598-018-25474-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalá L., Alonso C. A., Simón C., González-Esteban C., Orós J., Rezusta A., et al. (2016). Wild birds, frequent carriers of extended-spectrum β-lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 Types. Microb. Ecol. 72 861–869. 10.1007/s00248-015-0718-0 [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: https://github.com/s-andrews/FastQC (accessed on June 10, 2010). [Google Scholar]
- Athanasakopoulou Z., Tsilipounidaki K., Sofia M., Chatzopoulos D. C., Giannakopoulos A., Karakousis I., et al. (2021). Poultry and wild birds as a reservoir of CMY-2-producing Escherichia coli: the first large-scale study in Greece. J. Antibiot. 10:235. 10.3390/antibiotics10030235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atterby C., Ramey A. M., Hall G. G., Jarhult J., Borjesson S., Bonnedahl J. (2016). Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect. Ecol. Epidemiol. 6:32334. 10.3402/iee.v6.32334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez J., Hernández-Gracía M., Guamparito C., Díaz S., Olave A., Guerrero K., et al. (2015). Molecular characterization and genetic diversity of ESBL-producing Escherichia coli colonizing the migratory Franklin’s gulls (Leucophaeus pipixcan) in Antofagasta, North of Chile. Microb. Drug Resist. 21 111–116. 10.1089/mdr.2014.0158 [DOI] [PubMed] [Google Scholar]
- Batalha de Jesus A. A., Freitas A. A. R., de Souza J. C., Martins N., Botelho L. A. B., Girão V. B. C., et al. (2019). High-level multidrug-resistant Escherichia coli isolates from wild birds in a large urban environment. Microb. Drug Resist. 25 167–172. 10.1089/mdr.2018.0180 [DOI] [PubMed] [Google Scholar]
- Bélanger L., Garenaux A., Harel J., Boulianne M., Nadeau E., Dozois C. M. (2011). Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 62 1–10. 10.1111/j.1574-695X.2011.00797.x [DOI] [PubMed] [Google Scholar]
- BirdLife International (2021). Species factsheet: Fregata magnificens. Available online at: http://datazone.birdlife.org/species/factsheet/magnificent-frigatebird-fregata-magnificens (accessed on May 9, 2021). [Google Scholar]
- Bonnedahl J., Drobni M., Gauthier-Clerc M., Hernandez J., Granholm S., Kayser Y., et al. (2009). Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One 4:e5958. 10.1371/journal.pone.0005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmi S., Dunyach-Rémy C., Touati A., Lavigne J.-P. (2015). CTX-M-15-producing Escherichia coli and the pandemic clone O25b-ST131 isolated from wild fish in Mediterranean Sea. Clin. Microbiol. Infect. 21 e18–e20. 10.1016/j.cmi.2014.09.019 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2018). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. CLSI supplement VET08. Philadelphia: CLSI. [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2019). Performance Standards for Antimicrobial Susceptibility Testing CLSI supplement M100. Philadelphia: CLSI. [Google Scholar]
- Cunha M. P., Lincopan N., Cerdeira L., Esposito F., Dropa M., Franco L. S., et al. (2017). Coexistence of CTX-M-2, CTX-M-55, CMY-2, FosA3, and QnrB19 in extraintestinal pathogenic Escherichia coli from poultry in Brazil. Antimicrob Agents Chemother 61:e02474-16. 10.1128/AAC.02474-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho M. P. N., Fernandes M. R., Sellera F. P., Lopes R., Monte D. F., Hippólito A. G., et al. (2020). International clones of extended-spectrum β-lactamase (CTX-M)-producing Escherichia coli in peri-urban wild animals, Brazil. Transbound Emerg. Dis. 67 1804–1815. 10.1111/tbed.13558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Thoisy B., Lavergne A., Semelin J., Pouliquen J. F., Blanchard F., Hansen E., et al. (2009). Outbreaks of disease possibly due to a natural avian herpesvirus infection in a colony of young Magnificent Frigatebirds (Fregata magnificens) in French Guiana. J. Wildl. Dis. 45 802–807. 10.7589/0090-3558-45.3.802 [DOI] [PubMed] [Google Scholar]
- Dziva F., Stevens M. P. (2008). Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 37 355–366. 10.1080/03079450802216652 [DOI] [PubMed] [Google Scholar]
- EUCAST (2017). Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Sweden: European Comittee of Antimicrobial Susceptibility Testing. [Google Scholar]
- Ewbank A. C., Esperón F., Sacristán C., Sacristán I., Krul R., Cavalcante de Macedo E., et al. (2021). Seabirds as anthropization indicators in two different tropical biotopes: a One Health approach to the issue of antimicrobial resistance genes pollution in oceanic islands. Sci. Total Environ. 754:142141. 10.1016/j.scitotenv.2020.142141 [DOI] [PubMed] [Google Scholar]
- Ewbank A. C., Fuentes-Castillo D., Sacristán C., Cardoso B., Esposito F., Fuga B., et al. (2022). Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli survey in wild seabirds at a pristine atoll in the southern Atlantic Ocean, Brazil: first report of the O25b-ST131 clone harboring blaCTX–M–8. Sci. Total Environ. 806:150539. 10.1016/j.scitotenv.2021.150539 [DOI] [PubMed] [Google Scholar]
- Ewers C., Bethe A., Stamm I., Grobbel M., Kopp P. A., Guerra B., et al. (2014). CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrobial. Chemother. 69 1224–1230. 10.1093/jac/dkt516 [DOI] [PubMed] [Google Scholar]
- Ewers C., Li G., Wilking H., Kiessling S., Alt K., Antáo E. M., et al. (2007). Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297 163–176. 10.1016/j.ijmm.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Fernandes M. R., Sellera F. P., Moura Q., Esposito F., Sabino C. P., Lincopan N. (2020b). Identification and genomic features of halotolerant extended-spectrum-β-lactamase (CTX-M)-producing Escherichia coli in urban-impacted coastal waters, Southeast Brazil. Mar. Pollut. Bull. 150:110689. 10.1016/j.marpolbul.2019.110689 [DOI] [PubMed] [Google Scholar]
- Fernandes M. R., Sellera F. P., Cunha M. P. V., Lopes R., Cerdeira L., Lincopan N. (2020a). Emergence of CTX-M-27-producing Escherichia coli of ST131 and clade C1-M27 in an impacted ecosystem with international maritime traffic in South America. J. Antimicrob. Chemother. 75 1647–1649. 10.1093/jac/dkaa069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M. R., Sellera F. P., Esposito F., Sabino C. P., Cerdeira L., Lincopan N. (2017). Colistin-resistant mcr-1-positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob. Agents Chemother. 61:e00234-17. 10.1128/AAC.00234-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M. R., Sellera F. P., Moura Q., Gaspar V. C., Cerdeira L., Lincopan N. (2018). International high-risk clonal lineages of CTX-M-producing Escherichia coli F-ST648 in free-roaming cats, South America. Infect. Genet. Evol. 66 48–51. 10.1016/j.meegid.2018.09.009 [DOI] [PubMed] [Google Scholar]
- Franklin A. B., Ramey A. M., Bentler K. T., Barrett N. L., McCurdy L. M., Ahlstrom C. A., et al. (2020). Gulls as sources of environmental contamination by colistin-resistant bacteria. Sci. Rep. 10:4408. 10.1038/s41598-020-61318-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood K., Anderson P., Anderson V., Tan L., Nolan T. (2000). Twelve year outcomes following bacterial meningitis: further evidence for persisting effects. Arch. Dis. Child 83 111–116. 10.1136/adc.83.2.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther S., Grobbel M., Beutlich J., Bethe A., Friedrich N. D., Goedecke A., et al. (2010). CTX-M-15-type extended-spectrum beta-lactamases-producing Escherichia coli from wild birds in Germany. Environ. Microbiol. Rep. 2 641–645. 10.1111/j.1758-2229.2010.00148.x [DOI] [PubMed] [Google Scholar]
- Hasan B., Sandegren L., Melhus A., Drobni M., Hernandez J., Waldenström J., et al. (2012). Antimicrobial drug-resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg. Infect. Dis. 18 2055–2058. 10.3201/eid1812.120513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández J., González-Acuña D. (2016). Anthropogenic antibiotic resistance genes mobilization to the polar regions. Infect. Ecol. Epidemiol. 6:32112. 10.3402/iee.v6.32112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J., Bonnedahl J., Eliasson I., Wallensten A., Comstedt P., Johansson A., et al. (2010). Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX–M–15, found in Glaucous-winged gull at remote Commander Islands, Russia. Environ. Microbiol. Rep. 2 329–332. 10.1111/j.1758-2229.2010.00142.x [DOI] [PubMed] [Google Scholar]
- Hu Y. Y., Cai J. C., Zhou H. W., Chi D., Zhang X. F., Chen W. L., et al. (2013). Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 79 5988–5996. 10.1128/AEM.01740-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) (2017). Plano de Manejo da Estação Ecológica Tupinambás e Refúgio de Vida Silvestre do Arquipélago de Alcatrazes. Brasília, DF: Instituto Chico Mendes de Conservação da Biodiversidade. [Google Scholar]
- Jacoby G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamborova I., Dolejska M., Zurek L., Townsend A. K., Clark A. B., Ellis J. C., et al. (2017). Plasmid-mediated resistance to cephalosporins and quinolones in Escherichia coli from American crows in the USA. Environ. Microbiol. 19 2025–2036. 10.1111/1462-2920.13722 [DOI] [PubMed] [Google Scholar]
- Jobbins S. E., Alexander K. A. (2015). From whence they came: antibiotic-resistant Escherichia coli in African wildlife. J. Wildl. Dis. 51 811–820. 10.7589/2014-11-257 [DOI] [PubMed] [Google Scholar]
- Kemmett K., Williams N. J., Chaloner G., Humphrey S., Wigley P., Humphrey T. (2014). The contribution of systemic Escherichia coli infection to the early mortalities of commercial broiler chickens. Avian Pathol. 43 37–42. 10.1080/03079457.2013.866213 [DOI] [PubMed] [Google Scholar]
- Kim K. S. (2002). Strategy of Escherichia coli for crossing the blood-brain barrier. J. Infect. Dis. 1 S220–S224. 10.1086/344284 [DOI] [PubMed] [Google Scholar]
- Liakopoulos A., Olsen B., Geurts Y., Artursson K., Berg C., Mevius D. J., et al. (2016). Molecular characterization of extended-spectrum-cephalosporin-resistant Enterobacteriaceae from wild kelp gulls in South America. Antimicrob. Agents Chemother. 60 6924–6927. 10.1128/AAC.01120-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncaric I., Stalder G. L., Mehinagic K., Rosengarten R., Hoelzl F., Knauer F., et al. (2013). Comparison of ESBL and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS One 8:e84048. 10.1371/journal.pone.0084048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluta R. P., Logue C. M., Casas M. R., Meng T., Guastalli E. A., Rojas T. C., et al. (2014). Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One 9:e105016. 10.1371/journal.pone.0105016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhar S. H., Li X., Rashid A., Su J., Xu J., Brejnrod A. D., et al. (2021). Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 755:142702. 10.1016/j.scitotenv.2020.142702 [DOI] [PubMed] [Google Scholar]
- Melo L. C., Oresco C., Leigue L., Netto H. M., Melville P. A., Benites N. R., et al. (2018). Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet. Microbiol. 221 59–66. 10.1016/j.vetmic.2018.05.017 [DOI] [PubMed] [Google Scholar]
- Michael C. A., Dominey-Howes D., Labbate M. (2014). The antimicrobial resistance crisis: causes, consequences, and management. Front. Public Health 2:145. 10.3389/fpubh.2014.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughini-Gras L., Dorado-García A., van Duijkeren E., van den Bunt G., Dierikx C. M., Bonten M. J. M., et al. (2019). ESBL Attribution Consortium. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet Health 3 e357–e369. 10.1016/S2542-5196(19)30130-5 [DOI] [PubMed] [Google Scholar]
- Mukerji S., Stegger M., Truswell A. V., Laird T., Jordan D., Abraham R. J., et al. (2019). Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J. Antimicrob. Chemother. 74 2566–2574. 10.1093/jac/dkz242 [DOI] [PubMed] [Google Scholar]
- Niemeyer C., Favero C. M., Shivaprasad H. L., Uhart M., Musso C. M., Rago M. V., et al. (2017). Genetically diverse herpesviruses in South American Atlantic coast seabirds. PLoS One 12:e0178811. 10.1371/journal.pone.0178811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Potron A., de La Cuesta C., Cleary T., Nordmann P., Munoz-Price L. S. (2012). Wild coastline birds as reservoirs of broad-spectrum-β-lactamase-producing Enterobacteriaceae in Miami Beach, Florida. Antimicrob. Agents Chemother. 56 2756–2758. 10.1128/AAC.05982-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruden A., Pei R., Stortebomm H., Carlson K. H. (2006). Antibiotic resistance genes as emerging contaminants: studies in Northern Colorado. Environ. Sci. Technol. 40 7445–7450. 10.1021/es060413l [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B., Venkateswarlu K., Sethunathan N., Megharaj M. (2019). Local applications but global implications: can pesticides drive microorganisms to develop antimicrobial resistance? Sci. Total Environ. 654 177–189. 10.1016/j.scitotenv.2018.11.041 [DOI] [PubMed] [Google Scholar]
- Rocha F. R., Pinto V. P. T., Barbosa F. C. B. (2015). The spread of CTX-M-type extended-spectrum β-lactamases in Brazil: a systematic review. Microb. Drug Resist. 22 301–311. 10.1089/mdr.2015.0180 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Siek K. E., Giddings C. W., Doetkott C., Johnson T. J., Fakhr M. K., Nolan L. K. (2005). Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151 2097–2110. 10.1099/mic.0.27499-0 [DOI] [PubMed] [Google Scholar]
- Sacristaìn I., Esperoìn F., Acuna F., Aguilar E., Garciìa S., Loìpez M. J., et al. (2020). Antibiotic resistance genes as landscape anthropization indicators: using a wild felid as sentinel in Chile. Sci. Total Environ. 703:134900. 10.1016/j.scitotenv.2019.134900 [DOI] [PubMed] [Google Scholar]
- Sandegren L., Stedt J., Lustig U., Bonnedahl J., Andersson D., I, Järhult J. D. (2018). Long-term carriage and rapid transmission of extended spectrum beta-lactamase-producing E. coli within a flock of Mallards in the absence of antibiotic selection. Environ. Microbiol. Rep. 10 576–582. 10.1111/1758-2229.12681 [DOI] [PubMed] [Google Scholar]
- Sarowska J., Futoma-Koloch B., Jama-Kmiecik A., Frej-Madrzak M., Ksiazczyk M., Bugla-Ploskonska G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 21:10. 10.1186/s13099-019-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviolli J. Y., Cunha M. P., Guerra M. F., Irino K., Catão-Dias J. L., de Carvalho V. M. (2016). Free-ranging frigates (Fregata magnificens) of the southeast coast of Brazil harbor extraintestinal pathogenic Escherichia coli resistant to antimicrobials. PLoS One 11:e0148624. 10.1371/journal.pone.0148624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufler K., Semmler T., Wieler L. H., Trott D. J., Pitout J., Peirano G., et al. (2019). Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob. Agents Chemother. 63 e243–e219. 10.1128/AAC.00243-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellera F. P., Fernandes M. R., Moura Q., Carvalho M. P. N., Lincopan N. (2018a). Extended-spectrum-β-lactamase (CTX-M)-producing Escherichia coli in wild fishes from a polluted area in the Atlantic Coast of South America. Mar. Pollut. Bull. 135 183–186. [DOI] [PubMed] [Google Scholar]
- Sellera F. P., Fernandes M. R., Moura Q., Lopes R. B., Souza T. A., Cerdeira L., et al. (2018b). Draft genome sequence of a blaCMY–2/IncI1-harbouring Escherichia coli D:ST457 isolated from coastal benthic organisms. J. Glob. Antimicrob. Resist. 14 83–84. 10.1016/j.jgar.2018.06.010 [DOI] [PubMed] [Google Scholar]
- Sellera F. P., Fernandes M. R., Sartori L., Carvalho M. P., Esposito F., Nascimento C. L., et al. (2017). Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX–M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J. Antimicrob. Chemother. 72 1255–1256. 10.1093/jac/dkw543 [DOI] [PubMed] [Google Scholar]
- Stedt J., Bonnedahl J., Hernandez J., Waldenström J., McMahon B. J., Tolf C., et al. (2015). Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet. Scand. 57:74. 10.1186/s13028-015-0166-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 18 318–327. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Tenaillon O., Skurnik D., Picard B., Denamur E. (2010). The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8 207–217. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- West B. M., Liggit P., Clemans D. L., Francoeur S. N. (2010). Antibiotic resistance, gene transfer, and water quality patterns observed in waterways near CAFO farms and wastewater treatment facilities. Water Air Soil Pollut. 217 473–489. 10.1007/s11270-010-0602-y [DOI] [Google Scholar]
- Wright G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5 175–186. 10.1038/nrmicro1614 [DOI] [PubMed] [Google Scholar]
- Yang J., Wang C., Shu C., Liu L., Geng J., Hu S., et al. (2013). Marine sediment bacteria harbor antibiotic resistance genes highly similar to those found in human pathogens. Microb. Ecol. 65 975–981. 10.1007/s00248-013-0187-2 [DOI] [PubMed] [Google Scholar]
- Yang R. S., Feng Y., Lv X. Y., Duan J. H., Chen J., Fang L. X., et al. (2016). Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 60 6899–6902. 10.1128/AAC.01365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Meng J., McDermott P. F., Wang F., Yang Q., Cao G., et al. (2014). Presence of disinfectant resistance genes in Escherichia coli isolated from retail meats in the USA. J. Antimicrob. Chemother. 69, 2644–2649. 10.1093/jac/dku197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are deposited in the DDBJ/EMBL/GenBank repository, accession number NZ_JAGYFD010000000. The sequence has been released and is available at the repository: https://www.ncbi.nlm.nih.gov/nuccore/NZ_JAGYFD000000000.1.