Abstract
Karst caves are oligotrophic environments that appear to support a high diversity of fungi. Studies of fungi in Thailand’s caves are limited. During a 2019 exploration of the mycobiota associated with soil samples from a karst cave, namely, Phu Pha Phet in the Satun UNESCO Global Geopark in Satun Province, southern Thailand, two previously undescribed fungi belonging to Talaromyces (Trichocomaceae, Eurotiales, Eurotiomycetes) were studied using a polyphasic approach combining phenotypic and molecular data. Based on datasets of four loci (ITS, BenA, CaM, and RPB2), phylogenetic trees of the section Trachyspermi were constructed, and two new species—Talaromyces phuphaphetensis sp. nov. and T. satunensis sp. nov.—phylogenetically related to T. subericola, T. resinae, and T. brasiliensis, are described. Detailed descriptions and illustrations of the new species are provided. This study increases the number of cave-dwelling soil fungi discovered in Thailand’s Satun UNESCO Global Geopark, which appears to be a unique environment with a high potential for discovering fungal species previously undescribed.
Keywords: section Trachyspermi, polyphasic taxonomy, Trichocomaceae, cave-dwelling soil micro-fungi, Phu Pha Phet karst cave
1. Introduction
The genus Talaromyces was introduced [1] with Talaromyces vermiculatus (=T. flavus) as the type of species. Talaromyces taxa are classified into Aspergillaceae, Eurotiales, Eurotiomycetidae, Eurotiomycetes, Pezizomycotina, and Ascomycota (MycoBank. 2022; Species Fungorum. 2022; accessed on 1 June 2022). This genus is well-known and among the most prevalent groups of fungi, found in a range of habitats, including soil, vegetation, air, living or decaying plants, indoor environments, and a wide range of food products [2,3,4,5,6,7]. Phu Pha Phet Cave, a part of a mycological diversity project associated with Satun Geopark, Thailand’s first UNESCO Global Geopark, is also known as “Diamond Mountain Cave”. It is the fourth largest cavern on earth and the largest cave in Thailand, covering more than 80,000 m2. Based on estimated visitation, the cave has been opened as a tourist attraction and is regarded as an anthropogenic disturbance; nonetheless, some areas in the Phu Pha Phet Cave remain closed [8]. Research on fungal diversity and mycological systematics in karst caves has been scarce in Thailand’s Satun UNESCO Global Geopark.
In this study, soil samples randomly obtained from the Phu Pha Phet Cave were subjected to phenotypic examination and phylogenetic approaches, and two new cave-dwelling soil micro-fungi belonging to Talaromyces—T. phuphaphetensis and T. satunensis spp. nov.—were described and compared with similar taxa.
2. Materials and Methods
2.1. Collection, Isolation, and Morphology
On 3 December 2019, collections were performed during a fungal survey of Phu Pha Phet Cave. Two strains of Talaromyces were isolated from soil samples (110 m elevation; 7°07′35″ N 99°59′49″ E) in Thungwa, Manang District, La-Ngu, Satun Province, southern Thailand. Ten or twenty grams of soil were randomly collected at shallow depths (1–5 cm) after removing the surface layer, placed in zip lock bags, preserved at 4 °C in an ice box during collection, and transferred to the mycological laboratory at the National Center for Genetic Engineering and Biotechnology (BIOTEC).
The dilution plate technique was carried out using a modified version of the method of Zhang et al. [9], and 1 g of the sample was suspended in 9 mL of sterile distilled water and then serially diluted 10-fold. Dilutions from 10−1 to 10−5 were prepared, and 100 µL of each dilution was spread on potato dextrose agar (PDA; Difco, GA, USA) containing two antibiotics (50 μg/mL of ampicillin and 50 μg/mL of streptomycin) with three replicates. Plate cultures were incubated at room temperature for two–three days to allow fungal growth before subculture onto PDA without antibiotics for additional morphological investigation.
After seven days, macroscopic features and growth rates were examined on seven traditional culture media (Czapek yeast autolysate agar (CYA), Czapek’s agar (CZ), malt extract agar (MEA), yeast extract sucrose agar (YES), dichloran 18% glycerol agar (DG18), creatine sucrose agar (CREA), and oatmeal agar (OA, Difco)), as previously described [10]. Strains were inoculated with spore suspensions at three points and incubated in the dark at 25 °C, with additional temperatures of 30 and 37 °C for CYA. Extended incubation of MEA and OA plates for four weeks was performed to observe sexual reproduction. Microscopic observations were carried out on 7-day-old MEA, CZ, and CYA media. Ethanol (70%) and lactic acid (60%) were used to wash excess of conidia and mount slides, respectively.
Microscopic characters (i.e., conidiophores, conidiogenous cells, and conidia) were examined with a light microscope (OLYMPUS CX31; Olympus Corporation, Japan) and photographed using a Nomarski differential interference contrast microscope (OLYMPUS DP70). The Methuen Handbook of Color created color codes that were used to categorize the observed colors of the colonies [11]. The types and strains were deposited into the Thailand Bioresource Research Center (TBRC; https://www.tbrcnetwork.org, accessed on 21 July 2022) under the names Talaromyces phuphaphetensis sp. nov. (TBRC 16281) and T. satunensis sp. nov. (TBRC 16246). The type specimens are kept in the FUNGARIUM BIOTEC Bangkok Herbarium (BBH; https://www.nbt-microbe.org, accessed on 15 June 2022) as T. phuphaphetensis BBH 49306 (holotype) and T. satunensis BBH 49305 (holotype). The MycoBank numbers were registered as T. phuphaphetensis MB 844613 and T. satunensis MB 844614.
2.2. DNA Extraction, PCR Amplification, and Phylogenetic Analyses
Following the protocols of Sri-indrasutdhi et al. [12], genomic DNA was extracted from 7-day-old cultures grown on MEA using the cetyltrimethylammonium bromide (CTAB) method. The internal transcribed spacer (ITS) region, β-tubulin (BenA), calmodulin (CaM), and RNA polymerase II (RPB2) genes were amplified. The primers and amplification profiles used are shown in Table 1. PCR products were purified and sequenced by Macrogen Inc. (Seoul, South Korea) using the same PCR primers used for PCR amplification. The obtained sequences of ITS, BenA, CaM, and RPB2 were assembled and trimmed at both ends in BioEdit v.7.1.3 [13]. The newly generated sequences were deposited in GenBank (the National Centre for Biotechnology Information (NCBI)), and representative Talaromyces in the section Trachyspermi used in phylogenetic analyses, and their accession numbers are provided in Table 2.
Table 1.
Molecular markers, primers, and amplification profiles used and generated in this study.
| Molecular Locus | Primer Name | Direction | Reference | Amplification Profile | ||
|---|---|---|---|---|---|---|
| Denature | Repeat Step | Extension | ||||
| Internal transcribed spacers (ITS) | ITS1 | Forward | [17] | 94 °C (5 min) |
35 cycles, 94 °C (45 s), 55 °C (45 s), 72 °C (60 s) | 72 °C (7 min) |
| ITS5 | ||||||
| ITS4 | Reverse | |||||
| β-tubulin (BenA) | Bt2a | Forward | [18] | 94 °C (10 min) |
35 cycles, 94 °C (30 s), 57 °C (30 s), 72 °C (30 s) |
72 °C (10 min) |
| Bt2b | Reverse | |||||
| Calmodulin (CaM) | cmd5 | Forward | [19] | 94 °C (3 min) |
30 cycles, 94 °C (1 min), 57 °C (1 min), 72 °C (1 min) |
72 °C (10 min) |
| cmd6 | Reverse | |||||
| RNA polymerase II (RPB2) | 5F2 | Forward | [20] | 94 °C (3 min) |
34 cycles, 94 °C (1 min), 54 °C (1 min), 72 °C (1.30 min) |
72 °C (8 min) |
| 7cR | Reverse | |||||
Table 2.
Talaromyces species of sect. Trachyspermi used in phylogenetic analyses and their GenBank accession numbers.
| Taxon | Original Strain Number |
GenBank Accession Number | |||
|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | ||
| Talaromyces aerius | CBS 140611 T | KU866647 | KU866835 | KU866731 | KU866991 |
| T. affinitatimellis | CBS 143840 T | LT906543 | LT906552 | LT906549 | LT906546 |
| T. africanus | CBS 147340 T
= DTO 179-C5 |
OK339610 | OK338782 | OK338808 | OK338833 |
| T. albisclerotius | CBS 141839 T
= DTO 340-G5 |
MN864276 | MN863345 | MN863322 | MN863334 |
| T. albobiverticillius | CBS 133440 T | HQ605705 | KF114778 | KJ885258 | KM023310 |
| T. amyrossmaniae | NFCCI 1919 T | MH909062 | MH909064 | MH909068 | MH909066 |
| T. assiutensis | CBS 147.78 T | JN899323 | KJ865720 | KJ885260 | KM023305 |
| T. atroroseus | CBS 133442 T | KF114747 | KF114789 | KJ775418 | KM023288 |
| T. austrocalifornicus | CBS 644.95 T | JN899357 | KJ865732 | KJ885261 | MN969147 |
| T. basipetosporus | CBS 143836 T
= FMR 9720 |
LT906542 | LT906563 | - | LT906545 |
| T. brasiliensis | URM 7618 T | MF278323 | LT855560 | LT855563 | MN969198 |
| T. calidominioluteus | CBS 147313 T
= DTO 052-G3 |
OK339612 | OK338786 | OK338817 | OK338837 |
| T. catalonicus | CBS 143039 T
= FMR 16441 |
LT899793 | LT898318 | LT899775 | LT899811 |
| T. chongqingensis | CS26-67 T | MZ358001 | MZ361343 | MZ361350 | MZ361357 |
| T. clemensii | PPRI 26753 T | MK951940 | MK951833 | MK951906 | MN418451 |
| T. convolutus | CBS 100537 T | JN899330 | KF114773 | MN969316 | JN121414 |
| T. diversus | CBS 320.48 T | KJ865740 | KJ865723 | KJ885268 | KM023285 |
| T. erythromellis | CBS 644.80 T | JN899383 | HQ156945 | KJ885270 | KM023290 |
| T. gaditanus | CBS 169.81 T
= DTO 228-B8 |
MH861318 | OK338775 | OK338802 | OK338827 |
| T. germanicus | CBS 147314 T
= DTO 055-D1 |
OK339619 | OK338799 | OK338812 | OK338845 |
| T. guatemalensis | CCF 6215 T | MN322789 | MN329687 | MN329688 | MN329689 |
| T. halophytorum | KACC 48127 T | MH725786 | MH729367 | MK111426 | MK111427 |
| T. heiheensis | HMAS 248789 T = CGMCC 3.18012 | KX447526 | KX447525 | KX447532 | KX447529 |
| T. minioluteus | CBS 642.68 T | JN899346 | MN969409 | KJ885273 | JF417443 |
| T. minnesotensis | CBS 142381 T | LT558966 | LT559083 | LT795604 | LT795605 |
| T. pernambucoensis | URM 6894 T | LR535947 | LR535945 | LR535946 | LR535948 |
| T. phuphaphetensis | TBRC 16281 T | ON692803 | ON706960 | ON706962 | ON706964 |
| T. resinae | CBS 324.83 T
= IMI 080450 |
MT079858 | MN969442 | MT066184 | MN969221 |
| T. rubrifaciens | CGMCC 3.17658 T | KR855658 | KR855648 | KR855653 | KR855663 |
| T. samsonii | CBS 137.84 T = DTO 304-C3 = DTO 169-G6 | MH861709 | OK338798 | OK338824 | OK338844 |
| T. satunensis | TBRC 16246 T | ON692804 | ON706961 | ON706963 | - |
| T. solicola | DAOM 241015 T | FJ160264 | GU385731 | KJ885279 | KM023295 |
| T. speluncarum | CBS 143844 T
= FMR 16671 |
LT985890 | LT985901 | LT985906 | LT985911 |
| T. subericola | CBS 144322 T
= FMR 15656 |
LT985888 | LT985899 | LT985904 | LT985909 |
| T. systylus | BAFCcult 3419 T | KP026917 | KR233838 | KR233837 | - |
| T. trachyspermus | CBS 373.48 T
= IMI 040043 |
JN899354 | KF114803 | KJ885281 | JF417432 |
| T. ucrainicus | CBS 162.67 T
= FRR 3462 |
JN899394 | KF114771 | KJ885282 | KM023289 |
| T. udagawae | CBS 579.72 T
= IMI 197482 |
JN899350 | KF114796 | KX961260 | MN969148 |
| T. flavus | CBS 310.38 T | JN899360 | JX494302 | KF741949 | JF417426 |
New taxa proposed in this study are in bold. T, Ex-type strain. -, Data not available. Acronyms of culture collections: BAFC/BAFCcult, Culture Collection of the Department of Biological Sciences, Faculty of Exact and Natural Sciences, University of Buenos Aires, Argentina; BCC, BIOTEC Culture Collection, Pathum Thani, Thailand; CBS, Centraalbureau voor Schimmelcultures, CBS-KNAW Culture, Utrecht, Netherlands; CCF, Culture Collection of Fungi, Department of Botany, Faculty of Science, Charles University, Prague, Czech Republic; CGMCC, China General Microbiological Culture Collection Center, Beijing, China; DAOM, Canadian Collection of Fungal Cultures, Ottawa, Canada; DTO, culture collection of Food and Indoor Mycology Group of Westerdijk Institute, Utrecht, Netherlands; FMR, Faculty of Medicine in Reus, Spain; HMAS, Herbarium Mycologicum Academiae Sinicae, Beijing, China; IMI, International Mycological Institute (CABI Bioscience, Eggham), UK; KACC, Korean Agricultural Culture Collection, South Korea; NFCCI, National Fungal Culture Collection of India, India; PPRI, ARC-Plant Protection Research Institute, National Collection of Fungi: Culture Collection, Denmark; TBRC, Thailand Bioresource Research Center, Pathum Thani, Thailand; URM, Universidade Federal de Pernambuco Herbário, Brazil.
Multiple sequence alignments were performed separately using MAFFT v.7.490 [14] for each locus and adjusted manually. The four datasets were concatenated in BioEdit v.7.1.3 [13]. Maximum likelihood (ML) phylogenetic analyses, including 1000 bootstrap replicates, were performed using RAxML-NG [15] under the GTR + GAMMA model with default parameters on the Debian Linux operating system. Bayesian inference (BI) was carried out using MrBayes v.3.2.7 [16] with 5,000,000 Markov chain Monte Carlo (MCMC) generations, with the first 2,000,000 discarded as burn-in. The consensus tree was visualized and adjusted in Adobe Photoshop 2021 using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree, accessed on 10 September 2019).
3. Results
3.1. Phylogenetic Analysis
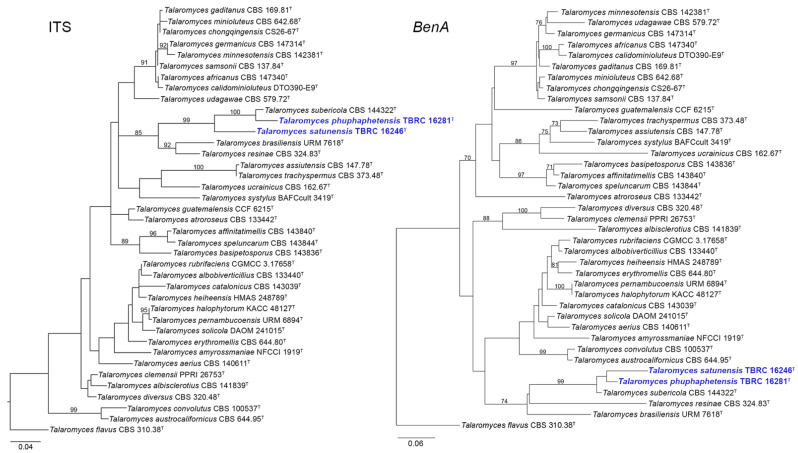
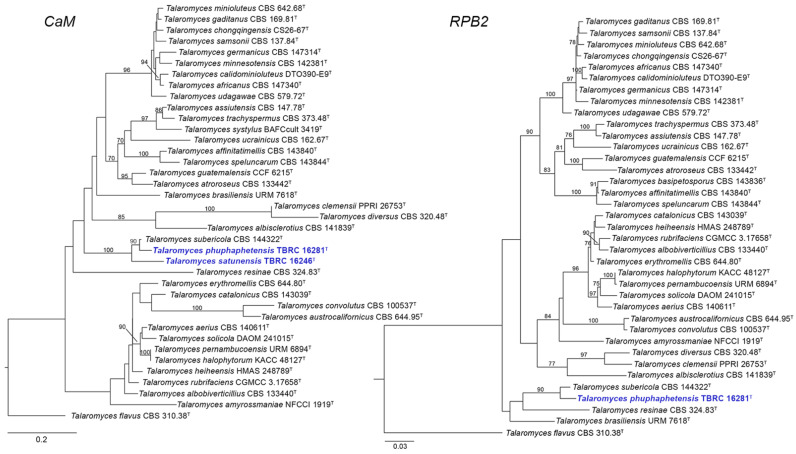
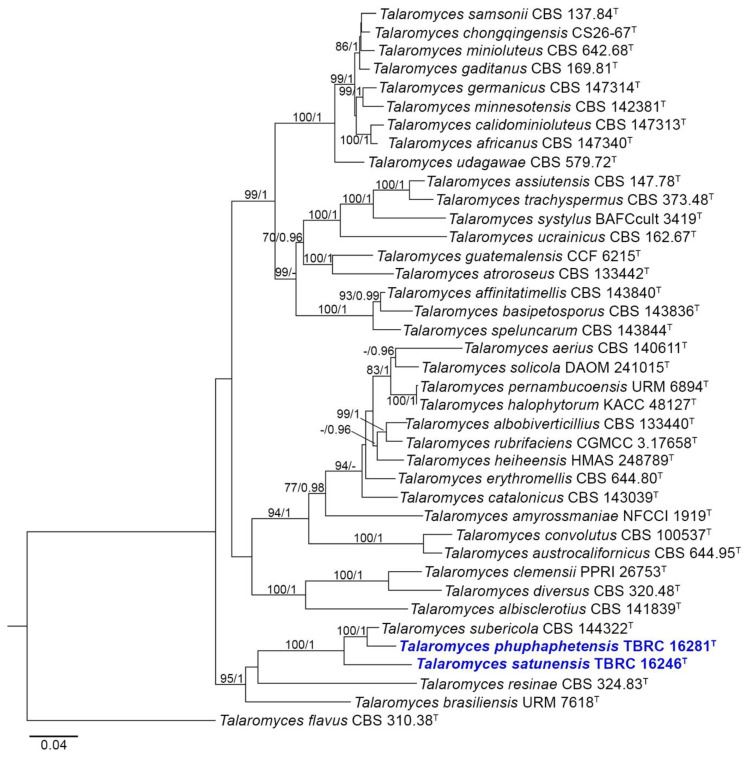
The phylogenetic trees of ITS, BenA, CaM, and RPB2 constructed separately using ML analyses and the concatenated datasets of four loci based on ML and Bayesian analyses revealed the relationships among the novel strains (TBRC 16281 and TBRC 16246) and Talaromyces species of the section Trachyspermi (Figure 1, Figure 2 and Figure 3). Based on the single-gene analyses, our two proposed new species, T. phuphaphetensis and T. satunensis, were clustered with T. brasiliensis URM 7618, T. resinae CBS 324.83, and T. subericola CBS 144322. The two new species and T. subericola formed a monophyletic group, and were revealed as phylogenetically related to T. brasiliensis and T. resinae (Figure 1 and Figure 2).
Figure 1.
Maximum likelihood phylogeny based on the ITS region (left) and the BenA gene (right) for the closely related species belonging to Talaromyces section Trachyspermi. Talaromyces flavus (CBS 310.38) was chosen as the outgroup. New species are indicated in blue. T = Ex-type strain. Bootstrap (BS) values ≥ 70% are indicated at the nodes.
Figure 2.
Maximum likelihood phylogeny based on the CaM (left) and RPB2 genes (right) for the closely related species belonging to Talaromyces section Trachyspermi. Talaromyces flavus (CBS 310.38) was chosen as the outgroup. New species are indicated in blue. T = Ex-type strain. Bootstrap (BS) values ≥ 70% are indicated at the nodes.
Figure 3.
Maximum likelihood phylogeny based on the combination of the ITS region and BenA, CaM, and RPB2 genes for the closely related species belonging to Talaromyces section Trachyspermi. Talaromyces flavus (CBS 310.38) was chosen as an outgroup taxon. New species are indicated in blue. T = Ex-type strain. Bootstrap (BS) values ≥ 70% (left) or posterior probability (PP) values ≥ 0.95 (right) are indicated at the nodes.
In the ITS and CaM phylograms, T. subericola was a sister taxon to T. phuphaphetensis, and these two lineages were closely related to T. satunensis with a good bootstrap support (Figure 1 and Figure 2). In the BenA phylogram, our two new species clustered together with a low support value (bootstrap value < 70%) and were closely related to T. subericola on a highly supported branch (99%). In the RPB2 analyses (no sequence data of T. satunensis), T. subericola was the closest sister taxon to T. phuphaphetensis, with good bootstrap support (90%).
Based on the combined datasets of ITS, BenA, CaM, and RPB2, the phylogenetic relationships showed a topology similar to those obtained from each gene individually (Figure 3). The two new species, T. phuphaphetensis and T. satunensis, formed two single branches and a well-supported clade with T. brasiliensis, T. resinae, and T. subericola (BS/PP = 95%/1.00). Talaromyces subericola was a sister taxon of T. phuphaphetensis, and these two species were closely related to T. satunensis in both fully supported subclades (BS/PP = 100%/1.00). Phylogenetically, T. resinae and T. brasiliensis were at a basal position, located on a single branch within the same clade as T. phuphaphetensis and T. satunensis.
3.2. Taxonomy
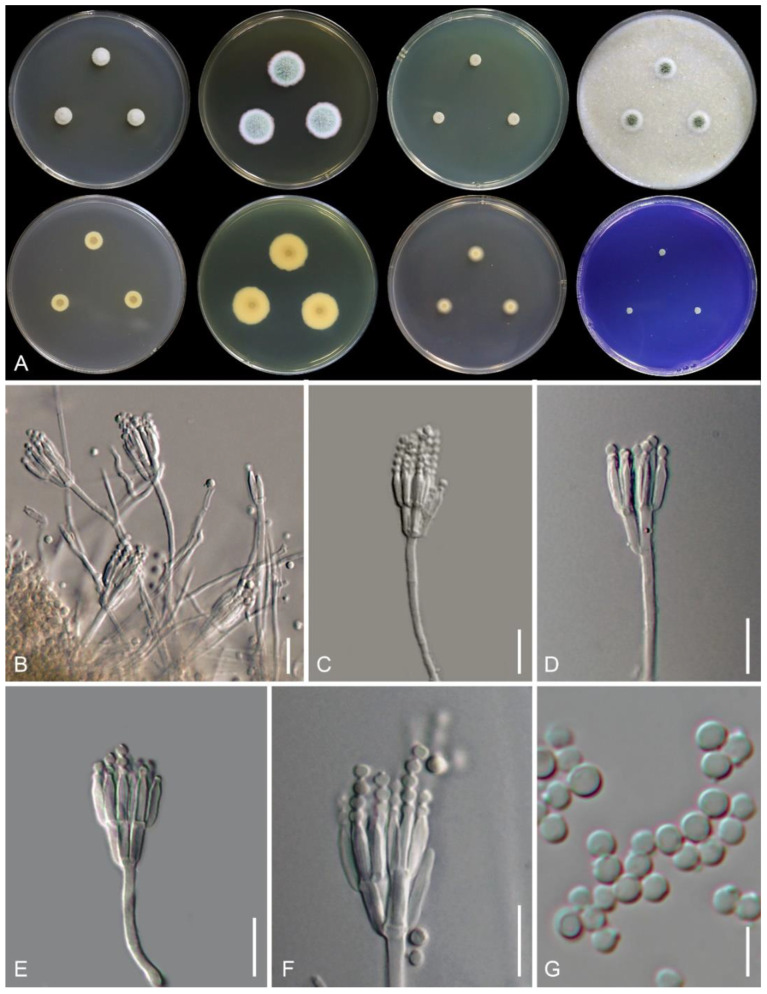
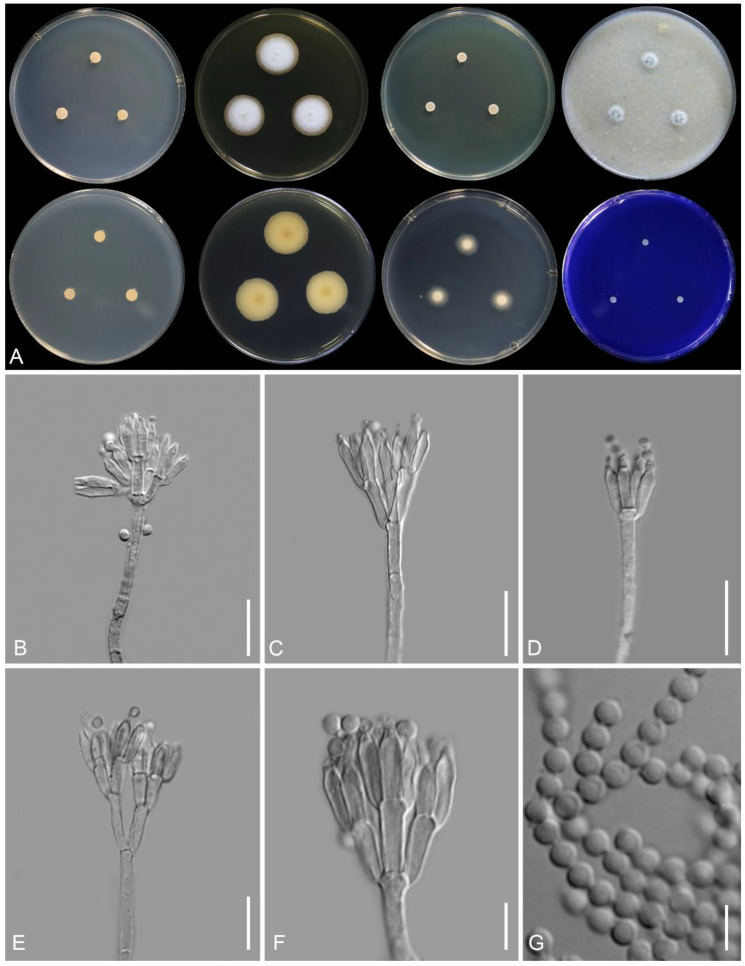
Talaromyces phuphaphetensis Nuankaew, Chuaseehar. & Somrith., sp. nov. is shown in Figure 4.
Figure 4.
Talaromyces phuphaphetensis TBRC 16281. (A) Colonies from left to right: (top row) CYA, MEA, YES, and OA, and (bottom row) CYA reverse, MEA reverse, DG18, and CREA. (B–F) Conidiophores. (G) Conidia. Scale bars: (B–F) = 10 µm, G = 5 µm.
MycoBank: 844613.
Etymology: The specific epithet refers to “Phu Pha Phet Cave”, where the type strain was first collected.
Typification: Thailand, Satun Province, Manang District, Satun UNESCO Global Geopark, Phu Pha Phet cave, from soil, 3 December 2019, Nattawut Boonyuen, Prasert Srikitikulchai and Sita Preedanon, culture, Sita Preedanon, CV00299 (holotype BBH 49306, ex-type strain TBRC 16281).
GenBank numbers: BenA = ON706960, CaM = ON706962, ITS = ON692803, RPB2 = ON706964.
In: Talaromyces sect. Trachyspermi.
Colony diameter (7 days, in mm): CYA 8–9; CYA 30 °C 3–5; CYA 37 °C 3–4; CZ 3–4; MEA 16–18; OA 10–12; DG18 8–9; YES 6–7; CREA 3–4.
Colony characteristics: CYA at 25 °C after 7 days: Colonies slightly raised at centers; margins low, entire (<1 mm); mycelia white; texture floccose; sparse to absent sporulation after 21 days; conidia en masse grayish green (25C4); soluble pigment light yellow (2A4); exudates absent; reverse center grayish yellow (2C4) and yellowish white (4A2). MEA at 25 °C after 7 days: Colonies slightly raised at centers; margins low, entire (<1 mm); mycelia white; texture loosely funiculose and floccose; sporulation strong; conidia en masse grayish green (27E4); soluble pigment absent; exudates absent; reverse center orange-gray (5B2) fading into orange-white (5A2). CZ at 25 °C after 7 days: Colonies low, slightly raised at centers; margin entire (2–3 mm); mycelia white; texture velvety; sporulation moderately; conidia en masse dull green (28D4); soluble pigment yellow (2A6) after 14 days of incubation; exudates absent; reverse yellowish gray (3B2). DG18 at 25 °C after 7 days: Colonies low, plane; margins low, plane, entire (2–3 mm); mycelia white; texture velvety; sporulation absent; soluble pigment yellow (2A6) after 14 days of incubation; exudates absent; reverse center orange-white (5A2) fading into pale orange (5A3). OA at 25 °C after 7 days: Colonies slightly raised at centers; margins low, plane, entire (3–4 mm); mycelia white; texture loosely funiculose; sporulation moderate; conidia en masse grayish green (26D3); soluble pigment absent; exudates absent. YES at 25 °C after 7 days: Colonies slightly raised at center, slightly concave, wrinkled; margins low, entire (<1 mm); mycelia white; texture floccose; sporulation absent; soluble pigment absent; exudates absent; reverse grayish yellow (2C4). CREA at 25 °C after 7 days: Acid production absent; poorly growing.
Micromorphology: On MEA, conidiophores mostly biverticillate, minor proportion monoverticillate; stipes finely tuberculate, non-vesiculate, 15–60 × 2.5–3 μm; metulae (2–) 3–6 per stipe, adpressed, 5–9 × 1.5–3 μm; phialides 3–5 per metula, acerose, 7–9.5 × 2–3 μm; conidia globose to sub-globose, smooth-walled, 2–3.5 μm in diameter. Ascomata absent.
Note: Phylogenetically, Talaromyces phuphaphetensis falls into a terminal clade of section Trachyspermi, including species T. brasiliensis, T. resinae, T. satunensis (described here), and T. subericola (Figure 3). Talaromyces phuphaphetensis and T. satunensis can be differentiated as having tuberculate stipes and smooth-walled conidia, whereas the other three related species have smooth-walled stipes and conidial ornamentation [4,7,21]. Talaromyces phuphaphetensis mainly differs from T. satunensis in having shorter stipes (15–60 × 2.5–3 μm in T. phuphaphetensis vs. 20–290 × 2–3.2 μm in T. satunensis), producing yellow diffusible pigments on CYA after 7 days, CZ and DG18 after 14 days, and possessing strong sporulation on MEA.
In addition, T. phuphaphetensis showed poor growth on CYA incubated at 37 °C (3–4 mm, 7 days), while T. brasiliensis, T. satunensis, and T. subericola had no growth on the medium. Morphological comparisons of T. phuphaphetensis, T. satunensis, and the three related species are shown in Table 3.
Table 3.
Comparisons of the morphological characteristics of T. phuphaphetensis sp. nov. (TBRC 16281), T. satunensis sp. nov. (TBRC 16246), and closely related species of Talaromyces section Trachyspermi based on the phylogeny in this study.
| Microscopic Characters |
T. brasiliensis [4] |
T. phuphaphetensis (This Study) |
T. resinae [21] |
T. satunensis (This Study) |
T. subericola [7] |
||
|---|---|---|---|---|---|---|---|
| On MEA | Conidiophore | stipes (μm) | 20–50 × 2.5–4 | 15–60 × 2.5–3 | N/A | 20–290 × 2–3.2 | N/A |
| branching | biverticillate | mostly biverticillate, monoverticillate | mostly biverticillate, monoverticillate, terverticillate | ||||
| ornamentation | smooth | finely tuberculate | tuberculate | ||||
| Metulae | size (μm) | 8–11 × 2.5–3.5 | 5–9 × 1.5–3 | 5.5–10 × 2–3.3 | |||
| per verticil | 5–6 | 2–6 | 2–5 | ||||
| Phialides | size (μm) | 7–11 (−14) × 2–3 | 7–9.5 × 2–3 | 6–9 × 2–3.2 | |||
| per metula | 3–4 | 3–5 | 2–5 | ||||
| Conidia | size (μm) | 2–3 | 2.0–3.5 | 2.5–3 | |||
| shape | globose | Globose to sub-globose | globose to sub-globose | ||||
| ornamentation | finely roughened | smooth | smooth | ||||
| On CZ | Conidiophore | stipes (μm) | N/A | 30 − 100 (–120) × 2 − 3 | 40 − 60 (−80) × 3 − 4 | 25 − 135 × 2 − 3.5 | N/A |
| branching | biverticillate, monoverticillate | most biverticillate, monoverticillate symmetric | biverticillate, monoverticillate | ||||
| ornamentation | tuberculate | smooth | tuberculate | ||||
| Metulae | size (μm) | 6 − 8 × 2.5 − 3 | 6–8 (–12) × 2.5–3.5 | 5−9 × 2−3.5 | |||
| per verticil | 2–4 | N/A | 2–3 | ||||
| Phialides | size (μm) | 5 − 11 × 2 − 3 | 6–8 (−12) × 2 − 3 | 6 − 9 × 2 − 3 | |||
| per metula | 3–4 | N/A | 3–5 | ||||
| Conidia | size (μm) | 3–4 | (3−) 3.5 − 4.5 (−5) | 3–4 | |||
| shape | globose to sub-globose | globose to sub-globose | globose to sub-globose | ||||
| ornamentation | smooth | tuberculate | smooth | ||||
| On CYA * | Conidiophore | stipes (μm) | N/A | 20 − 70 × 2 − 3 | N/A | 35 − 85 × 2 − 2.5 | 30 − 45 × 2 − 3 |
| branching | biverticillate, monoverticillate | biverticillate, monoverticillate | biverticillate | ||||
| ornamentation | finely tuberculate | finely tuberculate | smooth | ||||
| Metulae | size (μm) | 8 − 12 × 2 − 3 | 6 − 9 × 2 − 2.5 | 12 − 20 × 2 − 3 | |||
| per verticil | 2–6 | 2–4 | 2–3 | ||||
| Phialides | size (μm) | 8 − 11 × 2 − 3 | 6.5 − 10.5 × 2 − 2.5 | 7 − 10 × 2 − 3 | |||
| per metula | 3–5 | 3–4 | 2–4 | ||||
| Conidia | size (μm) | 2–2.5 | 2–3 | 3 | |||
| shape | globose to sub-globose | globose to sub-globose | ellipsoidal to globose | ||||
| ornamentation | smooth | smooth | smooth-walled but verruculose with age | ||||
N/A = data not available. * = Microscopic characters were derived after incubation for 2 to 3 weeks.
Talaromyces satunensis Nuankaew, Chuaseehar. & Somrith., sp. nov. is shown in Figure 5.
Figure 5.
Talaromyces satunensis TBRC 16246. (A) Colonies from left to right: (top row) CYA, MEA, YES, and OA, and (bottom row) CYA reverse, MEA reverse, DG18, and CREA. (B–F) Conidiophores. (G) Conidia. Scale bars: (B–F) = 10 µm, G = 5 µm.
MycoBank: 844614.
Etymology: The specific epithet refers to “Satun”, the name of the province where the species originated.
Typification: Thailand, Satun Province, Manang District, Satun UNESCO Global Geopark, Phu Pha Phet cave, from soil, 3 December 2019, Nattawut Boonyuen, Prasert Srikitikulchai and Sita Preedanon, culture, Sita Preedanon, CV00055 (holotype BBH 49305, ex-type strain TBRC 16246).
GenBank numbers: BenA = ON706961, CaM = ON706963, ITS = ON692804.
In: Talaromyces sect. Trachyspermi.
Colony diameter (7 days, in mm): CYA 5–6; CYA 30 °C 4–5; CYA 37 °C No growth; CZ 4–5; MEA 18–20; OA 8–10; DG18 12–13; YES 4–5; CREA 3–4.
Colony characteristics: CYA at 25 °C after 7 days: Colonies raised, sulcate; margins low, entire (<1 mm); mycelia pale gray to yellowish gray; texture floccose; sporulation poorly after 21 days; soluble pigment absent; exudates absent; reverse grayish yellow (4C3). MEA at 25 °C after 7 days: Colonies slightly raised at centers; margins low, plane, entire (1–2 mm); mycelia white; texture floccose; sporulation poor; conidia en masse grayish green (30C3); soluble pigment absent; exudates absent; reverse grayish yellow (3B3) with center fading into orange-white to pale orange (5A2–5A3). CZ at 25 °C after 7 days: Colonies low, plane; margin entire (<1 mm); mycelia white; texture velvety; sporulation moderately; conidia en masse dull green (30D5); soluble pigment absent; exudates absent; reverse yellowish gray (4B2). DG18 at 25 °C after 7 days: Colonies low, plane; margins low, plane, entire (2–3 mm); mycelia white; texture velvety; sporulation absent; soluble pigment absent; exudates absent; reverse yellowish white (2A2). OA at 25 °C after 7 days: Colonies slightly raised at centers; margins low, plane, entire (<1 mm); mycelia white; texture loosely funiculose; sporulation moderate; conidia en masse grayish gray (26D3); soluble pigment absent; exudates absent. YES at 25 °C after 7 days: colonies slightly raised at center, slightly concave; margins narrow (<1 mm); mycelia white; texture velvety; sporulation absent; soluble pigment absent; exudates absent; reverse pale orange (5A3) and grayish orange (5B3). CREA at 25 °C after 7 days: Acid production absent; poorly growing.
Micromorphology: On MEA, conidiophores mostly biverticillate, minor proportion monoverticillate and terverticillate; stipes tuberculate, non-vesiculate, 20–290 × 2–3.2 μm; metulae 2–5 per stipe, rather adpressed, 5.5–10 × 2–3.3 μm; phialides (2–)3–5 per metula, ampulliform to acerose, 6–9 × 2–3.2 μm; conidia globose to sub-globose, smooth-walled, 2.5–3 μm in diameter. Ascomata absent.
Note: Phylogenetically, T. satunensis is located within a terminal clade, and it is closely related to T. phuphaphetensis and T. subericola (Figure 3). Talaromyces subericola differs from our two new species in producing smooth-walled stipes and verruculose conidia. In comparison, T. satunensis differs from T. phuphaphetensis in the absence of diffusible pigments on CYA, lacking growth on CYA at 37 °C, and poor sporulation on MEA. In addition, T. satunensis has longer stipes and sometimes produces terverticillate branches (see Table 3).
4. Discussion
In this study, phylogenies and morphological characters supported the establishment of Talaromyces phuphaphetensis and T. satunensis as two new species belonging to Talaromyces section Trachyspermi. Phylogenetic analyses based on single loci (i.e., ITS, BenA, CaM, and RPB2) and the multi-locus approach showed that T. phuphaphetensis and T. satunensis are members of the Talaromyces clade composed of T. brasiliensis, T. resinae, and T. subericola. All phylogenetic trees also indicated that T. subericola has the closest relationship with our two new species described herein. Based on the combined dataset, the phylogenetic analyses revealed that T. brasiliensis and T. resinae are basal to T. phuphaphetensis, T. satunensis, and T. subericola (Figure 3).
The topology of the CaM tree for T. brasiliensis and T. resinae showed a slightly different position (Figure 2). In addition, the species relationships within the section Tra-chyspermi, as shown in the phylogenetic trees inferred from CaM, were different from those in the trees based on the ITS, BenA, and RPB2 genes. These data are congruent with the studies of Rajeshkumar et al. [5] and Zhang et al. [22]. However, the phylogenetic tree of CaM gene sequences could distinguish T. phuphaphetensis and T. satunensis from other species in the section. Although the RPB2 gene is formally accepted as a potential molecular locus for identifying Talaromyces species, it is often difficult to amplify the targeted DNA region [23,24,25]. Unfortunately, this study did not obtain RPB2 sequence data from T. satunensis, although we attempted with different PCR profiles. Nonetheless, the phylogenies based on single genes and the concatenated data also confirmed the taxonomic placements of T. phuphaphetensis and T. satunensis as two distinct species in the Trachyspermi section.
Both T. phuphaphetensis and T. satunensis are characterized by the production of biverticillate conidiophores, tuberculate-walled stipes, and smooth-walled conidia. They grow restrictedly on CYA, YES, and DG18, slightly faster on MEA, and poorly on CREA. These data are in agreement with the description of the section [25,26]. Colonies of T. phuphaphetensis produce yellow pigment on CYA, CZ, and DG18. Generally, Talaromyces species are reported to be good pigment producers [27,28,29]. Many species in the section Trachyspermi (such as T. albobiverticillius, T. atroroseus, and T. minioluteus) can produce a large amount of red pigment. However, only T. atroroseus produces pigments without known mycotoxins, which might be suitable for application in the food or healthcare industry as an alternative synthetic dye [27]. Likewise, the new species we propose can serve as an alternative source of natural pigments that need to be investigated for mycotoxin production, enhanced pigment production, and other testing for future research.
5. Conclusions
Two isolates of soil fungi were discovered in the Phu Pha Phet Cave of the Satun UNESCO Global Geopark in southern Thailand and identified as part of the genus Talaromyces in the section Trachyspermi. The two isolates are proposed as new species, namely Talaromyces phuphaphetensis and T. satunensis, based on their morphological and phylogenetic differences from the other species described in the section Trachyspermi. The discovery will support future evaluations of the unique species’ potential applications and functions. Information on the mycological biodiversity and habitat of UNESCO’s Satun cave would promote awareness of sustainable conservation and exploitation, supporting the future planning, monitoring, and management of Thai caves in achieving a balance between conservation and development. Furthermore, the results contribute to the knowledge of cave-dwelling soil fungi, their ecological uniqueness and diversity in Thailand, and their global geographical distribution. Interestingly, it is also possible that more new species will be discovered in this peculiar environment in Thailand’s Satun UNESCO Global Geopark.
Acknowledgments
We acknowledge Jennifer Luangsaard (APMT-BIOTEC-NSTDA) and Sissades Thongsima (NBT-NSTDA) for their continual mycological support in the study. Wonnop Visessanguan and Theerayut Toojinda are thanked for their research support at BIOTEC. We are grateful to Lily Eurwilaichitr and Supawadee Ingsriswang, who initiated the Thailand–China Joint Laboratory on Microbial Biotechnology under the collaboration between TBRC (BIOTEC) and CGMCC under the CAS-NSTDA Joint Research Program. We acknowledge Narongrit Thungprue, Director of Satun Geopark, and Bumrungrad Ploydam, head of Teagkkhaobanthad Wildlife Sanctuary, Department of National Park Wildlife and Plant Conservation, for sharing their facilities and permission to collect samples. We are indebted to Chaiyaporn Siripornpibul, ex-director of the Department of Groundwater Resources, and Chotika Muangsong at Mahidol University for speleological support and physical data collection during field trips. We thank Anupong Klaysuban, Charisa Srihom, and Boonchuai Chainuwong for their invaluable assistance in the fungal laboratory. The authors express gratitude to the anonymous reviewers and editors for their critical reviews of the manuscript.
Author Contributions
Conceptualization, S.S. (Satinee Suetrong), L.C. and N.B.; methodology, S.P. (Sita Preedanon), S.N., C.C. and S.S. (Sayanh Somrithipol); software, S.N.; visualization, S.N.; investigation, S.N., S.P. (Sita Preedanon), S.S. (Supicha Saengkaewsuk), and S.P. (Sarinya Phookongchai); morphological analysis, C.C., S.N., S.S. (Sayanh Somrithipol), and X.-C.W.; writing—original draft manuscript, S.N., C.C., S.P. (Sita Preedanon), and N.B.; review and editing, S.N., C.C., S.P. (Sita Preedanon), P.K., S.S. (Sayanh Somrithipol), X.-C.W., L.C., P.S., N.K., Z.-F.Z., S.S. (Satinee Suetrong), and N.B.; supervision, S.S. (Satinee Suetrong) and N.B.; project administration, S.S. (Satinee Suetrong) and N.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All newly generated sequences have been deposited to the GenBank.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This research was funded by research grant number P1951709 under the project titled “Diversity of rock-dwelling microbes in Satun UNESCO Global Geopark” and partially supported by BIOTEC-NSTDA (laboratory work). Lei Cai acknowledges the CAS-NSTDA Joint Research Program (NO. 153211KYSB20200039). The authors would like to thank Rungsima Tantalakha from RDI Management for National Strategic and Network Division at RNS-NSTDA for supporting this work under collaboration among BIOTEC-NSTDA-Thailand, NBT-NSTDA-Thailand, and CAS-China.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Benjamin C.R. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47:669–687. doi: 10.1080/00275514.1955.12024485. [DOI] [Google Scholar]
- 2.Guevara-Suarez M., Sutton D., Gené J., García D., Wiederhold N., Guarro J., Cano-Lira J.F. Four new species of Talaromyces from clinical sources. Mycoses. 2017;60:651–662. doi: 10.1111/myc.12640. [DOI] [PubMed] [Google Scholar]
- 3.Wang X.C., Chen K., Qin W.T., Zhuang W.Y. Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycol. Prog. 2017;16:73–81. doi: 10.1007/s11557-016-1251-3. [DOI] [Google Scholar]
- 4.Barbosa R.N., Bezerra J.D.P., Souza-Motta C.M., Frisvad J.C., Samson R.A., Oliveira N.T., Houbraken J. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek. 2018;111:1883–1912. doi: 10.1007/s10482-018-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajeshkumar K.C., Yilmaz N., Marathe S.D., Seifert K.A. Morphology and multigene phylogeny of Talaromyces amyrossmaniae, a new synnematous species belonging to the section Trachyspermi from India. MycoKeys. 2019;45:41–56. doi: 10.3897/mycokeys.45.32549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.You Y.H., Aktaruzzaman M., Heo I., Park J.M., Hong J.W., Hong S.B. Talaromyces halophytorum sp. nov. isolated from roots of Limonium tetragonum in Korea. Mycobiology. 2020;48:133–138. doi: 10.1080/12298093.2020.1723389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodríguez-Andrade E., Stchigel A.M., Guarro J., Cano-Lira J.F. Fungal diversity of deteriorated sparkling wine and cork stoppers in Catalonia, Spain. Microorganisms. 2020;8:12. doi: 10.3390/microorganisms8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yordkayhun S., Wattanasen K., Thungprue N. Geophysical investigation of the karst geosites in Satun UNESCO Global Geopark, Thailand: Implication for sinkhole hazard assessment. Geosci. J. 2022;26:249–266. doi: 10.1007/s12303-021-0025-3. [DOI] [Google Scholar]
- 9.Zhang Z.F., Liu F., Zhou X., Liu X.Z., Liu S.J., Cai L. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia. 2017;39:1–31. doi: 10.3767/persoonia.2017.39.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visagie C.M., Houbraken J., Frisvad J.C., Hong S.B., Klaassen C.H.W., Perrone G., Seifert K.A., Varga J., Yaguchi T., Samson R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornerup A., Wanscher J.H. Methuen Handbook of Colour. 2nd ed. Methuen; London, UK: 1967. pp. 1–243. [Google Scholar]
- 12.Sri-indrasutdhi V., Boonyuen N., Suetrong S., Chuaseeharonnachai C., Sivichai S., Gareth J.E.B. Wood-inhabiting freshwater fungi from Thailand: Ascothailandia grenadoidia gen. et sp. nov., Canalisporium grenadoidia sp. nov. with a key to Canalisporium species (Sordariomycetes, Ascomycota) Mycoscience. 2010;51:411–420. doi: 10.1007/S10267-010-0055-6. [DOI] [Google Scholar]
- 13.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 14.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlov A., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient bayesian phyloge-netic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White T.J., Bruns T., Lee S., Taylor J. PCR Protocols: A Guide to Methods and Applications. Elsevier; Amsterdam, The Netherlands: 1990. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenet-ics; pp. 315–322. [DOI] [Google Scholar]
- 18.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong S.B., Cho H.S., Shin H.D., Frisvad J.C., Samson R.A. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 2006;56:477–486. doi: 10.1099/ijs.0.63980-0. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 21.Qi Z.T., Kong H.Z. A new species of Penicillium. Acta Mycologica Sin. 1982;1:103–105. [Google Scholar]
- 22.Zhang Z.K., Wang X.C., Zhuang W.Y., Cheng X.H., Zhao P. New species of Talaromyces (Fungi) isolated from soil in southwestern China. Biology. 2021;10:745. doi: 10.3390/biology10080745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stošić S., Ristić D., Gašić K., Starović M., Grbić M.L., Vukojević J., Živković S. Talaromyces minioluteus: New postharvest fungal pathogen in Serbia. Plant Dis. 2020;104:656–667. doi: 10.1094/PDIS-08-19-1806-RE. [DOI] [PubMed] [Google Scholar]
- 24.Sun X.R., Xu M.Y., Kong W.L., Wu F., Zhang Y., Xie X.L., Li D.W., Wu X.Q. Fine identification and classification of a novel beneficial Talaromyces fungal species from masson pine phizosphere soil. J. Fungi. 2022;8:155. doi: 10.3390/jof8020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yilmaz N., Visagie C.M., Houbraken J., Frisvad J.C., Samson R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaguchi T., Someya A., Udagawa S.-I. A reappraisal of intrageneric classification of Talaromyces based on the ubiquinone systems. Mycoscience. 1996;37:55–60. doi: 10.1007/BF02461457. [DOI] [Google Scholar]
- 27.Frisvad J.C., Yilmaz N., Thrane U., Rasmussen K.B., Houbraken J., Samson R.A. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE. 2013;8:e84102. doi: 10.1371/journal.pone.0084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagashetti A., Dufossé L., Singh S.K., Singh P. Fungal pigments and their prospects in different industries. Microorganisms. 2019;7:604. doi: 10.3390/microorganisms7120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morales-Oyervides L., Oliveira J., Sousa-Gallagher M., Méndez-Zavala A., Montañez J.C. Assessment of the dyeing properties of the pigments produced by Talaromyces spp. J. Fungi. 2017;3:38. doi: 10.3390/jof3030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All newly generated sequences have been deposited to the GenBank.