Objective:
Although several studies have identified an association between the rs4784227-cancer susceptibility candidate gene 16 (CASC16) polymorphism and breast cancer, the results remain inconclusive. Therefore, we conducted a meta-analysis to assess the relationship between the rs4784227-CASC16 polymorphism and breast cancer risk.
Methods:
Studies were searched in the PubMed, Web of Science, Embase, Google Scholar, and Cochran Library databases until June 10, 2021, to identify all potential literature on rs4784227-CASC16 polymorphism and breast cancer risk association. Fixed-effect or random-effect models were used to calculate odds ratios (ORs) and their corresponding 95% confidence intervals (95% CIs). Subgroup analyses, publication bias, and sensitivity analyses were also conducted.
Results:
Seventeen eligible studies involving 34,719 subjects (18,445 cases and 16,274 healthy controls) from 7 articles were included in the current meta-analysis. The pooled ORs regarding the association between the rs4784227-CASC16 polymorphism and breast cancer risk were statistically significant [T vs C: OR = 1.244, 95% CI = 1.202–1.287; TT vs CT + CC: OR = 1.407, 95% CI = 1.296–1.528; CC vs CT + TT: OR = 0.777, 95% CI = 0.745–0.811; TT vs CC: OR = 1.544, 95% CI = 1.419–1.681; CT vs CC: OR = 1.244, 95% CI = 1.189–1.301]. On subgroup analysis, the rs4784227-CASC16 T/C gene has a certain correlation with breast cancer susceptibility in Asian and North American populations, but no significant risk in the Australian population.
Conclusion:
Our pooled analysis showed a significant association between the rs4784227- (T) allele and breast cancer susceptibility in Asian and North American populations, and intervention with this mutation might be a new therapeutic strategy for breast cancer. However, large-scale and well-designed studies are needed in different populations to further evaluate the role of the rs4784227-CASC16 polymorphism in breast cancer.
1. Introduction
Breast cancer is the most malignant neoplasm among females and the leading cause of death among women worldwide.[1] An estimated 2.1 million new cases of breast cancer were diagnosed worldwide in 2018.[2] The mechanism of breast carcinogenesis is still not fully understood. Environmental variables and germline mutations are 2 well-known risk factors that contribute to the development of breast cancer.[3] Epidemiological studies have indicated that age, age of menarche, obesity, family history, and menstrual history are associated with an increased susceptibility to breast cancer.[4,5] Many single-nucleotide polymorphisms(SNPs) in critical genes, such as BRCA1, BRCA2, TP53, and PTEN, have been identified to contribute to increased susceptibility to breast cancer.[6–8] However, the involvement of genes in breast cancer has not yet been fully elucidated.
In the past few years, several genome-wide association studies (GWAS) have identified numerous novel genetic susceptibility variants and loci that are independently associated with an increased risk of breast cancer.[9–11] Forkhead box A1 (FOXA1) and TOX high-mobility box protein group family member 3 (TOX3) are believed to be other probable candidates that cause breast cancer susceptibility.[12] Breast cancer risk-associated SNPs are enriched in the cistromes of FOXA1 in a cancer and cell-type–specific manner.[13] TOX3 is a nuclear protein that can modify chromatin structure. Its clinical implications and its role in tumor development and invasion have been shown in the risk of breast cancer.[12] Rs4784227 is a site of cancer susceptibility candidate gene 16 (CASC16). CASC16 is a noncoding RNA, located at chromosome 16q12, that may affect the DNA-binding sequence change on FOXA1 and subsequently, trigger the FOXA1-binding affinity to the TOX3 gene promoter.[13] Growing evidence suggests that rs4784227 (a C-to-T transition) is strongly correlated with the risk of breast cancer.[14–21] Considering the relatively small sample size in most studies, it is possible to perform a quantitative synthesis of the evidence for potential correlations with rigorous methods. Meta-analysis has been proven to be an effective statistical method combining available studies to produce a precise conclusion. A meta-analysis article was published in 2021 July and included 8284 subjects (4055 breast cancer cases and 4229 controls).[22] However, many studies were still excluded from that meta-analysis. Additionally, the study population of the previous meta-analyze was limited to the Asian population. It is undetermined whether sample sizes are sufficient to reach a definite conclusion. Therefore, we performed a meta-analysis of 17 published studies that included 34,719 subjects (18,445 breast cancer cases and 16,274 healthy controls) to identify the precise association between the rs4784227-CASC16 C/T polymorphism and breast cancer risk. Moreover, the association between the rs4784227-CASC16 C/T polymorphism and breast cancer risk in Asian, North American, and Australian populations was explored by subgroup analyses.
2. Materials and Methods
2.1. Search strategy
Search term combinations were keywords relating to rs4784227 (e.g., “rs4784227”, “CASC16”, “chromosome 16q12”, ”LOC643714”) and in combination with words related to breast cancer (e.g., “breast cancer”, “breast carcinoma”, “malignant breast neoplasm”) and polymorphism or variation. These keyword retrieval strategies were used in PubMed, Web of Science, Embase, Google Scholar, and Cochran Library databases for entries until June 2021.
2.2. Inclusion and exclusion criteria
All studies included in the meta-analysis met the following inclusion criteria:
(1) a case-control study;
(2) an investigation of the association between the rs4784227-CASC16 polymorphism and breast cancer risk;
(3) including sufficient data for calculating the odds ratios (ORs) and the corresponding 95% confidence intervals (CIs);
(4) The genotype distribution of the control group must be consistent with the Hardy–Weinberg equilibrium (HWE).
The exclusion criteria were as follows:
(1) duplicate publication;
(2) case reports, review articles, letters, comments, meta-analyses, irrelevant studies;
(3) not offering sufficient data for calculation of ORs with 95% CIs.
2.3. Data extraction and Synthesis
Information and data were extracted carefully from all qualified independent articles by 2 independent investigators (Liping Bao and Wenji Xu), based on the inclusion and exclusion criteria above. The data included the first author, publication year, country, source of controls, number of cases and controls for each genotype, and genotyping method. If genotype distributions were not given in the study, we calculated them from allele frequencies and number of cases and controls. When necessary, we wrote to the corresponding authors for extra information. Disagreements were resolved by discussion and consensus. If discussion and consensus were not achieved, a suggestion was offered by the third reviewer (Yao Zhong) to determine the correct selection.
2.4. Statistical analysis
We performed this meta-analysis based on published studies. So there is no need to conduct special ethic review, and the ethical approval is not necessary. The OR with 95% CI was used to assess the strength of the association between rs4784227-CASC16 C/T polymorphisms and breast cancer risk in 5 genetic models (T vs C, TT vs CT + CC, CC vs CT + TT, TT vs CC, and CT vs TT). The rs4784227-CASC16 polymorphism distribution in the control group was tested for HWE using the Pearson chi-square test.[23] The Newcastle–Ottawa Scale (NOS) was used to evaluate the quality of the eligible studies. Studies with NOS scores ≥ 6 were considered high quality. Between-study heterogeneities were assessed by Cochran chi-square-based Q-test and I2 test. A fixed-effect model was used for analyses if the heterogeneity was not significant (P > .1, I2 < 50.0%). Otherwise, a random-effect model was used. Subgroup analyses were performed based on ethnicity. Sensitivity analysis was conducted to identify the influence of the individual data on pooled results and test the reliability of the results. Begg funnel plots and Egger tests were used to assess the existence of publication bias. All statistical analyses were conducted by Stata version 12.0 (StataCorp LP, College Station, TX).
3. Results
3.1. Study characteristics
Figure 1 shows the literature search flowchart of our meta-analysis. A total of 22 potentially relevant citations were identified from the databases. Five duplicate records were removed. After we screened the titles and abstracts, 5 citations were removed due to irrelevant topics (not about breast cancer and rs4784227-CASC16 polymorphism). Then, the full text of the remaining 12 citations was downloaded for reading carefully; we removed 4 citations due to insufficient genotype data for extraction and 1 citation due to a P value for Hardy–Weinberg equilibrium < 0.05. All 17 case and control studies from 7 articles were included in our meta-analysis, incorporating 18,445 breast cancer cases and 16,274 controls. The populations were from Asia (China, Japan, and Iran), North America, and Australia. The characteristics of these studies and the quality scores are presented in Table 1.
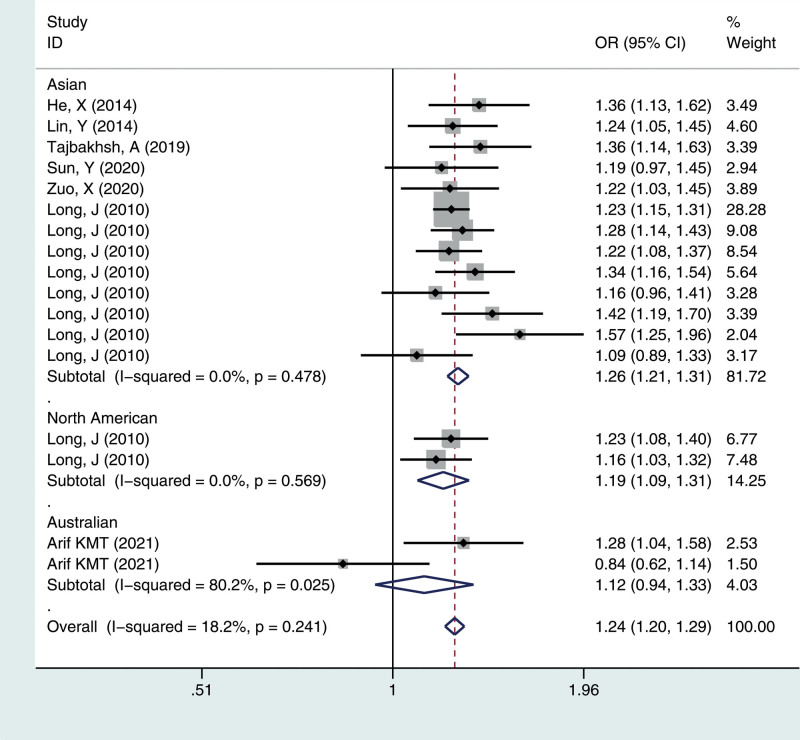
Figure 1.
Meta-analysis for the OR of breast cancer associated with the rs4784227-CASC16 polymorphism (T vs C). CASC16 = cancer susceptibility candidate gene 16, CI = confidence interval, OR = odds ratios.
Table 1.
Characteristics of the studies included in the meta-analysis.
| First author | Year | Country | Cases | Controls | Cases | Controls | P for HWE | NOS score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | |||||||
| He X[19] | 2014 | Chinese | 623 | 620 | 305 | 262 | 56 | 358 | 226 | 36 | .966 | 9 |
| Lin Y[18] | 2014 | Chinese | 701 | 794 | 331 | 302 | 68 | 424 | 313 | 57 | .941 | 9 |
| Fujian | ||||||||||||
| Tajbakhsh A[17] | 2019 | Iranian | 505 | 567 | 209 | 218 | 78 | 285 | 222 | 60 | .092 | 7 |
| Sun, Y[14] | 2020 | Chinese | 503 | 503 | 266 | 199 | 38 | 292 | 180 | 31 | .644 | 9 |
| Shanxi | ||||||||||||
| Zuo, X[16] | 2020 | Chinese | 675 | 675 | 353 | 270 | 52 | 394 | 240 | 41 | .581 | 9 |
| Shanxi | ||||||||||||
| Long J[20] | 2010 | Chinese | 6346 | 3921 | 3253 | 2581 | 512 | 2241 | 1447 | 233 | .977 | 7 |
| Shanghai | ||||||||||||
| Chinese | 1520 | 1583 | 747 | 637 | 136 | 890 | 594 | 99 | .993 | 7 | ||
| Tianjin | ||||||||||||
| Chinese | 1437 | 1437 | 726 | 591 | 120 | 808 | 539 | 90 | .993 | 7 | ||
| Nanjing | ||||||||||||
| Chinese | 1003 | 1010 | 500 | 416 | 87 | 587 | 366 | 57 | .996 | 7 | ||
| Taiwan | ||||||||||||
| Chinese | 456 | 644 | 231 | 187 | 38 | 354 | 247 | 43 | .992 | 7 | ||
| Hong Kong | ||||||||||||
| Japanese | 640 | 631 | 320 | 265 | 55 | 378 | 221 | 32 | .743 | 7 | ||
| Nagoya | ||||||||||||
| Japanese | 403 | 403 | 196 | 170 | 37 | 247 | 137 | 19 | .999 | 7 | ||
| Nagano | ||||||||||||
| Japanese | 531 | 511 | 288 | 206 | 37 | 289 | 191 | 31 | .940 | 7 | ||
| North American | 1145 | 1142 | 577 | 472 | 96 | 643 | 428 | 71 | .984 | 7 | ||
| North American | 1357 | 1148 | 704 | 547 | 106 | 646 | 430 | 72 | .960 | 7 | ||
| Arif KMT[21] | 2021 | Australian Caucasian | 369 | 484 | 173 | 159 | 37 | 262 | 188 | 34 | .972 | 7 |
| Australian Caucasian | 231 | 201 | 133 | 84 | 14 | 105 | 81 | 15 | .908 | 7 | ||
HWE = Hardy–Weinberg equilibrium, NOS = Newcastle–Ottawa Scale.
3.2. Overall and Subgroup analyses
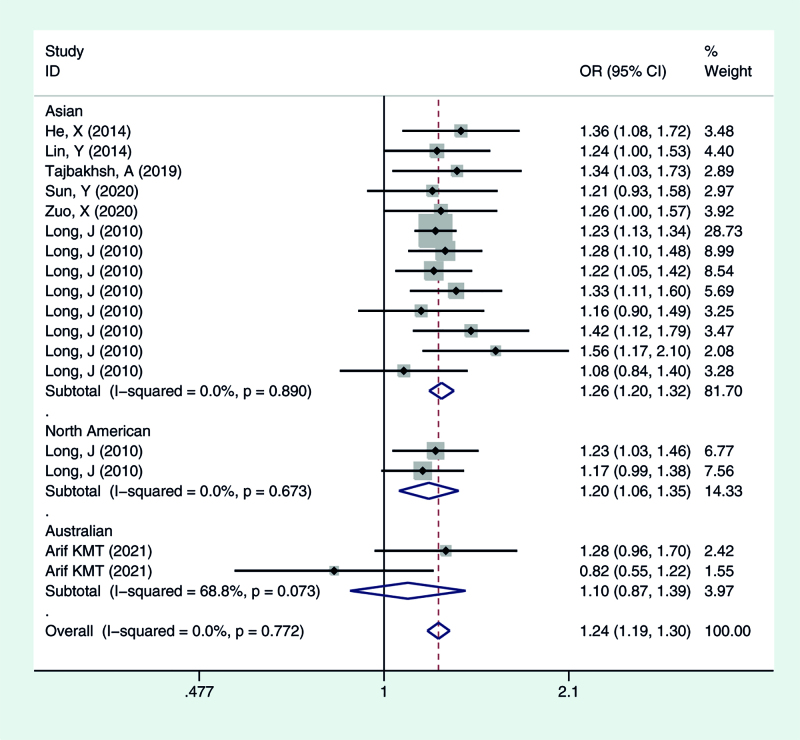
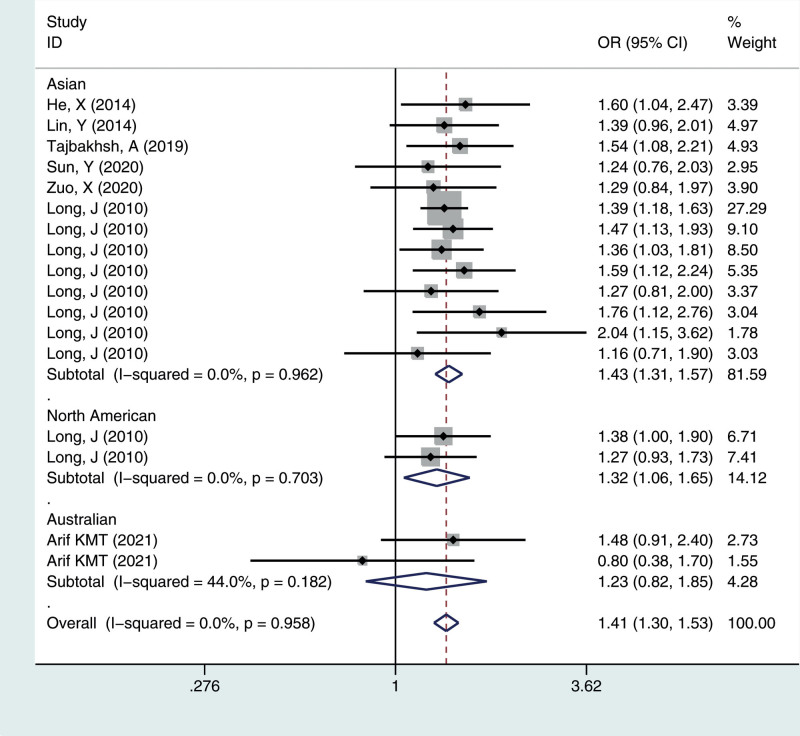
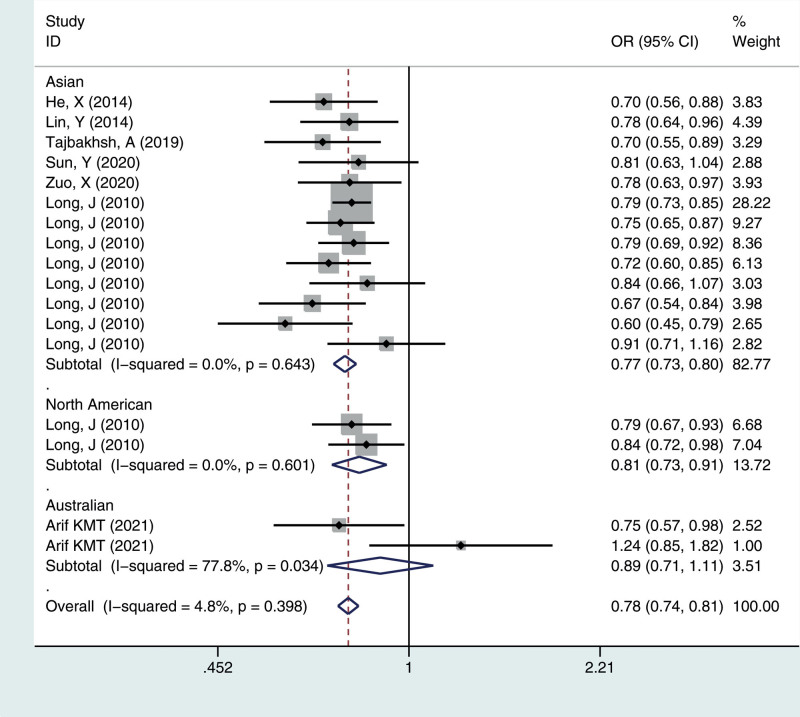
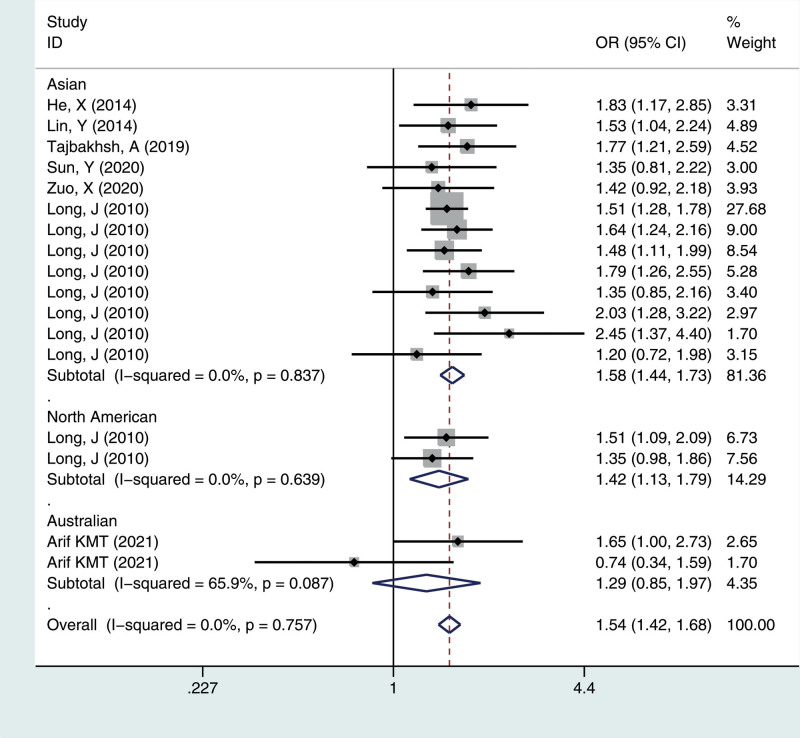
We calculated the summary ORs and their 95% CIs in 5 genetic models: the allelic contrast model (T vs C), the dominant model (CC vs CT + TT), the recessive model (TT vs TC + CC), and the additive model (TT vs CC), and the heterozygous model (CT vs CC). The evaluation of the association between the rs4784227-CASC16 C > T polymorphism and breast cancer risk is presented in Table 2. Overall, there was correlation between the prevalence of the rs4784227-CASC16 polymorphism and breast cancer, and the difference was statistically significant (T vs C: OR = 1.244, 95% CI = 1.202–1.287; TT vs CT + CC: OR = 1.407, 95% CI = 1.296–1.528; CC vs CT + TT: OR = 0.777, 95% CI = 0.745–0.811; TT vs CC: OR = 1.544, 95% CI = 1.419–1.681; CT vs CC: OR = 1.244, 95% CI = 1.189–1.301).
Table 2.
Summary of pooled ORs in the stratified analysis association between rs4784227-CASC16 and breast cancer risk.
| N | T vs C | TT vs CT + CC | CC vs CT + TT | TT vs CC | CT vs TT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | Ph | OR | Ph | OR | Ph | OR | Ph | OR | Ph | ||
| Total | 17 | 1.24 (1.20–1.29) | 0.241 | 1.41 (1.30–1.53) | 0.958 | 0.78 (0.74–0.81) | 0.398 | 1.54 (1.42–1.68) | 0.757 | 1.24 (1.19–1.30) | 0.772 |
| Asian | 13 | 1.26 (1.21–1.31) | 0.478 | 1.43 (1.31–1.57) | 0.962 | 0.77 (0.73–0.80) | 0.643 | 1.58 (1.44–1.73) | 0.837 | 1.26 (1.20–1.32) | 0.890 |
| North America | 2 | 1.19 (1.09–1.31) | 0.569 | 1.32 (1.06–1.65) | 0.703 | 0.81 (0.73–0.91) | 0.601 | 1.42 (1.13–1.79) | 0.639 | 1.20 (1.06–1.35) | 0.673 |
| Australian | 2 | 1.12 (0.94–1.33) | 0.025 | 1.23 (0.82–1.85) | 0.182 | 0.89 (0.71–1.11) | 0.034 | 1.29 (0.85–1.97) | 0.087 | 1.10 (0.87–1.39) | 0.073 |
CASC16 = cancer susceptibility candidate gene 16, N = number, OR = odds ratios, Ph = P value for heterogeneity.
To identify potential differences based on ethnicity, subgroup analysis was performed. In the stratified analysis by ethnicity, significant associations were found among different populations (Asian, North American, Australian) for the polymorphism in all genetic models. The results suggested that the rs4784227-CASC16 C > T gene has a certain correlation with breast cancer susceptibility in Asian and North American populations, but no significant risk in the Australian population, as shown in Figures 1–5.
Figure 5.
Meta-analysis for the OR of breast cancer associated with the rs4784227-CASC16 polymorphism (CT vs TT). CASC16 = cancer susceptibility candidate gene 16, OR = odds ratios.
Figure 2.
Meta-analysis for the OR of breast cancer associated with the rs4784227-CASC16 polymorphism (TT vs CT + CC). CASC16 = cancer susceptibility candidate gene 16, OR = odds ratios.
Figure 3.
Meta-analysis for the OR of breast cancer associated with the rs4784227-CASC16 polymorphism (CC vs CT + TT). CASC16 = cancer susceptibility candidate gene 16, OR = odds ratios.
Figure 4.
Meta-analysis for the OR of breast cancer associated with the rs4784227-CASC16 polymorphism (TT vs CC). CASC16 = cancer susceptibility candidate gene 16, OR = odds ratios.
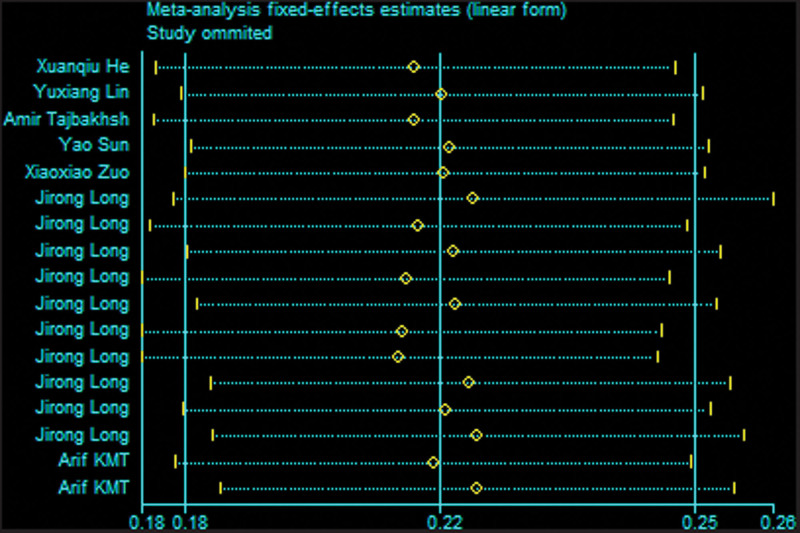
3.3. Sensitivity analysis
Sensitivity analysis was conducted by omitting a single study by turns to estimate the influence of the individual data on pooled results and test the reliability of the results (Fig. 6). The results of the sensitivity analysis showed that no individual study significantly affected the pooled OR, suggesting the stability of the meta-analyses.
Figure 6.
Sensitivity analysis of association between the rs4784227-CASC16 genetic variances and breast cancer. CASC16 = cancer susceptibility candidate gene 16.
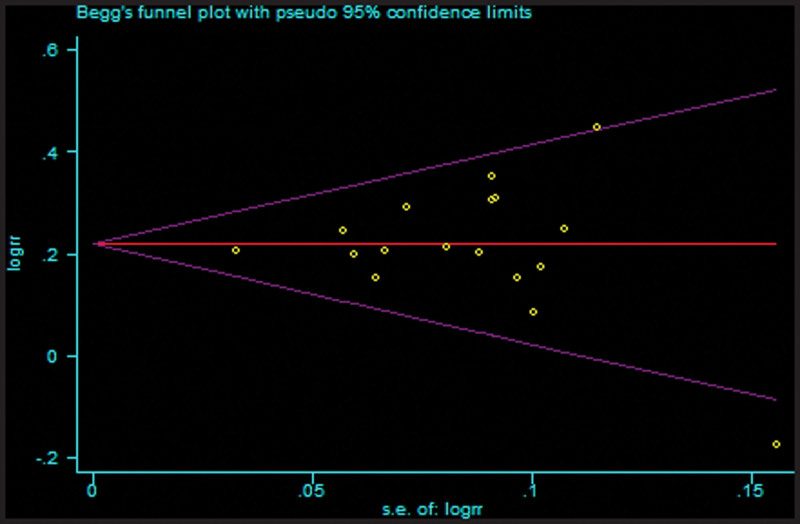
3.4. Publication bias
We used funnel plots and Egger test to evaluate potential publication biases. The shape of funnel plots was symmetrical for every comparison, thus suggesting no publication bias among the studies included. The results did not show any evidence of publication bias based on Begg funnel plot (PBegg = .902, T vs C, Fig. 7) or Egger regression test (PEgger = 0.982, T vs C). Similarly, there was no publication bias for the association between rs4784227-CASC16 polymorphism and breast cancer susceptibility under the other genetic models.
Figure 7.
Assessment of publication bias in the analysis of the association between the rs4784227-CASC16 gene polymorphism and breast cancer susceptibility. CASC16 = cancer susceptibility candidate gene 16.
4. Discussion
A variety of studies have focused on the association between the rs4784227-CASC16 gene polymorphism and breast cancer. However, the results obtained from such investigations have been inconclusive. Some studies have indicated that rs4784227-CASC16 may confer susceptibility to breast cancer by affecting the binding affinity of FOXA1 for the X3 gene promoter.[13] To derive a more precise estimation of the relationship, we performed this meta-analysis, combining data from similar studies to increase sample size and statistical power and achieve a more robust result.
The rs4784227-CASC16 SNP is an important SNP related to TOX3 and FOXA1, located upstream of the TOX3 gene. Meyer and Carroll suggested a tumor suppressor role for TOX3 in breast cancer.[24] FOXA1 is associated with ER and likely regulates TOX3 promoter activity. FOXA1-binding to DNA is crucial for the opening of chromatin and nucleosome positioning sequences for recruitment of transcription factors. Researchers have shown that rs4784227-CASC16 may disrupt enhancer function by FOXA1-binding affinity–modulation and, therefore, can change TOX3 expression.[13] Lupien et al[25] have demonstrated that the place for rs4784227-CASC16 on the FOXA1 genome for interaction is on the eighth position of the FKH motif recognized via FOXA1. Moreover, a study indicated that FOXA1 is modulated by T-rs4784227-CASC16 in vivo.[25] Katika et al[26] indicated that the affinity of the DNA site for the FOXA protein was enhanced for T-rs4784227-CASC16 compared with C-rs4784227-CASC16. Another study also showed that T rs4784227-CASC16 favors FOXA1-binding affinity over the C allele.[13] The Gro/transducin-like enhancer of split (TLE) protein, as a corepressor, is bound to the DNA sequence through DNA-binding repressor proteins. FOXA1 commonly stimulates gene expression, and cobinding to DNA sequences with Gro/TLE proteins, which leads to local chromatin condensation and transcriptional repression.[27] Cowper-Sal et al[13] showed that the risk allele T-rs4784227-CASC16 led to a reduction in TOX3 gene expression by decreasing the stability of the enhancer by increasing TLE repressor affinity recruitment. Additionally, a study showed that rs4784227-CASC16 affects the risk of breast cancer by regulating the sequence of RB transcriptional corepressor-like 2 gene expression.[28]
The objective of this meta-analysis was to explore the association between the CASC16 rs4784227 polymorphism and breast cancer risk. In this meta-analysis, a total of 7 articles including 17 case and control studies were used to evaluate the association between the rs4784227-CASC16 polymorphism and breast cancer risk. To eliminate heterogeneity, we established strict inclusion and exclusion criteria, and heterogeneity was not observed in the models in our meta-analysis. Therefore, the fixed-effect model was used in the genetic models. The results indicated that the rs4784227-CASC16 polymorphism significantly increased susceptibility to breast cancer. Considering that the polymorphism frequencies might differ among ethnic groups, we performed a subgroup analysis by ethnicity (Table 2). The results demonstrated that the rs4784227-CASC16 C/T polymorphism was associated with breast cancer risk in Asian and North American populations, but not in the Australian population. Since only 2 studies were performed and the total number of cases and controls is far lower in the Australian population, it should be noted that the ethnicity-based analysis may not be reliable in regard to the Australian subgroups. Our results indicated that the rs4784227-CASC16 gene polymorphism was associated with breast cancer risk in Asian and North American populations. The rs4784227-CASC16 T allele was a risk factor for breast cancer in Asian and North American populations.
Some potential limitations of the present meta-analysis should be considered. First, there were only 2 studies with a North American population and 2 studies with an Australian population, and the exploration of moderator variables was limited by the low number of studies. Further studies including a wider spectrum of subjects to investigate the role of this variant in other populations will be needed. Second, breast cancer is a complex disease with multiple determinants, and other risk factors were not well considered in the analysis, such as age and body mass index, which may affect the risk of breast cancer. We also need to consider the association between different types of breast cancer and the rs4784227-CASC16 polymorphism. As the limited original data contained in the study, we did not perform more hierarchical analysis, which could lead to a loss of significant evaluation subgroups.
Despite these limitations, this meta-analysis suggests that the rs4784227-CASC16 polymorphism was significantly associated with an increased risk of breast cancer in Asian and North American populations, particularly in Asian populations. Further studies including a wider spectrum of subjects in other populations and investigating multiple determinants and different types of breast cancer for breast cancer in large GWAS data are warranted.
Acknowledgments
The present study was supported by Kunming Medical University Joint Special Fund of Yunnan (grant number 202101AY070001-304 to Liping Bao) and High-level Health Technical Personnel Training Objects of Yunnan(grant number D-2018021 to Haihui Yang).
Abbreviations:
- CI =
- confidence interval
- FOXA1 =
- Forkhead box A1
- GWAS =
- Genome-wide association studies
- HWE =
- Hardy–Weinberg equilibrium
- NOS =
- Newcastle–Ottawa Scale
- ORs =
- odds ratios
- SNPs =
- single-nucleotidepolymorphisms
- TLE =
- Gro/transducin-like enhancer of split
- TOX3 =
- TOX high-mobility box protein group family member 3
Wenji Xu and Yao Zhong contributed equally to this work.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Xu W, Zhong Y, Yang H, Gong Y, Dao J, Bao L. Association between the rs4784227-CASC16 polymorphism and the risk of breast cance: A meta-analysis. Medicine 2022;101:34(e30218).
The authors have no funding and conflicts of interest to disclose.
References:
- [1].Winters S, Martin C, Murphy D, et al. Breast cancer epidemiology, prevention, and screening. Prog Mol Biol Transl Sci. 2017;151:1–32. [DOI] [PubMed] [Google Scholar]
- [2].Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–53. [DOI] [PubMed] [Google Scholar]
- [3].Barnard ME, Boeke CE, Tamimi RM. Established breast cancer risk factors and risk of intrinsic tumor subtypes. Biochim Biophys Acta. 2015;1856:73–85. [DOI] [PubMed] [Google Scholar]
- [4].McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ. 2000;321:624–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zaman K, Bodmer A, Pralong F, et al. Breast cancer and obesity, a dangerous relation. Rev Med Suisse. 2012;8:1101–4. [PubMed] [Google Scholar]
- [6].Shiovitz S, Korde LA. Genetics of breast cancer: a topic in evolution. Ann Oncol. 2015;26:1291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen Y, Pei J. Factors influencing the association between CYP17 T34C polymorphism and the risk of breast cancer: meta-regression and subgroup analysis. Breast Cancer Res Treat. 2010;122:471–81. [DOI] [PubMed] [Google Scholar]
- [8].Karami F, Mehdipour P. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int. 2013;2013:928562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Long J, Cai Q, Shu XO, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6:e1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mahfoudh W, Bouaouina N, Ahmed SB, et al. Hereditary breast cancer in Middle Eastern and North African (MENA) populations: identification of novel, recurrent and founder BRCA1 mutations in the Tunisian population. Mol Biol Rep. 2012;39:1037–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cowper-Sal LR, Zhang X, Wright JB, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun Y, Chen P, Wu J, et al. Association of polymorphisms in LOC105377871 and CASC16 with breast cancer in the northwest Chinese Han population. J Gene Med. 2020;22:e3131. [DOI] [PubMed] [Google Scholar]
- [15].Nourolahzadeh Z, Houshmand M, Mohammad FM, et al. Correlation between Lsp1 (Rs3817198) and Casc (Rs4784227) polymorphisms and the susceptibility to breast cancer. Rep Biochem Mol Biol. 2020;9:291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zuo X, Wang H, Mi Y, et al. The association of CASC16 variants with breast Cancer risk in a northwest Chinese female population. Mol Med. 2020;26:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tajbakhsh A, Farjami Z, Darroudi S, et al. Association of rs4784227-CASC16 (LOC643714 locus) and rs4782447-ACSF3 polymorphisms and their association with breast cancer risk among Iranian population. Excli J. 2019;18:429–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin Y, Fu F, Chen M, et al. Associations of two common genetic variants with breast cancer risk in a chinese population: a stratified interaction analysis. PLoS One. 2014;9:e115707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].He X, Yao G, Li F, et al. Risk-association of five SNPs in TOX3/LOC643714 with breast cancer in southern China. Int J Mol Sci . 2014;15:2130–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Long J, Cai Q, Shu XO, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6:e1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Arif K, Bradshaw G, Nguyen T, et al. Genetic association analysis implicates six MicroRNA-related SNPs with increased risk of breast cancer in Australian Caucasian Women. Clin Breast Cancer. 2021. [DOI] [PubMed] [Google Scholar]
- [22].Liang XS, Mo JL, Hu LM, et al. Association between CASC16 rs4784227 polymorphism and breast cancer susceptibility: a meta-analysis. Medicine (Baltim). 2021;100:e26215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chootrakool H, Shi JQ, Yue R. Meta-analysis and sensitivity analysis for multi-arm trials with selection bias. Stat Med. 2011;30:1183–98. [DOI] [PubMed] [Google Scholar]
- [24].Meyer KB, Carroll JS. FOXA1 and breast cancer risk. Nat Genet. 2012;44:1176–7. [DOI] [PubMed] [Google Scholar]
- [25].Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Katika MR, Hurtado A. A functional link between FOXA1 and breast cancer SNPs. Breast Cancer Res. 2013;15:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol. 2010;30:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Udler MS, Ahmed S, Healey CS, et al. Fine scale mapping of the breast cancer 16q12 locus. Hum Mol Genet. 2010;19:2507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]