Abstract
BrkA is a 103-kDa outer membrane protein of Bordetella pertussis that mediates resistance to antibody-dependent killing by complement. It is proteolytically processed into a 73-kDa N-terminal domain and a 30-kDa C-terminal domain as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. BrkA is also a member of the autotransporter family of proteins. Translocation of the N-terminal domain of the protein across the outer membrane is hypothesized to occur through a pore formed by the C-terminal domain. To test this hypothesis, we performed black lipid bilayer experiments with purified recombinant protein. The BrkA C-terminal protein showed an average single-channel conductance of 3.0 nS in 1 M KCl. This result strongly suggests that the C-terminal autotransporter domain of BrkA is indeed capable of forming a pore.
The autotransporters are a growing family of extracellular proteins, found in many gram-negative bacteria, that have many different functions but appear to have the same mechanism of export (19, 20, 25). Members of this diverse family include immunoglobulin A proteases from Neisseria gonorrhoeae (21) and Haemophilus influenzae (36); VacA (11), a vacuolating cytotoxin from Helicobacter pylori; the AIDA-I adhesin (28, 43) from Escherichia coli; IcsA (44) from Shigella flexneri, which is involved in intracellular spread; the ring-forming protein (32) from Helicobacter mustelae; Tsh (37), a temperature-sensitive hemagglutinin from an avian E. coli strain; EspP (8), an extracellular serine protease from enterohemorrhagic E. coli; and tracheal colonization factor (17), the adhesin pertactin (10), and serum resistance protein BrkA (15) from Bordetella pertussis.
These proteins are grouped together by the following three characteristics. (i) Most of the mature proteins are proteolytically processed into an approximately 30-kDa C-terminal domain and a much larger N-terminal domain, (ii) the C-terminal domains are predicted to form amphipathic β-barrels in the outer membrane, and (iii) export through the outer membrane does not require accessory proteins; hence, the name autotransporters.
In the proposed model of autotransporter secretion (22), an N-terminal signal sequence enables translocation across the cytoplasmic membrane. Once the protein is in the periplasm, the signal sequence is cleaved and the C-terminal domain then inserts itself into the outer membrane. It presumably forms a pore through which the N-terminal domain is exported by the formation of a hairpin loop. Cleavage of the N-terminal domain is thought to occur after translocation through the outer membrane, either autoproteolytically or by another protease (12).
In this study, we investigated the putative pore-forming ability of the C-terminal domain of the B. pertussis autotransporter BrkA through black lipid bilayer analysis. We found that the purified recombinant BrkA C-terminal protein forms channels in lipid bilayers whereas the BrkA N-terminal protein and the protein from a vector-only clone do not form channels in lipid bilayers.
Construction of clones.
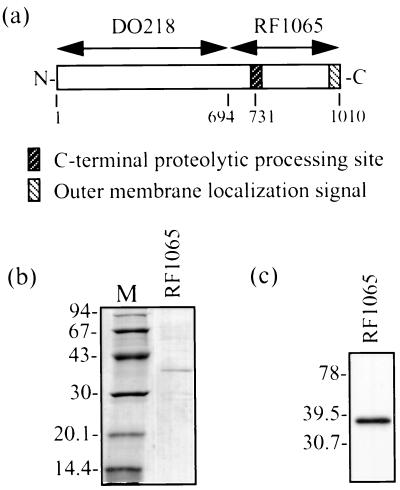
RF1065 and DO218 were subcloned from RF1066 (16). To construct RF1065 (Fig. 1a), brkA from the BamHI site to the HindIII site was ligated to pRSETb (Invitrogen, Carlsbad, Calif.), which represents amino acids (aa) 694 to 1010 of BrkA. This clone contains the C-terminal domain plus 37 aa that are upstream of the C-terminal processing site, as well as an N-terminal His tag. JS13 contains pRSETb (Invitrogen) without an insert. For DO218 (Fig. 1a), brkA from the AflIII site to the BamHI site was ligated to pET30b (Novagen, Madison, Wis.). This clone contains the first 693 aa of BrkA with N- and C-terminal His tags. All constructs were transformed into E. coli BL21 (DE3) pLysS cells (Novagen). Cultures were grown at 37°C in Luria broth or on Luria agar supplemented with 100 μg of ampicillin per ml and 34 μg of chloramphenicol per ml.
FIG. 1.
(a) Diagram of E. coli BrkA clones used in this study. Numbers below the boxes refer to amino acids. (b) SDS-PAGE and Coomassie blue staining showing pooled fractions of BrkA C-terminal protein obtained by denaturing Ni2+ chromatography after dialysis. Dialysis was performed slowly at 4°C against decreasing concentrations of urea and finally against 0.1% Triton X-100–10 mM Tris (pH 8.0). M, low-molecular-weight markers (Pharmacia) (molecular sizes are in kilodaltons). (c) Western immunoblot of BrkA C-terminal protein (same as in panel b). Detection was performed with an anti-BrkA C-terminal protein monoclonal antibody. Kaleidoscope Prestained Standards (Bio-Rad) were used for molecular size determination (molecular sizes are in kilodaltons).
Purification of the BrkA C-terminal domain from RF1065.
The RF1065 clone used as the source of our BrkA C-terminal protein is shown in Fig. 1a. Protein was purified from RF1065 by denaturing Ni2+-nitrilotriacetic acid purification using the Xpress System Protein Purification protocol (Invitrogen). In order to renature the protein, elution fractions containing the protein of interest were pooled and then slowly dialyzed against decreasing concentrations of urea in phosphate-buffered saline (PBS). Essentially, 8 M urea was diluted at a rate of 1 ml/min by PBS during the dialysis. The final dialysis was done overnight against 0.1% Triton X-100–10 mM Tris (pH 8.0). All dialysis was performed at 4°C.
After dialysis, pooled elution fractions were run on a sodium dodecyl sulfate (SDS)–11% polyacrylamide gel (23) and the proteins were visualized following staining with Coomassie brilliant blue. The Low Molecular Weight Electrophoresis Calibration Kit (Amersham Pharmacia Biotech, Baie d’Urfé, Quebec, Canada) was used to determine the molecular weight. SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 1b) revealed a 37-kDa band which corresponds in size to the BrkA C-terminal domain (30 kDa) along with the 37 aa upstream of the processing site (Fig. 1a) and the His tag.
Western blot analysis was also performed to confirm the identity of the protein. After electrophoresis was performed (23), the proteins were transferred to an Immobilon-P membrane (Millipore, Bedford, Mass.) at 100 V for 75 min by a wet transfer apparatus (Trans-Blot Electrophoretic Transfer Cell; Bio-Rad, Hercules, Calif.) in accordance with the manufacturer’s instructions. After transfer, the membrane was blocked with a 5% (wt/vol) skim milk solution in PBS for at least 1 h at room temperature. Washing and antibody incubation were carried out in a PBS solution containing 0.25% skim milk and 0.5% Tween 20. Membranes were incubated with a 1/30 dilution of mouse anti-BrkA C-terminal protein monoclonal antibody (a gift from Roger Parton, University of Glasgow) for 2 h at 37°C and then washed for 30 min. Secondary antibody incubation with a 1/10,000 dilution of goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Cappel, ICN Biomedicals, Costa Mesa, Calif.) was carried out for 1 h at room temperature and followed by 30 min of washing. Renaissance Western blot chemiluminescence reagent (NEN Life Science Products, Boston, Mass.) was used for detection. Kaleidoscope Prestained Standards (Bio-Rad) were used for molecular weight determination. The results of this Western blot analysis (Fig. 1c) confirmed that the BrkA C-terminal protein had been isolated.
Black lipid bilayer analysis of the BrkA C-terminal protein.
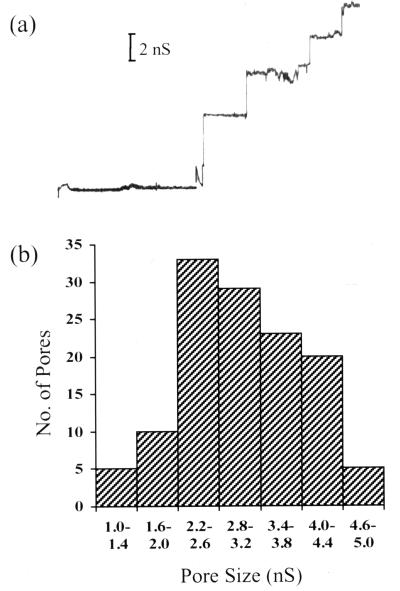
The pore-forming ability of the purified BrkA C-terminal protein was assessed through black lipid bilayer experiments, which were performed as previously described (3). Addition of the protein to a 1 M KCl solution bathing a membrane of 1.5% (wt/vol) oxidized cholesterol in n-decane with an applied voltage of 50 mV caused stepwise increases in conductance (Fig. 2a). This indicates that channels were being formed in the membrane. The distribution of these conductance measurements is shown in Fig. 2b. The average single-channel conductance of the BrkA C-terminal protein in 1 M KCl was found to be 3.0 nS. This average was calculated from 127 conductance increases obtained from two separate experiments. Similar-size channels were also observed when BrkA C-terminal protein from a second purification was used.
FIG. 2.
Black lipid bilayer analysis. (a) Single-channel conductance measurements after addition of the BrkA C-terminal protein from RF1065 to a 1 M KCl solution bathing a membrane of 1.5% oxidized cholesterol in n-decane. There was an applied voltage of 50 mV. (b) Histogram of single-channel conductance measurements showing pore size distribution.
To help rule out the possibility of contaminants in our protein preparation, we performed black lipid bilayer experiments with BrkA C-terminal protein that had undergone an additional gel purification step. In this case, the BrkA C-terminal protein was electrophoresed on an 11% polyacrylamide gel (23) and then a portion of the gel was stained with Coomassie brilliant blue so that it could be used as a guide for cutting of the protein out of the unstained segment of the gel. The protein was eluted from the gel overnight at room temperature with 0.1% Triton X-100–10 mM Tris (pH 8.0). Addition of this protein caused increases in conductance (Fig. 3) similar in size to those seen previously. This indicates that the channels seen were formed by the BrkA C-terminal protein and not by possible contaminants.
FIG. 3.
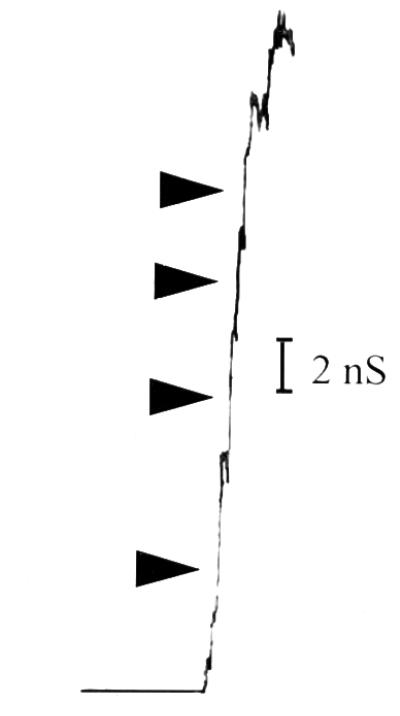
Black lipid bilayer analysis. Single-channel conductance measurements after addition of BrkA C-terminal protein that had been further purified by elution from an SDS-PAGE gel. Protein was added to a 1 M KCl solution bathing a membrane of 1.5% oxidized cholesterol in n-decane. There was an applied voltage of 50 mV. The arrows indicate stepwise increases in conductance.
Black lipid bilayer analysis of the BrkA N-terminal protein and protein from a vector-only clone.
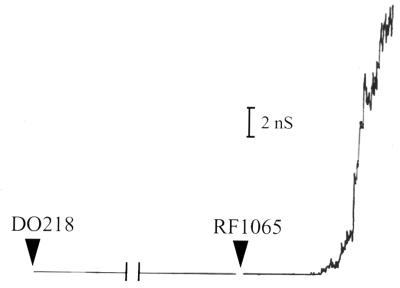
RF1065 overexpresses the BrkA C-terminal protein in the form of inclusion bodies, which necessitated purification under denaturing conditions. Porin proteins may contaminate preparations when proteins are produced in inclusion bodies (9a). In order to ensure that our results were not due to these possible contaminants, we used the BrkA N-terminal protein from DO218 (Fig. 1a) in black lipid bilayer experiments. This protein is also expressed in inclusion bodies, and we purified it in a way similar to that used for the C-terminal protein, except that the final dialysis was against PBS. Addition of the N-terminal protein diluted in 0.1% Triton X-100–10 mM Tris (pH 8.0) caused no increases in conductance (Fig. 4), even when large amounts of the protein were added. Upon addition of the BrkA C-terminal protein to the system (Fig. 4), channels were again observed.
FIG. 4.
Black lipid bilayer analysis. Single-channel conductance measurements were done after addition of first the BrkA N-terminal protein (DO218) and then the BrkA C-terminal protein (RF1065). Protein was added to a 1 M KCl solution bathing a membrane of 1.5% oxidized cholesterol in n-decane. There was an applied voltage of 50 mV. Protein from DO218 was purified in a way similar to that used for the protein from RF1065. The N-terminal protein was diluted in 0.1% Triton X-100–10 mM Tris (pH 8.0) before addition.
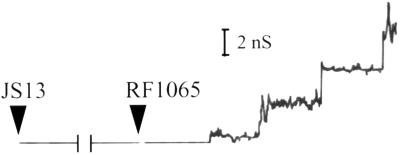
As another control for our protein purification method, we performed a black lipid bilayer experiment with protein that had been purified from a vector-only clone (JS13; does not contain a brkA insert) in the same manner as the BrkA C-terminal protein. Increases in conductance were not observed upon addition of protein from JS13 (Fig. 5). Addition of the BrkA C-terminal protein again caused the appearance of channels (Fig. 5). Both of these results help to rule out contaminants as the source of the channels seen with the BrkA C-terminal protein.
FIG. 5.
Black lipid bilayer analysis. Single-channel conductance measurements were done after addition of first protein from a clone containing the vector alone (JS13) and then the BrkA C-terminal protein (RF1065). Protein was added to a 1 M KCl solution bathing a membrane of 1.5% oxidized cholesterol in n-decane. There was an applied voltage of 50 mV. The protein from JS13 was purified in the same way as the protein from RF1065.
Discussion.
In this study, we demonstrated that the C-terminal autotransporter domain of BrkA is capable of forming a pore. Black lipid bilayer analysis showed the formation of channels upon addition of the BrkA C-terminal protein but not upon addition of the BrkA N-terminal protein or protein from a vector-only clone, all of which had been purified similarly. As well, the BrkA C-terminal protein that had been further purified by being cut out of an SDS-PAGE gel still formed channels.
As evidenced by our results, black lipid bilayer analysis can be used to determine the channel-forming capabilities not only of typical trimeric porins but also of a wide variety of proteins, including those involved in protein export (5) and those from mycobacterial (46, 47) and gram-positive (38) cell walls. Some examples of pore-forming proteins and their pore sizes are listed in Table 1.
TABLE 1.
Pore sizes of various proteins
| Organism and protein | Pore size determined by:
|
|||
|---|---|---|---|---|
| Black lipid bilayer analysis (nS) | Liposome analysis (nm) | Crystallography (nm) | Reference(s) or source | |
| Aeromonas salmonicida 28-kDa porin | 1.96 | 26 | ||
| Bordetella pertussis | ||||
| BrkA C-terminal domain | 3 | This study | ||
| MOMPd | 0.56 | 1 | ||
| Campylobacter coli MOMP | 0.53 | <<1 | 33 | |
| Escherichia coli | ||||
| OmpF | 2.1 | 1.2 | 6, 30 | |
| OmpC | 1.5 | 1.1 | 6, 30 | |
| PhoE | 1.8 | 2 | ||
| K | 1.5 | 6 | ||
| NmpC | 1.3 | 6 | ||
| LamB | 2.7/0.2 | 6 | ||
| OmpA | 1.2/0.18a | 1 | 39, 42 | |
| OmpG | 2 | 14 | ||
| Tsx | 0.01 | 7, 27 | ||
| FepA (ΔRV)b | 2 | 24 | ||
| TolC | 0.08 | 5 | ||
| PapC | 2 | 2–3 | 45 | |
| Haemophilus influenzae type b 40-kDa porin | 1.1 | 1.8 | 48, 49 | |
| Legionella pneumophila MOMP | 0.1c | 18 | ||
| Mycobacterium chelonae 59-kDa cell wall protein | 2.7 | 2.2 | 47 | |
| Mycobacterium smegmatis | ||||
| Cell wall extract | 4.1 | 46 | ||
| 40-kDa porin | 2 | 29 | ||
| Pseudomonas aeruginosa | ||||
| OprF | 5.6/0.36 | 2 | 3, 31, 50 | |
| OprP | 0.28 | 3 | ||
| Salmonella typhimurium | ||||
| OmpC (40K) | 2.4 | 4 | ||
| OmpF (39K) | 2.2 | 4 | ||
| OmpD (38K) | 2.5 | 4 | ||
| Treponema denticola 53-kDa outer sheath protein | 10.9 | 13 | ||
Measurements were made in 0.25 M KCl.
A portion of the N-terminus was deleted.
Measurements were made in 0.1 M NaCl.
MOMP, major outer membrane protein.
As can be seen in Table 1, the 3.0-nS pore size of the BrkA C-terminal protein is larger than those reported for the typical E. coli porins OmpF and OmpC, as well as many other proteins, but is not without precedent. OprF from Pseudomonas aeruginosa and the 53-kDa outer sheath protein of Treponema denticola were found to have single-channel conductances of 5.6 nS (3) and 10.9 nS (13), respectively. As well, a 59-kDa cell wall protein from Mycobacterium chelonae was found to have a channel size of 2.7 nS (47).
A larger pore size for the BrkA C-terminal domain would be expected based on its protein-exporting function. PapC, an outer membrane usher through which the subunits of P pili are exported in uropathogenic E. coli, has a pore diameter of at least 2 nm (45). This is large enough to allow the passage of unravelled pilus subunits. It appears that a similar requirement for unfolded passenger proteins also applies to autotransporters.
In order for the BrkA C-terminal protein to have consistently formed channels, one would expect it to be properly folded but the denaturing purification procedure that we used brings up the question of whether or not the protein should be capable of assuming its native conformation. In order to renature the protein, we slowly removed the urea and used a detergent. Examples of the renaturation of outer membrane proteins into a native conformation using related procedures have been shown previously. Outer membrane proteins extracted from inclusion bodies and renatured were found to have the same pore-forming characteristics as those obtained from the outer membrane (40, 41). As well, crystals of monomeric outer membrane protein A (OmpA) were obtained from inclusion bodies (35) and then used for structure determination by X-ray diffraction analysis (34).
The proposed native conformation of the autotransporter C-terminal domains, as appears to be the case for all outer membrane proteins, is a multistranded β-barrel. Even though all of the outer membrane proteins examined to date contain this same basic structure (34), they do not all form pores, which is one reason why the pore-forming ability of the BrkA C-terminal domain needed to be tested. The computer-predicted models of these proteins give a general idea of their structure, but as evidenced by the recently published structures of OmpA (34) and FepA (9), they can be wrong. The main structural features of autotransporters that need to be elucidated are the exact number of strands, whether the extreme N-terminal strand of the barrel faces in or out, and whether or not the pore is blocked after export of the N-terminal domain of the protein. We are currently addressing these questions by mapping the topology of the BrkA C-terminal domain.
In summary, we have shown that the BrkA C-terminal domain is capable of forming a pore, which supports the proposed model of autotransporter export. To our knowledge, this is the first time that pore-forming ability of the C-terminal domain has ever been demonstrated for a member of the autotransporter family.
Acknowledgments
This work was supported by Natural Sciences and Engineering Research Council grant OGP0194599. J.L.S. was a recipient of an NSERC PGS A scholarship.
We thank the R. E. W. Hancock laboratory for assistance with the black lipid bilayer experiments and helpful discussions of the data and, in particular, Fiona Brinkman for critical reading of the manuscript. We thank Roger Parton for the gift of the anti-BrkA C-terminal protein monoclonal antibody and Dave Oliver and Carrie Mathewson for the DO218 protein.
REFERENCES
- 1.Armstrong S K, Parr T R, Jr, Parker C D, Hancock R E W. Bordetella pertussis major outer membrane porin protein forms small, anion-selective channels in lipid bilayer membranes. J Bacteriol. 1986;166:212–216. doi: 10.1128/jb.166.1.212-216.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benz R, Darveau R P, Hancock R E W. Outer-membrane protein PhoE from Escherichia coli forms anion-selective pores in lipid-bilayer membranes. Eur J Biochem. 1984;140:319–324. doi: 10.1111/j.1432-1033.1984.tb08104.x. [DOI] [PubMed] [Google Scholar]
- 3.Benz R, Hancock R E W. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim Biophys Acta. 1981;646:298–308. doi: 10.1016/0005-2736(81)90336-9. [DOI] [PubMed] [Google Scholar]
- 4.Benz R, Ishii J, Nakae T. Determination of ion permeability through the channels made of porins from the outer membrane of Salmonella typhimurium in lipid bilayer membranes. J Membr Biol. 1980;56:19–29. doi: 10.1007/BF01869348. [DOI] [PubMed] [Google Scholar]
- 5.Benz R, Maier E, Gentschev I. TolC of Escherichia coli functions as an outer membrane channel. Zentbl Bakteriol. 1993;278:187–196. doi: 10.1016/s0934-8840(11)80836-4. [DOI] [PubMed] [Google Scholar]
- 6.Benz R, Schmid A, Hancock R E W. Ion selectivity of gram-negative bacterial porins. J Bacteriol. 1985;162:722–727. doi: 10.1128/jb.162.2.722-727.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benz R, Schmid A, Maier C, Bremer E. Characterization of the nucleoside-binding site inside the Tsx channel of Escherichia coli outer membrane: reconstitution experiments with lipid bilayer membranes. Eur J Biochem. 1988;176:699–705. doi: 10.1111/j.1432-1033.1988.tb14333.x. [DOI] [PubMed] [Google Scholar]
- 8.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 9.Buchanan S K, Smith B S, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 9a.Hancock, R. E. W. Personal communication.
- 10.Charles I, Fairweather N, Pickard D, Beesley J, Anderson R, Dougan G, Roberts M. Expression of the Bordetella pertussis P.69 pertactin adhesin in Escherichia coli: fate of the carboxy-terminal domain. Microbiology. 1994;140:3301–3308. doi: 10.1099/13500872-140-12-3301. [DOI] [PubMed] [Google Scholar]
- 11.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 12.Egile C, d’Hauteville H, Parsot C, Sansonetti P J. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol. 1997;23:1063–1073. doi: 10.1046/j.1365-2958.1997.2871652.x. [DOI] [PubMed] [Google Scholar]
- 13.Egli C, Leung W K, Müller K-H, Hancock R E W, McBride B C. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect Immun. 1993;61:1694–1699. doi: 10.1128/iai.61.5.1694-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fajardo D A, Cheung J, Ito C, Sugawara E, Nikaido H, Misra R. Biochemistry and regulation of a novel Escherichia coli K-12 porin protein, OmpG, which produces unusually large channels. J Bacteriol. 1998;180:4452–4459. doi: 10.1128/jb.180.17.4452-4459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez R C, Weiss A A. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect Immun. 1994;62:4727–4738. doi: 10.1128/iai.62.11.4727-4738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez R C, Weiss A A. Serum resistance in bvg-regulated mutants of Bordetella pertussis. FEMS Microbiol Lett. 1998;163:57–63. doi: 10.1111/j.1574-6968.1998.tb13026.x. [DOI] [PubMed] [Google Scholar]
- 17.Finn T M, Stevens L A. Tracheal colonization factor: a Bordetella pertussis secreted virulence determinant. Mol Microbiol. 1995;16:625–634. doi: 10.1111/j.1365-2958.1995.tb02425.x. [DOI] [PubMed] [Google Scholar]
- 18.Gabay J E, Blake M, Niles W D, Horwitz M A. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J Bacteriol. 1985;162:85–91. doi: 10.1128/jb.162.1.85-91.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 20.Jose J, Jähnig F, Meyer T F. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol Microbiol. 1995;18:377–382. doi: 10.1111/j.1365-2958.1995.mmi_18020378.x. [DOI] [PubMed] [Google Scholar]
- 21.Klauser T, Krämer J, Otzelberger K, Pohlner J, Meyer T F. Characterization of the Neisseria Igaβ-core, the essential unit for outer membrane targeting and extracellular protein secretion. J Mol Biol. 1993;234:579–593. doi: 10.1006/jmbi.1993.1613. [DOI] [PubMed] [Google Scholar]
- 22.Klauser T, Pohlner J, Meyer T F. The secretion pathway of IgA protease-type proteins in gram-negative bacteria. Bioessays. 1993;15:799–805. doi: 10.1002/bies.950151205. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Rutz J M, Feix J B, Klebba P E. Permeability properties of a large gated channel within the ferric enterobactin receptor, FepA. Proc Natl Acad Sci USA. 1993;90:10653–10657. doi: 10.1073/pnas.90.22.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loveless B J, Saier M H., Jr A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membr Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 26.Lutwyche P, Exner M M, Hancock R E W, Trust T J. A conserved Aeromonas salmonicida porin provides protective immunity to rainbow trout. Infect Immun. 1995;63:3137–3142. doi: 10.1128/iai.63.8.3137-3142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier C, Bremer E. Pore-forming activity of the Tsx protein from the outer membrane of Escherichia coli: Demonstration of a nucleoside-specific binding site. J Biol Chem. 1988;263:2493–2499. [PubMed] [Google Scholar]
- 28.Maurer J, Jose J, Meyer T F. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol. 1997;179:794–804. doi: 10.1128/jb.179.3.794-804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhopadhyay S, Basu D, Chakrabarti P. Characterization of a porin from Mycobacterium smegmatis. J Bacteriol. 1997;179:6205–6207. doi: 10.1128/jb.179.19.6205-6207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido H. Porins and specific channels of bacterial outer membranes. Mol Microbiol. 1992;6:435–442. doi: 10.1111/j.1365-2958.1992.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido H, Nikaido K, Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J Biol Chem. 1991;266:770–779. [PubMed] [Google Scholar]
- 32.O’Toole P W, Austin J W, Trust T J. Identification and molecular characterization of a major ring-forming surface protein from the gastric pathogen Helicobacter mustelae. Mol Microbiol. 1994;11:349–361. doi: 10.1111/j.1365-2958.1994.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 33.Page W J, Huyer G, Huyer M, Worobec E A. Characterization of the porins of Campylobacter jejuni and Campylobacter coli and implications for antibiotic susceptibility. Antimicrob Agents Chemother. 1989;33:297–303. doi: 10.1128/aac.33.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pautsch A, Schulz G E. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 35.Pautsch A, Vogt J, Model K, Siebold C, Schulz G E. Strategy for membrane protein crystallization exemplified with OmpA and OmpX. Proteins. 1999;34:167–172. [PubMed] [Google Scholar]
- 36.Poulsen K, Brandt J, Hjorth J P, Thøgersen H C, Kilian M. Cloning and sequencing of the immunoglobulin A1 protease gene (iga) of Haemophilus influenzae serotype b. Infect Immun. 1989;57:3097–3105. doi: 10.1128/iai.57.10.3097-3105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provence, D. L., and R. Curtiss III. Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect. Immun. 62:1369–1380. [DOI] [PMC free article] [PubMed]
- 38.Rieß F G, Lichtinger T, Cseh R, Yassin A F, Schaal K P, Benz R. The cell wall porin of Nocardia farcinica: biochemical identification of the channel-forming protein and biophysical characterization of the channel properties. Mol Microbiol. 1998;29:139–150. doi: 10.1046/j.1365-2958.1998.00914.x. [DOI] [PubMed] [Google Scholar]
- 39.Saint N, De E, Julien S, Orange N, Molle G. Ionophore properties of OmpA of Escherichia coli. Biochim Biophys Acta. 1993;1145:119–123. doi: 10.1016/0005-2736(93)90388-g. [DOI] [PubMed] [Google Scholar]
- 40.Saxena K, Drosou V, Maier E, Benz R, Ludwig B. Ion selectivity reversal and induction of voltage-gating by site-directed mutations in the Paracoccus denitrificans porin. Biochemistry. 1999;38:2206–2212. doi: 10.1021/bi982296f. [DOI] [PubMed] [Google Scholar]
- 41.Schmid B, Maveyraud L, Krömer M, Schulz G E. Porin mutants with new channel properties. Protein Sci. 1998;7:1603–1611. doi: 10.1002/pro.5560070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugawara E, Nikaido H. Pore-forming activity of OmpA protein of Escherichia coli. J Biol Chem. 1992;267:2507–2511. [PubMed] [Google Scholar]
- 43.Suhr M, Benz I, Schmidt M A. Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a β-barrel structure. Mol Microbiol. 1996;22:31–42. doi: 10.1111/j.1365-2958.1996.tb02653.x. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki T, Lett M, Sasakawa C. Extracellular transport of VirG protein in Shigella. J Biol Chem. 1995;270:30874–30880. doi: 10.1074/jbc.270.52.30874. [DOI] [PubMed] [Google Scholar]
- 45.Thanassi D G, Saulino E T, Lombardo M, Roth R, Heuser J, Hultgren S J. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trias J, Benz R. Permeability of the cell wall of Mycobacterium smegmatis. Mol Microbiol. 1994;14:283–290. doi: 10.1111/j.1365-2958.1994.tb01289.x. [DOI] [PubMed] [Google Scholar]
- 47.Trias J, Jarlier V, Benz R. Porins in the cell wall of mycobacteria. Science. 1992;258:1479–1481. doi: 10.1126/science.1279810. [DOI] [PubMed] [Google Scholar]
- 48.Vachon V, Kristjanson D N, Coulton J W. Outer membrane porin protein of Haemophilus influenzae type b: pore size and subunit structure. Can J Microbiol. 1988;34:134–140. doi: 10.1139/m88-027. [DOI] [PubMed] [Google Scholar]
- 49.Vachon V, Laprade R, Coulton J W. Properties of the porin of Haemophilus influenzae type b in planar lipid bilayer membranes. Biochim Biophys Acta. 1986;861:74–82. doi: 10.1016/0005-2736(86)90373-1. [DOI] [PubMed] [Google Scholar]
- 50.Woodruff W A, Parr T R, Jr, Hancock R E W, Hanne L F, Nicas T I, Iglewski B H. Expression in Escherichia coli and function of Pseudomonas aeruginosa outer membrane porin protein F. J Bacteriol. 1986;167:473–479. doi: 10.1128/jb.167.2.473-479.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]