Abstract
We have sequenced the entire region of DNA required for the biosynthesis of CS5 pili from enterotoxigenic Escherichia coli O115:H40 downstream of the major subunit gene, designated csfA (for coli surface factor five A). Five more open reading frames (ORFs) (csfB, csfC, csfE, csfF, and csfD) which are transcribed in the same direction as the major subunit and are flanked by a number of insertion sequence regions have been identified. T7 polymerase-mediated overexpression of the cloned csf ORFs confirmed protein sizes based on the DNA sequences that encode them. The expression of only the csf region in E. coli K-12 resulted in the hemagglutination of human erythrocytes and the cell surface expression of CS5 pili, suggesting that the cluster contains all necessary information for CS5 pilus biogenesis and function.
Enterotoxigenic Escherichia coli (ETEC) is a major cause of acute diarrhea in children in the developing world and in international travellers in these areas (24, 31). Specific colonization of the epithelium is mediated through the action of proteinaceous surface structures known as pili or fimbriae. In ETEC strains isolated from humans, such pili are referred to as colonization factor antigens (CFAs).
A number of the operons involved in CFA biosynthesis have been cloned and characterized, including CFA/I (17) and CFA/II isolates expressing the heterogeneous coli surface (CS) antigens CS1 (10, 32, 36), CS2 (11), and CS3 (16). CFA/IV is present on the surfaces of 10 to 20% of all ETEC strains. The pili produced are antigenically heterogeneous, consisting of CS4, CS5, and CS6 (39). CFA/IV ETEC strains express the nonpilus antigen CS6 with either CS5 or CS4 pili. CS5 and CS6 pili are found on strains of serotypes O6, O29, O92, O114, O115, and O167 (23, 39).
Members of our group previously described the molecular cloning of the region required for the biosynthesis of functional CS5 pili from ETEC strains belonging to the O115:H40 serotype, isolated originally during an outbreak among aboriginal children in Central Australia (15, 20). In subsequent nucleotide sequencing of the region encoding the major CS5 pilin subunit, significant homology within the signal sequence and at the carboxy terminus with the corresponding major subunit of F41 pili from porcine ETEC was shown (5).
In this study, we sequenced and analyzed the entire DNA region encoding the biosynthesis of CS5 pili immediately downstream of the major subunit gene, along with flanking regions of DNA. We have identified several insertion sequences (IS) and IS-related sequences surrounding five open reading frames (ORFs), designated csfB (for coli surface factor five B), csfC, csfE, csfF, and csfD, and the major subunit previously described (5), termed csfA. Through immunogold electron microscopy experiments, we also showed that the csf cluster, minus the flanking DNA regions, mediates the cell surface expression of CS5 pili in E. coli K-12, suggesting that all the information required for CFA biogenesis is contained within these ORFs.
Analysis of the DNA sequence downstream of the major subunit, csfA.
Previously, Clark et al. (5) showed that immediately downstream of the major subunit (csfA) is a proposed transcriptional attenuator sequence and the beginning of a second ORF with a good ribosome binding site (5′-GGAA-3′) located 8 nucleotides (nt) upstream of the AUG initiation codon. A predicted signal sequence cleavage site was between Phe25 and Ser26 (5).
To facilitate the sequencing of the region downstream from the beginning of this predicted ORF, designated csfB, pPM1312 (15) was digested with PstI and then cloned into pBS-SK+ (Stratagene) to yield pPM5303, which was digested with SacI and XbaI and then treated with exonuclease III, which deletes bases from the unprotected SacI overhang of DNA for a given period of time. The DNA was then treated with Klenow enzyme and S1 nuclease, ligated with T4 DNA ligase, and transformed into DH5α (Bethesda Research Laboratories). Chosen nested deletion products were then directly sequenced with the M13 forward and reverse sequencing primers. Sequencing kits for both dye primer and dye terminator reactions were purchased from Applied Biosystems (Foster City, Calif.), and the sequencing was carried out on a model 373A Applied Biosystems automated sequencer.
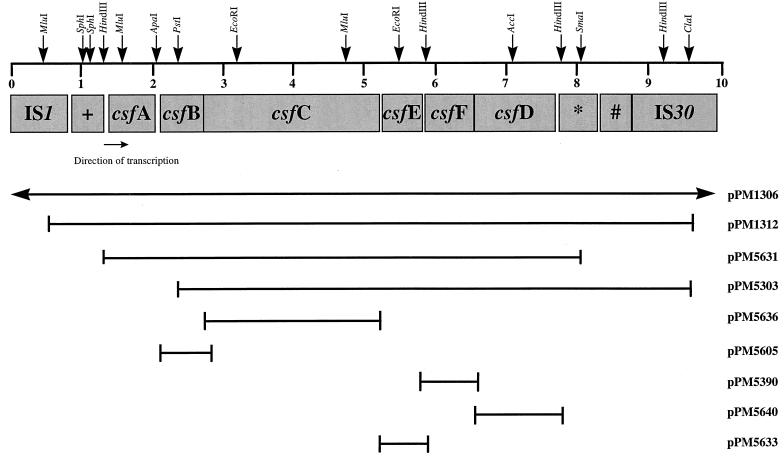
The csfB ORF extends from nt 2096 to 2770 (Fig. 1) and is predicted to encode a periplasmic protein of 24 kDa with a pI value of 6.42 and a mean hydrophobicity index of −0.41. The following ORF, termed csfC, which overlaps csfB by 4 nt, extends from nt 2767 to 5211 (Fig. 1). A potential ribosome binding site (5′-AAGA-3′) is located 8 nt upstream of the AUG start codon (37). The csfC ORF is predicted to encode a 90.3-kDa protein with a pI value of 8.61 and a mean hydrophobicity index of −0.44. Cleaving this protein at a potential signal sequence cleavage site between Ala21 and Asp22 (43) would result in a mature protein of 88 kDa in the periplasm. The overlap of reading frames csfB and csfC is suggestive of translational coupling (22). Immediately downstream of csfC, initiated within 17 nt, is a fourth ORF, denoted csfE, which extends from nt 5227 to 5838 (Fig. 1). csfE may encode a protein of 22.8 kDa with a pI value of 7.72 and a mean hydrophobicity index of −0.01. A potential signal sequence cleavage site exists between Leu20 and Gln21, giving rise to a mature protein of 20.3 kDa in the periplasm. The csfE reading frame is preceded by a potential ribosome binding site (5′-GAAGAG-3′) located at the optimal distance of 7 nt from the AUG start codon.
FIG. 1.
Genetic organization of the region required for the biosynthesis of CS5 pili in ETEC O115:H40. ORFs belonging to the CS5 cluster are designated csfA to csfF. The region is flanked by a variety of IS elements. +, region homologous to part of orfB in Tn21; ∗, region homologous to part of IS66; #, region homologous to part of IS911. IS1 and a defective IS30 are indicated. The upper line refers to the position of the genes relative to the sequence length, in kilobases, with relevant restriction sites indicated. Plasmids used in this study and the ORF(s) they encompass are indicated.
The fifth ORF in the cluster, termed csfF, lies 13 nt downstream of csfE and extends from nt 5852 to 6526 (Fig. 1), giving rise to a 25.5-kDa protein with a pI of 8.68 and a mean hydrophobicity index of −0.21. A potential signal sequence cleavage site located between Ala19 and Phe20 would give rise to a 23.3-kDa protein in the periplasm. A potential ribosome binding site (5′-GAGA-3′) is located 8 bases upstream from the AUG start codon.
The final ORF in the cluster, csfD, overlaps csfF by 4 nt (which is also indicative of translational coupling), extends from nt 6523 to 7644 (Fig. 1), and is predicted to encode a protein of 41.0 kDa with a pI value of 8.3 and a mean hydrophobicity index of −0.3. A mature 38.9-kDa protein in the periplasm would be produced from the potential signal sequence cleavage site located between Ala19 and Ala20. The potential ribosome binding site is located 10 nt upstream of the AUG start codon (5′-AAGG-3′), which is not optimal for efficient translation (13, 18) but may be offset by translational coupling with csfF.
Homologies.
DNA and amino acid sequence homologies were detected with the basic local alignment search tool (BLAST) (1), and the sequences were aligned with CLUSTAL W (40). A search of the databases with the derived sequence of the CsfC protein showed that the protein’s greatest homology is with the membrane usher protein CooC from CS1 pili (GenBank accession no. X76908) (10), with 19% protein identity and 38% similarity. Secondary-structure analysis with PHDSec (28–30) predicted a high β-sheet (44.5%) and low α-helix (9.8%) content for this protein, along with a high percentage of charged amino acids (18.4%) and no long regions of hydrophobic amino acids. Similar properties were defined for the CooC protein (10), and such features are commonly found in other known outer membrane proteins (25). Based on homology searches and secondary-structure analysis, csfC is predicted to encode an outer membrane usher protein.
The amino acid sequence of CsfB had 30% identity and 51% similarity with CsfF, while CsfA was homologous to CsfD, with 14% identity and 24% similarity. Although the percent identity between CsfA and CsfD is low, it is similar to that between the minor tip-associated pilin subunits CooD, PapG, and FimH and their cognate major pilins in CS1, Pap, and type I pili, respectively (12, 33). CsfE showed no homology to sequences in the database (P = 1.0).
DNA sequence upstream and downstream of the csf cluster.
DNA sequences upstream and downstream of the csf cluster from pPM1312 to pPM1306 (15) showed regions of homology with a number of IS elements, including orfB of transposon Tn21 from Shigella flexneri (GenBank accession no. U42226) (13a), IS66 (GenBank accession no. M10204) (19), and IS911 from Shigella dysenteriae (GenBank accession no. X17613) (27). Furthermore, a defective IS30 element in the transposase sequence was identified and determined to be 98% identical to the complete IS30 (6), along with an intact IS1 element that has 99% sequence identity and 98% protein identity with the published IS1 sequence (26) (Fig. 1).
Immediately upstream of csfA are two potential ς70 promoter sequences, from nt 1262 to 1291 and nt 1375 to 1402. The former has −35 (TTGACG) and −10 (TATTGT) sites with an 18-nt spacer region, and the latter has −35 (TTTTCA) and −10 (TATTGT) sites separated by a 16-nt spacer region. There is a high degree of identity for the −35 and −10 consensus sequences (TTGACA and TATAAT); however, the spacing between them is not optimal (17 nt), but it is acceptable (14).
Protein expression studies.
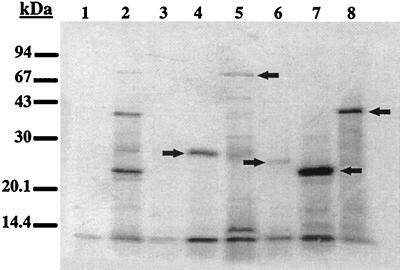
Plasmids were transformed into DH5α (Bethesda Research Laboratories) containing the plasmid pGP1-2 (38). Overexpression of the proteins was carried out essentially according to the method described by Tabor and Richardson (38). The csfB, csfC, csfD, csfE, and csfF ORFs were predicted to encode proteins of 25.8, 90.3, 41, 22.8, and 25.5 kDa, respectively, with the DNASIS program (LKB-Hitachi). In order to confirm this, each ORF was PCR amplified with Amplitaq polymerase (Roche) and gene-specific oligonucleotides and then cloned into pBS-SK+ (Stratagene) oriented downstream from the T7 promoter (Fig. 1). The generation of pPM5605 (csfB) utilized oligonucleotides 2944 (5′-GGGAATTCCCAGGCAGCTGCTGC-3′) and 2945 (5′-GGTCTAGAGCCAGCTCACTTTTATCAGC-3′), which included EcoRI and XbaI sites, respectively (underlined). pPM5636 (csfC) was constructed with oligonucleotides 2977 (5′-GGCTGCAGCGGACGCGAACGAAAACTG-3′) and 2978 (5′-GGTCTAGACCATATAGAAGACAAGTTTG-3′), which included PstI and XbaI sites, respectively (underlined). pPM5640 (csfD) was constructed with oligonucleotides 2979 (5′-GGCTGCAGGAGCATGTACAGACGCTT-3′) and 2980 (5′-GGTCTAGACCACTACAGGAGGTAATTG-3′), which included PstI and XbaI sites, respectively (underlined). pPM5633 (csfE) was constructed with oligonucleotides 2973 (5′-GGCTGCAGGGAAATATGTAAGCA-3′) and 2974 (5′-GGTCTAGACGATTAAAATCGCTCAAAAA-3′), which included PstI and XbaI sites, respectively (underlined). Finally, the construction of pPM5390 (csfF) required oligonucleotides 2971 (5′-GGGAATTCGAGGACTATGTAGGG-3′) and 2972 (5′-GGTCTAGATGAACCATAAAGGAAAAAAAAG-3′), which included EcoRI and XbaI sites, respectively (underlined). These plasmids were compared with pPM5631, which contained all of the csf cluster; constructed by the digestion of pPM1312 with SphI and SmaI; and then cloned into pGEM7-ZF+ (Promega) oriented from the T7 promoter. Overexpressed proteins were labelled with 10 μCi of l-[35S]methionine (Amersham) and separated by gel electrophoresis in sodium dodecyl sulfate on 15% polyacrylamide gels. Several protein bands in pPM5631 (Fig. 2, lane 2) that correlated well with the sizes of the individual overexpressed gene products (Fig. 2, lanes 4 to 8) were detected. The CsfE and CsfD proteins migrated with apparent molecular masses in approximate agreement with their respective DNA sequences (Fig. 2, lanes 7 and 8, respectively). A band corresponding to the expected size of CsfE in pPM5631 (Fig. 2, lane 2) may have been either CsfE or CsfA, which also migrated to approximately 23 kDa (5). CsfF migrated with an apparent molecular mass of 27 kDa, which was slightly higher than that predicted based on DNA sequence alone (Fig. 2, lane 4). Similarly, the apparent molecular mass of CsfB was 26 kDa, which was slightly higher than the molecular mass predicted in DNA sequence studies (Fig. 2, lane 6).
FIG. 2.
T7 RNA polymerase promoter expression of CsfB, CsfC, CsfD, CsfE, and CsfF clones in pBS-SK+. Proteins were labelled with [35S]methionine, and whole-cell fractions on 15% polyacrylamide gels were subjected to gel electrophoresis in sodium dodecyl sulfate, followed by autoradiography of the dried gels. Lane 1, pGEM7-ZF+; lane 2, pPM5631; lane 3, pBS-SK+; lane 4, pPM5390 (CsfF); lane 5, pPM5636 (CsfC); lane 6, pPM5605 (CsfB); lane 7, pPM5633 (CsfE); lane 8, pPM5640 (CsfD). Arrows indicate the positions of the overexpressed proteins.
The CsfC protein and the mature form were predicted to have molecular masses of 90.3 and 88 kDa, respectively; however, T7 analysis indicated an apparent molecular mass of only 71 kDa (Fig. 2, lane 5). The 71-kDa product may represent a degradation product which is readily observed in the E. coli K-12 background, while the full-length form is not detectable. Proteolytic degradation products are a common occurrence among other known outer membrane usher proteins, including CooC (42), PapC (8), FaeD (41), and FasD (35). However, it is worth noting that there is an unusual distribution of charged residues (18.4%) within the coding sequence for CsfC, which may also account for the 71-kDa band observed.
Expression of the csf cluster in E. coli K-12.
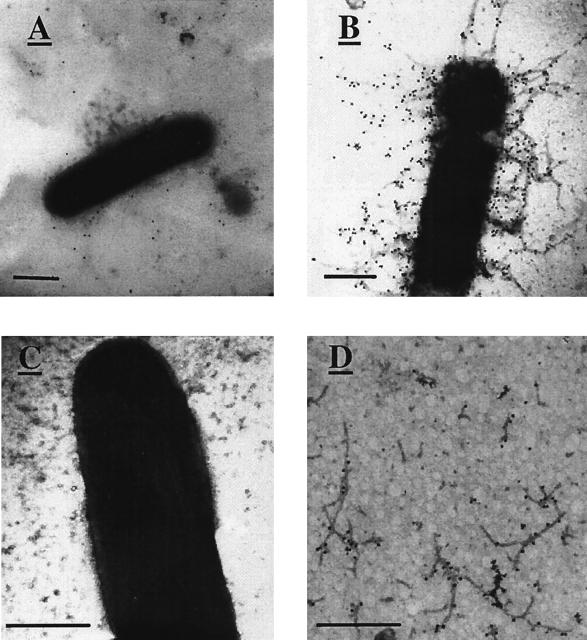
To examine whether pPM5631 contains all the information necessary for the biogenesis of functional CS5 pili, hemagglutinations and immunogold electron microscopy were performed. Bacterial strains were grown overnight on CFA agar (9) at 37°C, with ampicillin (100 μg/ml) as required. A sample of resuspended bacteria in phosphate-buffered saline was mixed with an equal volume of 5% human group A+ erythrocytes in the presence of 0.1 M d-(+)-mannose. Both wild-type strain PE423 (20) and E. coli K-12 strain DH5α containing pPM5631 readily agglutinated, while the negative control strain containing pGEM-7ZF+ was unable to do so. Immunogold electron microscopy with rabbit anti-native CS5 pilus antiserum (15) showed that E. coli K-12 containing pPM5631 exhibited peritrichous pili (Fig. 3B), some of which were found detached in the surrounding milieu (Fig. 3D), and they were in far greater amounts than pili of the corresponding wild-type strain, PE423 (Fig. 3A). E. coli K-12 containing pGEM-7ZF+ showed no cell surface pili, as expected (Fig. 3C). These data show that the csf cluster contains all information required for the expression of a functional CS5 pilus on the surface of E. coli K-12 strain DH5α.
FIG. 3.
Immunogold electron microscopy of PE423 and E. coli K-12 derivatives. (A) PE423; (B) E. coli K-12 containing pPM5631; (C) E. coli K-12 containing pGEM7-ZF+; (D) cell-free CS5 pili. Bars, 500 nm.
Conclusion.
CS5 pili may be representative of a distinct class of pili, since the protein products encoded by four of the six ORFs comprising the CS5 region show no significant homology (P > 0.65) to any sequence in the databases. Several of the products encoded by the csf ORFs are homologous to each other, namely, CsfA to CsfD and CsfB to CsfF. This may indicate that the protein products have similar functions in CS5 pilus biogenesis. A lack of homology to any of the known families of regulatory proteins suggests that the csf cluster does not encode a specific regulatory protein. Therefore, it is possible that a regulatory protein is not required for CS5 pilus expression in E. coli K-12, unless a homologous protein in E. coli K-12 is able to functionally replace a wild-type regulator. No specific regulatory gene has been identified within the cluster of CS3 pilus cst genes, which have been shown to readily produce CS3 pili in an E. coli K-12 background (16, 21). Conversely, the expression of CS1 and CS2 pili is dependent on a plasmid-borne positive regulatory gene, rns (3), which is a member of the araC family of regulators and highly homologous to cfaD and cfaR, which are required for CFA/I expression (4, 34). Another proposed plasmid-borne regulatory gene is csvR, originally cloned from E. coli O167:H5, a strain which produces not only CS5 pili but also CS6 pili (44). The csvR gene is 87% homologous to cfaR and is able to functionally replace cfaR to mediate CFA/I expression (7). It is currently unknown whether CsvR plays a role in CS5 biogenesis.
The average G+C content of the six ORFs comprising the csf cluster is only 37.1%, compared with the average G+C content of 50.8% in E. coli (2). The pattern of codon usage within the csf cluster also differs significantly from the codon usage of E. coli. The rare or modulator codons in the csf cluster of E. coli are used much more frequently than highly and minimally expressed genes of E. coli (data not shown). This may indicate that the csf cluster was recently introduced into E. coli from another organism by transposition, which may be supported by the presence of flanking IS regions. The identification of only one set of potential promoters immediately upstream of csfA, along with overlapping reading frames for four of the six csf ORFs, indicates that this region likely constitutes an operon.
Expression of the cloned csf region in E. coli K-12 produced cell surface CS5 pili far in excess of those observed in the wild-type strain, PE423. This may be due to a gene dosage effect, a consequence of the expression of the csf cluster from a high-copy-number plasmid versus expression from the very-low-copy-number virulence plasmids found in the wild-type ETEC strains. CS5 pilus expression from the high-copy-number plasmid in E. coli K-12 provides a useful system for investigating phenotypic changes in expression resulting from mutational and complementation experiments. Consequently, we are currently investigating the role of the individual ORFs in CS5 biogenesis.
Nucleotide sequence accession number.
The DNA sequence obtained in this study has been deposited into the EMBL database under accession no. AJ224079.
Acknowledgments
We thank the National Health and Medical Research Council (NH and MRC) of Australia for support and Chris Cursaro for excellent technical assistance.
T.G.D. is the recipient of a Northfield Laboratories University-Industry Scholarship.
Footnotes
Present address: Astra Research Center Boston, Inc., Cambridge, MA 02139-4239.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Caron J, Coffield L M, Scott J R. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc Natl Acad Sci USA. 1989;86:963–967. doi: 10.1073/pnas.86.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron J, Scott J R. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect Immun. 1990;58:874–878. doi: 10.1128/iai.58.4.874-878.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark C A, Heuzenroeder M W, Manning P A. Colonization factor antigen CFA/IV (PCF8775) of human enterotoxigenic Escherichia coli: nucleotide sequence of the CS5 determinant. Infect Immun. 1992;60:1254–1257. doi: 10.1128/iai.60.3.1254-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalrymple B, Caspers P, Arber W. Nucleotide sequence of the prokaryotic mobile genetic element IS30. EMBO J. 1984;3:2145–2149. doi: 10.1002/j.1460-2075.1984.tb02104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan L A, Willshaw G A, van der Zeijst B A, Gaastra W. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol Lett. 1991;67:341–346. doi: 10.1111/j.1574-6968.1991.tb04487.x. [DOI] [PubMed] [Google Scholar]
- 8.Dodson K W, Jacob-Dubuisson F, Striker R T, Hultgren S J. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans D G, Evans D J, Jr, Clegg S, Pauley J A. Purification and characterization of the CFA/I antigen of enterotoxigenic Escherichia coli. Infect Immun. 1979;25:738–748. doi: 10.1128/iai.25.2.738-748.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froehlich B J, Karakashian A, Melsen L R, Wakefield J C, Scott J R. CooC and CooD are required for assembly of CS1 pili. Mol Microbiol. 1994;12:387–401. doi: 10.1111/j.1365-2958.1994.tb01028.x. [DOI] [PubMed] [Google Scholar]
- 11.Froehlich B J, Karakashian A, Sakellaris H, Scott J R. Genes for CS2 pili of enterotoxigenic Escherichia coli and their interchangeability with those for CS1 pili. Infect Immun. 1995;63:4849–4856. doi: 10.1128/iai.63.12.4849-4856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girardeau J P, Bertin Y. Pilins of fimbrial adhesins of different member species of Enterobacteriaceae are structurally similar to the C-terminal half of adhesin proteins. FEBS Lett. 1995;357:103–108. doi: 10.1016/0014-5793(94)01340-7. [DOI] [PubMed] [Google Scholar]
- 13.Gold L, Pribnow D, Schneider T, Shinedling S, Singer B S, Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- 13a.Hall, R. M. Unpublished results.
- 14.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heuzenroeder M W, Neal B L, Thomas C J, Halter R, Manning P A. Characterization and molecular cloning of the PCF8775 CS5 antigen from an enterotoxigenic Escherichia coli O115:H40 isolated in Central Australia. Mol Microbiol. 1989;3:303–310. doi: 10.1111/j.1365-2958.1989.tb00175.x. [DOI] [PubMed] [Google Scholar]
- 16.Jalajakumari M B, Thomas C J, Halter R, Manning P A. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol Microbiol. 1989;3:1685–1695. doi: 10.1111/j.1365-2958.1989.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 17.Jordi B J, Willshaw G A, van der Zeijst B A, Gaastra W. The complete nucleotide sequence of region 1 of the CFA/I fimbrial operon of human enterotoxigenic Escherichia coli. DNA Sequence. 1992;2:257–263. doi: 10.3109/10425179209020811. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machida Y, Sakurai M, Kiyokawa S, Ubasawa A, Suzuki Y, Ikeda J E. Nucleotide sequence of the insertion sequence found in the T-DNA region of mutant Ti plasmid pTiA66 and distribution of its homologues in octopine Ti plasmid. Proc Natl Acad Sci USA. 1984;81:7495–7499. doi: 10.1073/pnas.81.23.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning P A, Higgins G D, Lumb R, Lanser J A. Colonization factor antigens and a new fimbrial type, CFA/V, on O115:H40 and H− strains of enterotoxigenic Escherichia coli in central Australia. J Infect Dis. 1987;156:841–844. doi: 10.1093/infdis/156.5.841. [DOI] [PubMed] [Google Scholar]
- 21.Manning P A, Timmis K N, Stevenson G. Colonization factor antigen II (CFA/II) of enterotoxigenic Escherichia coli: molecular cloning of the CS3 determinant. Mol Gen Genet. 1985;200:322–327. doi: 10.1007/BF00425443. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy J E, Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990;6:78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- 23.McConnell M M, Thomas L V, Day N P, Rowe B. Enzyme-linked immunosorbent assays for the detection of adhesion factor antigens of enterotoxigenic Escherichia coli. J Infect Dis. 1985;152:1120–1127. doi: 10.1093/infdis/152.6.1120. [DOI] [PubMed] [Google Scholar]
- 24.Merson M H, Black R E, Khan M U, Huq I. Enterotoxigenic Escherichia coli diarrhea: acquired immunity and transmission in an endemic area. In: Ouchterlony O, Holmgren J, editors. Cholera and related diarrheas: molecular basis of a global health problem. 43rd Nobel Symposium. S. Basel, Switzerland: Karger; 1980. pp. 34–45. [Google Scholar]
- 25.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtsubo H, Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci USA. 1978;75:615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prère M-F, Chandler M, Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990;172:4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 30.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 31.Sack R B. Human diarrheal disease caused by enterotoxigenic Escherichia coli. Annu Rev Microbiol. 1975;29:333–353. doi: 10.1146/annurev.mi.29.100175.002001. [DOI] [PubMed] [Google Scholar]
- 32.Sakellaris H, Balding D P, Scott J R. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol Microbiol. 1996;21:529–541. doi: 10.1111/j.1365-2958.1996.tb02562.x. [DOI] [PubMed] [Google Scholar]
- 33.Sakellaris H, Scott J R. New tools in an old trade: CS1 pilus morphogenesis. Mol Microbiol. 1998;30:681–687. doi: 10.1046/j.1365-2958.1998.01088.x. [DOI] [PubMed] [Google Scholar]
- 34.Savelkoul P H, Willshaw G A, McConnell M M, Smith H R, Hamers A M, van der Zeijst B A, Gaastra W. Expression of CFA/I fimbriae is positively regulated. Microb Pathog. 1990;8:91–99. doi: 10.1016/0882-4010(90)90073-y. [DOI] [PubMed] [Google Scholar]
- 35.Schifferli D M, Alrutz M A. Permissive linker insertion sites in the outer membrane protein of 987P fimbriae of Escherichia coli. J Bacteriol. 1994;176:1099–1110. doi: 10.1128/jb.176.4.1099-1110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott J R, Wakefield J C, Russell P W, Orndorff P E, Froehlich B J. CooB is required for assembly but not transport of CS1 pilin. Mol Microbiol. 1992;6:293–300. doi: 10.1111/j.1365-2958.1992.tb01471.x. [DOI] [PubMed] [Google Scholar]
- 37.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas L V, McConnell M M, Rowe B, Field A M. The possession of three novel coli surface antigens by enterotoxigenic Escherichia coli strains positive for the putative colonization factor PCF8775. J Gen Microbiol. 1985;131:2319–2326. doi: 10.1099/00221287-131-9-2319. [DOI] [PubMed] [Google Scholar]
- 40.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valent Q A, Zaal J, de Graaf F K, Oudega B. Subcellular localization and topology of the K88 usher FaeD in Escherichia coli. Mol Microbiol. 1995;16:1243–1257. doi: 10.1111/j.1365-2958.1995.tb02346.x. [DOI] [PubMed] [Google Scholar]
- 42.Voegele K, Sakellaris H, Scott J R. CooB plays a chaperone-like role for the proteins involved in formation of CS1 pili of enterotoxigenic Escherichia coli. Proc Natl Acad Sci USA. 1997;94:13257–13261. doi: 10.1073/pnas.94.24.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 44.Willshaw G A, Smith H R, McConnell M M, Rowe B. Cloning of regulator genes controlling fimbrial production by enterotoxigenic Escherichia coli. FEMS Microbiol Lett. 1991;66:125–129. doi: 10.1016/0378-1097(91)90320-a. [DOI] [PubMed] [Google Scholar]