Abstract
Abstract
This mini review focuses on the diagnosis and treatment of virus diseases using Crisper-Cas technology. The present paper describes various strategies involved in diagnosing diseases using Crispr-Cas-based assays. Additionally, CRISPR-Cas systems offer great potential as new therapeutic tools for treating viral infections including HIV, Influenza, and SARS-CoV-2. There are several major challenges to be overcome before this technology can be applied routinely in clinical settings, such as finding a suitable delivery tool, toxicity, and immunogenicity, as well as off-target effects. This review also discusses ways to deal with the challenges associated with Crisper-Cas technology.
Key points
• Crisper technology is being applied to diagnose infectious and non-infectious diseases.
• A new generation of CRISPR-Cas-based assays has been developed which detect pathogens within minutes, providing rapid diagnosis of diseases.
• Crispr-Cas tools can be used to combat viral infections, specifically HIV, influenza, and SARS-CoV-2.
Keywords: HIV, Influenza, SARS-CoV-2, Crispr-Cas
Introduction
The clustered regularly interspaced short palindromic sequence repeats-Cas (CRISPR-Cas) system has emerged as a promising tool for next-generation pathogen diagnosis, gene editing, drug discovery, and therapeutics. It forms a part of natural adaptive immune response in many species of archaea and bacteria, against foreign bacteriophage and plasmid infections by cleaving their nucleic acid (Brouns et al. 2008; Horvath et al. 2010; Garneau et al. 2010; Barrangou et al. 2007). Research investigations now focus on optimizing Crispr-Cas system to be utilized in Humans (Cebrian-Serrano et al. 2017; Hendel et al. 2015; Kumar et al. 2019; Moorthy et al. 2020; Naeem et al. 2020).
Rapid detection of disease-causing pathogens enables accurate and quick treatment and helps in preventing the spread of disease. While conventional diagnostic methods such as restriction enzymes, recombinases, nucleases, sequencing-based methods, PCR/qPCR-based methods, and isothermal amplification-based techniques (Yang and Rothman 2004; Zhao et al. 2015; Scheler et al. 2014) are time-consuming, have low specificity and sensitivity, and are expensive, requires technical expertise, and sophisticated devices, Crispr-Cas helps in nucleic acid detection (Khambhati et al. 2019; Li Y et al. 2019; Chertow 2018) with high sensitivity and specificity in comparatively less time. This makes Crispr-Cas system a powerful tool in next generation pathogen diagnosis. Crispr-Cas-based SHERLOCK technology allows detection of Zika virus and dengue virus in just 2 h with a sensitivity of about 1 copy per microliter of sample (Myhrvold et al. 2018). It has also enabled detection of West Nile, yellow fever virus, and SARS-CoV-2 (Gootenberg et al. 2017, 2018; Myhrvold et al. 2018; Zhang et al. 2020). Similarly, DETECTR technology has also been employed to detect SARS-CoV-2 (Broughton et al. 2020). Furthermore, human papilloma virus strains (HPV-16 and HPV-18) can be distinguished using DETECTR technology (Myhrvold et al. 2018). The development of Crispr-based chips has enabled detection of target genome without the need of amplification, providing quicker results (Hajian et al. 2019).
SARS COV-2 outbreak is severely contagious and has been gripping the world since December 2019, and has led to an alarming increase in worldwide mortality rate. According to WHO, 187,827,660 confirmed cases and 4,055,497 deaths have been reported worldwide by July 2021. Other viral infections such as HIV and Influenza are also a major public health issue worldwide. As per WHO, nearly 37.6 million people were living with HIV and 690,000 deaths have been reported due to HIV-related diseases in the year 2020. With advancement in medical research, HIV has become manageable and people are living with HIV and leading normal lives. However, there is no proper cure and vaccines available against it. Further, antiretroviral drugs have to be administered routinely which often cause adverse effects and do not target the latent reservoirs. Human Influenza viruses cause seasonal epidemics each year. According to WHO, influenza is responsible for 99% of deaths in children below the age of five years. WHO estimates that annual epidemics results in 3 to 5 million cases and 290,000 to 650,000 deaths worldwide due to severe illness from Influenza. Immunity provided from currently available vaccines against influenza diminishes over time, hence vaccines should be taken each year. Thus, devising an appropriate and effective treatment strategy is the need of the hour to fight against these disease (Figs. 1 and 2). In recent years, Crispr-Cas has unfolded as a promising tool for antiviral therapy. It comprises of two main components, the crRNA, designed to bind to the complementary sequence of the target DNA, and Cas proteins that cleaves the target DNA, producing a double stranded break which is repaired by cellular DNA repair mechanisms. The double-stranded breaks produced by Cas proteins disrupt the genes that are crucial for viral life cycle and proliferation, thereby providing an effective inhibition of viral replication within a cell. Studies show that Crispr Cas-mediated targeting of host genes essential for viral entry has conferred resistance to cells from viruses. Knockdown of CMP-sialic acid transporter SLC3A1 by Crispr-Cas in cells prevented IAV infection (Han et al. 2018). Furthermore, in vitro studies show that targeting highly conserved sequences of viral genome such as LTR sequences of HIV (Ebina et al. 2013; Liao et al. 2015), RdRP, and N gene of SARS-COV-2 and segment 6 and segment 4 of IAV decreased viral load in the cells (Abbott et al. 2020). However, several scientific limitations and ethical concerns need to be addressed before it can be approved for human clinical trails.
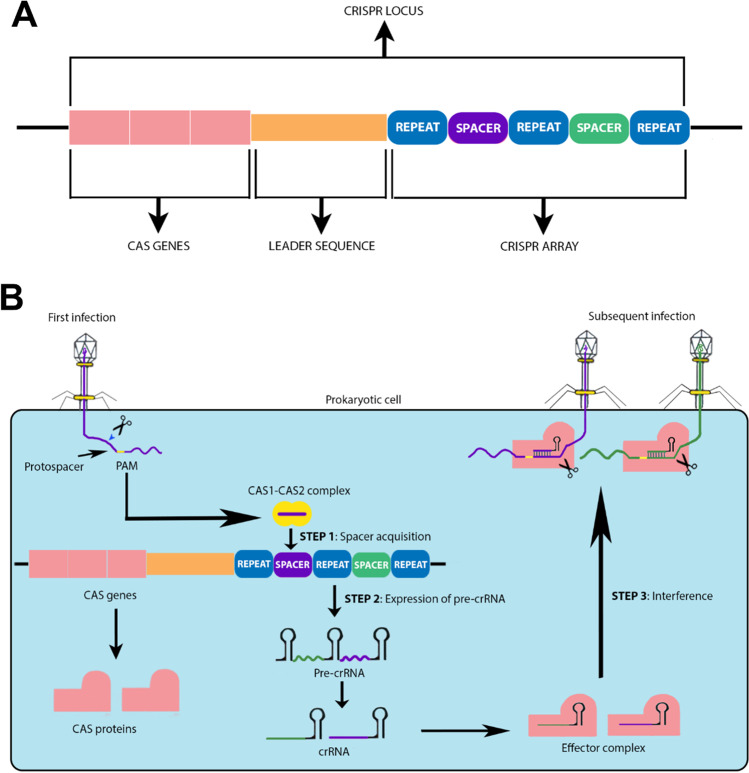
Fig. 1.
Schematic representation of the Crispr-Cas-adaptive immune response pathway in Prokaryotes. A Crispr locus showing crispr array, leader sequence, and Cas genes. B Three stages of Crispr-Cas pathway: as the viral genetic material is injected into the cell, cas1-cas2 complex identifies the PAM (protospacer adjacent motif) site and cleaves the protospacer sequence on the genetic material and integrates it into the host genome as spacer in Crispr loci (spacer acquisition). Pre-crRNA is transcribed from the Crispr array (expression) and processed to form mature crRNA, which binds to the Cas proteins forming an effector complex. When the same virus invades the cell, the crRNA-Cas protein recognizes and binds to the complementary sequence on the viral genome thereby cleaving it
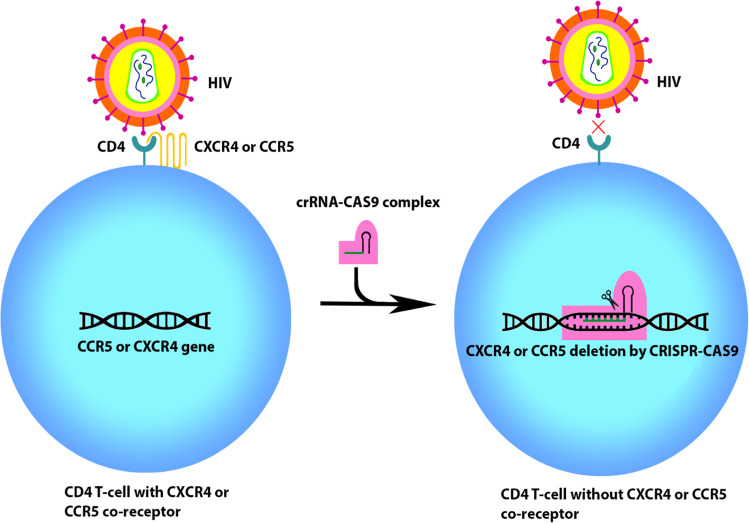
Fig. 2.
Receptor editing using crispr-cas9.HIV enters the cell by binding CD4 receptor and CXCR4 or CCR5 co-receptors. Crispr-Cas9 targeted against CXCR4 or CCR5 gene creates cells that are devoid of these co-receptors. HIV is unable to attack cells in the absence of co-receptor CXCR4 or CCR5, thereby providing resistance to the cell from HIV infection
This review aims to summarize how Crispr-Cas system can be used in the diagnosis of diseases and its potential as a treatment option for viral infections such as HIV, influenza, and COVID-19. While most of the recently published reviews discuss either Crispr based-detection of viruses (Yin et al. 2021) or Crispr-cas technology as an antiviral treatment option (Baddeley and Isalan 2021) our review looks at both aspects, from the basic functioning of the Crispr-Cas system to its detection and treatment. Therefore, it would help the readers to better understand Crispr-Cas system’s broad applicability and its advantages to healthcare. The review also highlights some of the latest detection methods, such as Crispr-Cas13a-based visual biosensors (Ma et al. 2022) and FELUDA (Azhar et al. 2021). In our paper, we offer therapeutic strategies to combat three of the most prevalent and lethal viral diseases in the world, viz., SARS-CoV2, HIV and influenza, by examining how these viruses hijack the human system, essential genes involved in their interaction with the host, and strategies used to combat them. Finally, we have not only highlighted some of the challenges associated with using Crispr-Cas in humans, but also provided some solutions to address them and directions for future research, including an anti-crispr strategy (Bondy-Denomy et al. 2015; Dolgin 2020; Aschenbrenner et al. 2020; Jain et al. 2021).
Basic anatomy and mechanism of Crispr-Cas system
Crispr-Cas system works by eliminating the viral infection by directly targeting the viral DNA or RNA. The bacterial genome consists of a CRISPR loci having a Crispr array. This array is formed by palindromic repeats intercalated by unique spacer sequences that is acquired from invading bacteriophage or viral genetic material (Bolotin et al. 2005; Mojica et al. 2005). These spacer sequences provide resistance by acting as genetic memory and prevents the host from being infected by virus comprising the same sequence (Brouns et al. 2008). The CRISPR array is flanked by a leader sequence and an operon consisting a group of Cas protein encoding genes (Makarova et al. 2013). These Cas proteins are necessary for adaptive immune response to function (Makarova et al. 2015). Amongst the diverse group of Cas proteins, Cas1 and Cas2 are universal and exists in all types of Crispr-Cas system and are involved in spacer integration (Haft et al. 2005; Makarova et al. 2006; Nunez et al. 2014).
Crispr-Cas system is divided into two major classes, viz., class I and class II and into six different types (I–VI) (Makarova et al. 2013, 2015, 2020) (Table 1). These types are further divided into subtypes and each type has a characteristic composition of functional modules. Those types which have multiunit crRNA-effector complex are categorized into class I and The types with single subunit crRNA-effector complex are grouped into class II (Makarova et al. 2015).
Table 1.
| Type | Functional modules | |||
|---|---|---|---|---|
| Adaptation | Expression/crRNA processing | Interference | ||
| Class I | Type-I | Cas1, Cas2, Cas4* | Cas6 | Cas7,Cas5, Cas8a, Cas11, Cas3′, Cas3″ |
| Type-III | Cas1,Cas2 | Cas6* | Cas7,Cas5, Cas11, Cas10 | |
| Type-IV | Cas1*,Cas2* | Cas6* | Cas7,C as5, Cas11, Csf1 (Cas8-like) | |
| Class II | Type-II | Cas1,Cas2, Cas4* | RNaseIII | Cas9 |
| Type-V | Cas1*,Cas2*, Cas4* | Cas12 | ||
| Type-VI | Cas1*, Cas2* | Cas13 | ||
*Modules missing in some subtypes
The adaptive immune response against foreign genetic material-mediated by Crispr-Cas system occurs mainly in three stages, viz., adaptation/spacer acquisition, expression of pre-crRNA, and interference.
Adaptation/spacer acquisition involves recognition of short stretch of conserved sequence of nucleotides known as to as PAM (protospacer adjacent motif) that is found in vicinity to the protospacer (foreign DNA sequence integrated as spacer) (Mojica et al. 2009), followed by its cleavage and acquisition by nuclease complex Cas1-Cas2 (Nunez et al. 2014; Wang, J. et al. 2015). The protospacer acquired from the invading viral genetic material is incorporated as spacer in between two repeats. The new spacer is integrated at the leader terminus of the Crispr array (Pourcel et al. 2005; Barrangou et al. 2007)
In second step, expression, Crispr locus is transcribed to form pre-crRNA, followed by its processing by Cas proteins which transform it into fully mature crRNA or guide RNA (Charpentier et al. 2015; Hochstrasser and Doudna 2015; Carte et al. 2008).
In the final step, interference, the mature crRNA/guide RNA, bound to a multiprotein processing complex, recognizes and base pairs with the complementary sequence of the invading viral genetic material which subsequently get cleaved by Cas proteins beside the PAM site (Brouns et al. 2008; Plagens et al. 2015). PAM sites on the viral genetic material are crucial for distinguishing self and non-self genetic materials. Studies show that mutations in PAM sites bypass the immunity conferred by the Crispr-Cas system (Deveau et al. 2008; Garneau et al. 2010).
Crispr-Cas system in next-generation pathogen diagnosis
The power of the Crispr-Cas system to detect a viral genetic material in a sample has revolutionized medical diagnostic field, enabling accurate and rapid detection of any pathogen within minutes with high sensitivity and specificity (Table 2).
Table 2.
Features of some Crispr-based diagnostic platforms
| Diagnostic platform | Cas protein involved | Approximate assay time | Preamplification method | Readout | Detection | References |
|---|---|---|---|---|---|---|
| NASBACC | Cas9 | 2-h amplification + 1 h (results) | NASBA | Colorimetry | Zika VIRUS | Pardee et al. 2016 |
| CAS-EXPAR | Cas9 | 1 h | EXPAR | Fluorescence | L.monocytogenes; DNA methylation | Huang et al. 2018 |
| CRISPR-Chip | Cas9 | 15 min | - | Electrochemical | DNA mutations in Duchenne muscular dystrophy | Hajian et al. 2019 |
| Paired dCAS9 (PC) reporter system | dCas9 | < 1-h ramplification + few minutes (results) | PCR | Luminescence | M. tuberculosis | Zhang et al. 2017; Zhang et al. 2021 |
| FELUDA | dFnCas9 | 1 h | RT-RPA, RT-PCR | Lateral flow assay, smartphone app | Detection of SNV, SARS-CoV2 | Azhar et al. 2021 |
| SHERLOCK | Cas13 | 2-h amplification + 30 min–3 h (results) | RPA, RT-RPA | Fluorescence | Zika virus, dengue virus, HIV, pathogenic bacteria | Gootenberg et al. 2017 |
| SHERLOCKv2 | Cas13 | 30 min amplification + 30 min–3 h (results) | RPA | Fluorescence, lateral flow assay | Zika virus, dengue virus, SNP, bacteria | Gootenberg et al. 2018 |
| DETECTR | Cas12a | 10-min amplification + 1 h (results) | RPA | Fluorescence | HPV16, HPV18 | Chen et al. 2018 |
| HOLMES | LbCas12a | 45-min amplification + 15-min results | PCR | Fluorescence | Japanese encephalitis virus | Li et al. 2018 |
| HOMESv2 | AacCas12b | 30-min amplification + 30-min (results) | LAMP | Fluorescence | Japanese encephalitis virus, DNA methylation | Li et al. 2019a, b |
| STOPcovid | Cas12b | 1 h | RT-LAMP | Fluorescence, lateral flow assay | SARS-CoV2 | Joung et al. 2020 |
| CARMEN | LwCas13a | 20-min amplification + 3 h (results) | PCR/RPA | Fluorescence | Drug-resistant HIV mutations, SARS-Cov2, subtyping influenza strain, human-associated viruses | Ackerman et al. 2020 |
| CRISPR-Cas12a-powered visual biosensor | Cas12a | 1-h amplification + 30 min (readout) | RT-PCR | Colorimetry, smartphone app | SARS-CoV2 | Ma et al. 2022 |
| E-CRISPR | Cas12a | 30 min–3 h | – | Electrochemical | HPV-16, PB19, TGF-β protein | Dai et al. 2019 |
Pathogen detection using Crispr-Cas9-based assay
Pathogen detection using Crispr-cas9, a class II protein, is based on the ability of the guide RNA to bind to the dsDNA with its subsequent cleavage by Cas9 endonuclease.
Zika virus has been detected through amplification of the viral RNA using nucleic acid sequence-based amplification (NASBA) with incorporation of a synthetic trigger sequence and T7 primer, followed by cleavage of sequence adjacent to the PAM site by Crispr-Cas9 (Pardee et al. 2016). This assay, developed by Pardee et al. (2016) and named as NASBA-CRISPR cleavage (NASBACC), involves the principle of toehold switch sensors which works by programming a synthetic riboregulator which binds to complementary trigger RNA. The start codon along with ribosome binding site is blocked by the riboregulator due to its hairpin structure and translation only occurs when the riboregulator binds to the complementary trigger RNA, freeing the ribosome binding site and start codon. The results are monitored by colorimetry. NASBACC involves amplification of target RNA by NASBA through reverse transcription followed by addition of T7 polymerase to form dsDNA. Since the amplified fragments are incorporated with trigger RNA sequence, cleavage by Cas9-mediated results in truncated trigger RNA products which is unable to activate the toehold switch. If PAM sequence is absent, trigger RNA is not cut and full-length trigger RNA is able to activate the switch (Pardee et al. 2016).
EXPAR is an emerging, highly efficient, and quick isothermal amplification technique that enables to work at a constant temperature without the need to exogenous primers unlike conventional amplification methods. CRISPR/Cas9 triggered isothermal exponential amplification reaction (CAS-EXPAR) and exploits the EXPAR amplification method, allowing the detection of pathogen with a sensitivity of around 1 amol–10 fmol (Huang et al. 2018). The target DNA is cleaved by Cas9/sgRNA to produce ssDNA substrates (acts as a primer for amplification). The cleaved fragment (X) binds to complementary sequence on the EXPAR template which has a central NEase (nicking enzyme) recognition sequence flanked by sequences complementary to X. After amplification by DNA polymerase forming DNA duplex, NEase nicks and releases dsDNA strand, displacing X which can the initiate new amplification cycle. The results are analyzed through real time fluorescence detection (Huang et al. 2018). This method was shown to detect methylated DNA as well because of its high specificity (Huang et al. 2018).
The cas9 that binds to the DNA but does not cleave, also referred to as nuclease dead cas9, has also been utilized in the field of diagnostics. The firefly enzymes luciferase is split into two and fused to two dCas9. Both dCas9 binds to the target sequence in such a way that the two halves of the enzymes are brought together and they catalyze a reaction, producing bioluminescent signal (Zhang et al. 2017). This technique was used to detect the Mycobacterium tuberculosis, which causes tuberculosis (Zhang et al. 2017).
To improve sensitivity, a Crispr-chip-based method has also been devised that does not require amplification. This method is able to diagnose DNA mutations in Duchenne muscular dystrophy. The chip is made up of a graphene-based field-effect transistor (gFET) which has dCas9-gRNA complex immobilized on it (Hajian et al. 2019). The characteristics of the gFET are modulated when dCas9 binds to the target DNA and are sensed by the biosensor (Hajian et al. 2019).
In the view of rising number of COVID-19 cases, Drug Controller General of India (DCGI) approved a crispr-cas9-based diagnostic assay named “FELUDA” (FNCAS9 Editor Linked Uniform Detection) which allows rapid detection of SARS-CoV2 within an hour at a low cost and without any complex instrumentation (Azhar et al. 2021). With an accuracy of 100%, FELUDA is able to detect single nucleotide variants and shows 97% specificity across all ranges of viral loads. The assay makes use of catalytically inactive dFnCas9 (from Francisella novicida) and results are interpreted via lateral flow assay readout. The target RNA is amplified by RT-PCR or RPA and biotinylated. dFnCas/sgRNA complex, labelled with FAM, is allowed to bind to the complementary sequence on the biotinylated target RNA. The test strip containing immobilized streptavidin captures the biotinylated RNA complexed with dFnCas/sgRNA (labelled with FAM) and is detected using Anti-Fam antibodies conjugated with gold nanoparticles, giving a visual signal (Azhar et al. 2021).
Pathogen detection using Crispr-Cas12/Cas13-based assay
Cas12 and Cas13 containing Crispr system make the use of its “collateral cleavage activity,” wherein, after the target nucleic acid is cleaved, a nearby non-specific ssDNA/RNA sequence is also cleaved. This principle has been exploited and used in SHERLOCK and DETECTR technology that uses fluorescently labelled ssDNA/RNA probes, which after cleavage by Cas protein would give a fluorescent signal once the fluorophore is separated from the quencher. (Chen et al. 2018; Gootenberg et al. 2017, 2018).
SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) technology harnesses collateral cleavage activity of Cas13a, a type VI class 2 Crispr-Cas protein (Gootenberg et al. 2018). The pathogenic DNA is amplified by RPA (recombinase polymerase amplification) or reverse transcriptase RPA if the genetic material is RNA (Piepenburg et al. 2006) followed by specific binding of crRNA to its cognate target, which switches on Cas13a. It then cleaves the target DNA and collaterally cleaves fluorescently tagged ssRNA reporter, separating fluorophore from the quencher which generates a fluorescent signal (Kellner et al. 2019). To enhance the efficiency of SHERLOCK, an improved method HUDSON (heating unextracted diagnostic samples to obliterate nucleases) was established, which enables direct diagnosis of pathogen from the sample without extraction of nucleic acid (Myhrvold et al. 2018). HUDSON-SHERLOCK enabled detection with 100% sensitivity and 100% specificity and allowed differentiation between 4 serotypes of dengue virus and Zika virus in samples obtained from different parts of the world. Crispr-Cas13-based SHERLOCK assay has been utilized to detect dengue, Zika, HIV, West Nile, and Yellow fever viruses (Gootenberg et al. 2017, 2018; Myhrvold et al. 2018). Recently, Zhang et al. (2020) devised a method to detect SARS-COV2 using Crispr-Cas13-based SHERLOCK assay, wherein two crRNA targeting S and ORF1ab gene of Covid-19 was harnessed, that provided a visual readout within an hour. A modified version of SHERLOCK assay, referred to as SHERLOCKv2, allows multiplexing such that in a single reaction three ssDNA targets and one dsDNA target can be detected by using combinations of Cas13 and Cas12 (Gootenberg et al. 2018).It also allows visual readouts in lateral flow strips with a greater sensitivity than SHERLOCK.
To reduce the number of fluid handling steps, a one-pot diagnostic test for SARS-Cov2 was devised known as STOPcovid (SHERLOCK Testing in One Pot) by Joung et al. (2020). It makes use of thermostable AacCas12b (from Alicyclobacillus acidiphilus) and preamplification by RT-LAMP. Results analyzed through lateral flow readout enables detection in 70 min; whereas with fluorescence readout, results are obtained in 40 min (Joung et al. 2020).
A combination of microfluidics and Crispr-Cas13a-based SHERLOCK system is exploited in CARMEN (combinatorial arrayed reactions for multiplexed evaluation of nucleic acids) by Ackerman et al. CARMEN allows, rapid, scalable, sensitive, high-through put, and low-cost detection of more than 4500 targets per chip. LwaCas13 (from Leptotrichia wadei) detection mixes containing cas13, crRNA, and reporters and samples amplified through PCR or RPA are combined with different solution-based fluorescent color code. After emulsification of the color-coded solutions in fluorous oil, the so formed droplets are pooled and added into microwell chips, each well containing a single pair, i.e., detection mixes and amplified samples. On applying electric field, the pair of droplets combine and fluorescence is detected by trans-cleavage activity of cas13a when it binds to complementary target sequence. CARMEN facilitated miniaturization of reaction, cutting down the cost and reducing the reagent and sample consumption. A total of 169 human-associated viruses, identification of drug-resistant HIV mutation, and subtyping of influenza A strains and coronaviruses were detected using this technique (Ackerman et al. 2020).
DETECTR (DNA endonuclease-targeted CRISPR trans-reporter) technology utilizes collateral cleavage activity of Cas12a, a class V Crispr-Cas protein (Chen et al. 2018). Similar to the SHERLOCK assay, in this method, the pathogenic genetic material is amplified through RPA (recombinase polymerase amplification) combined with reverse transcription if it involves RNA. The crRNA then guides Cas12a to dsDNA target sequence, where it binds and cleaves the target DNA and indiscriminately cleaves fluorescently tagged reporter ssDNA, thereby liberating the quenched fluorophore and generating a fluorescent signal (Chen et al. 2018). Human papillomavirus 16 (HPV16) and human papillomavirus 18 (HPV18) was distinguished using Crispr-Cas12-based DETECTR assay within an hour by creating crRNA targeted to hypervariable V loop of L1 gene, having a difference of only 6 nucleotides in the two strains of HPV (Chen et al. 2018; Myhrvold et al. 2018). Recently, Broughton et al. selected two crRNA targeting E and N genes specific to SARS-coV-2 and was able to diagnose COVID-19 within 30 min through DETECTR assay (Broughton et al. 2020). HOLMES (n one-HOur Low-cost Multipurpose highly Efficient System) uses LbCas12a and pre amplification through PCR unlike RPA in DETECTR (Li et al. 2018). A slightly variant version of HOLMES, known as HOLMESv2 enables one pot reaction and employs AacCas12b (from Alicyclobacillus acidiphilus) which is thermostable, using LAMP (loop-mediated isothermal amplification of DNA) for preamplification (Li et al. 2019a, b).
To provide rapid and ultrasensitive detection of SARS-CoV2, various biosensing techniques are being developed to allow timely detection of the disease, therapeutic interventions, and epidemiological surveillance, in the view of ongoing pandemic. Recently, CRISPR-Cas12a-powered visual biosensor with a smartphone readout has been developed by Ma et al. (2022). The technique was able to detect SARS-CoV2 gene in synthetic vectors, and SARS-CoV2 pseudo viruses with 1 µl/ml LOD of pseudo viruses (Ma et al. 2022). The time required for obtaining the results was about 90 min. It is based on trans-cleavage activity of cas12a and makes use of linker ssDNA that can hybridize with a pair of pre-made Au-DNA probe pairs. When target DNA is present in the sample, ssDNA linker is cleaved due to the trans-cleavage activity of the Cas12a leading to disaggregation of Au-DNA probes producing a color change which can be visualized by naked eyes or with a smartphone installed with the color picker app (Ma et al. 2022).
E-CRISPR employs electrochemical biosensor and uses trans-cleavage activity of Cas12a. A synthetic non-specific ssDNA reporter linked with methylene blue (MB) tag is immobilized on the gold electrode of the sensor. When target DNA is present, the Cas12a would cleave the ssDNA-MB probe due to the activation of its trans cleavage activity, resulting in lower electrochemical current of methylene blue. As a proof of concept, human papillomavirus 16 (HPV-16) and parvovirus B19 (PB-19) could be detected using E-CRISPR (Dai et al. 2019).
Crispr-Cas system as a potential antiviral treatment option
Along with timely detection of the disease, it is necessary to provide accurate treatment to control the spread of the disease and keep the mortality rate under control. The conventional antiviral drugs only keep the viral replication under control. Further, daily admiration of drugs causes a wide range of side effects and resistance in affected individuals. Crispr-Cas system can be an effective alternative which changes the host genes necessary for the survival of the virus or the viral genes crucial for viral life cycle (Jin et al. 2018).
Crispr-Cas9-based antiviral strategies against HIV-1
HIV infection is one of those viral infections that is highly prevalent worldwide and remains a global threat till the date with no vaccine and proper cure. Currently, antiretroviral therapy is used to limit the viral replication and to prevent its transmission; however, these therapies cannot cure the disease due to the formation of latent reservoirs consisting of HIV provirus in long-living resting T cells. The immune system does not attack the provirus residing in the reservoir cells, and when the antiretroviral therapy is interrupted, the virus rebounds from the inactive state (Siliciano et al. 2003; Blankson et al. 2002). Moreover, persistent use of drugs poses side effects and also leads to emergence of escape mutants.
HIV is a retrovirus that has RNA as its genetic material. The classical route of entry used by HIV to infect humans is through cells that express CD4 receptor and co-receptors CCR5 or CXCR4 recognized by HIV-1 gp120 (Maartens et al. 2014). Upon the viral entry into the cells having CD4, the viral RNA is reverse transcribed into DNA, followed by its integration into the host genome as Proviral DNA (Li and Clercq 2016). Subsequently, proviral DNA is transcribed using cellular RNA polymerase II transcriptional machinery. Occasionally, provirus becomes transcriptionally inactive and forms latent reservoir and escapes the attack from immune system. The antiretroviral therapy also fails to target the non-transcribing latent reservoirs.
Crispr-Cas9 has been widely exploited as genome editing tool to cleave HIV DNA. Cas9 obtained from Streptococcus pyogenes, designated as spCas9, is mainly used in crispr-cas9 system. A small guide RNA having complementarity to target DNA is allowed to bind and direct spCas9 to cleave the DNA sequence adjacent to the PAM site producing a double-stranded break in the DNA, which is repaired by cellular DNA repair mechanism, viz., non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ), whereby insertions and deletions (indels) and sometimes substitutions are introduced in the DNA at the cleavage site (Shen et al. 2018; Allen et al. 2019). Although most mutations inactivate the virus by disrupting the essential genes, however, sometimes virus may escape the inhibition due to the accumulation of mutations at the cleavage site which hampers the further binding of guide RNA (Wang et al. 2016a, 2016b).
Using this strategy, guide RNA targeting against LTR present at 3′ and 5′ ends of the provirus were harnessed which could block the expression of transcriptionally active provirus as well as latent reservoirs in cell lines (Ebina et al. 2013; Liao et al. 2015).It could also excise the proviral DNA from the host genome by targeting the LTRs at both terminals (Ebina et al. 2013; Kaminski et al. 2016). To improve the efficiency and to reduce the viral escape due to accumulation of mutations at the cleavage site, various combinations of two guide RNAs targeting different essential genes required for viral infectious cycle have also been utilized (Lebbink et al. 2017; Wang et al. 2016a). Experimental evidences show that combinations of guide RNAs that targets highly conserved sequences introduce large indels at the cleavage site, which is sufficient to inhibit the viral cycle (Lebbink et al. 2017; Wang et al. 2016b). However, further optimization is required for the efficient and safe use of Crispr-cas9 in HIV treatment.
In vitro studies show that mutant CD4+ T cells lacking CCR5 or CXCR4 co-receptors can prevent the HIV viral entry (Fig. 2). Crispr-Cas9 targeted against CCR5 gene created CD4+ T cells devoid of CCR5 receptors provided resistance to CCR5-utilizing HIV (Wang et al. 2014). Although only a minute fraction of HIV utilizes CXCR4 co-receptors, editing CCR5 does not confer resistance to CXCR4 harnessing HIV, and mutations might occur on HIV envelop, shifting the usage of CCR5 to CXCR4 co-receptor (Fatkenheuer et al. 2008; Verheyen, 2019). Recent in vitro studies reveal that it is possible to disrupt both CCR5 and CXCR4 receptors simultaneously with no cellular toxicity using combined sgRNA (small guide RNA) targeting both CCR5 and CXCR4 genes (Liu et al. 2017; Yu et al. 2018). For efficient use of Crispr-Cas9 in humans, validation using animal model is required. Crispr-Cas9-based genome editing along with combined antiviral retro therapy can be a potential treatment option for HIV after further research.
Combating SARS-Cov-2 using Crispr-Cas system
COVID-19 (SARS-CoV-2) emerged in China’s Wuhan market in 2019, and has spread worldwide since then. From past 2 year, the world has been suffering from this global pandemic, taking lives of millions with no effective treatment. Since SARS-CoV-2 is severely contagious, an accurate and effective treatment strategy is the need of the hour to fight against this pandemic.
SARS-CoV-2 is a positive single-stranded RNA virus (ssRNA) which belongs to coronaviridae family (Siddell et al. 2019). MERS-CoV and SARS-CoV are amongst the other viruses belonging to coronaviridae family. The RNA genome of SARS-CoV-2 comprises of 16 non-structural protein encoding genes, viz., nsp1-nsp16 and 4 structural protein encoding genes which encode for spike protein (S), membrane (M), nucleocapsid (N), and envelop (E), and approximately 8 genes coding for accessory proteins (Wu et al. 2020; Kandeel 2020). Spike protein is crucial for viral entry into the host cells by interacting with ACE2 receptor (Li et al. 2003), whose expression level is high in cells of the lungs, kidney, and heart (Donoghue et al. 2000).
While traditional vaccines work by generating an immune response by forming antibodies against foreign pathogen, Crispr-Cas system directly destroys the foreign genetic material by cleaving it. Crispr system using Cas13d, a class II type VI Crispr-Cas protein (Shmakov et al. 2017), has been harnessed to fight against COVID-19. Cas13d is advantageous in the sense that it is small in size, making it easier to be delivered using adeno-associated viruses (AAV) along with the guide RNA complementary to the target RNA and does not require protospacer adjacent motifs (PAM) for its cleavage activity thereby making it easier to design complementary guide RNA (Nguyen et al. 2020). Recently, Abbott et al. developed a Crispr-Cas13d-based strategy known as prophylactic antiviral CRISPR in huMAN cells (PAC-MAN) to provide pan-coronavirus protection by targeting multiple species within coronaviridae family. In this study, highly conserved regions of SARS-CoV-2 genome was targeted, viz., RNA-dependent RNA polymerase (RdRp) gene required for viral proliferation and nucleocapsid (N) gene coding for capsid protein (Abbott et al. 2020). Bioinformatic analysis of all the known genome of the coronavirus followed by analysis on human lung epithelial cells, it was observed that only 6 crRNA could target 91% of the known genomes of coronavirus and a total of 22 crRNA targeted all the known genomes of the coronavirus (Abbott et al. 2020). SARS-COV-2 inhibition was directly proportional to the crRNA concentration, hence high crRNA concentration is required for effective inhibition of SARS-COV-2 (Abbott et al. 2020). Nguyen et al. (2020) also harnessed Crispr-Cas13d system to inhibit SARS-COV-2 by targeting spike protein coding gene and ORF1ab. In this study, 10,333 guide RNAs were designed to specifically bind to 10 peptide-coding regions of viral genome (Nguyen et al. 2020).
For precise delivery of the Crispr-Cas system into the cell to avoid off-target effects, AntiBody And CAS fusion (ABACAS) was devised, which has Cas13 fused to an antibody against the SARS-COV-2 spike protein, crucial for viral entry into the cell (Yuan et al. 2020; Joyce et al. 2020). This facilitates the delivery of Crispr-cas13 directly into the cell along with the SARS-COV2 and ensures cleavage of the viral RNA before it could make further copies. However, there are not much efficient antibody developed against the spike protein, studies show that REGN-COV2 and LY-CoV555 can be potential candidates for the same (Joyce et al. 2020).
Crispr-Cas system to inhibit influenza A virus
Influenza A virus (IAV) is a negative sense single-stranded RNA virus comprising of 8 segments. IAV has a high pandemic potential in humans due to its ability to evolve rapidly. It has caused seasonal epidemics every year in various parts of the world, and therefore it is necessary to build strategies to inhibit IAV infection.
Abbott et al. (2020) extended the PAC-MAN strategy to inhibit IAV infection and succeed in providing a pan-IAV protection. In this study, highly conserved regions of all 8 segments of IAV genome was targeted. Studies show that the segment ends necessary for viral packaging are highly conserved in all the 8 segments of IAV genome (Desselberger et al. 1980; Muramoto et al. 2006; Skehel and Hay 1978), which makes it an ideal target. PAC-MAN targeting of segment 6(S6) which codes for neuraminidase (essential for releasing virus from the host) and segment 4 (S4) coding for hemagglutinin (essential for binding of the viral particle to the host cell) inhibited IAV, S6 being more potent than S4 that showed moderate inhibition of IAV (Abbott et al. 2020). Only 6 crRNA could target 92% of the IAV strains and 81 crRNA targeted all the 91,600 known IAV strains in lung epithelial cells (Abbott et al. 2020). Although PAC-MAN strategy could show satisfactory results in cell culture, it has to be validated in animal models and preclinical trials have to be conducted before it can be used in therapeutic treatment.
Freije et al. (2019) harnessed Cas13b derived from Prevotella sp. referred to as PspCas13b to check its antiviral activity in MDCK (Madin-Darby canine kidney) epithelial cells by designing crRNA targeted against mRNA in the cytoplasm and complementary viral RNA in the nucleus. IAV viral load was reduced by 7–22 folds, indicating that crispr-Cas13b could be a potential antiviral therapeutic option.
Cas13-assisted restriction of viral expression and readout (CARVER) enables rapid detection and destruction of the viral pathogen (Freije et al. 2019), using Crispr-Cas13 system. CARVER combines HUDSON (Myhrvold et al. 2018)-mediated RNA extraction and detection of viral RNA using SHERLOCK technology (Gootenberg et al. 2017, 2018), followed by Cas13-mediated cleavage of viral RNA. A combination of 4 crRNA against NP (nucleoprotein) and its cleavage by PspCas13b reduced the IAV viral load by eightfold (Freije et al. 2019) After proper optimization and further research, CARVER can be a promising platform for diagnostics as well as treatment of a wide variety mammalian viruses.
Analysis conducted using genome-wide Crispr knockout (GeCKO) on human lung epithelial cells reveal that a CMP-sialic acid transporter SLC3A1 is essential for IAV entry into the cell (Han et al. 2018). Knockdown of SLC3A1 hampered sialic acid biosynthesis and results in formation of cells devoid of sialic acid, thereby inhibiting viral entry and makes the cell resistant to IAV infection (Han et al. 2018). Crispr-Cas-targeted against SLC3A1 gene can be used to inhibit the interaction of viral hemagglutinin with host cell sialic acid and prevents viral entry. After validation in preclinical models, Crispr-Cas can be powerful antiviral tool against IAV infection.
Limitations of using Crispr-Cas system for diagnosis and in therapeutics
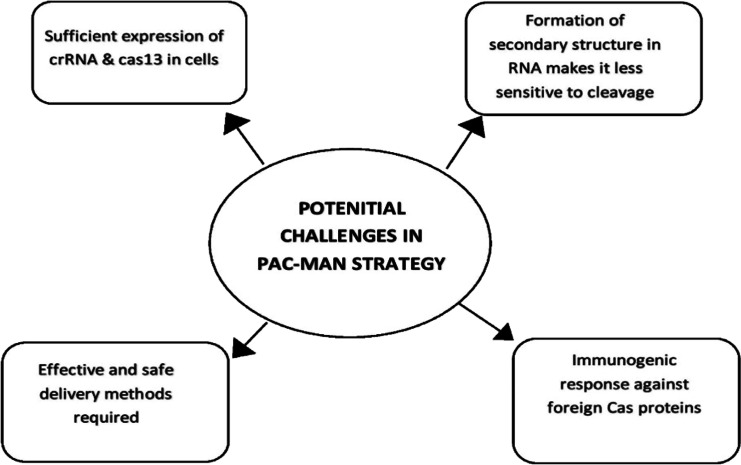
Crispr-Cas-based methods are highly versatile, cost effective, and does not require sophisticated devices and expertise. However, there are ethical concerns and scientific limitations that needs to be addressed before it could be used as a treatment option in humans (Fig. 3). One of the major challenges is the mode of delivery of Crispr-Cas system to the target cells. The choice of delivery method should allow transfer of large amount of Crispr-Cas proteins in the desired cell. Most widely used viral vectors are lentiviral vectors, adenovirus, and adeno-associated virus, but these vectors pose a limit to the size of Cas proteins being delivered. Lentiviral vectors and adenovirus vector allow larger insertions and better packing efficiency as compared to adeno-associated viral vectors. However, they often elicit immunological responses in the host (Ahi et al. 2011; Annoni et al. 2019). Lentiviral vector integrates into the host genome, increasing the potential of causing off-target site effects due to sustained release of gene. To avoid such adverse effects, several non-viral vectors have been developed amongst which lipid nanoparticles are the most widely used. Other nanoparticles such as gold nanoclusters (Ju et al. 2019), black phosphorus nanosheets (Zhou et al. 2018), and nanoscale zeolitic imidazole framework (Alsaiari et al. 2018) have also been used. Although nanoparticles have shown promising results in cell culture, they require complexing design and more research for safer use and their side effects need to be assessed carefully. Exosomes, a natural nanoparticle secreted by cells of the body such as immune cells, epithelial cells, have shown low immunogenicity and higher biocompatibility in transferring Crispr-Cas components; however, they have a lower packaging efficiency (Maas et al. 2017). The delivery of Crispr-Cas components should precisely target the desired cell to minimize off target effects. One of the major cause off target site effects is due to mismatch of gRNA. Off target site effects can cause loss of function or disruption of essential genes, leading to abnormalities in the host. A variety of in silico design tools such as DeepCRISPR and CRISPRitz (Cancellieri et al. 2020; Chuai et al. 2018) have been develop to predict these off target effects. Other in silico guided predication tools such as CHOPCHOP (https://chopchop.cbu.uib.no/), Cas-OFFinder (http://www.rgenome.net/cas-offinder/), E-Crisp (http://www.e-crisp.org/E-CRISP/), and Breaking-Cas (https://bioinfogp.cnb.csic.es/tools/breakingcas/) are also being widely used. Scientists are developing high fidelity Cas variants (Kleinstiver et al. 2016) and truncating the spacer region (Fu et al. 2014) as an alternative approach for improving the targeting specificity of Crispr-Cas system. Since Cas proteins are derived from prokaryotes, it can cause cell toxicity and immunogenic responses and can lead to formation of antibodies against them (Charlesworth et al. 2019). Several in silico prediction tools predict that some peptides sequences on the Cas proteins have affinity to bind to HLA class 1 and HLA class 2 alleles, eliciting a T cell response (Chew et al. 2018). Cas proteins may have B cell epitopes on them, hence neutralization mediated by antibodies is often observed (Chew et al. 2018). One of the approaches for preventing immune response involves modification of epitopes in such a way that the function of proteins remains intact. It has been reported that phosphatase treatment of in vitro transcribe guide RNA results in reduction of immune response (Kim et al. 2018). Co-administration of immunosuppressants and use of safer engineered delivery tools can also decrease the immune reactions. The RNA being targeted often forms secondary structures which decreases the efficiency of the Crispr Cas system. The occurrence of mosaicism is often observed when the transduced cells undergo division before the target nucleic acid is cleaved by Crispr Cas system. Furthermore, stability of RNA has to be maintained and should be prevented from attack by RNases. Sufficient level expression of Crispr-Cas in the desired cell and the duration of expression should be optimized in order to use Crispr-Cas as an antiviral therapy. Although Crispr-Cas system is a robust gene editing tool, its use in therapeutics had a long way to go.
Fig. 3.
Challenges that needs to be resolved before PAC-MAN could be used therapeutically. These challenges include a need suitable delivery tool, immunologic effects of cas proteins, interfere of secondary structure of RNA and expression
Conclusions and future prospects
Crispr-Cas-based methods are highly versatile, cost-effective, and do not require sophisticated equipment or expertise. There are, however, ethical concerns and scientific limitations that need to be addressed before it can be used as a treatment option in humans. In order to deliver Crispr-Cas to target cells, delivery mode is a major challenge. It is important that the delivery method chosen allows for the transfer of large amounts of Crispr-Cas proteins to the targeted cells. There is a limit to the size of Cas proteins that can be delivered via lentiviral vectors, adenoviruses, and adeno-associated viruses. Comparatively to adeno-associated viral vectors, lentiviral and adenovirus vectors allow larger insertions and better packing efficiency. However, they often elicit immunological responses in hosts (Ahi et al. 2011; Annoni et al. 2019). Since lentiviral vectors integrate into the host genome, they can cause off-target site effects due to sustained gene release. In order to avoid such adverse effects, several non-viral vectors have been developed, including lipid nanoparticles. Other nanoparticles such as gold nanoclusters (Ju et al. 2019), black phosphorus nanosheets (Zhou et al. 2018), and nanoscale zeolitic imidazole framework (Alsaiari et al. 2018) have also been used. Despite promising results in cell culture, nanoparticles require more research and complex designs to ensure safe use, and their side effects need to be assessed carefully. Exosomes, a natural nanoparticle secreted by cells of the body such as immune cells, epithelial cells have shown low immunogenicity and higher biocompatibility in transferring Crispr-Cas components; however, they have a lower packaging efficiency (Maas et al. 2017). A recent all-in-one AVV capable of expressing NmeCas9 with a single sgRNA with homology-directed repair template or dual sgRNA has shown promising in vivo results (Ibraheim et al. 2021). In order to minimize off-target effects, Crispr-Cas components should be delivered precisely to the desired cell. A mismatch between gRNA and target site is a major cause of off target site effects. As a result of off-target effects, essential genes can lose function or be disrupted, leading to abnormalities in the host. There have been a number of in silico design tools developed to predict these off target effects, including DeepCRISPR and CRISPRitz (Cancellieri and Chuai 2020; Chan et al. 2018). CHOPCHOP (https://chopchop.cbu.uib.no/), Cas-OFFinder (http://www.rgenome.net/cas-offinder/), E-Crisp (http://www.e-crisp.org/E-CRISP/), and Breaking-Cas (https://bioinfogp.cnb.csic.es/tools/breakingcas/) are also being widely used. Researchers have developed high fidelity Cas variants (Kleinstiver et al. 2016) and truncated the spacer region (Fu et al. 2014) to improve Crispr-Cas system specificity.
Anti-Crisprs, natural inhibitors of Crispr-Cas system, are being widely exploited to minimize the off target effects and improve target specificity (Bondy-Denomy et al. 2015; Dolgin 2020). Kinetic insulation mediated by anti-Crispr proteins that have been fused directly to the Cas9 or is co-expressed with Cas9 has enabled to fine tune the activity of Cas9 in mammalian cells (Aschenbrenner et al. 2020). A tuneable Crispr controller wherein anti-crispr and AcrIIA4 have been fused to ligand inducible destabilization domain, which can be induced by trimethoprim to turn off cas9 binding to DNA( Jain et al. 2021). Since Cas proteins are derived from prokaryotes, it can cause cell toxicity and immunogenic responses and can lead to formation of antibodies against them (Charlesworth et al. 2019). Several in silico prediction tools predict that some peptides sequences on the Cas proteins have affinity to bind to HLA class 1 and HLA class 2 alleles, eliciting a T cell response (Chew et al. 2018). Cas proteins may have B cell epitopes on them, hence neutralization mediated by antibodies is often observed (Chew et al. 2018). One of the approaches for preventing immune response involves modification of epitopes in such a way that the function of proteins remains intact. It has been reported that phosphatase treatment of in vitro transcribe guides RNA results in reduction of immune response (Kim et al. 2018). Co-administration of immunosuppressants and use of safer engineered delivery tools can also decrease the immune reactions. The RNA being targeted often forms secondary structure which decreases the efficiency of the Crispr Cas system. The occurrence of mosaicism is often observed when the transduced cells undergo division before the target nucleic acid is cleaved by Crispr Cas system. Furthermore, stability of RNA has to be maintained and should be prevented from attack by RNases. Sufficient level expression of Crispr-Cas in the desired cell and the duration of expression should be optimized in order to use Crispr-Cas as an antiviral therapy. Although Crispr-Cas system is Robust gene editing tool, its use in therapeutics have a long way to go.
Acknowledgements
PKA acknowledges the Department of Biotechnology, Government of India to provide him Ramalinga Re-entry Fellowship.
Author contribution
AR, SS, and JS collected data and wrote the manuscript. AK, PKA, and AS edited the manuscript. All authors read and approved the manuscript for submission.
Funding
This study was funded by the Department of Biotechnology, Government of India.
Data availability
Not applicable.
Declarations
Ethics statement
This article does not contain any studies with animals performed by any of the author.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Akhilesh Kumar, Email: akhilesh.kumar@bhu.ac.in.
Alok Kumar Singh, Email: alokjnu18@gmail.com.
Pankaj Kumar Arora, Email: arora484@gmail.com.
References
- Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R, Pande T. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman CM, Myhrvold C, Thakku SG, Freije CA, Metsky HC, Yang DK, Simon HY, Boehm CK, Kosoko-Thoroddsen TSF, Kehe J, Nguyen TG. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahi YS, Bangari S, D, Mittal KS, Adenoviral vector immunity: its implications and circumvention strategies. Curr Gene Ther. 2011;11:307–320. doi: 10.2174/156652311796150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, Páleníková P, Khodak A, Kiselev V, Kosicki M, Bassett AR. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol. 2019;37:64–72. doi: 10.1093/cid/ciy565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaiari SK, Patil S, Alyami M, Alamoudi KO, Aleisa FA, Merzaban JS, Li M, Khashab NM. Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework. J Am Chem Soc. 2018;140:143–146. doi: 10.1021/jacs.7b11754. [DOI] [PubMed] [Google Scholar]
- Annoni A, Gregori S, Naldini L, Cantore A. Modulation of immune responses in lentiviral vector-mediated gene transfer. Cell Immunol. 2019;342:103802. doi: 10.1016/j.cellimm.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner S, Kallenberger SM, Hoffmann MD, Huck A, Eils R, Niopek D (2020) Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Sci Adv 6(6):eaay0187 [DOI] [PMC free article] [PubMed]
- Azhar M, Phutela R, Kumar M, Ansari AH, Rauthan R, Gulati S, Sharma N, Sinha D, Sharma S, Singh S, Acharya S. Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens Bioelectron. 2021;183:113207. doi: 10.1016/j.bios.2021.113207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley HJE, Isalan M. The Application of CRISPR/Cas systems for antiviral therapy. Front Genome Ed. 2021;3:745559. doi: 10.3389/fgeed.2021.745559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med. 2002;53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR (2015) Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526(7571):136-9 [DOI] [PMC free article] [PubMed]
- Broughton JP, Deng X, Yu G, Fasching CL, Servellita SJ, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotech. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, Van Der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancellieri S, Canver MC, Bombieri N, Giugno R, Pinello L. CRISPRitz: rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing. Bioinformatics. 2020;36:2001–2008. doi: 10.1093/bioinformatics/btz867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian-Serrano A, Davies B. CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools. Mamm Genome. 2017;28:247–261. doi: 10.1007/s00335-017-9697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, Behlke MA. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbial Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsk JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow DS. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- Chew WL. Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip Rev Syst Biol Med. 2018;10:1408. doi: 10.1002/wsbm.1408. [DOI] [PubMed] [Google Scholar]
- Chuai G, Ma H, Yan J, Chen M, Hong N, Xue D, Zhou C, Zhu C, Chen K, Duan B, Gu F. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19:1–18. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R, Bushman FD. Hiv dna integration. Cold Spring Harb Perspect Med. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Somoza RA, Wang L, Welter JF, Li Y, Caplan AI, Liu CC. Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew Chem Int Ed. 2019;131:17560–17566. doi: 10.1002/ange.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U, Racaniello VR, Zazra JJ, Palese P. The 3'and 5'-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. The kill-switch for CRISPR that could make gene-editing safer. Nature. 2020;577:308–311. doi: 10.1038/d41586-020-00053-0. [DOI] [PubMed] [Google Scholar]
- Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circu Res. 2000;87:e1–e9. doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:1–7. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fätkenheuer G, Nelson M, Lazzarin A, Konourina I, Hoepelman AI, Lampiris H, Hirschel B, Tebas P, Raffi F, Trottier B, Bellos N. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. New Eng J Med. 2008;359:1442–1455. doi: 10.1056/NEJMoa0803154. [DOI] [PubMed] [Google Scholar]
- Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, Yozwiak NL. Programmable inhibition and detection of RNA viruses using Cas13. Mole Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comp Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajian R, Balderston S, Tran T, DeBoer T, Etienne J, Sandhu M, Wauford NA, Chung JY, Nokes J, Athaiya M, Paredes J. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat Biomed Engi. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Perez JT, Chen C, Li Y, Benitez A, Kandasamy M, Lee Y, Andrade J, Manicassamy B. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 2018;23:596–607. doi: 10.1016/j.celrep.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trend Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Huang M, Zhou X, Wang H, Xing D. Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal Chem. 2018;90:2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- Jain S, Shukla S, Yang C, Zhang M, Fatma Z, Lingamaneni M, Abesteh S, Lane ST, Xiong X, Wang Y, Schroeder CM, Selvin PR, Zhao H (2021) TALEN outperforms Cas9 in editing heterochromatin target sites. Nat Commun 12(1):606 [DOI] [PMC free article] [PubMed]
- Joyce MG, Sankhala RS, Chen WH, Choe M, Bai H, Hajduczki A, Yan L, Sterling SL, Peterson CE, Green EC, Smith C (2020) A cryptic site of vulnerability on the receptor binding domain of the SARS-CoV-2 spike glycoprotein. BioRxiv [Preprint]. 10.1101/2020.03.15.992883
- Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang ML, Kim NG, Yu X, Li J, Walker BD, Greninger A. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. MedRxiv. 2020 doi: 10.1101/2020.05.04.20091231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju E, Li T, Ramos da Silva S, Gao SJ. Gold nanocluster-mediated efficient delivery of Cas9 protein through pH-induced assembly-disassembly for inactivation of virus oncogenes. ACS Appl Mater Inter. 2019;11:34717–34724. doi: 10.1021/acsami.9b12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep. 2016;6:1–15. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M, Ibrahim A, Fayez M, Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020;92:660–666. doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Prot. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khambhati K, Bhattacharjee G, Singh V. Current progress in CRISPR-based diagnostic platforms. J Cell Biochem. 2019;120:2721–2725. doi: 10.1002/jcb.27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Koo T, Jee HG, Cho HY, Lee G, Lim DG, Shin HS, Kim JS. CRISPR RNAs trigger innate immune responses in human cells. Genome Res. 2018;28:367–373. doi: 10.1101/gr.231936.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Birnbaum MD, Moorthy BT, Singh J, Palovcak A, Patel DM, Zhang F. Insertion/deletion-activated frame-shift fluorescence protein is a sensitive reporter for genomic DNA editing. BMC Genom. 2019;20:609. doi: 10.1186/s12864-019-5963-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbink RJ, de Jong DC, Wolters F, Kruse EM, van Ham PM, Wiertz EJ, Nijhuis M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci Rep. 2017;7:1–10. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, De Clercq E. HIV genome-wide protein associations: a review of 30 years of research. Microbiology and Molecular Biol Rev. 2016;80:679–731. doi: 10.1128/MMBR.00065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL, Gao S, Cao RB, Zhao GP, Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discover. 2018;4:1–4. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li S, Wu N, Wu J, Wang G, Zhao G, Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synt Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li Y, Li S, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, Esteban CR. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Comm. 2015;6:1–10. doi: 10.1093/cid/ciy565. [DOI] [PubMed] [Google Scholar]
- Liu Z, Chen S, Jin X, Wang Q, Yang K, Li C, Xiao Q, Hou P, Liu S, Wu S, Hou W. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection. Cell Biosci. 2017;7:1–15. doi: 10.1186/s13578-017-0174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Yin L, Li X, Chen S, Peng L, Liu G, Ye S, Zhang W, Man S. A smartphone-based visual biosensor for CRISPR-Cas powered SARS-CoV-2 diagnostics. Biosens Bioelectron. 2022;195:113646. doi: 10.1016/j.bios.2021.113646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet. 2014;384:258–271. doi: 10.1093/cid/ciy565. [DOI] [PubMed] [Google Scholar]
- Maas SL, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:1–26. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR–Cas systems. Biochem Soc Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Moorthy BT, Kumar A, Lotenfoe LX, Zhang F. Evaluation of the efficiency of genome editing tools by a frameshift fluorescence protein reporter. Bio Protoc. 2020;10:e3622. doi: 10.21769/BioProtoc.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, Garcia KF. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem M, Majeed S, Hoque MZ, Ahmad I. Latest developed strategies to minimize the off-target effects in CRISPR-Cas-mediated genome editing. Cells. 2020;9:1608. doi: 10.3390/cells9071608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TM, Zhang Y, Pandolfi PP. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat Stru Mole Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Richter H, Charpentier E, Randau L. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol Rev. 2015;39:442–463. doi: 10.1093/femsre/fuv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Scheler O, Glynn B, Kurg A. Nucleic acid detection technologies and marker molecules in bacterial diagnostics. Expert Rev Mol Diagn. 2014;14:489–500. doi: 10.1586/14737159.2014.908710. [DOI] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, Liu DR, Gifford DK, Sherwood RI. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1093/cid/ciy565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell SG, Walker PJ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Harrach B, Harrison RL, Junglen S, Knowles NJ. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018) Arch Virol. 2019;164:943–946. doi: 10.1007/s00705-018-04136-2. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano SF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Skehel JJ, Hay AJ. Nucleotide sequences at the 5′ termini of influenza virus RNAs and their transcripts. Nucleic Acids Res. 1978;5:1207–1219. doi: 10.1093/nar/5.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen J, Thielen A, Lübke N, Dirks M, Widera M, Dittmer U, Kordelas L, Däumer M, de Jong DC, Wensing AM, Kaiser R. Rapid rebound of a preexisting CXCR4-tropic human immunodeficiency virus variant after allogeneic transplantation with CCR5 Δ32 homozygous stem cells. Clin Infect Dise. 2019;68:684–687. doi: 10.1093/cid/ciy565. [DOI] [PubMed] [Google Scholar]
- Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS ONE. 2014;9:e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell. 2015;163:840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhao N, Berkhout B, Das AT. A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep. 2016;17:2819–2826. doi: 10.1016/j.celrep.2016.11.057. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mole Ther. 2016;24:522–526. doi: 10.1093/cid/ciy565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1093/cid/ciy565. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas based antiviral strategies against HIV-1. Virus Res. 2018;244:321–332. doi: 10.1016/j.virusres.2017.07.020. [DOI] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004;4:337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Man S, Ye S, Liu G, Ma L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens Bioelectron. 2021;193:113541. doi: 10.1016/j.bios.2021.113541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Yao Y, Xiao H, Li J, Liu Q, Yang Y, Adah D, Lu J, Zhao S, Qin L, Chen X. Simultaneous knockout of CXCR4 and CCR5 genes in CD4+ T cells via CRISPR/Cas9 confers resistance to both X4-and R5-tropic human immunodeficiency virus type 1 infection. Hum Gene Ther. 2018;29:51–67. doi: 10.1089/hum.2017.032. [DOI] [PubMed] [Google Scholar]
- Yuan M, Wu NC, Zhu X, Lee CCD, So RT, Lv H, Mok CK, Wilson IA. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Abudayyeh OO, Gootenberg JS (2020) A protocol for detection of COVID-19 using CRISPR diagnostics. https://broad.io/sherlockprotocol [accessed 20th January 2022]
- Zhang Y. CRISPR-Cas13 as an Antiviral Strategy for Coronavirus Disease 2019. The CRISPR J. 2020;3:140–142. doi: 10.1089/crispr.2020.29094.yzh. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Qian L, Wei W, Wang Y, Wang B, Lin P, Liu W, Xu L, Li X, Liu D, Cheng S. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synt Biol. 2017;6:211–216. doi: 10.1021/acssynbio.6b00215. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Xu L, Lou C, Ouyang Q, Qian L. Paired dCas9 design as a nucleic acid detection platform for pathogenic strains. Methods. 2021 doi: 10.1016/j.ymeth.2021.06.003. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- Zhou W, Cui H, Ying L, Yu XF. Enhanced cytosolic delivery and release of CRISPR/Cas9 by black phosphorus nanosheets for genome editing. Angew Chem. 2018;130:10425–10429. doi: 10.1002/ange.201806941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.