Abstract
The diterpenoid triepoxides triptolide and triptonide from Tripterygium wilfordii (thunder god wine) exhibit unique bioactivities with potential uses in disease treatment and as a non-hormonal male contraceptives. Here, we show that cytochrome P450s (CYPs) from the CYP71BE subfamily catalyze an unprecedented 18(4→3) methyl shift required for biosynthesis of the abeo-abietane core structure present in diterpenoid triepoxides and in several other plant diterpenoids. In combination with two CYPs of the CYP82D subfamily, four CYPs from T. wilfordii are shown to constitute the minimal set of biosynthetic genes that enables triptonide biosynthesis using Nicotiana benthamiana and Saccharomyces cerevisiae as heterologous hosts. In addition, co-expression of a specific T. wilfordii cytochrome b5 (Twcytb5-A) increases triptonide output more than 9-fold in S. cerevisiae and affords isolation and structure elucidation by NMR spectroscopic analyses of 18 diterpenoids, providing insights into the biosynthesis of diterpenoid triepoxides. Our findings pave the way for diterpenoid triepoxide production via fermentation.
Subject terms: Natural product synthesis, Secondary metabolism, Metabolic engineering
How triptonide is made in the medicinal plant Tripterygium wilfordii is largely unknown. Here, the authors report the identification and characterization of a suite of cytochrome P450s and show their function in catalyzing the formation of triptonide from miltriadiene in tobacco and baker’s yeast.
Introduction
The Chinese medicinal plant Tripterygium wilfordii (léi gōng téng, Thunder god wine) is currently the exclusive source of the high value diterpenoids triptolide (1) and triptonide (2)1. These unique triepoxide diterpenoids (Fig. 1) have multiple applications as exemplified by the use of triptolide as the active component in new non-lethal rodent pest management products2 and the recent identification of triptonide as a promising non-hormonal male contraceptive agent that shows high efficacy, being reversible and safe. Biochemical analyses show that triptonide targets one of the final steps in sperm development resulting in loss of the motility required for egg fertilization and consequently reversible male infertility3. Widespread adoption of these novel applications for triepoxide diterpenoids is restricted by the high cost associated with triptolide extraction and purification from T. wilfordii or from established plant tissue cultures4,5. The establishment of a Saccharomyces cerevisiae production platform for T. wilfordii derived triepoxide diterpenoids has been hampered by insufficient knowledge about the biosynthetic pathway operating in the host plant6.
Fig. 1. 18(4→3) abeo-abietane methyl shift.
a Hypothesized scheme for the rearrangement of methyl groups on the A-ring of labdane type diterpenoids and examples of diterpenoids with the 18(4→3) abeo-abietane core structure. Examples of labdane type diterpenoids from the Lamiaceae species Coleus forskohlii and the Celastracae species T. wilfordii with an abeo-abietane core structure are shown. The rings of the tricyclic core structure are denoted A, B, and C. b Maximum-likelihood phylogenetic analysis of selected CYP genes from the 71 clan, including plant CYPs from the CYP82 and CYP71 subfamily, and NtCYP51 as the root. Shown CYPs are involved in either phenylpropanoid (purple) or terpenoid (grey) biosynthesis (Supplementary Table 22, Supplementary Fig. 1). TwCYP genes functionally characterized in this work highlighted in bold. Nodes marked * are supported by bootstrap values >85%. c Schematic overview of the genome location of the Tw genes used for heterologous biosynthesis of triptonide in the T. wilfordii genome6. Genes marked with * are found as tandem repeats with each copy having on average nucleotide sequence identity of 88.7% to the cDNA sequence of each of the genes used in this paper (Supplementary Table 23). TwCYP71BE84 and TwCYP71BE86 homologs are each found in tandem repeats in two different loci of the T. wilfordii genome.
Diterpenoid biosynthesis in plants is initiated by carbocation mediated carbon rearrangement of geranylgeranyl diphosphate (GGPP) into labdane type diterpenoids catalyzed by diterpene synthases (diTPS)7. In contrast to the canonical labdane type diterpenoids, T. wilfordii triepoxides harbors an unusual 18(4→3) abeo-abietane core structure (Fig. 1a). No diTPS has been linked to the formation of the abeo-abietane diterpene backbone directly from GGPP and previous studies support that the triepoxide diterpenoids found in T. wilfordii are derived from the abietane diterpene miltiradiene8,9. Thus, the biosynthesis of triptonide and triptolide must include enzymes that accept miltiradiene as a substrate, and catalyze reactions that can account for the unique positioning of the methyl groups on the A-ring of T. wilfordii triepoxides.
Here, we present the identification and characterization of a suite of T. wilfordii cytochrome P450s (CYPs) that in the heterologous hosts N. benthamiana and S. cerevisiae catalyze the formation of triptonide (2) from miltiradiene (3). To shed light on the biosynthetic processes leading to triptonide accumulation, 13 hitherto undescribed and 5 previously described diterpenoids are isolated and structure elucidated by NMR spectroscopy (Supplementary Figs. 8–53 and Supplementary Tables 1–19). In S. cerevisiae, the flux of diterpenoid through the cascade of orchestrated CYP oxygenations is increased by co-expression of a gene encoding a specific cytochrome b5 (cytb5). The functional expression of TwCYPs and Twcytb5 in S. cerevisiae constitute the foundation for the proof-of-concept strain for fermentation-based production of T. wilfordii derived triepoxide diterpenoids.
Results
Identification of TwCYP82s involved in miltiradiene oxygenation
To identify candidate CYPs capable of catalyzing oxygenation of diterpenes in T. wilfordii, homology searches were performed in publicly available T. wilfordii RNA sequencing data deposits (Supplementary Table 20). More than 61 plant CYPs mainly from the CYP71 and CYP85 clans have been shown to utilize diterpenes as substrate10,11 (Supplementary Fig. 1) and genes encoding CYPs from these two clans were used as queries in the homology-based search for candidate genes. In total, 68 candidate TwCYP-encoding transcripts (Supplementary Table 21) were identified and isolated from cDNA derived from root, stem and leaf tissue of T. wilfordii8,12. Three of these genes have previously been shown to be involved in terpenoid biosynthesis catalyzing key steps in the biosynthetic pathway of celastrol, a high value triterpenoid found in root extracts of T. wilfordii12.
For in planta characterization of the TwCYPs, each of the encoding genes were co-expressed separately in N. benthamiana plants13 together with CfTPS1 and CfTPS3, that catalyze the formation of 314, and with GGPP booster genes15,16. Combined, these are denoted as the miltiradiene biosynthetic genes. Metabolite extracts of the N. benthamiana leaf discs were analyzed by GC-MS and LC-qTOF-MS. Of the 68 candidate TwCYPs co-expressed individually with the miltiradiene biosynthetic genes, co-expression of TwCYP82D274 resulted in the depletion of 3 with an accompanied appearance of 14-hydroxy-dehydroabietadiene (5) (Fig. 2, and Supplementary Note 1), 3-epi-triptobenzene B (13)17 (Supplementary Fig. 29 and Table 9) and a number of additional compounds with monoisotopic masses (detected by LC-qTOF-MS) corresponding to oxygenated diterpenoids (Supplementary Fig. 2). Metabolites 5 and 13 have previously been isolated from T. wilfordii root tissue or from tissue cultures along with 1 and 217,18.
Fig. 2. Biosynthesis of triptonide from miltiradiene.

a Heterologously expressed genes constituting the minimal set of biosynthetic components required for heterologous triptonide biosynthesis. b quantity (bars) of putative key intermediates in the biosynthetic pathway of triptonide (right), when established in vivo via heterologous gene expression in N. benthamiana. c, quantification of intermediates from engineered S. cerevisiae (strains listed in Supplementary Table 28). The combination of genes co-expressed with the miltiradiene biosynthetic genes is indicated by black dots. Quantification was based on peak areas of signature m/z values for the putative intermediates, normalized to the peak area of signature m/z values for the internal standard (IS) used in the GC-MS (compounds 3–5) and LC-qTOF-MS (compounds 6–8 & 2). Signature m/z values are denoted in the header of each bar diagram. Bars represent the average of n = 3 [N. benthamiana] or n = 4 [S. cerevisiae] biological replicates, with error bars representing standard error of the mean (SEM). Values from each replicate is marked by black diamond squares. White- and grey fill color of the bars distinguishes compound quantity in relation to no expression (grey) or co-expression (white) of Twcytb5-A, respectively. Mass tolerance was ±0.1 m/z for GC-MS and ±0.005 m/z for LC-qTOF-MS for signature m/z values. Source data are provided as a Source Data file.
To assess whether enzymes native to N. benthamiana contributed to the turnover of miltiradiene-derived diterpenoids, we established an S. cerevisiae strain (NVJ0) capable of producing high levels of miltiradiene15. Furthermore, to support downstream CYP functionality, the gene encoding CYP reductase TwPOR1 (Supplementary Table 23) identified by BLAST searches in T. wilfordii transcriptomes, was integrated into the S. cerevisiae genome along with the TwCYPs. Interestingly, when TwCYP82D274 was genome integrated in the established S. cerevisiae strain, accumulation of 5 but not 13 was observed (Supplementary Fig. 2d), demonstrating consensus for formation of only 5 when comparing the plant and microbial host systems. The products specifically formed in the plant host could reflect intrinsic properties, such as the activity of endogenous enzymes, affecting the direct biosynthetic products of the co-expressed enzymes.
TwCYP71BE86 and TwCYP71BE85 oxygenate the A-ring of 14-hydroxy-dehydroabietadiene resulting in a C4→C3 methyl shift and lactone formation
We hypothesized that oxygenation of the A-ring of 3 could facilitate the 18(4→3) abeo-abietane methyl shift. Previously, CYPs from the CYP701A, CYP99A, CYP71Z, CYP71BE, and CYP71D subfamilies have been shown to be capable of oxygenating the A-ring of labdane type diterpenoids (Fig. 1 and Supplementary Fig. 1)10. CfCYP71D381 from Coleus forskohlii has been shown to oxygenate C-2 and C-20 of 13R-manoyl oxide when heterologous produced in N. benthamiana19, but to our knowledge, isolation of neither 2-hydroxy-13R-manoyl oxide nor 20-hydroxy-13-R-manoyl oxide from the host plant has been reported20. On the other hand, a number of abietane and 18(4→3) abeo-abietane diterpenoids have been isolated from C. forskohlii (Fig. 1a)20. We therefore hypothesized that CfCYP71D381 possibly accepts 3 and that members of the CYP71D subfamily could be involved in catalysis of reactions leading to the 18(4→3) methyl shift found in abeo-abietanes of both C. forskohlii and T. wilfordii. Accordingly, we tested whether 3 was a substrate for CfCYP71D381. Co-expression of miltiradiene biosynthetic genes14 and CfCYP71D381 in N. benthamiana resulted in new metabolites detected in leaf extracts by LC-qTOF-MS. In the mass spectra representing these new metabolites, the monoisotopic masses of likely parental ions supported the identification (<5 ppm) of oxygenated diterpenoids (Supplementary Fig. 3), showing that CfCYP71D381 can efficiently oxygenate 3 in planta. With this finding we searched the TwCYP library for CfCYP71D381 homologs and found four CYP-encoding genes of the CYP71BE subfamily. Recent expansion of the CYP71D subfamily by the identification of new CYP genes have caused the CYP71D subfamily to gain a phylogenetic overlap with the CYP71BE subfamily (Supplementary Fig. 1). These four CYP71BE subfamily encoding genes were selected for further studies.
To test for turnover of 3, the four candidates TwCYP71BE83, TwCYP71BE84, TwCYP71BE85, and TwCYP71BE86 were co-expressed individually in N. benthamiana, with the miltiradiene biosynthetic genes. Expression of TwCYP71BE85 and TwCYP71BE86 resulted in accumulation of oxygenated diterpenoids, with a larger number of new metabolites observed with TwCYP71BE86 expression in comparison to TwCYP71BE85 (Supplementary Fig. 3). We then tested whether the biosynthetic products of TwCYP82D274 could be utilized by TwCYP71BE85 or TwCYP71BE86 by co-expressing each of these genes together with TwCYP82D274. LC-qTOF-MS analysis of N. benthamiana leaf extracts revealed that co-expression of either TwCYP71BE85 or TwCYP71BE86 with TwCYP82D274 resulted in the accumulation of 13 in addition to other polyoxygenated diterpenoids (Fig. 2 and Supplementary Fig. 4). In leaves expressing TwCYP71BE86, we also observed formation of low amounts of additional metabolites, the structures of which were identified by NMR analyses or by comparison to authentic standards. One of these were identified as triptophenolide (8) (Fig. 2), a putative intermediate in the triptolide pathway that similar to compound 1 and 2 contains the 18(4→3) abeo-abietane core structure, and the lactone ring at the A-ring18 (Fig. 1b). Two other metabolites with similar core structures were 18(4→3) abeo-abietatrien-14,18-diol (6) and 14-hydroxy-18(4→3) abeo-abietatrien-18-al (7) (Fig. 2a, Supplementary Figs. 11–16 and Supplementary Tables 3–4). These results prompted us to co-express TwCYP71BE85 and TwCYP71BE86 in N. benthamiana leaves. Here we observed a significant increase in the accumulation of 8 (Fig. 2b, Supplementary Fig. 4), suggesting that both encoded enzymes can partake in the methyl rearrangement of the abietane core structure and in formation of the lactone moiety of 8.
Similar to the observations in the in planta expression host, co-expression of TwCYP82D274, TwCYP71BE85, TwCYP71BE86 and TwPOR1 in a miltiradiene producing strain of S. cerevisiae resulted in accumulation of 8 (Fig. 2c). However, in contrast to the results obtained using the N. benthamiana system, co-expression of TwCYP82D274 and TwCYP71BE86 did not result in production of 8 in S. cerevisiae. This supports that TwCYP71BE86 upon expression in N. benthamiana as well as in S. cerevisiae catalyzes the formation of the 18(4→3) abeo-abietane backbone and that TwCYP71BE85 expression in S. cerevisiae is required for catalysis of lactone ring formation and thus for formation of 8 (Fig. 2b, c).
Twcytb5-A enhances the capacity of TwCYPs to catalyze multiple oxygenations of their diterpenoid substrates
It is well established that cytb5 may serve as an additional electron donor in CYP catalyzed reactions21,22. To possibly improve the yield of the TwCYP products obtained in S. cerevisiae, the genes encoding six different Twcytb5’s were expressed individually in S. cerevisiae strains together with TwCYPs needed for biosynthesis of 8. Co-expression of Twcytb5-A resulted in a substantial increase of 8 in the S. cerevisiae extracts, while expression of the other Twcytb5’s did not have the same effect (Supplementary Fig. 5). Co-expression of Twcytb5-A with the different combinations of TwCYPs both in N. benthamiana and in S. cerevisiae (Fig. 2) revealed that the levels of CYP products were predominantly improved in the fungal host. Quantitative data for the accumulation of miltiradiene-derived diterpenoids by transient expression in leaves of N. benthamiana showed substantially higher variability across replicates when compared to production in engineered S. cerevisiae. This is consistent with the variability observed in other N. benthamiana heterologous biosynthesis studies16. Still, it is to be noticed that co-expression of Twcytb5A and TwCYP82D274 in N. benthamiana increased the accumulation of 5 when compared to N. benthamiana only expressing TwCYP82D274 alone. The opposite was observed in relation to the amount of 8, 6 and 7 (Fig. 2b) detected in leaf extracts co-expressing TwCYP82D274 and TwCYP71BE86, in which Twcytb5-A co-expression had a negative impact of accumulation of these compounds. This observation renders it possible that presence of Twcytb5-A effect the catalytic rate of TwCYP82D274 and TwCYP71BE86, differently in planta23. In contrast, the level of all intermediates identified in the S. cerevisiae extracts increased with Twcytb5-A co-expression, with the most pronounced effect seen on the levels of intermediates associated with expression of TwCYP71BE86 (Fig. 2c).
TwCYP82D213 completes the biosynthetic pathway for triptonide
The uniqueness of T. wilfordii triepoxide diterpenoids as pharmaceutical agents are attributed to the distinct configuration of epoxides on the B- and C-rings (Fig. 1a). These epoxides are essential for bioactivity, e.g., anticancer activity of triptolide towards multiple cancer cell lines24,25. CYPs have previously been shown to catalyze epoxide formation in plant specialized metabolism26. TwCYP82D274 catalyzed oxygenation of the C-ring in 3, resulting in accumulation of 5, but also oxygenations resulting in the accumulation of other miltiradiene derived compounds with more than two oxygens (Supplementary Fig. 2). Accordingly, we hypothesized that TwCYP82D274 in combination with other CYP82D homologs identified from T. wilfordii cDNA could catalyze oxygenations leading to the formation of the epoxides on the B- and C-ring of 2. Among the collection of TwCYP82D encoding genes isolated from T. wilfordii cDNA (Supplementary Table 21), TwCYP82D213 was selected and co-expressed with TwCYP82D274, TwCYP71BE86 and TwCYP71BE85 in N. benthamiana and subsequently in S. cerevisiae. In both expression hosts, co-expression of these four TwCYP encoding genes resulted in the accumulation of 2 (Fig. 2).
Based on these findings we conclude that the four TwCYPs together with the miltiradiene biosynthetic genes constitute a minimal set of triptonide biosynthetic genes in N. benthamiana and S. cerevisiae expression hosts (Figs. 2 and 3).
Fig. 3. Accumulation in fed-batch fermentation.
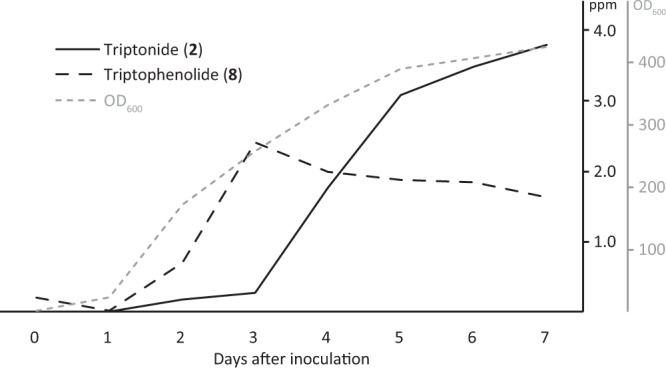
The accumulation of 8 (dotted black) and 2 (solid black) from NVJ8.5 (Supplementary Table 28) grown in a fed-batch fermentation. Density of the S. cerevisiae culture was based on the optical density of daily culture samples.
The route towards triepoxide formation by TwCYP catalyzed oxygenations in two orthologous biosynthesis hosts
A number of miltiradiene derived products including compounds with monoisotopic masses corresponding to glucosides and glutathione diterpenoid conjugates were identified by LC-qTOF-MS analysis of extracts of N. benthamiana leaves expressing the triptonide biosynthetic genes (Supplementary Fig. 6, Supplementary Table 24). Glucosylation and glutathionylation of oxygenated terpenoids heterologously produced in N. benthamiana tissues have previously been observed27,28 and their accumulation is likely caused by native N. benthamiana glucosyltransferases and glutathione S-transferases29.
From the engineered biosynthetic hosts, 23 GGPP derived compounds were identified, including 18 which were structure elucidated by use of 1D and 2D NMR spectroscopy in this work (Supplementary Figs. 9–53, Supplementary Table 1–20). Besides isocopal-13(16)-en-3,12,15-triol (19), labda-8(17),13-E−15-O-acetate-dien-3-ol (20), 14,15-epoxygeranylgeraniol (21), and trimethylcyclohexane-11,14-diolgeraniol (22) (Supplementary Note 3, Supplementary Figs. 21–53 and Supplementary Tables 15–19), all compounds (including the lactam-diterpenoid (23) (Supplementary Note 2, Supplementary Figs. 51–53 and Supplementary Table 19) were considered to be derived from 3 (see below). Of these, 2, 3, 13, 8, and triptobenzene I (16) have previously been isolated from species within the Tripterygium genus18,30,31 (Supplementary Table 1). Furthermore, 4-epi-triptobenzene J (14) and 4-epi-tripquinone C (18), diastereomers of compounds found in Tripterygium species were identified30,32. Hence, a substantial fraction of the molecular diversity of diterpenoid compounds present in Tripterygium spp. can be found in the engineered S. cerevisiae strain (NJV11.11, Supplementary Table 28) extracts. This demonstrates that the concerted enzymatic capacity of only four TwCYP enzymes enables biosynthesis of a number of abietane/abeo-abietane compounds that constitute a major part of the diterpenoid compound diversity found in the Tripterygium species.
Alternative biosynthetic routes for triepoxide aboe-abietane diterpenoids
In previous work, it has been proposed that dihydroabietic acid is an intermediate in the biosynthetic pathway for abeo-abietanes including the triepoxide terpenoids from T. wilfordii6,18,33. A possible route for dihydroabietic acid biosynthesis in T. wilfordii could be through TwCYP728B70 catalyzed oxygenation of miltiradiene and a subsequent spontaneous oxidation6. Here we find that CYPs from the CYP71BE and CYP82D subfamilies are sufficient for triptonide biosynthesis in both N. benthamiana and S. cerevisisae. Furthermore, none of the miltiradiene-derived compounds identified here carried a carboxyl group at C-18.
Genomic organization of triptonide specific TwCYP genes from T. wilfordii
Recently a high quality chromosome level genome of T. wilfordii have become available6. To our surprise genes from the CYP82D and CYP71BE subfamilies could not be identified by BLAST searches on the T. wilfordii RNA library based on the genome annotation. However, BLAST searches with the triptonide biosynthetic genes as queries on the assembled genome enabled us to identify the genome position of all (Supplementary Table 23). Interestingly, similar to TwTPS9 and TwTPS27, close homologs of TwCYP82D274 and TwCYP71BE86 were found in tandem repeats on chromosome CM023879 and CM23888, respectively (Fig. 1c). The genes encoding TwTPS9 and TwTPS27 involved in miltiradiene biosynthesis8 were identified positioned approximately 3 Mbp downstream from the TwCYP71BE86 tandem repeats (Supplementary Table 23). In contrast, the genome location of, the majority of genes enabling heterologous triptonide biosynthesis reveal that they are situated on different chromosomes in the T. wilfordii genome (Fig. 1c).
S. cerevisiae as a heterologous fermentation host for triptonide production
Co-expression of the triptonide biosynthetic genes including Twcytb5-A in S. cerevisiae (NJV11.11) resulted in 0.081 (n = 3, SD: 0.004) mg triptonide/L in 0.5 mL cultures using a 96-well plate as fermentation vessel. Substantial quantities of other GGPP derived compounds were also identified as being produced in the triptonide biosynthesizing strains. It remains to be determined which of these may function as intermediates in the triptonide biosynthesis pathway or as shunt products arising from an inefficient, imbalanced or incomplete triptonide pathway (see below).
Based on the established triptonide biosynthetic pathway, we sought to optimize the biosynthetic output by stable integration of additional copies of the triptonide biosynthetic genes in the S. cerevisiae genome. Following successful integration of one additional copy of the four TwCYP encoding genes, the yield of triptonide doubled to 0.20 (n = 3, SD: 0.06) mg triptonide/L in S. cerevisiae (NJV 8.15) extracts, making it our elite strain. To explore the use of S. cerevisiae as a production host for 2, the elite strain was grown in a 1 L fed-batch fermentation with daily monitoring of OD600 and quantification of 8 and 2. The level of 8 reached a maximum of 2.4 mg/L on day 3, whereas the highest level of 3.79 mg/L of 2 was at day 7 (Fig. 3). The reason for the observed differences between the accumulation of the compounds over time is unclear. It could be speculated that 8 is a rate limiting intermediate in the triptonide biosynthetic pathway. OD600 level at the end of the fermentation was high as a result of considerable water evaporation occurring during the fermentation run. Still, the fed-batch fermentation of NJV8.5 enabled us to obtain production titers supporting proof-of-concept for S. cerevisiae fermentation-based production of 2.
Discussion
By co-expression of T. wilfordii biosynthetic genes in two orthogonal heterologous host organisms, we demonstrate that four genes from the CYP71BE and the CYP82D subfamilies constitute a minimal set of genes required for formation of triptonide (2) from miltiradiene (3). The distinct nature of the two chosen production systems minimizes the likelihood of endogenous enzyme activities in the heterologous hosts being contributors to the formation of 2. Production in S. cerevisiae was more easily scaled and offered a cleaner background for purification of the intermediates selected for NMR analysis and structure elucidation.
In prior work, a majority of the functionally characterized genes from the CYP82D subfamily have been associated with enzymes catalyzing steps in flavonoid biosynthesis11 (Fig. 1b). No gene from this subfamily has to our knowledge been shown to be involved in oxygenation of diterpenoids11. A reaction similar to the one catalyzed by TwCYP82D274 is carried out by SmCYP76AH1 and homologs from Lamiaceae species resulting in formation of 11-hydroxy-dehydroabietadiene from 3 as demonstrated by expression studies in S. cerevisiae34. However, in many cases it remains to be determined whether conversion of 3 to dehydroabietadiene (4) is a spontaneous reaction or whether this happens in conjunction with oxidation of 3 resulting in aromatization of the C-ring (Fig. 2)35.
TwCYP71BE86 was shown to catalyze an unprecedented 18(4→3) abeo-abietane methyl shift within the abietane type scaffold found in 3, 4 and 5 when its encoding gene was expressed in N. benthamiana as well as in S. cerevisiae. This demonstrates that TwCYP71BE86 catalyzes a carbon rearrangement of labdane type diterpenoids, a highly unusual CYP reaction not previously assigned to any identified plant CYP36. Interestingly, ElCYP71D445 from the phylogenetically overlapping subfamily (Fig. 1b, Supplementary Fig. 1) has been shown in combination with ElCYP726A27 and ElADH1 to catalyze oxygenations facilitating carbon rearrangement of its macrocyclic diterpenoid substrate possibly via an aldol reaction37. SmCYP71D375 from the same CYP family has been shown to catalyze the formation of the heterocycle in Tanshinone IIA from miltirone possibly via a P450 mediated carbocation reaction mechanism38. Thus, three plant CYPs from the CYP71D/BE subfamily have been shown to be involved in rearrangement of diterpenoid core structures.
Carbocation mediated carbon rearrangements are the common mechanism in terpenoid synthase (TPS) catalyzed formation of cyclic terpenoids from prenyldiphosphates39. In contrast, the reported additional ability of plant CYPs to catalyze the formation of carbocations mediating terpenoid carbon rearrangements opens up a new route to the formation of complex natural products. Thus, the formation of 5(12)-oxa-3(11)-cyclotaxane upon heterologous expression of TbCYP725A from Taxus brevifolia in N. benthamiana, has been argued to be the result of a carbocation mediated carbon rearrangement of taxadiene40. TbCYP725A is part of the CYP85 clan. From the same clan, detailed studies of members of the CYP88A subfamily support that they catalyze ring-contraction of ent-kaurenoic acid via formation of a carbocation and a pinacol type rearrangement41. The bacterium Streptomyces arenae synthesizes the terpenoid pentanolactone. The final step of its biosynthesis proceeds via a CYP161C2 catalyzed formation of a carbocation, resulting in a Wagner-Meerwein type methyl shift and formation of a cyclo-pentene moiety42,43. A similar catalytic mechanism has to our knowledge never been reported for plant CYPs. CYP161C2 is distantly related to TwCYP71BE86 but renders it possible that TwCYP71BE86 catalyzes the 18(4→3) abeo-abietane methyl shift using a Wagner-Meerwein type reaction mechanism (Fig. 4b). This is further supported by the observation that the compounds 13, 6, 8, 18 R(4→3) abeo-abietatrien-19-ene-14,18,20-triol (9), 18 R(4→3) abeo-abietatrien-14,18-diol (10), 18-S-(4→3) abeo-abietatrien-14,18-diol (11), 18 S(4→3) abeo-abietatrien-14,18,20-triol (12), 18(4→3) abeo-abietatrien-14,18,20-triol (15), (Fig. 4a, b) were identified in extracts of S. cerevisiae expressing the genes encoding TwCYP82D274 and TwCYP71BE86. Compounds 9-12 all lack the C-3(C-4) double bond which could possibly cause stereochemical constraints of the C-3 and C-4 methyl’s preventing lactone formation (Fig. 4a). Assuming that the 18(4→3) abeo-abietane methyl shift is facilitated by a TwCYP71BE86 catalyzed formation of a carbocation, these compounds could be considered shunt products from an early quenching of the carbocation leading to a failure of establishing the C-3(C-4) double bond and the lactone (Fig. 4b). In this scenario TwCYP71BE86 would go through multiple catalytic cycles, by initially catalyzing formation of the carbocation of C-3 causing the 18(4→3) abeo-abietane methyl shift and subsequent oxygenations of C-18 resulting in the formation of 6, and possibly also 9, 10, 11, and 12. If TwCYP71BE86 utilizes a mechanism similar to CYP161C243, failure to inhibit the O-rebound at the TwCYP71BE86 active site could explain the formation of 13, that subsequently is oxygenated causing the accumulation of 14, 17, and 18 (Fig. 4a).
Fig. 4. Proposed biosynthetic pathway of triptonide (2) from 14-hydroxy-dehydroabietadiene (5) in S. cerevisiae.
a The biosynthetic pathway from 5 to 2 illustrating the formation of oxygenated miltiradiene derived intermediates catalyzed by the action of TwCYP82’s (blue arrows) and TwCYP71BE’s (red arrows) on the A- and C-ring of the abietane diterpene backbone, respectively. Underlined compounds or diastereomers hereof have previously been identified in plant extracts from Tripterygium species (Supplementary Table 1). b a hypothetical Wagner-Meerwein rearrangement reaction (W.-M.) to account for the methyl shift of C-18 to C-3 in the abietane carbon backbone. Cpd I and Cpd II represents states of the heme in the CYP catalytic cycle. Red line at heme bound hydroxyl symbolizes inhibition of oxygen rebound facilitating e- transfer as proposed by43 for CYP mediated carbocation formation. Compound 13 and oxygenated compounds hereof including 14, 17, and 18, would be products caused by the failure to inhibit oxygen rebound and additional oxygenation likely to be catalyzed by the TwCYPs heterolgously expressed in S. cerevisiae. Alternatively, the C18(4→3) could be mediated via a mechanism relying on the radical formed by the Compound II (CpdII) state of the involved CYP enzyme (Supplementary Fig. 7).
Cationic rearrangements facilitated by CYP catalysis have previously been suggested44 and substantial experimental support has been provided in43 to show that CYPs can catalyze a Wagner-Meerwein shift. Nevertheless, it cannot be completely excluded that the C18(4→3) shift proceeds via a free radical formed at CpdII (Supplementary Fig. 7), or that both cationic and free radical based reactions are occurring in parallel. If so, this may represent an intriguing evolutionary strategy offering the opportunity to increase the structural diversification of natural products in the plant kingdom by the ability of some enzymes to convert a specific substrate into different products by using different types of catalytic reaction mechanisms.
Out of the five T. wilfordii cytochrome b5 enzymes tested, only Twcytb5-A stimulated the activity of TwCYP71BE86 in S. cerevisiae45. A similar cytb5 mediated stimulation has been observed in artemisinic acid biosynthesis catalyzed by AaCYP71AV1. While data on appropriate POR/cytb5 expression ratios for heterologous biosynthesis of plant diterpenoids in S. cerevisiae has not been reported45, we hypothesize that differences in expression ratios could explain the disparate effects of Twcytb5-A expression in S. cerevisiae and N. benthamiana. In cytb5 titration studies with mammalian CYPs, CYP activity was inhibited at specific ratios46. Alternatively, or in addition, differences in ER membrane properties, native POR and cytb5 enzymes, or unknown factors in the two distinct hosts used for heterologous triptonide biosynthesis could influence Twcytb5-A functionality. The different effects of Twcytb5 co-expression show the advantage of using orthogonal systems for in vivo characterization of biosynthetic genes involved in plant specialized metabolism.
The TwCYP71BE86 associated formation of 6 and 7 requires a multiplicity of consecutive catalytic cycles. To facilitate these, electrons from TwPOR and/or Twcytb5-A need to be readily available. Presence of CYP reducing partners oxidizing NADPH and NADH, respectively, could promote the availability of electrons for an orchestrated series of CYP oxygenations. Failure to complete these concerted oxygenations could account for the accumulation of a number of the diterpenoid products formed in the S. cerevisiae triptonide biosynthesis strain.
In this study we demonstrate that biosynthesis of 2 from 3 can be catalyzed by CYPs from the CYP71BE and CYP82D subfamilies involving unique reactions including a methyl shift on the A-ring and multiple epoxidations of the B/C-ring system, respectively. Epoxides are generally highly reactive and typically labile moieties. LC-qTOF-MS analysis of extracts and NMR analysis of purified diterpenoids from S. cerevisiae afforded 2 as the only epoxide carrying compound identified. A triepoxide with a conformation of epoxide rings identical to 2 is the only epoxide containing diterpenoid observed in T. wilfordii tissue cultures18. Thus, derivatives of 3 with one or two epoxide rings have neither been identified in the native plant nor in the heterologous expression hosts described here. Instead, two abietane quinones were identified, whereof 18 and 16 have previously been isolated from Tripterygium hyoglaucoma and T. wilfordii32,47. To achieve efficient formation of the unique triepoxide configuration, TwCYP82D274 and TwCYP82D213 could be required to pass through multiple highly coordinated consecutive catalytic cycles, possibly facilitated by metabolon formation48. Intermediates or products escaping the concerted oxygenations including diterpenoids with one or two epoxide rings are possibly less stable compounds that rearrange into quinone diterpenoids. Further studies including evaluation of the stability of mono- and diepoxide abeo-abietane diterpenoids possibly derived by chemical synthesis would be required to clarify some of these questions and hypotheses.
In addition to being organized in metabolons, orchestration of plant biosynthetic pathways may also be controlled at the genome level e.g. by localization of the biosynthetic genes in gene clusters49. The enzymes identified here, including close orthologs of TwCYP82D274 and TwCYP71BE86, were found in tandem repeats on the chromosomes CM023878, CM023886 and CM023888 (Fig. 1c and Supplementary Table 23). These tandem repeats could have emerged from gene duplication throughout the evolution of this plant species50. Multiple copies of these genes could enable T. wilfordii to achieve high levels of the CYP enzymes via simultaneous transcription from multiple biosynthetic genes in the genome. With the diTPS genes encoding the first committed step in biosynthesis of 2 being co-localized on the same chromosome as TwCYP71BE86/85, the biosynthetic pathway of 2 in T. wilfordii could be considered clustered51. Still, while TwCYP71BE86 can accept 3 as substrate (Supplementary Fig. 3) in planta, it remains to be determined how the derived products contribute to the biosynthesis of 2 or whether miltiradiene derived products from TwCYP82D274 catalysis are more appropriate substrates. Thus, it is unclear how and in which order the TwCYP71BE86 and TwCYP82D274 contribute to the biosynthetic step(s) following the formation of 3.
Considering that co-expression of four CYPs in two orthologous host systems resulted in the formation of several miltiradiene derived compounds also observed in Tripterygium species, a linear pathway might be considered a too simplistic model for the biosynthesis of 2 in T. wilfordii. Instead, broad substrate and product promiscuity of the TwCYPs could suggest that 2 is only one out of many miltiradiene derived products in Tripterygium species that emerges from a grid-type biosynthetic pathway with involvement of a limited number of biosynthetic enzymes (Fig. 4a).
To conclude, the identification of CYPs catalyzing unprecedented types of reactions in plant diterpenoid biosynthesis significantly advances our understanding about their diverse catalytic capacities. This finding will guide future efforts in assigning CYPs to biosynthetic pathways with reaction steps not easily explained as classical CYP catalyzed reactions. Diterpenoid scaffold diversity have until now mainly been associated with the catalytic capacity of diTPS while CYPs have been associated with further decoration of the core structures by monooxygenation reactions52. Our data demonstrate that plant CYPs may also play a key role in the modification of the basic diterpenoid core structures. In the case of the biosynthesis of 2, these unique CYPs are harbored within the CYP71BE subfamily possibly catalyzing a Wagner-Meerwein reaction resulting in scaffold diversification of labdane diterpenoids, including formation of the 18(4→3) abeo-abietane backbone essential for biosynthesis of 2.
By introducing the genes shown to be involved in formation of 2 into yeast, we have established proof-of-concept for an on-demand and scalable production platform for 2 as a replacement for the current plant extraction-based production of this high value triepoxide diterpenoid.
Methods
Establishing constructs for high yield miltiradiene biosynthesis in N. benthamiana
For enhanced biosynthesis of miltiradiene, and to employ a gene assembly system that was compatible with the EasyCloni system53, we designed a new plasmid system for transient expression in N. benthamiana. Promoters for the new vector were sourced from literature and obtained via DNA synthesis (pCm9, pSGT54,55), from Addgene (pSIM24 – pM24 promoter56) or from in-house DNA template (p35S - pLIFE3357). A synthetic DNA Gblock was ordered from Integrated DNA Technologies (IDT, USA) containing the terminator tOCS58 and t3A terminator59 connected by the USER cassette 5´-CAACGGAATGCGTGCGATCGCGTGCATTC-3′. The new N. benthamiana expression vector was established by introducing the tOCS - USER cassette - t3A construct into the pLIFE33 backbone amplified with primer pairs GBA31 + GBA32, by InFusion cloning (Takara Bio, USA). Assembled plasmid was transformed into E. Cloni 10 G cells (Lucigen, USA). Plasmid sequence was verified by Sanger sequencing (Macrogen, South Korea) and named New_pLIFE. Genes selected for transient expression in N. benthamiana were cloned into the established plasmids by using methods described in the EasyCloni system. While working on the data presented here, it was shown that transient co-expression of GGPPS and diTPS encoding genes targeted to the cytosol, together with HMGR in N. benthamiana provides enhanced diterpenoid accumulation16. Employing a similar strategy, we constructed two dual expression constructs by cloning ScHMGR, together with a bidirectional promotor and SpGGPPS in the New_pLIFE plasmid. On another New_pLIFE plasmid tCfTPS1, was assembled with a bidirectional promotor and tCfTPS360. The bidirectional promotor consisted of pM24 and Cm9. Combined the four genes in the two expression construct are denoted the miltiradiene biosynthetic genes.
Isolation of T. wilfordii CYP genes and expression in Nicotiana benthamiana
Tripterygium wilfordii CYP genes (TwCYPs) were cloned from cDNA synthesized using the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher) from RNA isolated from either root, stem or leaf material from T. wilfordii grown at the green house facilities at the University of Copenhagen. TwCYPs were cloned into pLIFE33 or New_pLIFE33 by USER cloning57. A complete list of tested TwCYPs is provided in Supplementary Table 21.
Full length TwCYP genes were transiently co-expressed with the miltiradiene biosynthetic genes in N. benthamiana using agrobacteria mediated transfection13. Briefly, binary vectors each containing the miltiradiene biosynthetic genes or TwCYPs were transformed by electroporation into agrobacteria. Liquid cultures of transformed agrobacteria each containing specific plasmids were mixed for co-expression. Leaf material of N. benthamiana co-expressing specific combinations of TwCYPs together with the miltiradiene biosynthetic genes was harvested 7 d after agrobacterial infiltration. 1 mL MeOH was added to two leaf disks (Ø = 2 cm). Extraction was done at room temperature at 200 rpm orbital shaking. 200 μL of extract were filtered by using a 0.22 μm 96-well filter plate (Merck Millipore, Darmstadt, Germany) and at stored at 4 °C prior to LC-MS analysis.
Saccharomyces cerevisiae growth media
YPD media: 20 g/L Bacto™ Peptone, 10 g/L Bacto™ Yeast extract, 20 g/L glucose.
Synthetic complete (SC) meda without uracil: 1.92 g/L Yeast Synthetic Drop-out Media Supplements without uracil (Sigma-Aldrich Co. LLC. Catalog number Y1501), 6.7 g/L Yeast Nitrogen Base Without Amino Acids (Sigma-Aldrich Co. LLC. Catalog number Y0626), 20 g/L glucose. Feed-In-Time (FIT) was based on EnPump200 (Enpresso GmbH), and made according to protocol enclosed with the product. Agar plates: SC media including agar (15 g/L).
Uracil auxotrophy in parent strains was introduced by selecting for lack of URA3 function on agar plates of SC medium without uracil containing also 5-fluoroorotic acid (5-FOA, 0.74 g/L) and uracil (30 mg/L). Yeast transformants were isolated on SC without uracil agar plates.
Assembly of genetic constructs for S. cerevisiae genome engineering
All plasmids were generated by USER cloning57. Also, parent vectors named assembler −1, −2 and −3, for simultaneous genome integration of up to six gene constructs, and harboring AsiSI/Nb.BsmI USER-cassettes, were prepared for USER cloning53. Primers used for PCR amplification with USER compatible PfuX7 polymerase61 are listed in Supplementary Table 25. Vectors used and generated in this work is listed in Supplementary Table 26.
S. cerevisiae strain construction
Parent yeast strain was S. cerevisiae S288C (NCYC 3608; National Collection of Yeast Cultures, Norwich, UK). Genotypes and source of strains are listed in Supplementary Table 28. Biosynthesis of the triptonide precursor miltiradiene was established in S. cerevisiae as described in ref. 60. Codon optimized versions of TwCYP71BE85, TwCYP71BE86 and TwCYP82D213 (TWIST bioscience) were used (Supplementary Table 27). Other genes used for S. cerevisiae triptonide biosynthesis originated from T. wilfordii cDNA.
Constructed yeast strains were made using the lithium acetate transformation method62. Parent strains without functional URA3 were made competent by the following procedure: Inoculation from a glycerol stock into 5 ml YPD medium and growing at 30 °C O/N. Then, transfer of 3 mL of O/N culture to 50 mL YPD medium and continued growing for 4–5 h followed by centrifugation at 3500 × g for 10 min, then discarding the supernatant. Cells were then ready for transformation after 2 washes in sterile water (1st in 25 mL, 2nd in 1 mL) and resuspension in 0.4 mL of sterile water.
Transformation of competent yeast cells was carried out as follows: Mixes of designated NotI digested plasmids (2 µL of each) were each added 10 µL competent yeast cells and mixed with 60 µL PEG 3350 (50% w/v), 9 µL LiAc (1 M) and 12.5 µL preboiled salmon sperm DNA. The resulting mixes were next incubated at 42 °C for 40 min before cells were collected by centrifugation (2000 × g for 5 min) and removal of supernatant. Cells were then resuspended in 100 µL sterile water and spread on SC without uracil agar plates. Isolated transformants appeared as single colonies after 2 d of incubation at 30 °C. Insertion of gene constructs was confirmed by colony PCR, using the gene and construct specific primers found in Supplementary Table 25. For colony PCR, colonies were resuspended in 50 µL 20 mM NaOH and incubated at 99 °C for 15 min. 1 µL colony suspension was used for PCR.
Extraction and metabolite analysis
Genetically engineered S. cerevisiae strain was transferred into 0.5 mL media in a 96-well plate and grown for 3 d at 30 °C with orbital shaking at 350 rpm. For extraction, 0.1 mL of S. cerevisiae culture was transferred to 1.5 mL glass vials. 0.4 mL MeOH uHPLC grade was added. S. cerevisiae extracts were filtered by using a 0.22 μm 96-well filter plate (Merck Millipore, Darmstadt, Germany) and at stored at 4 °C prior to LC-MS analysis.
LC-MS analysis
MeOH extracts were analysed using an Ultimate 3000 UHPLC + Focused system (Dionex Corporation, Sunnyvale, CA) coupled to a Bruker Compact ESI-QTOF-MS (Bruker) system. Samples were separated on a Kinetex XB-C18 column (100 × 2.1 mm ID, 1.7 μm particle size, 100 Å pore size; Phenomenex Inc., Torrance, CA) maintained at 40 °C with a flow rate of 0.3 mL min−1 and mobile phase consisting of 0.05% (v/v) formic acid in water (solvent A) and 0.05% (v/v) formic acid in acetonitrile (solvent B).
Three LC protocols were used:
LC method 1: 0–0.5 min, 10 % B; 0.5–21 min, linear increase from 10 to 80% B; 21–31 min, to 90% B; 31–34 min, to 100% B; 34–39 min 100% B; 39–40 min linear decrease from 100 to 10% B. Isocratic 20%, B 41–45.5 min.
LC method 2: 0–0.5 min, 20% B; 0.5–11 min, linear increase from 20 to 80% B; 11–20 min, to 90% B; 20–22 min, to 100% B; 22–27 min 100% B; 27–28 min linear decrease from 100 to 20% B. Isocratic 20%, B 28–32 min.
LC method 3: 0–0.5 min, 20 % B; 0.5–9 min, linear increase from 20 to 100% B; 9–11 min, 100% B; 11–11.5 min, linear decrease from 100 to 20% B; 11.5–15 min, 20% B.
Mass spectra were acquired in positive ion mode over a scan range of m/z 50–1200 with the following ESI and MS settings: capillary voltage, 4000 V; end plate offset, 500 V; dry gas temperature, 220 °C; dry gas flow of 8 L min−1; nebulizer pressure, 2 bar; in source CID energy, 0 eV; hexapole RF, 50 Vpp; quadrupole ion energy, 4 eV; collision cell energy, 7 eV. Raw chromatogram data was calibrated using an internal sodium formate standard and subsequently exported as *.mzML format using DataAnalysis 4.3 (Bruker). MZmine ver 2.53 was used for visualizing the LC-MS chromatograms.
GC-MS analysis
GC-MS analysis was carried out on a Shimadzu GCMS-QP2010 Ultra (Shimadzu Corp.) with an Agilent HP-5MS column (Agilent Technologies) 20 m × 0.18 mm i.d., 0.18 µm film thickness). Hydrogen was used as a carrier gas at a constant linear velocity of 50 cm s−1, and the injection volume was 1 µL at 250 °C (splitless mode). The oven program was 80 °C for 2 min, ramp at rate 20 °C/min to 180 °C, ramp at rate 10 °C/min to 300 °C, ramp at rate 20 °C/min to 310 °C, hold for 3 min. Data was stored in.CDF format and processed in MZmine2.
Relative quantification of miltiradiene derived diterpenoids
Relative compound quantities in yeast cultures were based on normalized peak areas of characteristic ions (data obtained using targeted feature detection in the MZmine2 software). The signal for the following ions were quantified, 3: m/z 255 (GC-MS), 5: m/z 189 (GC-MS), 13: m/z 303.2318, 6: m/z 283.2059, 7: m/z 299.2002, 8: m/z 313.1794, 2: m/z 359.1481. For LC-qTOF-MS and GC-MS data a mass deviation of 5 ppm and 100 ppm, respectively, was tolerated.
For LC-qTOF-MS, the peak area of the base peak ion (m/z 315.1947) for the internal standard andrographolide was used for normalization. Additional data analysis of normalized peak areas were done with Microsoft Excel for Mac Ver. 16.59 (Microsoft Inc.) and SigmaPlot Version 14.5 (Systat Software Inc.).
Absolute quantifications of 2 and 6 from fed-batch fermentation
Absolute quantifications of 8 (FT65732, CarboSynth) and 2 (FT65197, CarboSynth) were carried out by co-analysis of authentic standards prepared in MeOH and a final concentration of 5 ppm internal standard (andrographolide). Quantification was based on normalized peak area and calculated from the slopes of linear extrapolations of the standards response curve (triptophenolide 0.05, 0.5, 1, 2 ppm; triptonide 0.5, 1, 2, 10, 20 ppm).
Metabolomics, identification of peaks observed in N. benthamiana and S. cerevisiae expressing the triptonide biosynthetic genes
LC-MS data from duplicate samples of the negative control and triptonide biosynthesis samples were analyzed using MZmine ver2.5363. Briefly, noise level was set to 1.5E4, ADAP Chromatogram builder was used for feature detection, while chromatographic peaks were detected by Chromatogram deconvolution using the local minimum search algorithm. Monoisotopic masses were identified using the Isotope peak grouper with a tolerance of m/z 0.01 or 10 ppm, while peaks in the N. benthamiana and S. cerevisiae extract samples, respectively, were aligned using a m/z and retention time tolerance of 5 ppm and 0.15 min. All peaks not identified in both of the duplicate samples were removed. Peaks exclusively identified the triptonide biosynthesis samples while not appearing in the background samples were retained. Identification of oxygenated diterpenoids or conjugates thereof was based on the molecular formula predicted from the accurate mass of monoisotopic peaks (<5 ppm). Putative monoisotopic peaks resulting from in source fragmentation of diterpenoid conjugates were removed.
Feed-batch fermentation of engineering S. cerevisiae strains for isolation of miltiradiene derived diterpenoids
All engineered S. cerevisiae strains were cultivated in 96-deepwell plates using a Feed-In-Time (FIT; m2p-labs)60. For isolation and purification of key intermediates in the triptonide pathway selected engineered S. cerevisiae strains were cultivated in feed batch fermentor using a 2 L Biostat® A bioreactor (Sartorius AG).
Batch media contained: 55 g/L glucose · H2O, 25 g/L (NH4)2SO4, 1.7 g/L MgSO4 · 7H2O, while feed media consisted of: 880 g/L glucose · H2O, 21.6 g/L KH2PO4, 24.24 g/L MgSO4 · 7H2O, 8.4 g/L K2SO4, and 0.672 g/L Na2HSO4. Batch- and feed salt mixes as well as batch and feed glucose were prepared separately by dissolving components in Milli-Q water and sterilizing by autoclavation. The feeding solution was made by mixing 500 mL of feed glucose with 500 mL of feed salt mix, 10 mL of vitamin mix (0.64 g/L D-biotin, 3 g/L nicotinic acid, 10 g/L thiamin HCl, 4 g/L D-pantothenic acid hemicalcium salt, 8 g/L myo-inositol, 2 g/L pyridoxine HCl), 10 mL microelements (6.7 g/L Titriplex III, 6.7 g/L (NH4)2Fe(SO4)2 · 6H2O, 0.55 g/L CuSO4 · 5H2O, 2 g/L ZnSO4 · 7H2O, and MnSO4 · H2O), and 1 mL of trace elements solution (1.25 g/L NiSO4 · 6H2O, 1.25 g/L CoCl2 · 6H2O, 1.25 g/L, boric acid, 1.25 g/L Kl, and 1.25 g/L Na2MoO4 · 2H2O).
Fed batch fermentation was initiated by addition of a 100 mL starter culture to the reactor tank (with impellers), which in turn was prepared earlier by autoclaving while containing 200 mL Batch glucose and 300 mL Batch salt mix. Also 5 mL vitamin mix, 5 mL micro elements and 0.5 mL trace elements, were added. Cultivation in the bioreactor was started under the following conditions (monitored and automatically controlled): pH = 5, temperature = 30 °C, dissolved oxygen (DO) = 20%. While pH was controlled by feeding of ammonium hydroxide (32%) and sulfuric acid (10 %), dissolved oxygen was controlled by air supply combined with stirring. Also foam levels were adjusted by addition of anti-foam emulsion (35119, Serva Electrophoresis GmbH). After 18 h of initial cultivation in the bioreactor, feeding with Feeding solution at a rate of 1.3% was started. The fermentation process continued for 7 d with daily sampling of the culture.
Isolation and purification of miltiradiene derived compounds from engineered S. cerevisiae strain for NMR analysis
Compounds in this invention were isolated from bioreactor cultured yeast strains NVJ8.15, and NVJ3.10, and structurally elucidated by NMR. The combined ethyl acetate extracts of broth and MeOH-lysed cells (cells:MeOH = 1:4, v/v) were initially dried in presence of Celite S® (06858, Sigma-Aldrich) via rotary evaporation. Compounds were subsequently isolated by successive fractionations using a puriFlash® 5.250 (Interchim, Montluçon, France) instrument with detection by UV absorbance and Evaporative Light-Scattering Detection (ELSD). This was equipped with a s PF-15SIHP-F0025 (C1) column (OV002A, Interchim) for normal phase and a US5C18HQ-100/300 (C2) column (SSP750, Interchim) - for reverse phase separation.
An initial pre-fractionation of the dry mix of Celite S®/crude extract was achieved using column (C1) with loading from a manually packed dry-loading column. Separation was obtained using mobile phases hexane (A) and ethyl acetate (B), a constant flow rate of 15 mL/min, followed by a final washing step with 100% MeOH. Compounds of interest were detected by UV and ELSD and collected. Collected fractions were continuously evaluated by LCMS using LC-MS method 3 and TLC analysis prior to further fractionation or NMR studies. Additional purification of compounds of interest from fractions containing multiple compounds was carried out by an additional normal phase fractionation using C1 or a reverse phase column fractionation using C2.
For reverse phase purification with C2, sample solvents were evaporated using rotor evaporation and the residue resuspended in 2 mL MeOH. Sample was injected directly onto the pre conditioned column C2. Mobile phases for C2 consisted of solvent C: deionized water and solvent D acetonitrile each acidified with 0.05% (v/v) formic acid. A constant flow rate of 32 mL/min was used, with a linear solvent gradient with increasing concentration of solvent D. Compounds of interest were detected by ELSD and UV and collected.
Additional reverse phase purification was done by multiple injections of 100 μL onto a semi-prep Phenomenex Luna 5 µm C18(2) 100 Å 250 × 10 mm i.d. (fully porous) (Phenomenex, Inc., Torrance, CA, USA) column on a Shimadzu HPLC (SPD-M20A diode array detector, FRC-10A fraction collector, DGU-20A5 degasser, LC-20AT pump, CBM-20A System controller, CTO-10AS VP column oven, SIL-10AP autosampler). Mobile phase was a linear gradient between C and D with an increasing amount of D going from 50–100%. Compounds of interest were detected by UV absorbance at 210 nm and collected.
NMR analysis
NMR experiments were acquired on a Bruker Avance III HD 600 MHz NMR spectrometer (1H operating frequency 599.85 MHz) equipped with a 5-mm cryogenically cooled DCH cryoprobe optimized for 13C and 1H or a Avance III 600 MHz spectrometer (1H operating frequency 600.13 MHz) equipped with a 1.7-mm TCI cryoprobe (Bruker Biospin, Karlsruhe, Germany). NMR data was recorded in 1.7- or 5 mm tubes in CDCl3 or CD3OD (Euriso-top, 99.8 atom % D) with temperature equilibration to 300 K, optimization of lock parameters, gradient shimming, and setting of receiver gain, all automatically controlled by Topspin ver. 3.2 or 3.5 and IconNMR ver. 4.7.5 or 5.0.7 (Bruker Biospin, Karlsruhe, Germany). 1H and 13C chemical shifts were referenced to the residual solvent signals at respectively δH 7.26 ppm and δC 77.16 ppm (CDCl3) and δH 3.31 ppm and δC 49.00 ppm (CD3OD). 1D 1H and 13C NMR spectra were acquired with 30° pulses and 64k data points and zero-filled to 128k data points, 1H spectra were acquired with a spectral width of 12 kHz, a relaxation delay of 1 s and an acquisition time of 2.7 s. 13C spectra were 1H-decoupled using the Waltz-16 composite pulse decoupling scheme. 2D homo- and heteronuclear experiments were acquired with 4096 (HMBC), 2048 (DQF-COSY and ROESY), or 1024 (multiplicity edited HSQC) data points in the direct dimension and 256 (DQF-COSY, HMBC and ROESY) or 256/128 (multiplicity edited HSQC) data points in the indirect dimension. 2D NMR data was zero-filled to 1k in F1 and zero-filled to twice the number of points in F2, employing forward linear prediction in F1 (LPBIN = 0). The 2D experiments supported the results from the 1D-experiments and structures of the identified molecules in Supplementary Figs. 8–53 and Supplementary Table 1–19. Processing of NMR data was done using Topspin ver. 4.0.9 (Bruker Biospin, Karlsruhe, Germany).
Optical rotation was conducted on a ADP410 from Bellingham and Stanley, using MeOH as a reference. Column length was 20 cm and pure compound was dissolved in 10 mL of MeOH. See Supplementary Note 4 for optical rotation measurement values and the comments.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The authors would like to thank Fernando Geu-Flores for his input on the hypothesized mechanisms for the CYP catalyzed C18(C4→C3). This research has been funded by grants from the Novo Nordisk foundation (NNF18OC0031974 [J.A.R.], NNF20OC0061048 [J.A.R.], and NNF16OC0021616 [B.L.M, D.S.]), The Danish Innovation Fund (0160-00016B) [J.A.R] and the Lundbeck foundation (R223-2016-85 [B.L.M., D.S.]).
Author contributions
N.L.H, B.L.M., and J.A.R. conceived and initiated the study. N.L.H., and J.A.R. Identified, and isolated genes from plant cDNA, performed N. benthamiana expression studies, metabolite analysis and isolation of compounds for NMR analysis. N.L.H., V.F., Q.H. and J.A.R. established yeast strains used. L.K. and D.S. conducted the NMR analysis. B.L.M. and J.A.R. wrote the manuscript. N.L.H., V.F., Q.H., D.S., L.K., B.L.M. and J.A.R. revised the manuscript.
Peer review
Peer review information
Nature Communications thanks Shengying Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Sequence information data for TwCYP71BE85 (accession ON375998), TwCYP71BE86 (accession ON375999), TwCYP82D213 (accession ON376000), TwCYP82D274 (accession ON376001), Twb5-A (accession ON376002), Twb5-B (accession ON376003), Twb5-C (accession ON376004), Twb5-D (accession ON376005), Twb5-E (accession ON376006), Twb5-F (accession ON376007), and TwPOR1 (accession ON376008) are available in the NCBI GenBank. Source data are provided with this paper.
Competing interests
N.L.H., V.F., and J.A-R. are inventors of the patent entitled “Production of oxygenated diterpenoid compounds” (application number: PCT/EP2021/073656) related to the cytochrome P450 enzymes described in the paper. J.A-R. has established TriptoBIO to commercialize the patented technology. Other authors claim no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/24/2025
A Correction to this paper has been published: 10.1038/s41467-025-62209-8
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-32667-5.
References
- 1.Kupchan, S. M., Court, W. A., Dailey, R. G., Gilmore, C. J. & Bryan, R. F. Tumor inhibitors. LXXIV. Triptolide and tripdiolide, novel antileukemic diterpenoid triepoxides from Tripterygium wilfordii. J. Am. Chem. Soc.94, 7194–7195 (1972). [DOI] [PubMed] [Google Scholar]
- 2.Dyer, C. A. et al. Accelerated follicle depletion in vitro and in vivo in sprague-dawley rats using the combination of 4-vinylcyclohexene diepoxide and triptolide. J. Zoo. Wildl. Med.44, S9–S17 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Chang, Z. et al. Triptonide is a reversible non-hormonal male contraceptive agent in mice and non-human primates. Nat. Commun.12, 1253 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutney, J. P. et al. Cultivation of Tripterygium wilfordii tissue cultures for the production of the cytotoxic diterpene tripdiolide. Planta Med48, 158–163 (1983). [DOI] [PubMed] [Google Scholar]
- 5.Kutney, J. P. et al. Cytotoxic diterpenes triptolide, tripdiolide, and cytotoxic triterpenes from tissue cultures of Tripterygium wilfordii. Can. J. Chem.59, 2677–2683 (1981). [Google Scholar]
- 6.Tu, L. et al. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat. Commun.11, 971 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters, R. J. Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep.27, 1521–1530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen, N. L. et al. The terpene synthase gene family in Tripterygium wilfordii harbors a labdane-type diterpene synthase among the monoterpene synthase TPS-b subfamily. Plant J.89, 429–441 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Su, P. et al. Identification and functional characterization of diterpene synthases for triptolide biosynthesis from Tripterygium wilfordii. Plant J.93, 50–65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bathe, U. & Tissier, A. Cytochrome P450 enzymes: A driving force of plant diterpene diversity. Phytochemistry161, 149–162 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Hansen, C. C., Nelson, D. R., Møller, B. L. & Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant14, 1244–1265 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Hansen, N. L. et al. Integrating pathway elucidation with yeast engineering to produce polpunonic acid the precursor of the anti-obesity agent celastrol. Microb. Cell Factories.19, 15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.S. Bach, S. et al. High throughput testing of terpenoid biosynthesis candidate genes using transient expression in Nicotiana benthamiana, Methods in Molecular Biology: Plant isoprenoids. Vol. 1153, 245–255 (2014). [DOI] [PubMed]
- 14.Pateraki, I. et al. Manoyl Oxide (13 R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii. Plant Physiol.164, 1222–1236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen-Ranberg, J. et al. Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew. Chem. Int. Ed.55, 2142–2146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De La Peña, R. & Sattely, E. S. Rerouting plant terpene biosynthesis enables momilactone pathway elucidation. Nat. Chem. Biol.17, 205–212 (2021). [DOI] [PMC free article] [PubMed]
- 17.Gao, C., Wang, D., Zhang, Y., Huang, X.-X. & Song, S.-J. Kaurane and abietane diterpenoids from the roots of Tripterygium wilfordii and their cytotoxic evaluation. Bioorg. Medicinal Chem. Lett.26, 2942–2946 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Kutney, J. P. & Han, K. Studies with plant-cell cultures of the Chinese herbal plant, Tripterygium wilfordii. Isolation and characterization of diterpenes. Recl. des. Trav. Chimiques des. Pays-Bas115, 77–93 (1996). [Google Scholar]
- 19.Pateraki, I. et al. Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii. eLife6, e23001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alasbahi, R. H. & Melzig, M. F. Plectranthus barbatus: a review of phytochemistry, ethnobotanical uses and pharmacology – Part 1. Planta Med.76, 653–661 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Bhatt, M. R., Khatri, Y., Rodgers, R. J. & Martin, L. L. Role of cytochrome b5 in the modulation of the enzymatic activities of cytochrome P450 17α-hydroxylase/17,20-lyase (P450 17A1). J. Steroid Biochem. Mol. Biol.170, 2–18 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Hildebrandt, A. & Estabrook, R. W. Evidence for the participation of cytochrome b5 in hepatic microsomal mixed-function oxidation reactions. Arch. Biochem. Biophys.143, 66–79 (1971). [DOI] [PubMed] [Google Scholar]
- 23.Gou, M. et al. Cytochrome b5 is an obligate electron shuttle protein for syringyl lignin biosynthesis in Arabidopsis. Plant Cell.31, 1344–1366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, Z.-L., Yang, Y.-X., Ding, J., Li, Y.-C. & Miao, Z.-H. Triptolide: structural modifications, structure–activity relationships, bioactivities, clinical development and mechanisms. Nat. Prod. Rep.29, 457–475 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Kupchan, S. M. & Schubert, R. M. Selective alkylation: a biomimetic reaction of the antileukemic triptolides? Science185, 791–793 (1974). [DOI] [PubMed] [Google Scholar]
- 26.Geisler, K. et al. Biochemical analysis of a multifunctional cytochrome P450 (CYP51) enzyme required for synthesis of antimicrobial triterpenes in plants. Proc. Natl Acad. Sci.110, E3360–E3367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Herpen, T. W. J. M. et al. Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS ONE.5, e14222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Q. et al. Reconstitution of the costunolide biosynthetic pathway in yeast and Nicotiana benthamiana. PLoS ONE.6, e23255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geu-Flores, F. et al. Glucosinolate engineering identifies a γ-glutamyl peptidase. Nat. Chem. Biol.5, 575–577 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Duan, H., Kawazoe, K., Bando, M., Kido, M. & Takaishi, Y. Di- and triterpenoids from Tripterygium hypoglaucum. Phytochemistry46, 535–543 (1997). [Google Scholar]
- 31.Li, K., Duan, H., Kawazoe, K. & Takaishi, Y. Terpenoids from Tripterygium wilfordii. Phytochemistry45, 791–796 (1997). [Google Scholar]
- 32.Shishido, K. et al. Tripterygium wilfordii var. regelii which are interleukin-1 inhibitors. Phytochemistry35, 731–737 (1994). [Google Scholar]
- 33.Forman, V., Callari, R., Folly, C., Heider, H. & Hamberger, B. Production of putative diterpene carboxylic acid intermediates of triptolide in yeast. Molecules22, 981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo, J. et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl Acad. Sci.110, 12108–12113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zi, J. & Peters, R. J. Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org. Biomol. Chem.11, 7650–7652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, X. & Li, S. Expansion of chemical space for natural products by uncommon P450 reactions. Nat. Prod. Rep.34, 1061–1089 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Luo, D. et al. Oxidation and cyclization of casbene in the biosynthesis of Euphorbia factors from mature seeds of Euphorbia lathyris L. Proc. Natl Acad. Sci.113, E5082–E5089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma, Y. et al. Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza. Nat. Commun.12, 685 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesburg, C. A., Zhai, G., Cane, D. E. & Christianson, D. W. Crystal Structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science277, 1820–1824 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Rontein, D. et al. CYP725A4 from yew catalyzes complex structural rearrangement of taxa-4(5),11(12)-diene into the cyclic ether 5(12)-Oxa-3(11)-cyclotaxane. J. Biol. Chem.283, 6067–6075 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Citron, C. A., Brock, N. L., Tudzynski, B. & Dickschat, J. S. Labelling studies on the biosynthesis of terpenes in Fusarium fujikuroi. Chem. Commun.50, 5224–5226 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Zhu, D., Seo, M.-J., Ikeda, H. & Cane, D. E. Genome mining in Streptomyces. Discovery of an unprecedented P450-catalyzed oxidative rearrangement that is the final step in the biosynthesis of pentalenolactone. J. Am. Chem. Soc.133, 2128–2131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan, L., Jogl, G. & Cane, D. E. The Cytochrome P450-catalyzed oxidative rearrangement in the final step of pentalenolactone biosynthesis: substrate structure determines mechanism. J. Am. Chem. Soc.138, 12678–12689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ortiz de Montellano, P. R. Hydrocarbon hydroxylation by cytochrome P450 enzymes. Chem. Rev.110, 932–948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paddon, C. J. et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature496, 528–532 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Im, S.-C. & Waskell, L. The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch. Biochem. biophysics507, 144–153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan, H. et al. Immunosuppressive diterpenoids from Tripterygium wilfordii. J. Nat. Products62, 1522–1525 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Laursen, T. et al. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science354, 890–893 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Field, B. & Osbourn, A. E. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science320, 543–547 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Dutartre, L., Hilliou, F. & Feyereisen, R. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: gene duplications and origin of the Bx cluster. BMC Evolut. Biol.12, 64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nützmann, H.-W., Huang, A. & Osbourn, A. Plant metabolic clusters – from genetics to genomics. N. Phytologist211, 771–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boutanaev, A. M. et al. Investigation of terpene diversification across multiple sequenced plant genomes. Proc. Natl Acad. Sci.112, E81–E88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen, N. B. et al. EasyClone: method for iterative chromosomal integration of multiple genes Saccharomyces cerevisiae. FEMS Yeast Res.14, 238–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sahoo, D. K., Sarkar, S., Raha, S., Maiti, I. B. & Dey, N. Comparative analysis of synthetic DNA promoters for high-level gene expression in plants. Planta240, 855–875 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Kumar, D. et al. Development of useful recombinant promoter and its expression analysis in different plant cells using confocal laser scanning microscopy. PLOS ONE.6, e24627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahoo, D. K., Dey, N. & Maiti, I. B. pSiM24 is a novel versatile gene expression vector for transient assays as well as stable expression of foreign genes in plants. PLOS ONE.9, e98988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nour-Eldin, H. H., Hansen, B. G., Nørholm, M. H. H., Jensen, J. K. & Halkier, B. A. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucl. Acids Res.34, e122 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Greve, H. et al. Nucleotide sequence and transcript map of the Agrobacterium tumefaciens Ti plasmid-encoded octopine synthase gene. J. Mol. Appl. Genet.1, 499–511 (1982). [PubMed] [Google Scholar]
- 59.Coruzzi, G., Broglie, R., Edwards, C. & Chua, N. H. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J.3, 1671–1679 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forman, V., Bjerg-Jensen, N., Dyekjær, J. D., Møller, B. L. & Pateraki, I. Engineering of CYP76AH15 can improve activity and specificity towards forskolin biosynthesis in yeast. Microb. Cell Factories.17, 181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nørholm, M. H. H. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol.10, 21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc.2, 31–34 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Pluskal, T., Castillo, S., Villar-Briones, A. & Oresic, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinforma.11, 395–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence information data for TwCYP71BE85 (accession ON375998), TwCYP71BE86 (accession ON375999), TwCYP82D213 (accession ON376000), TwCYP82D274 (accession ON376001), Twb5-A (accession ON376002), Twb5-B (accession ON376003), Twb5-C (accession ON376004), Twb5-D (accession ON376005), Twb5-E (accession ON376006), Twb5-F (accession ON376007), and TwPOR1 (accession ON376008) are available in the NCBI GenBank. Source data are provided with this paper.




