Abstract
The Saccharomyces cerevisiae open reading frame YLL057c is predicted to encode a gene product with 31.5% amino acid sequence identity to Escherichia coli taurine/α-ketoglutarate dioxygenase and 27% identity to Ralstonia eutropha TfdA, a herbicide-degrading enzyme. Purified recombinant yeast protein is shown to be an Fe(II)-dependent sulfonate/α-ketoglutarate dioxygenase. Although taurine is a poor substrate, a variety of other sulfonates are utilized, with the best natural substrates being isethionate and taurocholate. Disruption of the gene encoding this enzyme negatively affects the use of isethionate and taurine as sulfur sources by S. cerevisiae, providing strong evidence that YLL057c plays a role in sulfonate catabolism.
In the absence of sulfate, a number of yeasts can use aliphatic sulfonates, such as taurine, cysteate, and isethionate, as alternative sulfur sources (11). Sulfonate utilization by Saccharomyces cerevisiae occurs only under aerobic conditions, is independent of sulfate-utilizing enzymes, and requires sulfite reductase, consistent with the formation of sulfite prior to assimilation. This pattern of sulfonate utilization is similar to that in Escherichia coli and other enteric bacteria (10, 12), suggesting the presence of a common pathway in these diverse organisms. Recently, the enzyme responsible for the degradation of taurine (2-aminoethanesulfonate) in E. coli was described (4). Taurine/α-ketoglutarate (α-KG) dioxygenase (TauD) hydroxylates the carbon atom in the C-S bond of taurine to give an unstable intermediate that spontaneously decomposes to sulfite and aminoacetaldehyde. The cosubstrate, α-KG, is oxidatively decarboxylated to form succinate in a reaction that requires Fe(II) and O2. TauD is mechanistically similar to TfdA, a Ralstonia eutropha enzyme that metabolizes the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) (5). The YLL057c open reading frame in S. cerevisiae (GenBank accession no. Z73162) encodes a protein with 27% identity to TfdA, 31.5% identity to TauD, and significant identity to several uncharacterized TauD homologs from a variety of different bacteria (8). Several regions are highly conserved among these proteins, including the H-X-D-X53–57-H motif common to α-KG-dependent dioxygenases (2). Here, we report both the characterization of the S. cerevisiae YLL057c gene product and studies with the YLL057c deletion mutant to determine if this enzyme plays a role that is analogous to TfdA in 2,4-D-degrading microorganisms or to TauD in E. coli.
Cloning and expression of YLL057c.
Open reading frame YLL057c was PCR amplified and cloned into pET23a for expression in E. coli. The 1,236-bp YLL057c sequence, which contains no introns, was amplified from λPM-5392 (ATCC) containing a 30-kb fragment of S. cerevisiae chromosome XII, with primers 5′-CAT ATG TAC AGA GGA CGT CGT CGA G-3′ and 5′-CTA CAA CAC TTT TCG TCT CCG AGG-3′. The forward primer contained a 5′ extension that introduced an NdeI restriction site directly upstream of the start ATG codon. Template DNA was added by transferring a small amount of material from a λPM-5392 plaque, obtained by plating on E. coli Y1090 (9), directly to the PCR mix. The PCR mix included 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 25 μM dNTPs (Gibco), 1 μM each primer, and 0.05 U of Taq polymerase (Gibco). The temperature program was comprised of an initial denaturation step at 95°C for 2 min 10 s; 35 cycles of a denaturation step at 95°C for 1 min, an annealing step at 55°C for 1 min, and a 2-min-10-s extension step at 68°C; and a final extension at 68°C for 6 min. The resulting 1,734-bp PCR product (which included an additional downstream sequence) was cloned into pCR2.1 TOPO vector (Invitrogen) in accordance with the manufacturer’s instructions to give plasmid pCR2.1-YDO. The cloned gene was cut from pCR2.1-YDO with NdeI and SacI and inserted into pET23a cut with the same enzymes to create pB10. To create an N-terminal fusion of the YLL057c gene product to the maltose binding protein (MBP), YLL057c was amplified from pB10 using the protocol described above with primers 5′-TCT CTA GAA TGT CTC CTG CAG CAG C-3′ and 5′-GTC AAG CTT AAA GAA GTG TTG TCG CCG-3′. The forward primer introduced an XbaI site directly upstream of the start site for in-frame insertion into pMAL-c2 (New England Biolabs). The amplification product was directly cloned into pGEM-T (Promega) to give pGTYDO16. YLL057c was cut from pGTYDO16 with XbaI and HindIII and ligated to pMAL-c2 prepared with the same enzymes to create pMBPYDO, a fusion of the malE gene to YLL057c.
Production and purification of YLL057c gene product.
Various efforts to directly express the yeast gene in E. coli DH5α from pB10 failed to achieve high-level production of the desired protein. In contrast, an MBP fusion of this product was produced in E. coli DH5α(pMBPYDO), consistent with an increased stability and decreased toxicity imparted by the N-terminal extension. Cultures were grown to an A600 of 0.4 to 0.6 at 30°C in Luria broth containing 100 μg of ampicillin/ml, induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubated for 3 h. Harvested cells were suspended in 20 mM Tris buffer (pH 7.5) containing 1 mM EDTA, 200 mM NaCl, and 1 mM dithiothreitol and lysed by two passages through a French pressure cell at 120 MPa. The cell lysates were spun at 100,000 × g to yield the clarified cell extracts. The 89.7-kDa fusion protein was readily purified by passing cell extracts over an amylose column in accordance with the manufacturer’s instructions, followed by elution in the above buffer amended with 10 mM maltose.
Characterization of MBP-dioxygenase fusion protein.
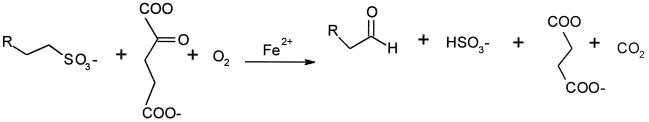
The product of YLL057c was found to be an Fe(II)- and α-KG-dependent dioxygenase capable of utilizing a variety of sulfonates, but was inactive on 2,4-D. Since sulfite was a common product (Fig. 1) from sulfonate degradation, activity was determined by quantifying the amount of sulfite released using Ellman’s reagent [5,5′-dithiobis(2-nitrobenzoic acid)] (4). Standard assay conditions were defined as the use of 10 mM dimethylglutarate (pH 7.0), 1 mM α-KG, and 50 μM Fe(II)-ascorbic acid salt (Sigma) and a 5-min incubation at 30°C in the presence of varied substrate concentrations. Maximum rates (Vmax), affinities (Km), and catalytic efficiencies (kcat/Km) for various substrates are compared in Table 1. Whereas taurine was a relatively poor substrate for the yeast sulfonate/α-KG dioxygenase (with a moderate reaction rate and high Km), isethionate (2-hydroxyethanesulfonate) and taurocholate were turned over more rapidly and bound with much higher affinity by the enzyme. According to their catalytic efficiencies, the best substrates were the synthetic compounds MOPS and N-phenyltaurine. Several of the better substrates for this enzyme had large adducts on the amino group, indicating a relaxed substrate specificity for this portion of the molecule. Of the substrates listed in Table 1, only taurine, MOPS (4), and taurocholate (1) were also substrates of E. coli TauD. Notably, TauD activity toward taurine exhibits a kcat of 133 min−1, which is the same order of magnitude as found here for the yeast enzyme.
FIG. 1.
Reaction mechanism of sulfonate/α-KG dioxygenase.
TABLE 1.
Sulfonate specificity and kinetic parameters for sulfonate/α-KG dioxygenase
| Substratea | Vmax (μmol/min/mg) | Km (mM) | kcat/Km (min−1/mM) | Ki (mM) |
|---|---|---|---|---|
| 3-N-Morpholinopropanesulfonic acid (MOPS) (0–20 mM) | 1.80 ± 0.06 | 0.24 ± 0.02 | 670.9 | 34.3 ± 5.8b |
| N-Phenyltaurine (0–3 mM) | 0.81 ± 0.03 | 0.12 ± 0.01 | 606.4 | 1.5 ± 0.1b |
| 2-Hydroxyethanesulfonate (isethionate) (0–20 mM) | 1.26 ± 0.05 | 0.27 ± 0.05 | 418.4 | 20.3 ± 2.5b |
| 3-Morpholino-2-hydroxypropanesulfonic acid (MOPSO) (0–20 mM) | 1.51 ± 0.03 | 0.33 ± 0.02 | 411.2 | 28.4 ± 2.4b |
| Taurocholate (0–10 mM) | 0.99 ± 0.05 | 0.42 ± 0.05 | 210.7 | 8.2 ± 1.1b |
| 2-(4-Morpholino)ethanesulfonic acid (MES) (0–20 mM) | 1.48 ± 0.05 | 1.94 ± 0.14 | 68.4 | 45.2 ± 6.6b |
| 2-(Cyclohexylamino)ethanesulfonic acid (CHES) (0–20 mM) | 0.56 ± 0.02 | 2.57 ± 0.34 | 19.6 | —c |
| N-(Tris[hydroxymethyl]methyl)-2-amino-ethanesulfonic acid (TES) (0–15 mM) | 0.75 ± 0.01 | 3.60 ± 0.12 | 18.7 | —c |
| N-[2-Hydroxyethyl] piperazine-N′-[2-hydroxy-propanesulfonic acid] (HEPPSO) (0–20 mM) | 1.01 ± 0.08 | 5.04 ± 1.07 | 18.0 | —c |
| N-(Tris(hydroxymethyl)methyl)-3-aminopropane sulfonic acid (TAPS) (0–20 mM) | 1.61 ± 0.06 | 10.38 ± .80 | 13.9 | —c |
| 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (0–20 mM) | 0.82 ± 0.02 | 16.62 ± 0.80 | 4.4 | —c |
| 2-Aminoethanesulfonic acid (taurine) (0–20 mM) | 0.46 ± 0.04 | 10.37 ± 1.82 | 4.0 | —c |
The concentrations of substrates examined are shown in parentheses.
Data for these substrates were fit to the equation given in the text.
No inhibition was detected. Kinetic parameters were fit to the Michaelis-Menten equation.
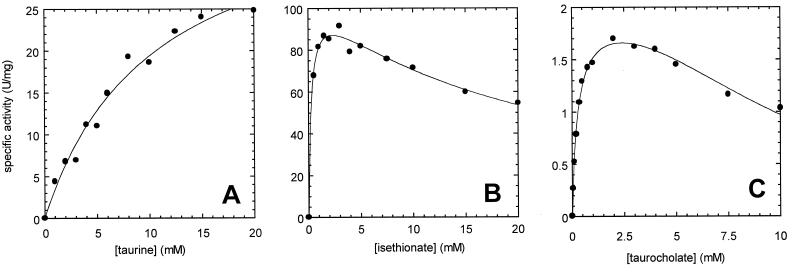
While taurine utilization followed simple Michaelis-Menten kinetics (Fig. 2A), the kinetic properties for several other substrates were complicated by apparent inhibition of the enzyme at elevated substrate concentrations (Fig. 2B and 2C). In those cases, the data were fit to the standard equation for substrate inhibition: v = Vmax[S]/([S] + Km + [S]2/Ki). Inhibition constants (Ki) are listed in Table 1. TauD exhibited similar substrate inhibition profiles for certain substrates other than taurine (1). Activity towards the following compounds was not detected: cysteate, aniline-2-sulfonic acid, homotaurine, picrylsulfonic acid, sulfosalicylic acid, 3-[(3-cholamidopropyl)-dimethylammonio]propanesulfonic acid (CHAPS), 4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid (EPPS), piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), and 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS).
FIG. 2.
Substrate concentration dependence of sulfonate/α-KG dioxygenase with taurine (A), isethionate (B), and taurocholate (C). The data were fit to standard Michaelis-Menten kinetics for taurine and to the standard equation for substrate inhibition (see text) for the other substrates. One unit is defined as 1 μmol of sulfite released per min under the standard assay conditions.
Several additional properties of the MBP-dioxygenase fusion protein were characterized. The Km for α-KG was determined to be 30 ± 16 μM by a previously described method using α-[14C-1]-KG (5). The approximate Kd for Fe was found to be 44 ± 5 μM. Temperature stability experiments, conducted by incubating the protein at varying temperatures for 1 h prior to measuring activity at 30°C, showed that the enzyme lost 77% activity after incubation at 37°C and was completely inactivated at higher temperatures. The fusion protein was shown to be cleaved by Factor Xa (New England Biolabs) to yield active 47-kDa enzyme. There was no significant difference in kcat towards MOPS for the fusion protein and the free enzyme.
Role of sulfonate/α-KG dioxygenase in S. cerevisiae.
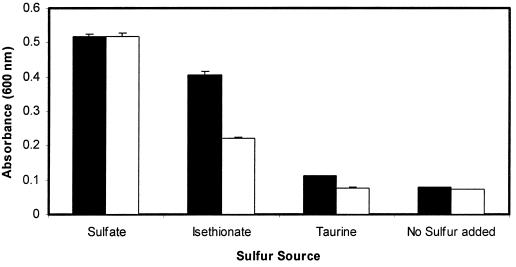
Studies with a S. cerevisiae YLL057c deletion mutant, BY4742-11545 (Research Genetics, Huntsville, Ala.), demonstrated the importance of sulfonate/α-KG dioxygenase for growth on alternative sulfur sources. Both S. cerevisiae BY4742-11545 and the corresponding wild-type parent strain, BY4742, were grown aerobically in defined medium (3) with isethionate (250 μM), taurine (250 μM), or sulfate (250 μM), or with no sulfur source added. Comparison of the growth profiles for these two strains clearly showed that deletion of the sulfonate/α-KG dioxygenase gene decreased the extent to which S. cerevisiae could use taurine and isethionate as the sole source of sulfur (Fig. 3). The rate at which BY4742 grew in medium with isethionate was also affected (data not shown). The growth rate and final yield were identical for the two strains in medium containing sulfate. The fact that S. cerevisiae BY4742-11545 could grow to some degree on isethionate suggests that an additional pathway exists for sulfonate utilization. Examples of enzymes capable of the liberation of sulfite from sulfonates include a flavin mononucleotide-dependent monooxygenase from Pseudomonas aeruginosa (6) and a sulfolyase from an Acinetobacter isolate (7). Redundancy of sulfonate catabolic pathways accentuates the importance of these compounds as sources of cell sulfur.
FIG. 3.
Utilization of different sulfur sources by wild-type S. cerevisiae BY4742 (solid bars) and the YLL057c knockout strain BY4742-11545 (open bars). Growth in medium containing either sulfate, isethionate, taurine, or no added sulfur source was measured as absorbance at 600 nm of stationary-phase cultures. Error bars represent the standard deviations between replicate cultures.
Acknowledgments
This work was supported by NSF grants MCB9603520 (R.P.H.) and DEB9120006 (Center for Microbial Ecology) and by the Michigan State University Agricultural Experiment Station. Partial support of D.A.H. was also provided by National Institutes of Health Biotechnology Training Grant T32-GM08350.
We thank Larry Snyder and Stephen Ekunwe for providing selected E. coli strains.
REFERENCES
- 1.Auchtung, T. A. 1999. Unpublished observations.
- 2.Borovok I, Landman O, Kreisberg-Zakarin R, Aharonowitz Y, Cohen G. Ferrous active site of isopenicillin N synthase: genetic and sequence analysis of the endogenous ligands. Biochemistry. 1996;35:1981–1987. doi: 10.1021/bi951534t. [DOI] [PubMed] [Google Scholar]
- 3.Cherest H, Surdin-Kerjan Y. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics. 1992;130:51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichhorn E, van der Ploeg J R, Kertesz M A, Leisinger T. Characterization of α-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J Biol Chem. 1997;272:23031–23036. doi: 10.1074/jbc.272.37.23031. [DOI] [PubMed] [Google Scholar]
- 5.Fukumori F, Hausinger R P. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J Biol Chem. 1993;268:24311–24317. [PubMed] [Google Scholar]
- 6.Kertesz M, Schmidt-Larbig K, Wüest T. A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J Bacteriol. 1999;181:1464–1473. doi: 10.1128/jb.181.5.1464-1473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King J E, Jaouhari R, Quinn J P. The role of sulfoacetalaldehyde sulfolyase in the mineralization of isethionate by an environmental Acinetobacter isolate. Microbiology. 1997;143:2339–2343. doi: 10.1099/00221287-143-7-2339. [DOI] [PubMed] [Google Scholar]
- 8.Microbial Genomes Blast Databases Website. 27 July 1999, revision date. [Online.] National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html. [11 August 1999, last date accessed.]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 10.Uria-Nickelsen M R, Leadbetter E R, Godchaux W., III Comparative aspects of utilization of sulfonate and other sulfur sources by Escherichia coli K12. Arch Microbiol. 1994;161:434–438. doi: 10.1007/BF00288955. [DOI] [PubMed] [Google Scholar]
- 11.Uria-Nickelsen M R, Leadbetter E R, Godchaux W., III Sulfonate-sulfur assimilation by yeasts resembles that of bacteria. FEMS Microbiol Lett. 1993;114:73–78. doi: 10.1016/0378-1097(93)90144-q. [DOI] [PubMed] [Google Scholar]
- 12.Uria-Nickelsen M R, Leadbetter E R, Godchaux W., III Sulphonate utilization by enteric bacteria. J Gen Microbiol. 1993;139:203–208. doi: 10.1099/00221287-139-2-203. [DOI] [PubMed] [Google Scholar]