Graphical abstract
Keywords: Arrhenius, Temperature compensation, Splicing, Translation, Heat shock, Arabidopsis, Saccharomyces cerevisiae, Mouse
Abstract
Temperature is an environmental condition that has a pervasive effect on cells along with all the molecules and reactions in them. The mechanisms by which prototypical RNA molecules sense and withstand heat have been identified mostly in bacteria and archaea. The relevance of these phenomena is, however, broader, and similar mechanisms have been recently found throughout the tree of life, from sex determination in reptiles to adaptation of viral RNA polymerases, to genetic disorders in humans. We illustrate the temperature dependence of RNA metabolism with examples from the synthesis to the degradation of mRNAs, and review recently emerged questions. Are cells exposed to greater temperature variations and gradients than previously surmised? How do cells reconcile the conflicting thermal stability requirements of primary and tertiary structures of RNAs? To what extent do enzymes contribute to the temperature compensation of the reaction rates in mRNA turnover by lowering the energy barrier of the catalyzed reactions? We conclude with the ecological, forensic applications of the temperature-dependence of RNA degradation and the biotechnological aspects of mRNA vaccine production.
1. Introduction
The information RNA molecules transmit from each gene varies depending on the cell type, time and environmental conditions, such as temperature. All types of macromolecules, proteins, RNAs, lipids and sugars, play a role in the sensing and adaptation to temperature; however, the physicochemical constraints and the molecular mechanisms differ for each class of molecules. The sensing of temperature by RNA molecules and their heat resistance has mostly been studied in bacteria [1], [2], but more and more cases have been identified in higher organisms.
The range of temperature to which microorganisms, plants and poikilothermic animals can adapt is quite broad, but even mammals face temperature gradients larger than often assumed. As an introduction, we review the relevant ranges of temperatures cells face. Subsequently, we focus on how these temperatures affect the integrity, folding and structure of RNAs and the strategies how organisms maintain RNA integrity and folding at extreme temperatures.
In the following sections, we will describe the physical principles underlying the temperature-dependence of enzyme reactions that help to understand how specific steps of RNA synthesis, processing and degradation are coordinated. Instead of a comprehensive overview, we present a few examples in particular model organisms at each step between the birth and death of RNA molecules (Sections 6-10). Since all reactions in RNA metabolism – transcription, splicing, RNA degradation etc. – are catalyzed by enzymes, we will discuss the Arrhenius relation to understand how the temperature dependence of enzymatic activities affects RNA abundances (Section 5). Some of these processes, such as transcription, are deeply intertwined with protein-based heat- and cold-shock regulation, which have been extensively reviewed. Therefore, we do not cover upstream transcriptional regulation and will focus on general processes related to mRNA synthesis, the temperature dependence of transcriptional initiation and elongation.
We highlight new developments in areas that await resolution, in the hope that the interactions of these areas will contribute to a better understanding of RNA-mediated temperature adaptation. We conclude the review with the biotechnological applications of the temperature control of RNA-based processes (Section 11).
2. The relevant range of temperatures
2.1. Temperature ranges compatible with cellular growth
The growth range of some extremophiles overlaps with the freezing or boiling points of water. The cold adapted (psychrophilic) microorganism Methanococcoides burtonii, an archaeon isolated from a lake in Antarctica, grows between −2 °C and 28 °C [3], [4]. On the other end, Pyrococcus furiosus has an optimum growth temperature at around 100 °C. Discovered on the Volcano island in Italy, this hyperthermophilic archaeon can multiply rapidly even at the boiling point of water, with a doubling time of 37 min [5].
Whereas special forms of organisms, such as spores, can survive freezing and boiling temperatures [6], the temperature range in which individual microorganisms can grow, and not just survive, span at most 45 °C. Cells mount a cold or heat shock response when exposed to temperatures at the lower or upper limit of their growth range, respectively. Importantly, the temperature that evokes stress responses is not uniform across the tree of life. In fact, the cold shock temperature of a microorganism can be deadly hot for another. For example, a lukewarm environment (23 °C) evokes heat stress in M. burtonii, [3], whereas P. furiosus experiences cold shock at 75 °C [7].
Interestingly, the heat shock response of M. burtonii at 23 °C coincides with the optimal growth temperature, indicating that growth at optimal temperature is stressful for this cold-adapted organism. However, such an overlap is not specific to psychrophilic (cold-loving) organisms. In Saccharomyces cerevisiae (yeast), a mesophilic model organism, transcription driven by heat shock response elements is activated already at optimal growth temperature (30 °C). The intensity of this response depends on how cold the prior environment was and how long the temperature was raised [8], [9]. Furthermore, the heat shock response is transient at 37 °C but long lasting at 42 °C [10]. Thus, no single temperature threshold can be defined for cold or heat shock responses. Similarly, the temperature optimum can vary depending on the particular trait or function, as exemplified by the model organism Drosophila, which can develop from egg to adult fruit fly in the range between 12 and 32 °C. Maximum values are observed at 22 °C for ovariole number but at 16 °C for wing lengths of females [11], indicating that the particular optimum temperature can differ as much as 6 °C. The response can even become binary: the temperature of eggs determines the sex of the developing reptiles [12].
In summary, the temperature optimum of growth or specific organismic activity may not coincide with the temperature at which cold and heat shock responses are minimal, and a certain degree of stress response may be part of normal physiology.
2.2. Basal metabolism in mammals and the contribution of RNA turnover to it
All homeotherms, which regulate their body temperatures, such as birds and mammals, maintain high body temperatures in the range of 36 to 42 °C [13]. When low ambient temperatures threaten to overcool the body, muscles generate heat through shivering as do mitochondria in brown adipose tissue or beige cells through proton leak [14]. For example, the body temperature of men with brown adipose tissue raises in response to cold environment by 0.4 °C more than of those without [15]. At intermediate temperatures, also known as the thermoneutral zone, energy expenditure is the lowest and corresponds to the basal metabolic rate. Counterintuitively, the metabolic rate increases again above the thermoneutral zone, which is attributed to the acceleration of reactions with temperature (see 5.1.). Interestingly, several mammals, such as mice, react biphasically. Mice exposed to high ambient temperatures (39.5 °C) increase their core body temperature to 42-43 °C. Upon returning mice to 25 °C, the core temperature declines rapidly to around 28-30 °C [16], which is clearly below the temperature that evokes cold shock (less than 32–34 °C) [17], [18]. Thus, cells experience both heat and cold shock in close succession before returning to the thermoneutral zone.
The major source of heat generated by basal metabolic rate, which is typically measured by oxygen consumption, is the proton leak in mitochondrial respiration. In addition, other ATP consuming processes, especially ion pumps (Na+/K+ ATPase and Ca2+ pump), contribute significantly. Similarly, transcription consumes ATP, CTP GTP and UTP as RNA is synthesized, which is likely to contribute between 1 % and 5 % of basal metabolism and hence heat generation in some cells, particularly in the liver [19].
2.3. Temperature gradients in the cells and the body, and their potential impact on RNA
In homeotherms, even small temperature variations can be lethal. Athletes can tolerate core temperatures as high as 41.7 °C without adverse effects. The critical thermal maximum in humans is a body temperature of 42 °C lasting for few hours. Due to the ongoing climate change, the mortality due to heat strokes is expected to increase [20]. At extreme temperatures (49 °C to 50 °C), all cellular structures are destroyed and cellular necrosis occurs in less than five minutes. At lower temperatures, cell death is largely due to apoptosis. Apoptosis affects dividing cells in the developing brain but not in postmitotic neurons [21].
Despite the relative constancy of core body temperatures, gradients are common in the bodies of most homeotherms. For example, the skin, has lower temperature (33–35 °C), especially on the extremities [22], which can further decrease to 29 °C when the ambient temperature drops from 25 °C to 15 °C [23]. Thus, cells in the skin and most cells in the extremities experience lower temperatures than the standard 37 °C.
In addition to gradients within the body, there are also gradients at the cellular scale. In particular, the nuclei and the mitochondria may have higher temperatures than the rest of the cells [15], [24]. Some studies suggest that mitochondria can be as hot as close to 50 °C based on measurements with a fluorescent protein thermosensor, whose fluorescence emission changes in response to temperature [25]. If mitochondria are so hot, the intramitochondrial transcription, ribosomes and other RNA mediated processes will be affected by the high temperature. There is evidence that another feature of mitochondria, the strong oxidizing environment in the mitochondrial matrix, affects RNAs. For example, oxidatively damaged RNA is nearly exclusively observed in the mitochondria of retinal ganglion cells [26]. Furthermore, guanine, which is the most sensitive base to oxidative damage, are predominantly localized in solvent inaccessible parts of ribosomal RNA in mitochondrial ribosomes, a sign of RNA adaptation to the mitochondrial environment [27]. It remains to be determined whether RNAs in the mitochondria show also signs of adaptation to higher temperature. An example has been recently suggested: the mitochondrial isoform of an RNase in Arabidopis, AtPRORP1, has a 12 °C higher melting temperature than the nuclear isoforms [28].
This raises the question of how steep the intracellular temperature gradients can be. Steep gradients do not easily arise because water is an excellent heat conductor. Computational models based on Fourier’s law of heat conduction suggest that at the scale of a cell, temperature gradients of most 10−5C can arise, which is considerably surpassed by the 1 to 10 °C difference detected in the above studies. This discrepancy may be explained by the spatial and temporal averaging through which fluorescent reporters reflect the proton spikes near the ATP synthase [29], [30]. In this case, the proteins would nonlinearly overestimate the actual temperature in the mitochondrial matrix. On the other hand, the heat conduction models should incorporate endothermic reactions in the cell, which can steepen the gradients because they absorb heat [24]. Consequently, the high temperature may be limited to the respiratory chain or the immediate vicinity of the inner mitochondrial membrane. This hypothesis is compatible with the results of a systematic study on the melting temperature of proteins; this study found that mitochondrial proteins were not substantially more stable than others were, but the members of the mitochondrial respiratory chain have robustly higher melting temperatures [31].
Nuclei also appear to have higher temperature than the cytoplasm [24], possibly due to the strongly exothermic RNA synthesis (2.2). The faster the RNA turnover, the larger the heat production.
3. RNA folding and temperature
The characteristics of RNA folding is easy to understand when contrasted to protein folding. The secondary structure motifs (e.g. alpha-helices, beta-sheets) of proteins are contextual, i.e. they are stable in the context of the rest of the protein, but they may not form when they are isolated [32]. Such a strong dependence on tertiary structures is typically absent in RNAs, because secondary structures (e.g. stem-loops, see Fig. 1) are highly autonomous, largely due to the stronger interactions between the base pairs. Magnesium promotes the appearance of tertiary structure. This dominance of secondary structures in RNAs may explain the relatively high accuracy of structure prediction models based on physical rules, which reaches around 80 % (the fraction of correctly predicted base pairs in all predicted base pairs) [33], [34], [35].
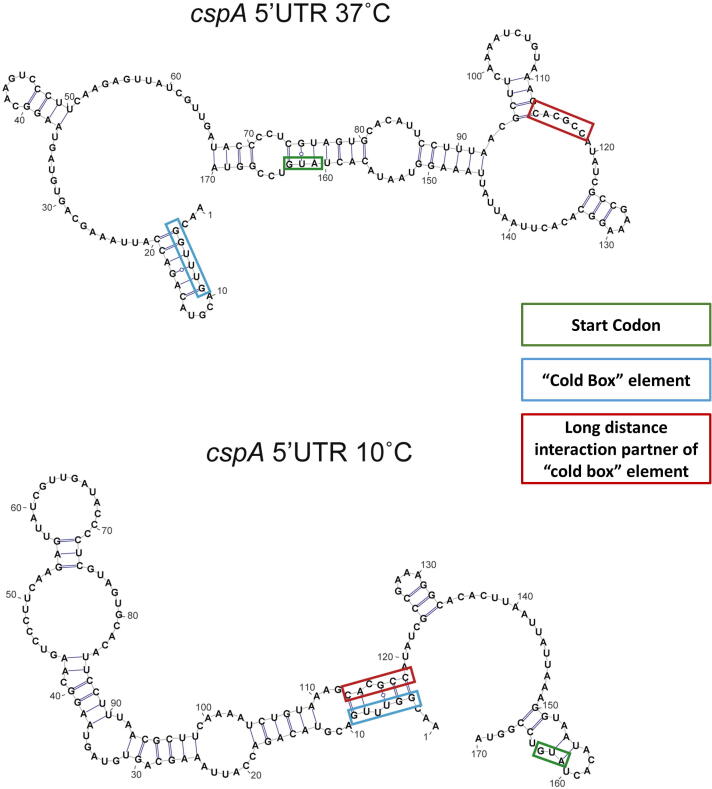
Fig. 1.
The structure of the 5′UTR of the cspA mRNA in E.coli at 37 and 10 °C. Reproduced from Zhang et al [45].
Two questions arise regarding the temperature dependence of RNA folding, which will be addressed in the next subsections. How can RNAs sense changes in ambient temperatures? How can RNAs maintain folded structure at extreme temperatures?
3.1. Temperature sensing by RNA thermosensors
RNAs, just like proteins, contain a very large number of secondary bonds, mostly hydrogen bonds [36], which are sensitive to changes in ambient temperature. Consequently, both proteins and mRNAs can undergo substantial conformational changes with temperature. When a conformational change alters the protein output, such as DNA binding, ion channel activity or inhibition of translation, we talk about protein thermosensors [37]. Many protein thermosensors have been described in both in prokaryotes and eukaryotes. In comparison, RNA thermosensors affect a narrower range of processes, typically splicing and translation, because they act within the same RNA molecule. For example, an RNA stem-loop close to the translation start site inhibits translation of the same RNA (in cis) [1], [38]. As always in biology, there are exceptions to the rule: a recent study unveiled that the pairing of two different mRNA molecules controls the expression of a bacterial cold-shock protein (in trans) [39].
Even in non-bacterial organisms, stem-loops can effectively tune translation. A stem consisting of 6 G-C pairs inserted upstream of a start codon decreases translation by roughly 100-fold in yeast cells. When the stem is shortened to 5 base pairs, consisting of three A-T the two G-C pairs, the number of possible hydrogen bonds is reduced from 18 to 12, and consequently translation is inhibited much less, only fourfold [40]. Analogously, a temperature-induced reduction of base pairing is expected to alleviate the inhibition. Indeed, increasing temperatures shift the equilibrium from the folded to the unfolded state of RNA stem loops [41]. The shift between folded and unfolded structures provides a simple intuitive view, but there may be many more structures in equilibrium [42].
Most biophysical studies on RNA structures are performed in vitro, raising the question of how they reflect in vivo conditions. For example, a comparative study did not find a significant difference in the computed melting temperature between the bacterial thermosensor and non-thermosensor RNAs [43]. Can this surprising observation be explained by differences between in vivo and in vitro structures? This question can be answered using methods capable of detecting in vivo structures, such as DMS-seq. Dimethyl sulfate (DMS) labels unpaired C and this modification can be detected with RNA sequencing (DMS-seq) [44]. Much fewer in vivo RNA structures were found in yeast cells (around 4 % of the mRNA regions) with DMS-seq in comparison to isolated RNA in vitro (24 %) This reduction of structures is energy-dependent, as evidenced by ATP depletion experiments. The scarcity of RNA structures is not restricted to the coding regions and extend to the untranslated regions (UTRs). Structures appear only much below the optimal growth temperature of yeast cells.
With the same method (DMS-seq), a similar increase in RNA folding was found at low temperatures in E. coli [45]. At 10 °C, RNA translation decreases due to the increased RNA folding; however, the abundance of the cold shock proteins (csp) increases after the cold shock. This is possible in part due to the major rearrangement of the structure of the 5′UTR of the RNA chaperone cspA (Fig. 1). While it is easy to understand how high temperatures can increase abundance of protein by disinhibiting translation that is under the control of a stem-loop, it is less clear how cold can increase protein abundance via RNA structures. At 37 °C, there is a 6 bp stem loop the 5′end of the cspA 5′UTR, which contains the cold box sequence. At 10 °C, the cold box sequence pairs with a more distant downstream sequence, resulting in a long tandem of stems, which is likely to prevent the degradation of the mRNA. The cspA mRNA has a much longer half-life at 10 °C than at 37 °C, resulting in higher cspA RNA abundance after cold shock [46]. At the same time, the structure around the start codon becomes looser, which facilitates the translation of Csp. Thus, a real-life RNA thermosensor can undergo complex rearrangements, affecting multiple functions. Well-designed experiments tare required to decipher the structure-function relationship in these molecules. The increased abundance of the cspA protein leads to a genome-wide reduction of RNA structures due to its RNA chaperone activity in the later phases of the adaptation to the cold shock.
The strong folding of RNAs in the cold, observed in prokaryotes and eukaryotes, and their ATP dependent, active unfolding may explain why RNA helicases, RNA chaperones and nucleases play a prominent role in the cold shock in both bacteria and plants [2], [47].
Bacterial RNA thermosensors can detect not only cold, but also warmth [48]. Bacteria often respond when temperature reaches or exceeds 37 °C, as this threshold signals them they are likely to have entered the host organism. A lead (Pb)-based method was used to monitor RNA structures in Yersinia bacteria when temperature was shifted from 25 °C to 37 °C. Slightly more unfolding was observed in the UTRs than in the coding regions. Confirming some well-known riboswitches, this study also identified new mRNAs that contain structures in the UTR that can alter translation in a temperature dependent way [49].
3.2. RNA folding at extreme temperatures
The structural stabilization of ribonucleic acids in hyperthermophiles is particularly important in transfer RNAs (tRNAs), which must maintain a complex three-dimensional structure. Covalent modification of nucleotides seem to provide the answer [50]. The analysis of ribonucleosides from P. furiosus grown at 70, 85 and 100 °C revealed that the relative abundance of the nucleoside 5-methyl-2-thiouridine and several other acetylated and methylated nucleotides increased as a function of cell culture temperature [51]. In addition to covalent modification of nucleotides, macromolecular associations can promote tRNA folding. Polyamines are cations that bind to nucleic acids, which were shown to increase the melting temperature of tRNAs. The number of polyamine species is higher in hyperthermophilic microorganisms, exceeding the three standard polyamines: spermin, spermidine and putrescine [52]. The polyamines specific to hyperthermophilic organisms have complex, branched structures.
On the other hand, tRNAs in the psychrophilic archaea, M. burtonii, have very few modifications. One of them is dihydrouridine, molecule associated with maintenance of polynucleotide flexibility at low temperatures [53].
4. Integrity of RNA molecules at high temperatures
High temperatures do not only unfold RNAs but can also affect their primary bonds by hydrolyzing the RNA, irreversibly damaging RNA integrity. RNA is more susceptible to hydrolysis than DNA since the 2′OH group, present only in RNAs, can interact with the phosphate group, which results in breaking up of the bonds through transesterification. Whereas magnesium promotes tertiary structure of RNA, it does also efficiently catalyze transesterification, especially at high pH. Thus, the primary and tertiary structures have conflicting requirements for magnesium. In contrast to magnesium, monovalent cations oppose the transesterification. Taking into account realistic solvent properties, the expected half-life of an mRNAs consisting of 1000 or more nucleotides (nt) is expected to be ∼1 min or less at 100 °C due to the chemical instability of RNA at such high temperatures [54]. Even smaller RNAs such as tRNA would have a half-life of only ∼10 min at 100 °C.
The above in vitro studies on RNA hydrolysis raise the question of how RNAs withstand the high temperatures in hyperthermophilic organisms. Interestingly, recent studies indicate that the intracellular milieu can resolve the opposing effect of magnesium: the detrimental effect on RNA hydrolysis and stabilizing effect on tertiary structure promotion. The free amino acids in the cell weakly chelate magnesium. The amino acid-chelated Mg2 + retains its capacity to enhance RNA folding, by shifting the equilibrium distribution toward the folded structure, but at the same time prevents RNA degradation [55]. Other chelators, such as citrate may further decrease the free magnesium, which may explain how RNA integrity is maintained at high temperatures [56]. Furthermore, the concentration of potassium, a monovalent cation, is often higher in thermophilic than in mesophilic organisms [57], which further enhances the integrity of RNAs.
Potassium may play a role also in the thermotolerance of mesophilic organisms. Intracellular potassium concentration as well as thermotolerance increase in proportion to extracellular osmolality [58], [59], [60]. It remains to be determined if the enhanced thermotolerance is mediated by potassium through the stabilization of RNA primary structure.
5. Temperature effect on reactions
5.1. The Arrhenius equation and the catalysis of RNA turnover
Changes in RNA abundance in response to temperature are determined by both the RNA structures and the reactions catalyzed by the enzymes in RNA metabolism. Since these two factors can have different contributions, the response of the reactions has to be quantified separately. Several empirical relations were suggested by the end of 19th century for the temperature dependence of reaction rate constants. van’t Hoff observed that the rate constants (k) of most reactions increase by a factor Q10 = 2 to 3 when the temperature is increased by 10 °C in the range of ambient temperatures [61], [62], [63], [64]. Arrhenius provided a physical explanation for this relation, which is similar to but not identical with the van’t Hoff’s functional form (Fig. 2A), stating that a reaction can occur only if the preceding molecular collision has a sufficient energy (activation energy. Ea):
| (1) |
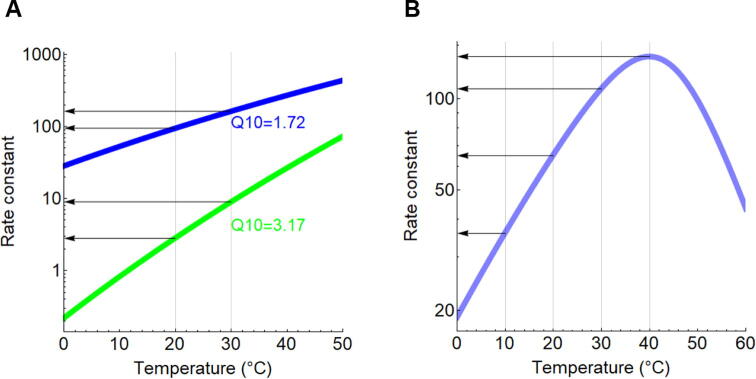
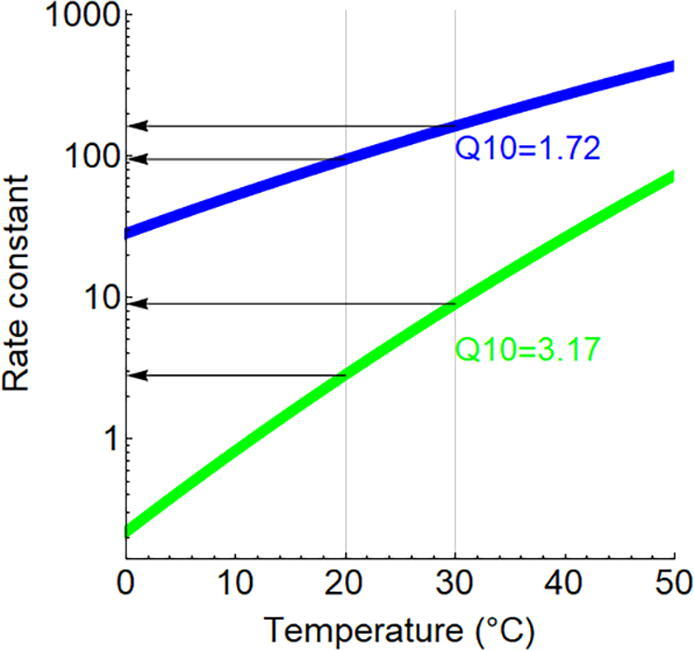
Fig. 2.
Temperature dependence of reaction rates (A) Reaction rates calculated according to Arrhenius formula. The blue curve represents an enzymatic reaction (Ea = 40 kJ/mol [9.5 kcal/mol]), whereas the green curve denotes the uncatalyzed counterpart with higher activation energy (Ea = 85 kJ/mol [20.3 kcal/mol]). The corresponding Q10 values are shown for a temperature shift between 20 and 30 °C. (B) Temperature dependence of an imaginary enzymatic reaction rate based on Peterson et al that takes enzyme denaturation into account, with a melting temperature of 45 °C, in equilibrium condition [66]. The Q10 values indicate a gradual decrease with temperature: Q10(10/20 °C) = 1.81, Q10(20/30 °C) = 1.65 and Q10 (30/40 °C) = 1.27. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Measurement of the reaction rate constants at two (or more) temperatures permits the determination of Ea from equation 1:
| (2) |
The Arrhenius relation (equation 1) closely approximates the observations with most elementary uni- and bimolecular reactions, but deviations occur when the relation is examined over a broad temperature range [65].
Most biochemical reactions are not elementary reactions and the reaction rates do not increase monotonically but reach a maximum because the enzymes that catalyze the reaction undergo a phase transition (i.e. denature) at high temperatures (Fig. 2B). Consequently, the temperature at which the reaction rate reaches a maximum has been often termed optimal temperature [66]. It is difficult to determine this value because enzyme denaturation is often slow and thus the temperature optimum depends on the duration of temperature shift before starting the kinetic experiment [67]. Therefore, the optimum temperature is now rarely used but it can be helpful when similar enzymes are directly compared, as illustrated by the following example. It has been shown that a miniaturized ribozymes can be assembled from the fragments of a parent ribozyme. The smaller, fragmented ribozyme did not only have lower optimal temperature but also a higher catalytic activity at lower temperatures in comparison to the parent ribozyme [68]. This shift in the optimal temperature and the improved activity enables ribozymes to function at lower temperatures, which is relevant in a a hypothetical primordial RNA world.
A systematic analysis showed that the mean Q10 of mesophilic enzymes is 1.8 [69]. This value is equivalent to Ea = 10.3 kcal / mol (assuming temperature is shifted between 25 and 35 °C, equation (2)). At the same time, Q10 for the same reactions without enzymatic catalysis was found to be 3.4 (n = 9 reactions), i.e. Ea = 20.7 kcal / mol. In other words, when an enzyme lowers the energy barrier of the reaction, the temperature dependence curve of the reaction rate becomes flattened (see also 5.2).
Interestingly, the Q10 of thermophilic and psychrophilic enzymes is similar to that of mesophilic enzymes [69]. This also means that compared to the non-catalyzed reaction, the enzymatic reaction rate increases less with temperature (Fig. 2A). Nevertheless, the absolute rates are very high even at high temperatures, which allows fast kinetics of enzymatic reactions.
In order to assess the activation energies in mRNA metabolism, we collected Ea for RNA hydrolysis (degradation). For the enzymatic reactions Ea = 13.1–16 kcal/mol (hammerhead ribozyme, ribonuclease T1) [54], [70], whereas Ea = 23.9, 28.5 and 29 kcal/mol for the non-enzymatic reactions [54], [71], [72]. The ∼ 10 kcal reduction of the activation energies by ribonucleases and ribozymes is comparable to the recent observations with other mesophilic enzymes [69]. It also underscores the observation the Q10 has a lower value for enzymatic reactions than for the uncatalyzed counterparts, a distinction not made until the second half of the 20th century [64], [69]. The determination of Ea of RNA hydrolysis can help also the improvement of RNA vaccine formulation (see 11.2).
Enzymes can reduce the activation energy by multiple mechanisms, among which the stabilization of the transient state stands out. The transient state has an intermediate configuration between the substrate and product. The pre-organized polar environment of the enzyme stabilizes the transition state by electrostatic interactions much more than the corresponding environment in water [73]. Metal cations are key contributors to electrostatic interactions in many enzymes. The importance of metals in DNA and RNA processing enzymes is underscored by the observation that metal cations are coordinated by acidic amino acids to relatively fixed positions within the active site in a large number of enzymes [74]. Interestingly, no major rearrangement was observed in the RNase H1during the transition state formation; instead, cation trafficking was found: magnesium and potassium ions enter and exit the active site at specific reaction steps [75].
5.2. Temperature compensation in reaction networks
In biochemical networks, the primary source of temperature compensation is simply catalysis by enzymes, which lower the activation energy of the reaction, thereby making the temperature dependence of the reaction rate less steep (Fig. 2A, section 5.1).
If the individual reactions in RNA metabolism had the same Q10 values, the RNA would be expressed at a constant level under temperature fluctuations, since the faster RNA synthesis rate is compensated by the equally faster rate of degradation.
Q10 was measured in Arabidopis upon shifting the temperature from 17 °C to 27 °C. The mean (and median) Q10 calculated from transcriptome-wide measurement of RNA synthesis rate was 3.6 (3.4). For the degradation rate, Q10 = 3.3 (2.8). These Q10 values were relatively high in comparison to average Q10 of enzymes (see sections 5.1, 6). Since the Q10 of RNA synthesis and degradation are similar, only a small average (10 %) change is expected in the steady-state expression upon the temperature shift, implying the two processes are reasonably well compensated.
Even if the Q10 values differ among the reactions in a reaction network, gene or biochemical networks can display adaptation, compensating the differences in the Q10 values by various network motifs [76], [77]. Time-varying expression, such as oscillations are particularly sensitive to temperature-dependent changes in reactions [78], [79]. They are more dependent on active temperature compensation by tuning of activation energies and network motifs, such as negative feedback [79].
6. Transcription
In this section, we review the temperature dependence of transcription (6.1) and transcriptional fluctuations (6.2), and the relevant genetic diseases (6.3).
6.1. Effect of temperature on transcriptional initiation and elongation
RNA synthesis occurs in three main steps: transcriptional initiation, elongation, and termination. Elongation can be interrupted by backtracking of RNAP, a phenomenon when the 3′ end of RNA disengages from the template DNA and the polymerase shifts backward along the DNA. Backtracking can occur due to misincorporation of nucleotides. The resulting pause ends mostly with the correction of the mutant RNA sequence. Longer pauses, on the other hand, are usually caused by RNA secondary structures, such as hairpins [80], [81].
The effect of temperature on transcriptional elongation can be monitored in vitro by heating single molecules with infrared laser. In such an assay, the rate of elongation of the E. coli RNA polymerase increases with temperature. Conversely, little change was found in the frequency and the lifetime of the paused states [82]. In a related study, which examined the RNA polymerase over broader temperature range, both elongation and pausing was found to be temperature dependent [83]. The (pause-free) velocity increased from 3 nt/s at 7 °C to 22 nt/s at 45 °C, implying Ea = 9.7 kcal/mol. However, Ea of the entry into the paused state was only 1.3 kcal/mol, which is much smaller than the activation energy of the elongation, suggesting that the RNA polymerase is prone to pause, which may actually be beneficial for the correction of misincorporated nucleotides.
In yeast cells, the analysis of RNA polymerase II with a genomic run-on assay yielded a somewhat lower activation energy of the elongation (6.6 kcal/mol), with velocities from 11.6 nt/s (23 °C) to 21.8 nt/s (37 °C). The faster elongation, however, did not result in larger transcription rate because the polymerase density across the genes reduced, possibly due to the lower initiation rate at higher temperatures [84]. Transcription initiation rate displays also a very complex temperature dependence in E.coli [85], suggesting that the reaction is not elementary enough to follow the Arrhenius relation.
Different viral RNA-dependent RNA polymerases show strikingly different temperature dependence [86]. Whereas some human viruses, the poliovirus and the human rhinovirus C, show temperature dependence similar to that of the E. coli polymerase, the RNA polymerase of the phage Φ6 has nearly constant elongation speed of ∼ 20 nt/s with minimal increase between 25 and 45 °C, implying a low activation energy (4.1 kcal/mol). This unusual property may be related to the bacterial host of this phage, a plant pathogen, which grows over a broad range of temperatures.
Fever-like temperatures have been shown to reduce transcription of SARS-CoV-2 and hence their replication in respiratory epithelium. This phenomenon was specifically due to inhibition of transcription since the virus entry into the cells was intact [87]. The mechanism of the temperature dependence remains to be determined.
6.2. Temperature dependence of transcriptional fluctuations
Messenger RNAs are typically present at low copy numbers [88], which results in substantial fluctuations in molecule numbers. Consequently, RNA copy numbers are distributed in a cell population. A typical distribution is the Poisson distribution, with a characteristic an inverse relationship between the coefficient of variation (CV) and the mean copy number (CV2 = 1/µ). These fluctuations due to the low discrete number of molecules can be supplemented by fluctuations in epigenetic changes, gene regulation, chromosomal neighborhood, RNA synthesis and RNA degradation [89], [90], [91], [92], [93], which can increase noise above the Poisson level. Such an increase is prominent when transitions between active and inactive states of a promoter occur rarely and active promoter initiates the transcription of multiple mRNAs in bursts, resulting in a large burst size. While most additional regulation increases noise, it can be also reduced: a posttranscriptional RNA negative feedback in HIV reduces noise and stabilizes the viral decision [94].
The temperature results in a velocity distribution of molecular velocities, the Maxwell-Boltzmann distribution, the effect of which can be observed in liquid water, as well [95]. The velocity distribution is taken into account by widely used stochastic simulation algorithm (SSA) [96]. Thus, temperature-related distribution is inherent in this algorithm.
In mammalian cells, the burst size of reporter gene expression was inversely proportional with temperature [97]. Some heat shock genes associate with the nuclear speckle compartment after heat shock, which is associated with a higher transcription rate, which may result in a burst-like gene expression [98]. These two studies indicate that heat shock genes and other genes may react differently to temperature shifts in their stochastic behavior.
6.3. Genetic disorders due to temperature sensitive alleles of general transcription factors
Temperature can play a role in diseases by an inverse mechanism when certain gene alleles render a protein temperature sensitive. Temperature-sensitive alleles have played an important role in molecular biology as tools to inhibit a process when temperature is raised. For example, such methods can be used to inhibit transcription to study mRNA half-lives. A similar mechanism was found to underlie genetically inherited diseases, affecting specific components of transcriptional initiation. Specific mutations of the xeroderma pigmentosum group D, XPD, one of the subunits of the transcription/DNA repair complex TFIIH, affect transcription and DNA repair in a temperature-dependent way. These mutations lead to a fever-dependent reversible deterioration of the disease symptom brittle hair. This symptom (also known as trichothiodystrophy) arises due to the transient reduction of cross-linking proteins during the febrile periods, which introduces a fragile point at the base at the hair, resulting in complete hair loss [99], [100]. Whereas transcription is similar at 37 °C and 40 °C in WT cells, cells expressing the relevant mutation have less than half of the original transcription upon this temperature shift.
Mutations have been described that can deteriorate cell function already at physiological temperatures, causing a permanent dysfunctionality in the BRCA1 protein. BCRA1 combines the functions transcription and DNA repair, just like XPD [101], [102]. Carriers of BRCA1 mutations are predisposed to breast and ovarian cancer. One of these mutants yields a functional protein at 30 °C but fails to do so at 37 °C [103] making it constitutively inactive at the normal body temperature.
7. Splicing
Thermosensitive alternative splicing can alter the distribution of protein isoforms or suppress the expression of some mRNA isoforms. The simplest form of alternative splicing is intron retention. mRNAs with retained introns contain a premature stop codon which targets the mRNA for rapid degradation. An intron retention based mechanism was found to be associated with the temperature-dependent sex determination in reptiles [104]. Thermosensitive alternative splicing has been recently identified even in mouse, a homoeothermic animal. A protein thermosensor, the clock kinase protein (CLK1/4) phosphorylates the serine arginine rich SR proteins, which executes the actual alternative splicing event [105]. The mRNA encoding the SR proteins is itself subject to temperature-dependent alternative splicing in both mice and plants, indicating a conservation of the process [106]. This process may sense smaller, diurnal variations in body temperature.
Heat shock typically acts instantaneously but it also elicits a longer-term memory. Priming of plants with a non-lethal heat stress results in de-repression of splicing after a second exposure to heat stress. In contrast, non-primed plants showed significant repression of splicing. This “splicing memory” helps plants to survive subsequent and otherwise lethal heat stress [107].
8. RNAi-mediated gene silencing
RNAi-mediated gene silencing acts as an antiviral mechanism in plants, which is triggered by double-stranded viral RNA (dsRNA). Viral dsRNA is cleaved into virus-derived small interfering RNAs (vsiRNAs) with lengths of 21–24 nucleotides by the dsRNA-specific DICER-LIKE (DCL) endonuclease. The short length siRNAs are then loaded into ARGONAUTE (AGO), which silences the target RNAs, including viral RNA by cleaving target viral RNAs or by repressing their translation.
Several studies suggested that vsiRNAs amounts and silencing increased in strength with temperature in some systems, resulting in a better antiviral defense at higher temperature [108], [109]. In some systems, however, increases in temperature was associated with larger viral titers [110]. A strong temperature dependence of AGO activity was observed also in animals [111]. Furthermore, base pairing strength may affect the functionality of the miRNA-target duplex. The temperature-dependence of this pairing is also reflected in the significant correlation between the average GC content of miRNAs and the physiological temperature of a given organism [112].
9. RNA degradation and its measurement across temperatures
9.1. Nonlinear temperature dependence in RNA degradation in yeast cells
Being a first order reaction, RNA degradation is well suited to study the van’t Hoff law. A shift of temperature from 20 °C to 30 °C increases mRNAs degradation rate in yeast cells Q10(20/30 °C) = 1.82 times on average [113]. This value is in close agreement with the mean Q10 = 1.8 calculated for different mesophilic enzymes (section 5.1). The half-life of mRNAs is further shortened when the cells are exposed to heat shock at 42 °C. However, the extent of this shortening is smaller, Q(30/42 °C) = 1.58. This indicates a plateauing of decay rates possibly due to the partial denaturing of the RNA degradation enzymes at higher temperatures. Indeed, the melting temperature of Xrn1, which degrades mRNAs in the 5′ to 3′ direction, is 44 °C, implying that 50 % of this protein is denatured at this temperature [31].
The median mRNA half-lives determined by a gene control method are 3.4 and 1.5 min at 20 and 42 °C, respectively, which indicates that the mRNA turnover is very rapid throughout temperature range that permits cell growth. Short half-lives are important because they allow oscillatory transcription to propagate between the stages of the cell-cycle [114], which would be impossible with longer mRNA half-lives, as suggested by earlier studies [115]. This brings us to the next topic, how the methods affect the measurement of mRNA half-life, which will be discussed in the next section.
9.2. Impact of the methods on the measurement of RNA degradation at high temperatures
For many years, different methods and sometimes even different studies using the very same method had yielded uncorrelated mRNA half-lives [116]. However, datasets obtained with different methods were published in the past decade that displayed higher correlation, putting an end to the inconsistencies at the standard growth temperature (30 °C). On the other hand, exposing the cells to heat shock resulted in uncorrelated half-lives even between methods that otherwise show a high correlation at the standard temperature [113]. Thus, the measurement of half-lives under heat shock is particularly challenging, and displays strong method-dependence, which is partly caused by the interference from the different methods [116].
This raises the question of whether the heat shock results described in the previous section reflect the degradation rates faithfully. The reasoning applied at the standard temperature gives the way out: if two different methods yield consistent results, it is likely that the half-lives will be reflected with little distortion or completely faithfully. The gene control method used in the above study correlates highly (rs = 0.75) with one of the datasets that use transcriptional inhibition with the temperature sensitive allele of RNA polymerase II, rbp1-1 at 37 °C [117], which suggests that consistent results can be indeed achieved even at heat shock (Fig. 3). Since the rbp1-1 allele can be used to measure half-lives only at high temperatures, Q10 cannot be measured by this method.
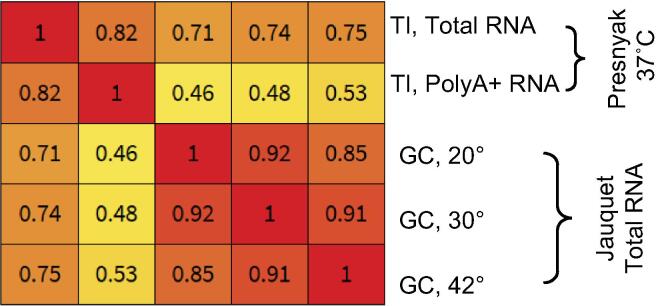
Fig. 3.
Correlation of RNA half-lives at high temperatures in budding yeast measured with gene control (GC) and transcriptional inhibition (TI) methods. The Spearman rank correlation (rs) is calculated from half-lives of mRNAs common in all indicated half-life datasets (n = 75). The correlation between the GC (Jaquet el, 42 °C) [113] and TI (Presnyak et al, 37 °C) [117] datasets is 0.75. The respective median half-lives are 1.5 and 6.4 min.
Q10 was higher in Arabidopsis than in yeast (3.3, see section 5.1) when the temperature was shifted from 17 to 27 °C. It remains to be determined if the large value is due to a molecular mechanism, such as temperature induced unfolding of RNA structures in the UTR, or due to method-dependence. In Arabidopsis, various protocols of transcriptional inhibition and metabolic labelling have been used and the correlations between different the various studies are low to intermediate (Fig. 4). Datasets with good correlations (>0.7) are yet to come. The application of a third, independent method can also help the verification of the findings.
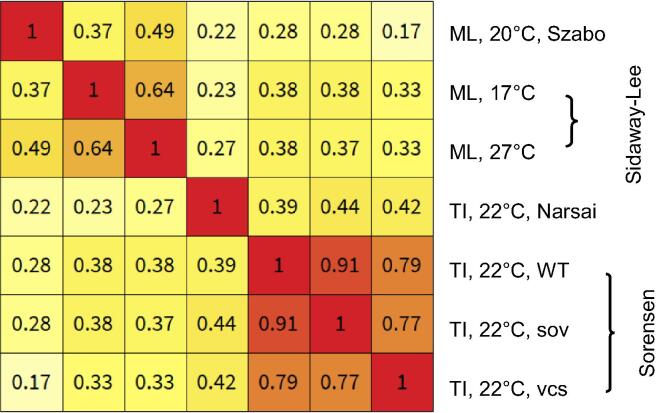
Fig. 4.
Correlation of RNA half-lives in Arabidopis thaliana measured with metabolic labelling (ML) or transcriptional inhibition (TI). The Spearman rank correlation is calculated between datasets for genes common in all studies (n = 5049). The following median half-lives were obtained for these common genes: 1.61 h in Szabo et al [147], 3.99 h (17 °C) and 1.35 h (27 °C) in Sidaway-Lee et al [148], 3.70 h in Narsai et al [149], and 1.83 (WT), 1. 64 (sov) and 2.42 (vcs) in Sorensent et al [150].
10. Regulation of translation by RNAs
In mammals, the cold shock response is triggered at and below 32 °C. A major mediator of this response is the cold inducible RNA-binding protein (CIRP or CIRBP). CIRBP is upregulated in various organs upon mild hypothermia or cold stress, in different species from amphibians to humans. It typically binds to the 5′UTRs of mRNAs and can affect their stability and translation [118]. In mouse fibroblasts, CIRP selectively binds to the 5′ untranslated region of the CDK (cyclin dependent kinase) inhibitor p27Kip1 mRNA and enhances its translation, which in turn reduces cell division [119]. p27Kip1 is central for the decreased proliferation at lower temperature, since mouse embryonic fibroblasts lacking this protein did not reduce their doubling time in hypothermic conditions.
In keratinocytes, the induction of CIRP at 32 °C is dependent on a protein thermosensor, the transient receptor potential vanilloid 4 (TRPV4) channel protein [120]. Since keratinocytes in the skin are frequently exposed to lower temperatures (2.3), cold shock proteins may affect skin physiology [121]. Interestingly, most experiments with keratinocytes were performed at 37 °C despite the fact that dermis regenerates and keratinocytes differentiate better at 31–32 °C than at 37 °C [122], [123].
In turtles, the expression of CIRBP is allele specific and temperature dependent: one of the alleles is induced in embryos exposed to a female-producing temperature, while expression of the other allele does not differ between female- and male-producing temperatures [124]. The SNP underlying the alleles is associated with temperature-dependent sex determination. Thus, thermosensitive splicing and CIRBP may cooperate in temperature-dependent sex determination [104], [124].
11. Biotechnological applications of temperature control
11.1. Ribozymes, riboswitches and RNA thermosensors
An outstanding achievement in combining temperature control with the synthesis of nucleic acids polymers was the discovery of PCR. The DNA conformational alterations induced by temperature cycling (i.e. denaturation and hybridization) are coupled with alternating DNA synthesis rates, leading to an amplification of DNA. Temperature cycling has been also applied to RNA synthesis [125]. A ribozyme catalyzed ligation process was developed to understand RNA amplification in a possible primordial RNA world in the early terrestrial history. The biotechnological application was not the primary aim of RNA amplification in this study, since RNA can be transcribed from DNA. It is important, however, to note that temperature cycling is not required for RNA replication per se [126]. However the temperature alterations may enhance ribozyme reactions [125], [127], with possible applications in biotechnology.
RNA folding is dominated by secondary structures with less contextual effects; this modularity permits thermosensing stem-loops to be easily inserted to the RNAs of interests [128]. RNA thermometers can be coupled to riboswitches, which bind and sense small molecules, and this combination permits the sensing of small molecules at different temperatures. However, some riboswitches, for example the add riboswitch, which senses both adenine and temperature, requires the interaction with a specific protein subunit of the ribosome, which may limit the transmission between different species [129]. Additional engineering of RNA sequence may be required to facilitate their use in different species. This riboswitch illustrates that many RNA structures cannot be analyzed in isolation. As an alternative to pure RNA modules, RNA and protein thermosensors can be combined to amplify the the temperature response. For this purpose, the temperature-sensitive coiled-coil domain of the Tlpa protein can be fused to the protein of interest [130], [131].
11.2. Role of temperature in the manufacturing and delivery of mRNA vaccines
Whereas mRNA vaccines have only been developed recently, they have proven to be the most effective vaccines against the coronavirus in the Covid pandemics (2019–2022) [132], [133]. An important step in this development was the incorporation of modified nucleosides, such as pseudouridine, in the mRNA, which reduces the undesired immune response and enhances translation [134], [135], [136]. One of the main causes of the undesired immune response is the double stranded RNA, which can arise as a byproduct of the vitro transcription. An extra HLPC purification of the in vitro transcribed RNA was shown to reduce this immune response [137]. An alternative method based on the thermostable T7 RNA polymerase has been developed recently. This thermostable polymerase permits the synthesis of the mRNA at high temperature (50 °C), preventing the formation of double stranded RNAs [138]. The mutation rate in the transcription by the T7 RNA polymerase does not increase when the temperature is raised from 30 to 42 °C [139]. Whether this finding can be extended to the transcription at 50 °C by the thermostable T7 RNA polymerase variants remains to be determined.
The degradation of the mRNA is an important practical aspect of the vaccine since RNAs require storage at very low temperatures. The activation energy of 17.9 kcal/mol (74.8 kJ/mol) has been found for mRNA degradation in lipid particles, which is less than the previous estimates for nonenzymatic hydrolysis (see section 5.1). These data suggest that the degradation of mRNAs encapsulated in lipid nanoparticles requires low activation energy, making it susceptible to degradation [140]. Using different materials for encapsulation has been shown to change the activation energies for degradation of different compounds [141]. Thus, the modification of encapsulation technology may permit mRNA vaccines to be stored at higher temperatures by raising the activation energy of degradation.
11.3. Forensic and ecological applications of the temperature dependence of RNA degradation
RNA degradation plays an important role in diagnostic and forensic science [142]. Evidence of degradation of some mRNAs suggest that the 5′ end of an mRNA transcript degrades in dried stains faster than the 3′ end. Statistical analysis of degradation kinetics suggests, depending upon the age of the sample, the age of bloodstains can be accurately estimated to within 2–4 weeks for stains less than 6 months of age and 4–6 weeks for stains 6 months to 1 year old [143]. The RNA half-life is dependent on temperature and air humidity [144].
RNAs can be also used to assess the past microbial communities in environmental samples [145]. The temperature dependence of RNA degradation affects the interpretation of microbiome since RNA persists much longer in low temperature soil [146].
12. Conclusions
Our review highlighted recent developments in temperature control of RNA biology. There are many phenomena that are more closely connected than commonly assumed, whose understanding requires both empirical observations and computational modeling, summarized briefly below.
Due to the biphasic temperature response observed in mouse, a succession of high and low temperature pulses may be more appropriate to study cell adaptation instead of monophasic heat or cold shock. In human biology, it remains to be seen how often SNPs encode temperature-sensitive enzymes, from RNA polymerases to processing enzymes, that display temperature-dependent symptoms.
New imaging results revealed the existence of intracellular temperature gradients, the magnitude of which is a subject of a lively debate at the interface of physical modelling and experimental observations. RNA turnover may contribute to these gradients through basal metabolism, and vice versa, RNA may experience temperature-induced changes in the nuclei and mitochondria.
RNA is prone to hydrolysis at high temperatures a typical hyperthermophile is exposed to, but the intracellular milieu seems to employ tricks to mitigate the detrimental effect of magnesium. If RNA is sufficiently stabilized by proper cation composition and chelation, the main source of degradation is likely to be enzymatic just like in most other organism. This hypothesis remains to be tested. The Arrhenius plot can help distinguishing enzymatic degradation from non-enzymatic hydrolysis since enzymes lower the activation energy of a reaction (Fig. 2A).
On the other hand, the appearance of strong RNA structures in cold environment may reduce the relevant reaction rates more than expected from a typical Q10 value of around two, which may contribute to a cessation of microorganism growth at low temperatures above the freezing point. Measurement of activation energies in these processes can help to understand the complex interplay between RNA structures, RNA degradation and translation.
CRediT authorship contribution statement
Attila Becskei: Writing – original draft, Writing – review & editing, Conceptualization, Supervision, Visualization. Sayanur Rahaman: Writing – original draft, Writing – review & editing, Conceptualization, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Mandin P., Johansson J. Feeling the heat at the millennium: thermosensors playing with fire. Mol Microbiol. 2020;113:588–592. doi: 10.1111/mmi.14468. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Gross C.A. Cold shock response in bacteria. Annu Rev Genet. 2021;55:377–400. doi: 10.1146/annurev-genet-071819-031654. [DOI] [PubMed] [Google Scholar]
- 3.Williams T.J., Lauro F.M., Ertan H., Burg D.W., et al. Defining the response of a microorganism to temperatures that span its complete growth temperature range (-2 degrees C to 28 degrees C) using multiplex quantitative proteomics. Environ Microbiol. 2011;13:2186–2203. doi: 10.1111/j.1462-2920.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- 4.Cavicchioli R. On the concept of a psychrophile. ISME J. 2016;10:793–795. doi: 10.1038/ismej.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiala G., Stetter K.O. Pyrococcus-Furiosus Sp-Nov represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100-degrees C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 6.Margosch D., Ehrmann M.A., Buckow R., Heinz V., et al. High-pressure-mediated survival of Clostridium botulinum and Bacillus amyloliquefaciens endospores at high temperature. Appl Environ Microbiol. 2006;72:3476–3481. doi: 10.1128/AEM.72.5.3476-3481.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg M.V., Schut G.J., Brehm S., Datta S., Adams M.W. Cold shock of a hyperthermophilic archaeon: pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J Bacteriol. 2005;187:336–348. doi: 10.1128/JB.187.1.336-348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonner J.J., Carlson T., Fackenthal D.L., Paddock D., et al. Complex regulation of the yeast heat shock transcription factor. Mol Biol Cell. 2000;11:1739–1751. doi: 10.1091/mbc.11.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hjorth-Sorensen B., Hoffmann E.R., Lissin N.M., Sewell A.K., Jakobsen B.K. Activation of heat shock transcription factor in yeast is not influenced by the levels of expression of heat shock proteins. Mol Microbiol. 2001;39:914–923. doi: 10.1046/j.1365-2958.2001.02279.x. [DOI] [PubMed] [Google Scholar]
- 10.Muhlhofer M., Berchtold E., Stratil C.G., Csaba G., et al. The heat shock response in yeast maintains protein homeostasis by chaperoning and replenishing proteins. Cell Rep. 2019;29(4593–4607):e4598. doi: 10.1016/j.celrep.2019.11.109. [DOI] [PubMed] [Google Scholar]
- 11.DeWitt T.J., Scheiner S.M. Oxford University Press; 2004. Phenotypic plasticity: functional and conceptual approaches. [Google Scholar]
- 12.Martinez-Juarez A., Moreno-Mendoza N. Mechanisms related to sexual determination by temperature in reptiles. J Therm Biol. 2019;85 doi: 10.1016/j.jtherbio.2019.102400. [DOI] [PubMed] [Google Scholar]
- 13.Akin J.A. Homeostatic processes for thermoregulation. Nature Education Knowledge. 2011:3. [Google Scholar]
- 14.Nayak G., Zhang K.X., Vemaraju S., Odaka Y., et al. Adaptive thermogenesis in mice is enhanced by opsin 3-dependent adipocyte light sensing. Cell Rep. 2020;30:672–686. doi: 10.1016/j.celrep.2019.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chondronikola M., Volpi E., Borsheim E., Chao T., et al. Brown adipose tissue is linked to a distinct thermoregulatory response to mild cold in people. Front Physiol. 2016;7 doi: 10.3389/fphys.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leon L.R., DuBose D.A., Mason C.W. Heat stress induces a biphasic thermoregulatory response in mice. Am J Physiol Regul Integr Comp Physiol. 2005;288:R197–R204. doi: 10.1152/ajpregu.00046.2004. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko T., Kibayashi K. Mild hypothermia facilitates the expression of cold-inducible RNA-binding protein and heat shock protein 70.1 in mouse brain. Brain Res. 2012;1466:128–136. doi: 10.1016/j.brainres.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Masuda T., Itoh K., Higashitsuji H., Higashitsuji H., et al. Cold-inducible RNA-binding protein (Cirp) interacts with Dyrk1b/Mirk and promotes proliferation of immature male germ cells in mice. Proc Natl Acad Sci U S A. 2012;109:10885–10890. doi: 10.1073/pnas.1121524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolfe D.F., Brown G.C. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 20.Bouchama A., Abuyassin B., Lehe C., Laitano O., et al. Classic and exertional heatstroke. Nat Rev Dis Primers. 2022;8:8. doi: 10.1038/s41572-021-00334-6. [DOI] [PubMed] [Google Scholar]
- 21.Khan V.R., Brown I.R. The effect of hyperthermia on the induction of cell death in brain, testis, and thymus of the adult and developing rat. Cell Stress Chaperones. 2002;7:73–90. doi: 10.1379/1466-1268(2002)007<0073:teohot>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor N.A., Tipton M.J., Kenny G.P. Considerations for the measurement of core, skin and mean body temperatures. J Therm Biol. 2014;46:72–101. doi: 10.1016/j.jtherbio.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 23.van der Lans A.A., Vosselman M.J., Hanssen M.J., Brans B., van Marken Lichtenbelt W.D. Supraclavicular skin temperature and BAT activity in lean healthy adults. J Physiol Sci. 2016;66:77–83. doi: 10.1007/s12576-015-0398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okabe K., Uchiyama S. Intracellular thermometry uncovers spontaneous thermogenesis and associated thermal signaling. Commun Biol. 2021;4:1377. doi: 10.1038/s42003-021-02908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chretien D., Benit P., Ha H.H., Keipert S., et al. Mitochondria are physiologically maintained at close to 50 degrees C. PLoS Biol. 2018;16:e2003992. doi: 10.1371/journal.pbio.2003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu L., Kwong J.M., Caprioli J., Piri N. DNA and RNA oxidative damage in the retina is associated with ganglion cell mitochondria. Sci Rep. 2022;12:8705. doi: 10.1038/s41598-022-12770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosseini M., Roy P., Sissler M., Zirbel C.L., et al. How to fold and protect mitochondrial ribosomal RNA with fewer guanines. Nucleic Acids Res. 2018;46:10946–10968. doi: 10.1093/nar/gky762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T.H., Sotomayor M., Gopalan V. Biochemical studies provide insights into the necessity for multiple arabidopsis thaliana protein-only RNase P isoenzymes. J Mol Biol. 2019;431:615–624. doi: 10.1016/j.jmb.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fahimi P., Matta C.F. The hot mitochondrion paradox: reconciling theory and experiment. Trends Chem. 2022;4:96–110. [Google Scholar]
- 30.Macherel D., Haraux F., Guillou H., Bourgeois O. The conundrum of hot mitochondria. Bba-Bioenergetics. 2021;1862:148348. doi: 10.1016/j.bbabio.2020.148348. [DOI] [PubMed] [Google Scholar]
- 31.Jarzab A., Kurzawa N., Hopf T., Moerch M., et al. Meltome atlas-thermal proteome stability across the tree of life. Nat Methods. 2020;17:495–503. doi: 10.1038/s41592-020-0801-4. [DOI] [PubMed] [Google Scholar]
- 32.Tinoco I., Jr., Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 33.Xu X., Chen S.J. Physics-based RNA structure prediction. Biophys Rep. 2015;1:2–13. doi: 10.1007/s41048-015-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh J., Hanson J., Paliwal K., Zhou Y.Q. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nature Commun. 2019;10:5407. doi: 10.1038/s41467-019-13395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fox D.M., MacDermaid C.M., Schreij A.M.A., Zwierzyna M., Walker R.C. RNA folding using quantum computers. PLoS Comput Biol. 2022;18:e1010032. doi: 10.1371/journal.pcbi.1010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halder A., Data D., Seelam P.P., Bhattacharyya D., Mitra A. Estimating strengths of individual hydrogen bonds in RNA base pairs: toward a consensus between different computational approaches. ACS Omega. 2019;4:7354–7368. doi: 10.1021/acsomega.8b03689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sengupta P., Garrity P. Sensing temperature. Curr Biol. 2013;23:R304–R307. doi: 10.1016/j.cub.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoe N.P., Goguen J.D. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ignatov D., Vaitkevicius K., Durand S., Cahoon L., et al. An mRNA-mRNA interaction couples expression of a virulence factor and its chaperone in listeria monocytogenes. Cell Rep. 2020;30(4027–4040):e4027. doi: 10.1016/j.celrep.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu C., Jaquet V., Gencoglu M., Becskei A. Protein dimerization generates bistability in positive feedback loops. Cell Rep. 2016;16:1204–1210. doi: 10.1016/j.celrep.2016.06.072. [DOI] [PubMed] [Google Scholar]
- 41.Stephenson W., Keller S., Santiago R., Albrecht J.E., et al. Combining temperature and force to study folding of an RNA hairpin. Phys Chem Chem Phys. 2014;16:906–917. doi: 10.1039/c3cp52042k. [DOI] [PubMed] [Google Scholar]
- 42.Meyer S., Carlson P.D., Lucks J.B. Characterizing the Structure-Function Relationship of a Naturally Occurring RNA Thermometer. Biochemistry. 2017;56:6629–6638. doi: 10.1021/acs.biochem.7b01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah P., Gilchrist M.A. Is thermosensing property of RNA thermometers unique? PLoS ONE. 2010;5:e11308. doi: 10.1371/journal.pone.0011308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Burkhardt D.H., Rouskin S., Li G.W., et al. A Stress Response that Monitors and Regulates mRNA Structure Is Central to Cold Shock Adaptation. Mol Cell. 2018;70(274–286):e277. doi: 10.1016/j.molcel.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldenberg D., Azar I., Oppenheim A.B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 47.Pandey S., Prasad A., Sharma N., Prasad M. Linking the plant stress responses with RNA helicases. Plant Sci. 2020;299 doi: 10.1016/j.plantsci.2020.110607. [DOI] [PubMed] [Google Scholar]
- 48.Somero G.N. RNA thermosensors: how might animals exploit their regulatory potential? J Exp Biol. 2018;221 doi: 10.1242/jeb.162842. [DOI] [PubMed] [Google Scholar]
- 49.Twittenhoff C., Brandenburg V.B., Righetti F., Nuss A.M., et al. Lead-seq: transcriptome-wide structure probing in vivo using lead(II) ions. Nucleic Acids Res. 2020;48:e71. doi: 10.1093/nar/gkaa404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniel R.M., Cowan D.A. Biomolecular stability and life at high temperatures. Cell Mol Life Sci. 2000;57:250–264. doi: 10.1007/PL00000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalak J.A., Dalluge J.J., McCloskey J.A., Stetter K.O. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- 52.Hori H. Regulatory Factors for tRNA Modifications in Extreme- Thermophilic Bacterium Thermus thermophilus. Front Genet. 2019;10:204. doi: 10.3389/fgene.2019.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noon K.R., Guymon R., Crain P.F., McCloskey J.A., et al. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23 degrees C) and Stetteria hydrogenophila (Topt, 95 degrees C) J Bacteriol. 2003;185:5483–5490. doi: 10.1128/JB.185.18.5483-5490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y.F., Breaker R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2 '-hydroxyl group. J Am Chem Soc. 1999;121:5364–5372. [Google Scholar]
- 55.Yamagami R., Bingaman J.L., Frankel E.A., Bevilacqua P.C. Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis. Nat Commun. 2018;9:2149. doi: 10.1038/s41467-018-04415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.AbouHaidar M.G., Ivanov I.G. Non-enzymatic RNA hydrolysis promoted by the combined catalytic activity of buffers and magnesium ions. Z Naturforsch C J Biosci. 1999;54:542–548. doi: 10.1515/znc-1999-7-813. [DOI] [PubMed] [Google Scholar]
- 57.Grosjean H., Oshima T. How nucleic acids cope with high temperature. Extremophiles. 2007:39–56. [Google Scholar]
- 58.Tesone S., Hughes A., Hurst A. Salt extends the upper temperature limit for growth of food-poisoning bacteria. Can J Microbiol. 1981;27:970–972. doi: 10.1139/m81-154. [DOI] [PubMed] [Google Scholar]
- 59.Richey B., Cayley D.S., Mossing M.C., Kolka C., et al. Variability of the intracellular ionic environment of Escherichia coli. Differences between in vitro and in vivo effects of ion concentrations on protein-DNA interactions and gene expression. J Biol Chem. 1987;262:7157–7164. [PubMed] [Google Scholar]
- 60.Kataoka N., Matsutani M., Murata R., Koga R., et al. Potassium ion leakage impairs thermotolerance in Corynebacterium glutamicum. J Biosci Bioeng. 2022;133:119–125. doi: 10.1016/j.jbiosc.2021.10.005. [DOI] [PubMed] [Google Scholar]
- 61.Logan S.R. The origin and status of the Arrhenius equation. J Chem Educ. 1982;59:279–281. [Google Scholar]
- 62.Lloyd J., Taylor J.A. On the temperature-dependence of soil respiration. Funct Ecol. 1994;8:315–323. [Google Scholar]
- 63.Wolfenden R., Snider M., Ridgway C., Miller B. The temperature dependence of enzyme rate enhancements. J Am Chem Soc. 1999;121:7419–7420. [Google Scholar]
- 64.Wolfenden R., Snider M.J. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 65.Smith I.W. The temperature-dependence of elementary reaction rates: beyond Arrhenius. Chem Soc Rev. 2008;37:812–826. doi: 10.1039/b704257b. [DOI] [PubMed] [Google Scholar]
- 66.Peterson M.E., Eisenthal R., Danson M.J., Spence A., Daniel R.M. A new intrinsic thermal parameter for enzymes reveals true temperature optima. J Biol Chem. 2004;279:20717–20722. doi: 10.1074/jbc.M309143200. [DOI] [PubMed] [Google Scholar]
- 67.Cornish-Bowden A. John Wiley & Sons; 2013. Fundamentals of enzyme kinetics. [Google Scholar]
- 68.Akoopie A., Muller U.F. Lower temperature optimum of a smaller, fragmented triphosphorylation ribozyme. Phys Chem Chem Phys. 2016;18:20118–20125. doi: 10.1039/c6cp00672h. [DOI] [PubMed] [Google Scholar]
- 69.Elias M., Wieczorek G., Rosenne S., Tawfik D.S. The universality of enzymatic rate-temperature dependency. Trends Biochem Sci. 2014;39:1–7. doi: 10.1016/j.tibs.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi K. Effects of temperature, salts, and solvents on the enzymatic activity of ribonuclease T1. J Biochem. 1974;75:201–204. doi: 10.1093/oxfordjournals.jbchem.a130377. [DOI] [PubMed] [Google Scholar]
- 71.Fabre A.L., Colotte M., Luis A., Tuffet S., Bonnet J. An efficient method for long-term room temperature storage of RNA. Eur J Hum Genet. 2014;22:379–385. doi: 10.1038/ejhg.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawamura K. Kinetic analysis of the cleavage of the ribose phosphodiester bond within guanine and cytosine-rich oligonucleotides and dinucleotides at 65–200 C and its implications concerning the chemical evolution of RNA. Bulletin of the Chemical Society of Japan. 2003;76:153–162. [Google Scholar]
- 73.Warshel A., Sharma P.K., Kato M., Xiang Y., et al. Electrostatic basis for enzyme catalysis. Chem Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 74.Genna V., Colombo M., De Vivo M., Marcia M. Second-Shell Basic Residues Expand the Two-Metal-Ion Architecture of DNA and RNA Processing Enzymes. Structure. 2018;26(40–50):e42. doi: 10.1016/j.str.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Samara N.L., Yang W. Cation trafficking propels RNA hydrolysis. Nat Struct Mol Biol. 2018;25:715–721. doi: 10.1038/s41594-018-0099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruoff P., Zakhartsev M., Westerhoff H.V. Temperature compensation through systems biology. FEBS J. 2007;274:940–950. doi: 10.1111/j.1742-4658.2007.05641.x. [DOI] [PubMed] [Google Scholar]
- 77.Krantz M., Legen J., Gao Y., Zoschke R., et al. Modeling indicates degradation of mRNA and protein as a potential regulation mechanisms during cold acclimation. J Plant Res. 2021;134:873–883. doi: 10.1007/s10265-021-01294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chandraseelan J.G., Oliveira S.M., Hakkinen A., Tran H., et al. Effects of temperature on the dynamics of the LacI-TetR-CI repressilator. Mol Biosyst. 2013;9:3117–3123. doi: 10.1039/c3mb70203k. [DOI] [PubMed] [Google Scholar]
- 79.Ruoff P., Loros J.J., Dunlap J.C. The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proc Natl Acad Sci U S A. 2005;102:17681–17686. doi: 10.1073/pnas.0505137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qian J., Dunlap D., Finzi L. Basic mechanisms and kinetics of pause-interspersed transcript elongation. Nucleic Acids Res. 2021;49:15–24. doi: 10.1093/nar/gkaa1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mosaei H., Zenkin N. Two distinct pathways of RNA polymerase backtracking determine the requirement for the Trigger Loop during RNA hydrolysis. Nucleic Acids Res. 2021;49:8777–8784. doi: 10.1093/nar/gkab675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abbondanzieri E.A., Shaevitz J.W., Block S.M. Picocalorimetry of transcription by RNA polymerase. Biophys J. 2005;89:L61–L63. doi: 10.1529/biophysj.105.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mejia Y.X., Mao H., Forde N.R., Bustamante C. Thermal probing of E. coli RNA polymerase off-pathway mechanisms. J Mol Biol. 2008;382:628–637. doi: 10.1016/j.jmb.2008.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miguel A., Monton F., Li T., Gomez-Herreros F., et al. External conditions inversely change the RNA polymerase II elongation rate and density in yeast. Biochim Biophys Acta. 2013;1829:1248–1255. doi: 10.1016/j.bbagrm.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 85.Plaskon D.M., Henderson K.L., Felth L.C., Molzahn C.M., et al. Temperature effects on RNA polymerase initiation kinetics reveal which open complex initiates and that bubble collapse is stepwise. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2021941118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seifert M., van Nies P., Papini F.S., Arnold J.J., et al. Temperature controlled high-throughput magnetic tweezers show striking difference in activation energies of replicating viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2020;48:5591–5602. doi: 10.1093/nar/gkaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herder V., Dee K., Wojtus J.K., Epifano I., et al. Elevated temperature inhibits SARS-CoV-2 replication in respiratory epithelium independently of IFN-mediated innate immune defenses. PLoS Biol. 2021;19:e3001065. doi: 10.1371/journal.pbio.3001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lahtvee P.J., Sanchez B.J., Smialowska A., Kasvandik S., et al. Absolute Quantification of Protein and mRNA Abundances Demonstrate Variability in Gene-Specific Translation Efficiency in Yeast. Cell Syst. 2017;4(495–504):e495. doi: 10.1016/j.cels.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 89.Baudrimont A., Jaquet V., Wallerich S., Voegeli S., Becskei A. Contribution of RNA Degradation to Intrinsic and Extrinsic Noise in Gene Expression. Cell Rep. 2019;26:3752–3761. doi: 10.1016/j.celrep.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 90.Lannan R., Maity A., Wollman R. Epigenetic fluctuations underlie gene expression timescales and variability. Physiol Genomics. 2022;54:220–229. doi: 10.1152/physiolgenomics.00051.2021. [DOI] [PubMed] [Google Scholar]
- 91.Iakovlev M., Faravelli S., Becskei A. Gene Families With Stochastic Exclusive Gene Choice Underlie Cell Adhesion in Mammalian Cells. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.642212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wada T., Wallerich S., Becskei A. Stochastic Gene Choice during Cellular Differentiation. Cell Rep. 2018;24:3503–3512. doi: 10.1016/j.celrep.2018.08.074. [DOI] [PubMed] [Google Scholar]
- 93.Griego A., Douche T., Gianetto Q.G., Matondo M., Manina G. RNase E and HupB dynamics foster mycobacterial cell homeostasis and fitness. iScience. 2022;25 doi: 10.1016/j.isci.2022.104233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hansen M.M.K., Wen W.Y., Ingerman E., Razooky B.S., et al. A Post-Transcriptional Feedback Mechanism for Noise Suppression and Fate Stabilization. Cell. 2018;173(1609–1621):e1615. doi: 10.1016/j.cell.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mo J.Y., Simha A., Kheifets S., Raizen M.G. Testing the Maxwell-Boltzmann distribution using Brownian particles. Opt Express. 2015;23:1888–1893. doi: 10.1364/OE.23.001888. [DOI] [PubMed] [Google Scholar]
- 96.Gillespie D.T., Petzold L.R., Seitaridou E. Validity conditions for stochastic chemical kinetics in diffusion-limited systems. J Chem Phys. 2014;140 doi: 10.1063/1.4863990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arnaud O., Meyer S., Vallin E., Beslon G., Gandrillon O. Temperature-induced variation in gene expression burst size in metazoan cells. BMC Mol Biol. 2015;16:20. doi: 10.1186/s12867-015-0048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim J., Venkata N.C., Hernandez Gonzalez G.A., Khanna N., Belmont A.S. Gene expression amplification by nuclear speckle association. J Cell Biol. 2020;219 doi: 10.1083/jcb.201904046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vermeulen W., Rademakers S., Jaspers N.G., Appeldoorn E., et al. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat Genet. 2001;27:299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- 100.Theil A.F., Mandemaker I.K., van den Akker E., Swagemakers S.M.A., et al. Trichothiodystrophy causative TFIIEbeta mutation affects transcription in highly differentiated tissue. Hum Mol Genet. 2017;26:4689–4698. doi: 10.1093/hmg/ddx351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Compe E., Egly J.M. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 102.Zhang X., Li R. BRCA1-Dependent Transcriptional Regulation: Implication in Tissue-Specific Tumor Suppression. Cancers (Basel) 2018;10 doi: 10.3390/cancers10120513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Worley T., Vallon-Christersson J., Billack B., Borg A., Monteiro A.N. A naturally occurring allele of BRCA1 coding for a temperature-sensitive mutant protein. Cancer Biol Ther. 2002;1:497–501. doi: 10.4161/cbt.1.5.164. [DOI] [PubMed] [Google Scholar]
- 104.Deveson I.W., Holleley C.E., Blackburn J., Marshall Graves J.A., et al. Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci Adv. 2017;3:e1700731. doi: 10.1126/sciadv.1700731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haltenhof T., Kotte A., De Bortoli F., Schiefer S., et al. A Conserved Kinase-Based Body-Temperature Sensor Globally Controls Alternative Splicing and Gene Expression. Mol Cell. 2020;78(57–69):e54. doi: 10.1016/j.molcel.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 106.Neumann A., Meinke S., Goldammer G., Strauch M., et al. Alternative splicing coupled mRNA decay shapes the temperature-dependent transcriptome. EMBO Rep. 2020;21:e51369. doi: 10.15252/embr.202051369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ling Y., Serrano N., Gao G., Atia M., et al. Thermopriming triggers splicing memory in Arabidopsis. J Exp Bot. 2018;69:2659–2675. doi: 10.1093/jxb/ery062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim Y., Kim Y.J., Paek K.H. Temperature-specific vsiRNA confers RNAi-mediated viral resistance at elevated temperature in Capsicum annuum. J Exp Bot. 2021;72:1432–1448. doi: 10.1093/jxb/eraa527. [DOI] [PubMed] [Google Scholar]
- 109.Szittya G., Silhavy D., Molnar A., Havelda Z., et al. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 2003;22:633–640. doi: 10.1093/emboj/cdg74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tenllado F., Canto T. Effects of a changing environment on the defenses of plants to viruses. Curr Opin Virol. 2020;42:40–46. doi: 10.1016/j.coviro.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 111.Park J.H., Shin C. Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res. 2015;43:9418–9433. doi: 10.1093/nar/gkv937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carmel I., Shomron N., Heifetz Y. Does base-pairing strength play a role in microRNA repression? RNA. 2012;18:1947–1956. doi: 10.1261/rna.032185.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jaquet V., Wallerich S., Voegeli S., Turos D., et al. Determinants of the temperature adaptation of mRNA degradation. Nucleic Acids Res. 2022;50:1092–1110. doi: 10.1093/nar/gkab1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Becskei A., Boselli M.G., van Oudenaarden A. Amplitude control of cell-cycle waves by nuclear import. Nat Cell Biol. 2004;6:451–457. doi: 10.1038/ncb1124. [DOI] [PubMed] [Google Scholar]
- 115.Baudrimont A., Voegeli S., Viloria E.C., Stritt F., et al. Multiplexed gene control reveals rapid mRNA turnover. Sci Adv. 2017;3:e1700006. doi: 10.1126/sciadv.1700006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wada T., Becskei A. Impact of Methods on the Measurement of mRNA Turnover. Int J Mol Sci. 2017;18:2723. doi: 10.3390/ijms18122723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Presnyak V., Alhusaini N., Chen Y.H., Martin S., et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhong P., Huang H. Recent progress in the research of cold-inducible RNA-binding protein. Future Sci OA. 2017;3:FSO246. doi: 10.4155/fsoa-2017-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roilo M., Kullmann M.K., Hengst L. Cold-inducible RNA-binding protein (CIRP) induces translation of the cell-cycle inhibitor p27Kip1. Nucleic Acids Res. 2018;46:3198–3210. doi: 10.1093/nar/gkx1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fujita T., Higashitsuji H., Higashitsuji H., Liu Y., et al. TRPV4-dependent induction of a novel mammalian cold-inducible protein SRSF5 as well as CIRP and RBM3. Sci Rep. 2017;7:2295. doi: 10.1038/s41598-017-02473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liao Y., Feng J., Zhang Y., Tang L., Wu S. The mechanism of CIRP in inhibition of keratinocytes growth arrest and apoptosis following low dose UVB radiation. Mol Carcinog. 2017;56:1554–1569. doi: 10.1002/mc.22597. [DOI] [PubMed] [Google Scholar]
- 122.Borowiec A.S., Delcourt P., Dewailly E., Bidaux G. Optimal differentiation of in vitro keratinocytes requires multifactorial external control. PLoS ONE. 2013;8:e77507. doi: 10.1371/journal.pone.0077507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Frese L., Darwiche S.E., von Rechenberg B., Hoerstrup S.P., et al. Thermal conditioning improves quality and speed of keratinocyte sheet production for burn wound treatment. Cytotherapy. 2021;23:536–547. doi: 10.1016/j.jcyt.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 124.Schroeder A.L., Metzger K.J., Miller A., Rhen T. A Novel Candidate Gene for Temperature-Dependent Sex Determination in the Common Snapping Turtle. Genetics. 2016;203:557–571. doi: 10.1534/genetics.115.182840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim K.S., Oh S., Yea S.S., Yoon M.Y., Kim D.E. Amplification of an RNA ligase ribozyme under alternating temperature conditions. FEBS Lett. 2008;582:2745–2752. doi: 10.1016/j.febslet.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 126.Vaidya N., Manapat M.L., Chen I.A., Xulvi-Brunet R., et al. Spontaneous network formation among cooperative RNA replicators. Nature. 2012;491:72–77. doi: 10.1038/nature11549. [DOI] [PubMed] [Google Scholar]
- 127.Dropulic B., Lin N.H., Jeang K.T. A method to increase the cumulative cleavage efficiency of ribozymes: thermal cycling. Nucleic Acids Res. 1993;21:2273–2274. doi: 10.1093/nar/21.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rossmanith J., Narberhaus F. Exploring the modular nature of riboswitches and RNA thermometers. Nucleic Acids Res. 2016;44:5410–5423. doi: 10.1093/nar/gkw232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.de Jesus V., Qureshi N.S., Warhaut S., Bains J.K., et al. Switching at the ribosome: riboswitches need rProteins as modulators to regulate translation. Nat Commun. 2021;12:4723. doi: 10.1038/s41467-021-25024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chee W.K.D., Yeoh J.W., Dao V.L., Poh C.L. Highly Reversible Tunable Thermal-Repressible Split-T7 RNA Polymerases (Thermal-T7RNAPs) for Dynamic Gene Regulation. ACS Synth Biol. 2022;11:921–937. doi: 10.1021/acssynbio.1c00545. [DOI] [PubMed] [Google Scholar]
- 131.Naik R.R., Kirkpatrick S.M., Stone M.O. The thermostability of an alpha-helical coiled-coil protein and its potential use in sensor applications. Biosens Bioelectron. 2001;16:1051–1057. doi: 10.1016/s0956-5663(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 132.Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin Microbiol Infect. 2022;28:202–221. doi: 10.1016/j.cmi.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Premikha M., Chiew C.J., Wei W.E., Leo Y.S., et al. Comparative Effectiveness of mRNA and Inactivated Whole Virus Vaccines against COVID-19 Infection and Severe Disease in Singapore. Clin Infect Dis. 2022 doi: 10.1093/cid/ciac288. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., et al. Nucleoside modifications in RNA limit activation of 2'-5'-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dolgin E. The tangled history of mRNA vaccines. Nature. 2021;597:318–324. doi: 10.1038/d41586-021-02483-w. [DOI] [PubMed] [Google Scholar]
- 137.Kariko K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu M.Z., Asahara H., Tzertzinis G., Roy B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA. 2020;26:345–360. doi: 10.1261/rna.073858.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Herman C, Bradley C, Gordon A, Wang C, et al., RNA polymerase inaccuracy underlies SARS-CoV-2 variants and vaccine heterogeneity. 2022.
- 140.Raffaele J., Loughney J.W., Rustandi R.R. Development of a microchip capillary electrophoresis method for determination of the purity and integrity of mRNA in lipid nanoparticle vaccines. Electrophoresis. 2022;43:1101–1106. doi: 10.1002/elps.202100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lavelli V., Sereikaite J. Kinetic study of encapsulated beta-carotene degradation in dried systems: a review. Foods. 2022;11:437. doi: 10.3390/foods11030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Takeuchi K., Yanagisawa H., Kurosawa Y., Iida Y., et al. Degradation of SARS-CoV-2 specific ribonucleic acid in samples for nucleic acid amplification detection. PLoS ONE. 2022;17:e0264541. doi: 10.1371/journal.pone.0264541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fu J., Allen R.W. A method to estimate the age of bloodstains using quantitative PCR. Forensic Sci Int Genet. 2019;39:103–108. doi: 10.1016/j.fsigen.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 144.Heneghan N., Fu J., Pritchard J., Payton M., Allen R.W. The effect of environmental conditions on the rate of RNA degradation in dried blood stains. Forensic Sci Int Genet. 2021;51 doi: 10.1016/j.fsigen.2020.102456. [DOI] [PubMed] [Google Scholar]
- 145.Malmstrom C.M., Martin M.D., Gagnevin L. Exploring the Emergence and Evolution of Plant Pathogenic Microbes Using Historical and Paleontological Sources. Annu Rev Phytopathol. 2022:60. doi: 10.1146/annurev-phyto-021021-041830. [DOI] [PubMed] [Google Scholar]
- 146.Schostag M.D., Albers C.N., Jacobsen C.S., Prieme A. Low Turnover of Soil Bacterial rRNA at Low Temperatures. Front Microbiol. 2020;11:962. doi: 10.3389/fmicb.2020.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Szabo E.X., Reichert P., Lehniger M.K., Ohmer M., et al. Metabolic Labeling of RNAs Uncovers Hidden Features and Dynamics of the Arabidopsis Transcriptome. Plant Cell. 2020;32:871–887. doi: 10.1105/tpc.19.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sidaway-Lee K., Costa M.J., Rand D.A., Finkenstadt B., Penfield S. Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biol. 2014;15:R45. doi: 10.1186/gb-2014-15-3-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Narsai R., Howell K.A., Millar A.H., O'Toole N., et al. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sorenson R.S., Deshotel M.J., Johnson K., Adler F.R., Sieburth L.E. Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc Natl Acad Sci U S A. 2018;115:E1485–E1494. doi: 10.1073/pnas.1712312115. [DOI] [PMC free article] [PubMed] [Google Scholar]