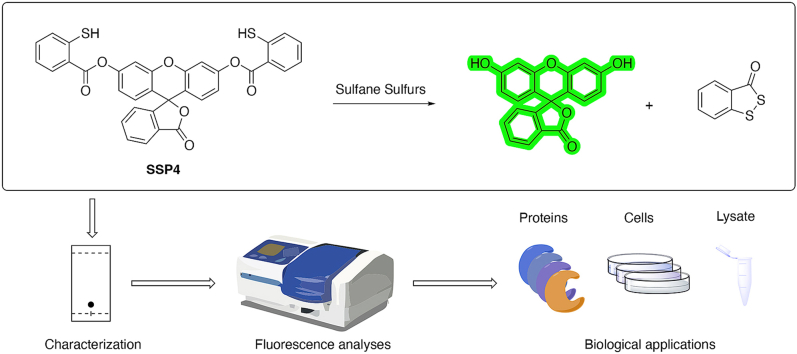
Abstract
Fluorescent probes are useful tools for the detection of sulfane sulfurs in biological systems. In this work, we report the development of SSP4, a widely used probe generated in our laboratory. We describe its evolution, preparation, and physical/chemical properties. Fluorescence analyses of SSP4 determined its high selectivity and sensitivity to sulfane sulfurs, even with the interfering presence of other species, such as amino acids and metal ions. Protocols for using SSP4 in a relatively quick and simple manner for the detection of persulfidated proteins, including papain, BSA, and GAPDH were developed. The method was then applied to human protein disulfide isomerase (PDI), leading to the discovery that persulfidation can occur at PDI's non-active site cysteines, and that PDI reductase activity is affected by sulfane sulfur treatment. Protocols for using SSP4 for the bioimaging of exogenous and endogenous sulfane sulfurs in different -cell lines were also established. These results should guide further applications of SSP4.
Keywords: SSP4, Sulfane sulfurs, Fluorescent probe, S-persulfidation
Graphical abstract
1. Introduction
Reactive sulfur species (RSS) are sulfur-containing molecules found in biological systems and include thiols, disulfides, hydrogen sulfide (H2S), and a variety of H2S-related sulfane sulfurs. Sulfane sulfurs (S0) refer to sulfur atoms with six valence electrons and no charge (typically bound covalently to other sulfur atoms). Biologically important sulfane sulfurs include persulfides (RSSH), hydrogen persulfide (H2S2), polysulfides (R-S-Sn-S-R), and protein-bound elemental sulfur (S8). While many studies have established that H2S is involved in multiple physiological processes, such as the mediation of neurotransmission, regulation of inflammation, relaxation of vascular smooth muscles, and cytoprotection against oxidative stress, sulfane sulfurs are less well understood [[1], [2], [3]]. H2S and sulfane sulfurs coexist, and recent work suggests that sulfane sulfur species may be the actual mediators in some biological phenomena previously thought to be mediated by H2S [[4], [5], [6]]. Thus, the development of chemical tools to detect sulfane sulfurs is essential to understanding their production, roles, and mechanisms of action in biological systems [[7], [8], [9]].
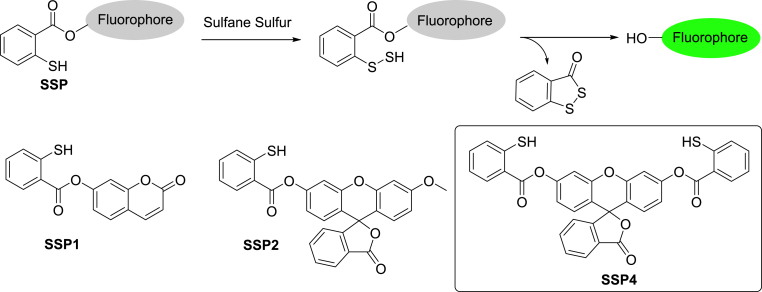
Techniques for sulfane sulfur detection have evolved over time. The first known method was based on the reaction of sulfane sulfur with cyanide ions to form thiocyanate, which can then be measured by absorption spectroscopy as ferric thiocyanate [10]. However, this method cannot be applied towards real-time detection in biological samples. As such, the development of sensitive and non-invasive fluorescent sensors for sulfane sulfurs in biological samples has emerged [9,11]. In 2013, our lab reported the first fluorescent probes for sulfane sulfurs: SSP1 and SSP2 [12]. Our design utilized the nucleophilic thiophenol moiety on the probe to attack the electrophilic sulfane sulfur, forming an –SSH intermediate that then rapidly underwent an intramolecular cyclization to release the fluorophore and turn on fluorescence (Scheme 1). While SSP1 and SSP2 showed promising activity for sulfane sulfurs, the low stability of SSP1 and moderate sensitivity of SSP2 warranted further optimizations. We envisioned that the sulfane sulfur recognition site was appropriate and a more suitable fluorophore was needed. We then selected free fluorescein as the fluorophore due to its excellent photophysical property and the high stability of its benzoate esters. This new probe, named SSP4, was first made in 2014 and used that same year by Ichinose et al. in lipopolysaccharide-treated human umbilical vein endothelial (HUVEC) cells to identify sulfide metabolites increased by sodium thiosulfate and in SH-SY5Y cells to determine that increased intracellular sulfur levels may be useful in protecting against neurodegenerative diseases [13,14]. Due to its excellent performance, SSP4 was later acquired by many researchers and successfully used in different biological settings (e.g. plants, bacteria, mammalian cells, tissues, human platelets, etc.), and those studies led to many peer-reviewed journal articles [15]. The popularity of SSP4 also made it the only commercialized sulfane sulfur probe by Dojindo Molecular Technologies, Inc.
Scheme 1.
The structures and reaction of SSP fluorescent probes.
Unlike traditional works on fluorescent probes (normally a paper reporting the development of the probe will be published first, and then the probe will be used in different systems), a comprehensive study describing the development and basic properties of SSP4 with evaluation of its applications under diverse conditions has never been reported, despite the fact that it is already a successful probe in the field. Herein, we report the rational design, preparation, and chemical/biochemical evaluation of SSP4. Protocols for using SSP4 for the detection of persulfidated proteins and to conduct bioimaging in cells are also provided. We expect this work will serve as a general guide for the use of SSP4.
2. Materials and methods
2.1. Materials and general methods
All reagents and solvents were of the highest grade available. Sodium disulfide was from Dojindo. Cell culture media (DMEM) and phenol-red free media (Fluorobrite DMEM) were from Gibco (Invitrogen; Darmstadt, Germany). Papain (from Carica papaya, #P76220), bovine serum albumin (BSA, # A2153), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, from rabbit muscle, #G2267) were obtained from Sigma-Aldrich. Recombinant human protein disulfide isomerase was provided by the Flaumenhaft Lab [16].
Chemical reactions were magnetically stirred and monitored by thin layer chromatography (TLC) with 0.25 mm pre-coated silica gel plates. Flash chromatography was performed with silica gel 60 (particle size 0.040–0.062 mm). 1H NMR and 13C NMR spectra were recorded by a Bruker 600 MHz NMR spectrometer. UV–vis spectra were recorded by a Thermo UV–Vis spectrophotometer. Fluorescence spectra were recorded by a Cary Eclipse spectrofluorometer or by a Molecular Devices SpectraMax iD3 Multi-Mode Microplate Reader.
2.2. Chemical synthesis and characterization of SSP4
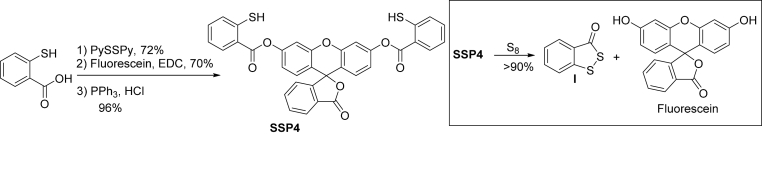
SSP4 was prepared in 3 steps from commercially available 2-mercapto benzoic acid (as shown in Scheme 2). Detailed synthetic protocols are provided in the SI. SSP4 was obtained as a white solid. m.p. 211–214 °C. 1H NMR (600 MHz, CDCl3) δ 8.28 (d, J = 8.0 Hz, 2H), 8.08 (d, J = 7.7 Hz, 1H), 7.88–7.58 (m, 2H), 7.42 (dt, J = 13.7, 7.6 Hz, 4H), 7.30–7.23 (m, 5H), 7.09–6.86 (m, 4H), 4.68 (s, 2H); 13C NMR (150 MHz, CDCl3) δ 169.2, 164.6, 153.1, 152.1, 151.6, 139.8, 135.3, 133.4, 132.3, 131.2, 130.1, 129.0, 126.0, 125.2, 124.9, 124.5, 124.1, 118.0, 116.7, 110.7, 81.6; HRMS calculated for C34H21O7S2 [M+H]+ 605.0729, found 605.0702.
Scheme 2.
Preparation and reaction mechanism of SSP4.
2.3. General fluorescence measurements
Probes were dissolved in DMSO to obtain a 1 mM stock solution. CTAB was dissolved in ethanol to yield a 5 mM stock solution. Analytes were dissolved in MilliQ water, 50 mM PBS (pH 7.4), or THF to make 10 mM stock solutions. Buffer was added to a 4.5 mL vial, followed by surfactant (25 μM), SSP4 (5 μM), and other species to a final volume of 4 mL. Samples were vortexed briefly and incubated in the dark at room temperature (rt) for 20 min before 3 mL was added to a 1 cm quartz cuvette and measured on a Cary Eclipse fluorescence spectrophotometer (emission 514 nm; excitation 482 nm). Samples measured on a Molecular Devices SpectraMax iD3 Multi-Mode Microplate Reader (emission 525 nm; excitation 485 nm; integration time 400s, low PMT sensitivity, attenuation 1, read height 1.00 mm) followed the same general procedure as above, though with a final volume of 200 μL added to a black polystyrene, flat-bottomed clear 96-well plate.
2.4. General protein-SSH measurements
Proteins were treated with Na2S, H2O2, or Na2S2 and purified by gel filtration on a PD-10 or Zeba 7K MWCO column. 50 mM PBS (pH 7.4) was added to a vial, followed by the protein and SSP4 (5 μM) to a final volume of 230–250 μL. Samples were mixed by pipetting, incubated for 30 min in the dark at rt, and then 200 μL was transferred to a black polystyrene, flat-bottomed clear 96-well plate. Samples were measured on a Molecular Devices SpectraMax iD3 Multi-Mode Microplate Reader (emission 525 nm; excitation 485 nm; integration time 400s, low PMT sensitivity, attenuation 1, read height 1.00 mm) to obtain relative fluorescence units (RFU).
2.5. General cell culturing and imaging protocol
2.5.1. HeLa cells
HeLa cells were seeded in a 48-well clear flat-bottomed plate at a density of 4 × 105 cells per well and cultured in DMEM supplemented with 10% FBS at 37 °C under 5% CO2 for 24 h. Culturing media was then aspirated, and live cells were washed twice with PBS. SSP4 (5 μM, 0.5% DMSO in serum-free Fluorobrite DMEM) was then added to the wells, and cells were incubated for 20 min at 37 °C under 5% CO2. The solution was removed post-incubation, and cells were washed once with PBS. Cetyltrimethylammonium bromide (CTAB) (100 μM) and Na2S2 (50 μM) in serum-free Fluorobrite DMEM were then added to the cells. Cells not treated with Na2S2 were used as negative controls. After 20 min incubation at 37 °C under 5% CO2, cells were washed once with PBS and then taken for fluorescence imaging using the Keyence All-in-One Fluorescence Microscope (BZ-X810) (excitation: 470/40 nm; emission: 525/50 nm).
2.5.2. COS-7 and HEK293T cells
COS-7 and HEK293T cells were seeded in eight-well glass chamber slides at a density of 4 × 105 cells per well. In some experiments, cells were pre-treated with cystathionine γ-lyase (CSE)-expression plasmid. The CSE overexpression and cysteinyl-tRNA synthetase 2 (CARS2) KO cells were produced according to the methods reported previously [17,18]. The cells were washed once with serum-free DMEM, followed by incubation with 20 μM SSP4 in serum-free DMEM containing 100 μM CTAB at 37 °C for 30 min. Cells not treated with probe were used as negative controls. After removing the excess probes from the cells and washing them with PBS, they were incubated in PBS for 30 min at 37 °C. Cells were then washed twice with PBS, followed by the measurement of fluorescence in images (excitation wavelength: 488 nm) with a confocal laser scanning microscope (Nikon C2 plus, NIS elements Version 5.01 software). Fluorescence intensity of images was calculated as fluorescence intensity per cell (a.u./cell) by using Image J software (National Institutes of Health) and dividing the intensity value by the number of cells.
2.6. Statistical analyses
Data are expressed as mean ± standard error of the mean (SEM) unless otherwise stated. Statistical evaluations were performed on GraphPad Prism with either one-way ANOVA or Student's t-test.
3. Results and discussion
3.1. Design, preparation, and mechanism evaluation of SSP4
Sulfane sulfur species are reactive, labile, and electrophilic. Our probe design exploits these properties with the inclusion of a thiophenol moiety, ester linker, and hydroxyl-containing fluorophore in all SSPs. As the electrophilic sulfane sulfurs have a special affinity to thiol-based nucleophiles, the nucleophilic thiophenol will attack the sulfane sulfur, forming an –SSH intermediate. This intermediate will then rapidly attack the pendant ester moiety, releasing the OH-containing fluorophore. Xanthene-based fluorophores were selected for this design as the –OH group in the xanthene core is an excellent optically tunable group. The acylation of –OH would lead to fluorescence quenching while the removal of the acyl group should restore the fluorescence. SSP1 and SSP2 used 7-hydroxycoumarin and 3-O-methylfluorescein as the fluorophores, respectively. SSP1 was found to be somewhat sensitive to hydrolysis while the sensitivity of SSP2 was moderate due to the release of partially quenched fluorescein. To improve these properties, we decided to use free fluorescein as the fluorophore due to its excellent photophysical properties including high quantum yield in aqueous solutions and photostability. Fluorescein-derived probes are widely used in cell imaging. Moreover, fluorescein-derived benzoate esters are quite stable even in the presence of esterases [19]. Since fluorescein contains two free hydroxyl groups, two sulfane sulfur recognition sites were needed, leading to the structure of SSP4. The preparation of SSP4 was straightforward (Scheme 2). The treatment of thiosalicylic acid and pyridyl disulfide followed by esterification with fluorescein and then disulfide reduction with PPh3 provided the desired SSP4 in excellent yields. The structure of SSP4 was fully characterized by 1H, 13C NMR and MS (see Supplementary).
With SSP4 in hand, we first tested its reaction with sulfane sulfurs to validate the reaction mechanism. Elemental sulfur (S8) was used as the model sulfane sulfur. The reaction was monitored by TLC and ESI-MS. When SSP4 was treated with an excess of S8, we observed the clean and almost quantitative formation of the cyclization product benzodithiolone and free fluorescein. These results indicated that both thiophenol sites of SSP4 could rapidly react with S8 and confirmed the mechanism shown in Scheme 1.
3.2. Fluorescence evaluation of SSP4
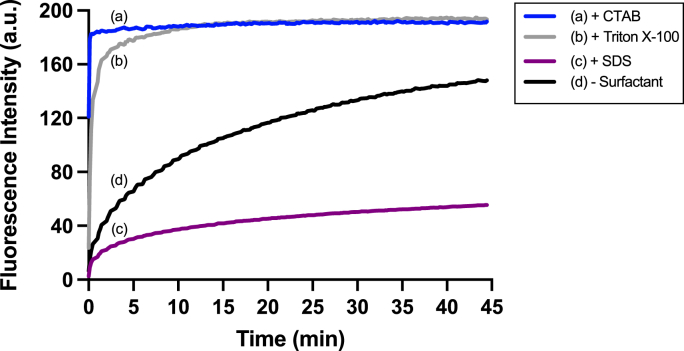
The fluorescence responses of SSP4 toward sulfane sulfurs were next evaluated in PBS buffer solution (50 mM, pH 7.4). Na2S2 was used as the equivalent of hydrogen persulfide (H2S2), a well-known sulfane sulfur model. The time-dependent response of SSP4 (5 μM) to Na2S2 (25 μM) was first assessed to determine optimal incubation times and fluorescence intensities. As shown in Fig. 1-d, SSP4 showed a quick and significant fluorescence ‘turn-on’ in the presence of H2S2, and the intensity reached a plateau at ∼45 min. Our previous work on fluorescent sensors revealed that the use of small amounts of surfactants as co-solvents in fluorescence measurements often significantly enhanced reaction rates and fluorescence intensity [12,19,20]. Thus, we then tested three surfactants: cationic CTAB, anionic SDS, and non-ionic Triton X-100 as co-solvents, all at 25 μM. CTAB was found to be the most effective and promoted a complete and high fluorescence turn-on in less than 1 min (Fig. 1-a). Triton X-100 was also effective, but the reaction was slightly slower (∼5–10 min) (Fig. 1-b). SDS, however, gave the opposite effect as it decreased the reaction rate and fluorescence intensity (Fig. 1-c). The effects of CTAB and Triton X-100 might be due to their ability to increase the solubility of SSP4 in aqueous buffers and to absorb or capture the target (HSS−) in micelles, thus facilitating the reaction. The anionic surfactant SDS may have repelled the HSS− from the probe and therefore, lowered the reactivity and fluorescence intensity. It should be noted that we also tested different concentrations of surfactants in this fluorescence turn-on process. SDS always yielded the lowest fluorescence intensities while CTAB and Triton X-100 worked similarly. Based on these results, we selected 25 μM CTAB and a 20 min incubation time as the optimal conditions for the remaining experiments for the purpose of reproducibility.
Fig. 1.
Time-dependent response of SSP4 (5 μM) to Na2S2 (25 μM) in 50 mM PBS buffer (pH 7.4) and (a) CTAB, (b) Triton X-100, (c) SDS (25 μΜ), or (d) no surfactant at room temperature. Emission: 514 nm, excitation: 482 nm.
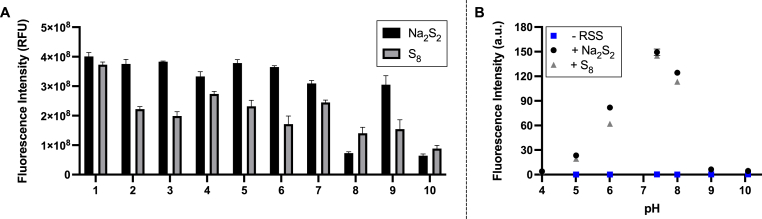
As SSP4 is expected to be applied in different solvent systems for biological tests, the responses of the probe in the presence of various buffers toward both Na2S2 and S8 were assessed (Fig. 2A). These included 50 mM PBS (pH 7.4), 10 mM PBS (pH 7.4), 50 mM Tris buffer (pH 7.4), 50 mM sodium phosphate buffer (pH 7.4), 50 mM potassium phosphate buffer (pH 7.0), 50 mM HEPES, Fluorobrite DMEM, Fluorobrite DMEM + 10% fetal bovine serum (FBS), phenol red-containing DMEM, and phenol red-containing DMEM + 10% FBS. Understandably, solvents with phenol red and FBS yielded the lowest fluorescence responses. The former is known to interfere with and quench fluorescence signals while the latter observably lowered the solubility of the sulfane sulfurs and probe. The lower fluorescence intensities of S8 in comparison with Na2S2 were also noted. S8 is the most stable sulfane sulfur compound, so it should have weaker reactivity to the probe as compared to H2S2. Solubility is another issue. S8 is insoluble in water, so it was dissolved in THF first and then diluted in the desired buffers. Precipitate was observed after the dilution and incubation with non-50 mM PBS (pH 7.4) buffers. 50 mM PBS (pH 7.4) was determined to be the ideal buffer for both Na2S2 and S8, so this buffer was used for additional evaluations.
Fig. 2.
Fluorescence responses of SSP4 (5 μM) toward Na2S2 or S8 (25 μM) in (A) different buffers: (1) 50 mM PBS, pH 7.4; (2) 10 mM PBS, pH 7.4; (3) 50 mM Tris buffer, pH 7.4; (4) 50 mM sodium phosphate buffer, pH 7.4; (5) 50 mM potassium phosphate buffer, pH 7.0; (6) 50 mM HEPES (7) Fluorobrite DMEM; (8) Fluorobrite DMEM + 10% FBS; (9) DMEM; (10) DMEM + 10% FBS. (B) different pHs in 50 mM PBS (pH 7.4). CTAB (25 μM) was added to all reactions, and reactions were carried out for 20 min at room temperature. Results are expressed as mean ± SEM (n = 3).
To evaluate the potential applications of SSP4 in different biological environments, we studied the effects of pH on the probe (Fig. 2B). The probe was less effective under acidic pHs (4–6), and this can be explained by the decreased nucleophilicity of the –SH residue on the probe under lower pHs. Interestingly, more basic conditions (pH 9–10) also yielded very low fluorescence. This might be due to the decreased electrophilicity of the sulfane sulfur species under basic conditions, which led to weak or no reaction with the probe. SSP4 worked optimally under normal biological pH ranges (ca. 6.5–8). The probe also appeared to be quite resistant to hydrolysis even under pH 9–10 as no fluorescence was observed by itself under those conditions.
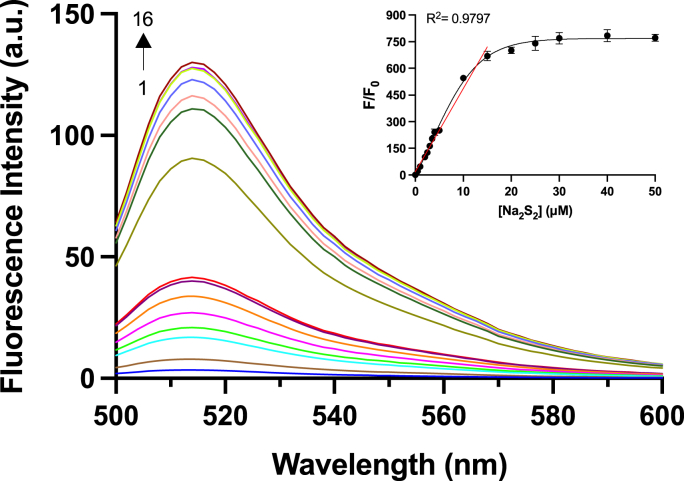
To further understand the efficiency of SSP4 in determining sulfane sulfur concentration, varied concentrations of Na2S2 (0–50 μM) were tested with SSP4 (5 μM) (Fig. 3). The fluorescence intensity increased linearly up to 15 μM before reaching a steady state. The detection limit of SSP4 was calculated to be 28 nM. This confirms that SSP4 is highly sensitive and suitable for detecting sulfane sulfurs in biological systems.
Fig. 3.
The fluorescence emission spectra of SSP4 (5 μM) with varied concentrations of Na2S2 (0, 0.5, 1, 1.5, 2, 3, 5, 10, 15, 25, 30, 50 μM) for curves 1–16, respectively. Results are expressed as mean ± SEM (n = 3).
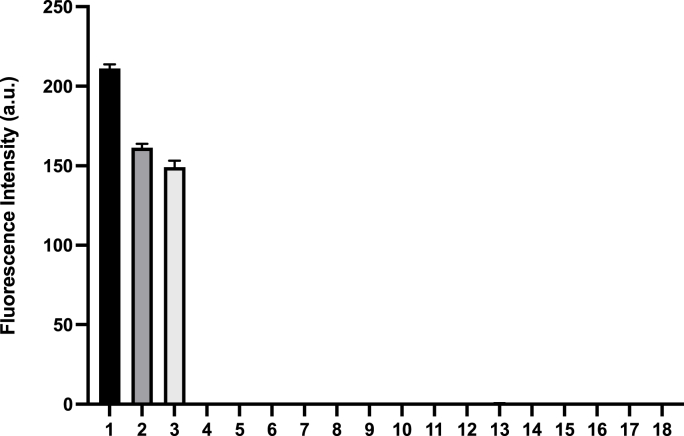
Next, the selectivity of SSP4 for sulfane sulfurs over other species was evaluated (Fig. 4). Three representative sulfane sulfur species were Na2S2, S8, and cysteine-polysulfide (Cys-polysulfide), and they all gave greater than 900-fold response. In contrast, other biologically relevant sulfur species tested, including cysteine (Cys), glutathione (GSH), homocysteine (Hcy), methionine (Met), and N-acetyl-l-cysteine (NAC), oxidized glutathione (GSSG), Na2SO4, Na2S2O3, Na2SO3, and Na2S, yielded no significant fluorescence enhancement. We also tested amino acids such as glycine (Gly), tyrosine (Tyr), tryptophan (Trp), and arginine (Arg) and did not observe any response. These results indicated the high selectivity of SSP4 to sulfane sulfurs.
Fig. 4.
Mean fluorescence response of SSP4 (5 μM) in the presence of: (1) 25 μM cysteine polysulfide (Cys-polysulfide); (2) 25 μM sodium disulfide (Na2S2); (3) 25 μM elemental sulfur (S8); (4) 1 mM l-cysteine (Cys); (5) 1 mM glutathione (GSH); (6) 1 mM homocysteine (Hcy); (7) 30 μΜ methionine (Met); (8) 100 μM N-acetyl-l-cysteine (NAC); (9) 100 μM oxidized glutathione (GSSG); (10) 100 μM sodium sulfate (Na2SO4); (11) 100 μM sodium thiosulfate (Na2S2O3); (12) 100 μM sodium sulfite (Na2SO3); (13) 100 μM sodium sulfide (Na2S); (14) 150 μM glycine (Gly); (15) 55 μM tyrosine (Tyr); (16) 40 μM tryptophan (Trp); (17) 100 μM arginine (Arg); (18) probe only. Results are expressed as mean ± SEM (n = 3).
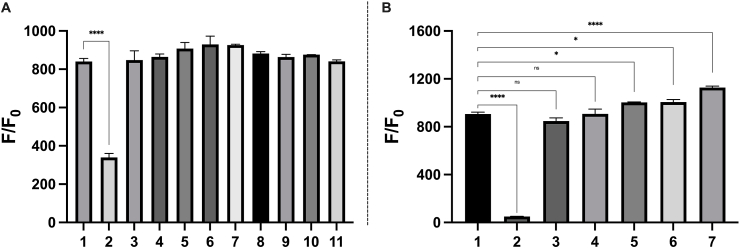
While SSP4 showed high selectivity for sulfane sulfurs presented individually, real detection systems would likely include other species. Thus, interference assays were conducted to investigate the specificity of SSP4 for sulfane sulfurs in the presence of selected other species at biologically relevant concentrations (Fig. 5A). As expected, there was no statistically significant difference between the SSP4 and S8 positive control condition with the non-sulfur containing amino acids: Gly, Arg, Trp, Tyr. Except for 1 mM GSH, there was also no statistical significance in fluorescence intensities between the control and the sulfur-containing amino acids. To investigate whether the significant interference, or decrease, in fluorescence intensity due to the presence of GSH was attributable to concentration, the experiment was repeated with 100 μM GSH and resulted in a strong fluorescence, comparable to that of the positive control. The issue caused by the high concentration of GSH can be explained by two possibilities: 1) GSH at high concentrations could rapidly react with the small amounts of S8 and thus consume the analyte, 2) even when SSP4 reacts with S8, the persulfide intermediate could be depleted by the large excess of GSH and would not lead to productive fluorophore release. Yet, despite the differences in fluorescence enhancement between the S8 only and S8 with 1 mM GSH conditions, >300-fold fluorescence enhancement was still observed in the latter condition, demonstrating that SSP4 is still highly sensitive to sulfane sulfurs.
Fig. 5.
Mean fluorescence enhancements of SSP4 (5 μM) with 25 μM S8 in the presence of (A) various analytes: (1) control; (2) 1 mM GSH; (3) 200 μΜ Ηcy; (4) 30 μM methionine (Met); (5) 100 μM Cys; (6) 100 μM GSH; (7) 100 μM N-acetyl-l-cysteine (NAC); (8) 150 μM glycine (Gly); (9) 55 μM tyrosine (Tyr); (10) 40 μM tryptophan (Trp), (11) 100 μM arginine (Arg). (B) metal ions: 1) control; (2) CuSO4; (3) FeCl2; (4) FeCl3; (5) MgCl2; (6) CaCl2; (7) ZnCl2 at 20 equivalents (500 μΜ) to the 25 μM S8. Results are expressed as mean ± SEM (n = 3). Statistical analyses were performed using one-way ANOVA. ****P < 0.0001, *P = 0.0326–0.0431, ns = not significant.
Besides organic molecules, many biological models also contain metal ions. Metal ions are essential components with catalytic or structural functions in more than one-third of all cellular proteins [21]. To examine their possible interference with SSP4 detection, essential metal cations (Cu2+, Fe2+, Fe3+, Mg2+, Ca2+, Zn2+) were tested at relevant concentrations (500 μM) to S8 (Fig. 5B). As expected, Cu2+ yielded a significantly inhibitory effect, possibly because Cu2+ can coordinate with the thiol groups on SSP4, preventing SSP4 from reacting with the electrophilic sulfane sulfur. Fe2+ and Fe3+ did not exhibit inhibitory or enhancement effects. Interestingly, the additions of Mg2+, Ca2+, and Zn2+ resulted in an increased fluorescence response, suggesting that SSP4 may fluoresce more brilliantly in biological systems that naturally have higher concentrations of these metals.
Our results thus far have demonstrated the sensitivity and specificity of SSP4 for small molecule sulfane sulfurs. Whether or not SSP4 is capable of detecting protein-bound sulfane sulfurs is less clear, and the protocol for such measurements has not been established. We next turned to address this question. S-Persulfidated papain was prepared following a published protocol and used as the first model in our studies [22]. After monitoring the time-dependent fluorescence response of SSP4 with different concentrations of persulfidated papain over time, we found that 30 min was sufficient for the enhanced fluorescence of the probe. As proteins possess different micro-environments from small molecules in aqueous solutions, their reactions with SSP4 might be different as well. We tested if CTAB was necessary for protein assays. Interestingly, the fluorescence intensity of SSP4 in the presence of persulfidated papain and with CTAB resulted in a 5-fold fluorescence enhancement, but the condition without CTAB yielded a 15-fold fluorescence enhancement (Fig. S1). While the use of surfactants like CTAB at high concentrations (∼500 μM) can denature proteins [23], this may not happen at the low concentrations used in our studies. If denaturation occurred, the persulfide residue would be more accessible for SSP4, which could promote the reaction. However, denaturation would also destabilize the persulfide residue, thus preventing the reaction. On the other hand, proteins in their natural structures may have suitable hydrophobic microenvironments that could 1) stabilize the persulfides and 2) facilitate the reactions with SSP4. Thus, we decided not to use CTAB in the following protein-based studies.
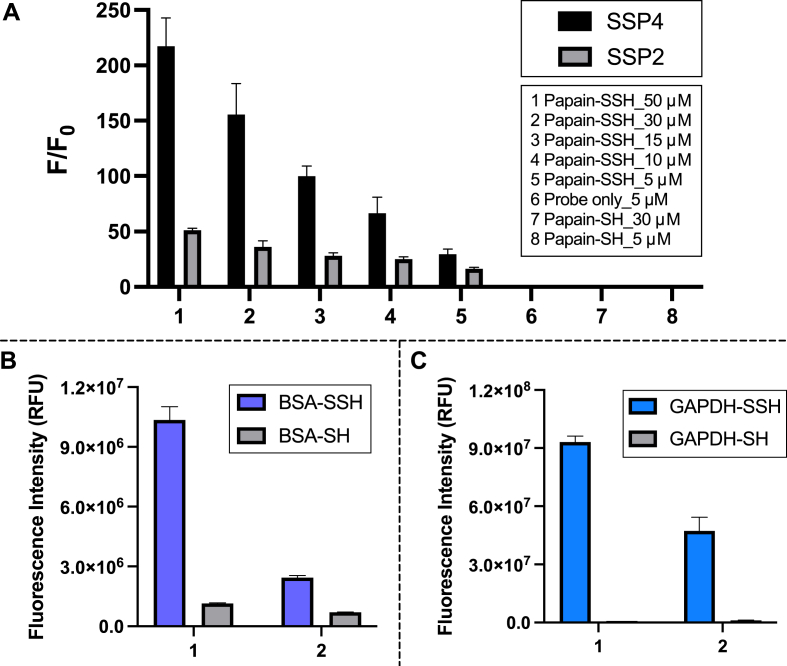
Considering SSP4 contains two sulfane sulfur reaction sites, it theoretically needs to react with two molecules of sulfane sulfurs to fully release the fluorophore. This should be easily achievable with small molecule sulfane sulfurs, but it was unclear if protein-based sulfane sulfurs could do the same. To address this question, we compared the fluorescence responses of SSP4 with SSP2 (which only contains one reaction site). Various concentrations of persulfidated papain and unpersulfidated papain (5–50 μM) were incubated with SSP4 or SSP2 (5 μM), and their fluorescence intensities were recorded (Fig. 6A). Interestingly, the fluorescence enhancement from SSP4 was still much higher than that of SSP2, and no fluorescence was detected in reduced protein. These results demonstrated that SSP4 is more sensitive than SSP2 for protein substrates.
Fig. 6.
(A) Fluorescence enhancement (F/F0) of 5 μM SSP4 or SSP2 in the presence of varied concentrations of persulfidated papain. (B)/(C) Fluorescence intensity of 5 μM SSP4 for the detection of various concentrations of persulfidated and reduced BSA or GAPDH at (1) 30 μM and (2) 5 μM. Results are expressed as mean ± SEM (n = 3).
Using this protocol, two other protein models (BSA and GAPDH) were also examined to encompass proteins of different sizes and functions. As expected, persulfidated BSA and GAPDH yielded significantly higher fluorescence signals than their reduced forms, though persulfidation efficacy was different between the models (Fig. 6B–C). BSA treated with H2O2 and Na2S yielded ∼10-fold and 4-fold fluorescence enhancement at 30 μM and 5 μM relative to untreated BSA, respectively. GAPDH treated with Na2S2 yielded ∼100-fold and ∼40-fold fluorescence enhancement at 30 μM and 5 μM protein, respectively, relative to the untreated protein.
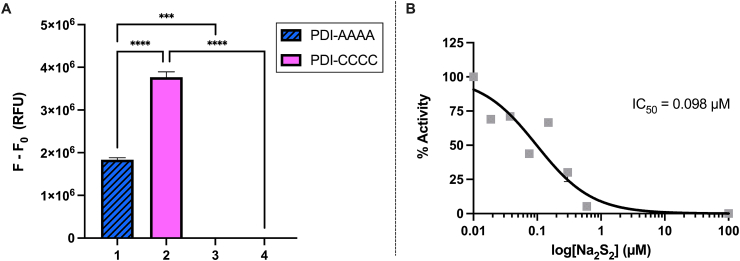
The successful application of our protocol on three protein models established that SSP4 is effective at detecting protein persulfidation and prompted us to study a protein with reactive cysteines that has not previously been evaluated for persulfidation. Protein disulfide isomerases (PDIs) are thiol isomerases that promote proper protein folding in the endoplasmic reticulum by catalyzing disulfide bond oxidation, reduction, and isomerization in proteins and acting as chaperones to aid in folding in the absence of catalytic activity [24]. The archetypical PDI, human PDIA1 (encoded by the P4HB gene), has a domain structure of a-b-b’-a’ and is of particular interest due to its role in the formation and rearrangement of disulfide bonds in cells [25]. However, it is not known whether or not cellular sulfane sulfurs cause PDI persulfidation or affect its activity. PDI possesses two CGHC motifs that contain active site cysteines that are sensitive to redox environments and undergo a variety of cysteine modifications [24,26]. PDI functions in both normal physiology and in several disease states, including thrombosis, cancer, Parkinson's disease, Alzheimer's disease, amyotrophic lateral sclerosis (ALS), prion related disorders, and Huntington's disease [24,27,28]. A better understanding of the post-translational modifications of PDI will lead to a greater understanding of potential treatments for these biological problems. The SSP4 protocol was applied to recombinant PDI (Fig. 7A). Reduced wild type human PDI (PDI-CCCC) has free thiols at C53 and C56 within the CGHC of the a domain and at C397 and C400 of the a’ domain. The mutant PDI (PDI-AAAA) has C53A, C56A, C397A, and C400A mutations. Both PDI-CCCC and PDI-AAAA were treated with Na2S2. SSP4 was then added to the treated and untreated (reduced) samples. Only samples exposed to Na2S2 showed a fluorescence signal. Interestingly, the mutant PDI in which all active site cysteines of its two CGHC motifs were mutated to alanine showed a significant fluorescence response (as compared to its control), suggesting that persulfidation is not restricted to active site cysteines and may occur at free cysteines such as C312 and C343 that are located in the b’ domain [29]. Nevertheless, Na2S2 treatment of wild type (PDI-CCCC) yielded much stronger fluorescence than that of the mutant (PDI-AAAA), indicating that active site cysteines were also persulfidated. Since active site cysteines were persulfidated, we evaluated whether this modification affected PDI enzymatic activity. A di-eosin-GSSG assay was conducted to monitor PDI reductase activity and demonstrated potent, dose-dependent inhibition by Na2S2 (Fig. 7B), confirming PDI persulfidation and demonstrating its effect on PDI reductase activity. Overall, these results demonstrate a relatively simple and quick method of detecting persulfidated proteins using SSP4 (see Fig. 8).
Fig. 7.
A) Fluorescence intensity difference between 97 μM Na2S2-treated (1) PDI-AAAA or (2) PDI-CCCC and untreated (3) PDI-AAAA or (4) PDI-CCCC. SSP4 (5 μM) was incubated with the proteins (5 μM) for 30 min at room temperature in the dark. Results are expressed as mean ± SEM (n = 3). Statistical analyses were performed using one-way ANOVA. ****P < 0.0001, ***P = 0.0001. B) Inhibition of PDI reductase activity by Na2S2. PDI (0.2 uM) was incubated with the indicated concentration of Na2S2 and assayed for reductase activity using a di-eosin-GSSG assay.
Fig. 8.
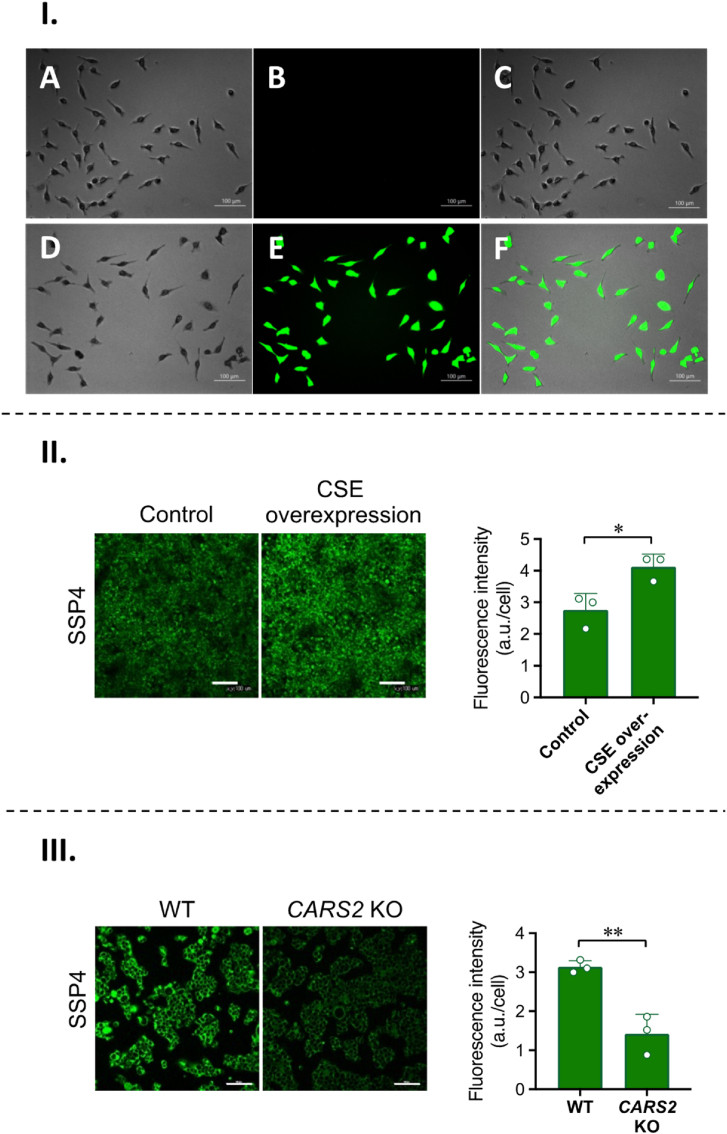
Representative cell images with SSP4. (I) Brightfield (A, D), fluorescence (B, E), and overlaid images (C, F) of Η2S2 in HeLa cells. Cells were first treated with 5 μΜ of SSP4, then washed and subjected to (A–C) No Na2S2 added, (D–F) 50 μM Na2S2. (II) COS-7 cells overexpressing CSE and WT and (III)CARS2 KO HEK293T were subjected to fluorescence staining. Fluorescence images (left panel) and fluorescence intensities (right panel) of SSP4 staining. Scale bars, 100 μm. Data are mean values ± S.D (from three different images, each containing more than 100 cells in a representative experiment). *P < 0.05, **P < 0.01; Student's t-test.
Finally, we tested conditions for the detection of sulfane sulfurs in cells using SSP4. The cytotoxicity of SSP4 was first evaluated by the CCK-8 assay. HeLa cells were incubated with various concentrations (5, 20, 50 100, 150 μM) of SSP4 for 4 h. This time was chosen because SSP4 applications typically do not exceed 1 h. Results demonstrated that SSP4 at these concentrations and for this duration in cells was not cytotoxic. As 5 μM SSP4 was used throughout the previous investigations, cell viability was also assessed at this concentration over 24 h. SSP4 did not significantly affect cell viability either (Fig. S2). We next imaged HeLa cells following exposure to 5 μM SSP4. After a 20 min incubation, cells were washed with PBS and 50 μM Na2S2 was added (Fig. 8-I). After another 20 min incubation, cells were washed with PBS before imaging on the Keyence All-in-One Fluorescence Microscope (BZ-X810). The resulting data indicates that a low concentration of SSP4 generates strong fluorescence in the presence of Na2S2, demonstrating that the probe is highly sensitive for the intracellular detection of sulfane sulfurs.
After determining that SSP4 could be used for exogenous sulfane sulfur detection, SSP4 was also applied to detect the endogenous production of sulfane sulfurs. COS-7 cells overexpressing CSE, CARS2 KO HEK293T cells, and wild type controls were subjected to SSP4 treatment (Fig. 8-II and III). As expected, the overexpression of CSE led to greater fluorescence than that of the control due to the increased production of persulfides, and the knockout of CARS2 reduced fluorescence intensities due to the decreased production of CysSSH [18]. This demonstrates that SSP4 is able to sensitively detect the differences in sulfane sulfur production within cells. Moreover, we also evaluated the application of SSP4 in sensing sulfane sulfurs in cell lysates. Briefly, HeLa cell lysates spiked with different concentrations of Na2S2 (1–100 μM) were treated with SSP4 (5 μM). Concentration-dependent fluorescence increases were noted in the range from 1 to 50 μM (Fig. S3).
4. Conclusions
We report in this study the design, preparation, characterization, fluorescence analysis, and applications evaluation of SSP4. The sulfane sulfur-mediated benzodithiolone formation and fluorescein release reaction proved to be highly selective for small molecule sulfane sulfur species including persulfide, polysulfide, and elemental sulfur. The reaction did not proceed with other biologically relevant sulfur species, such as cysteine, glutathione (both reduced and oxidized), homocysteine, methionine, N-acetyl-l-cysteine, sulfate, thiosulfate, sulfite, and H2S, nor with non-sulfur-containing species, such as glycine, tyrosine, tryptophan, and arginine. SSP4 also demonstrated high sensitivity for detecting sulfane sulfurs even with the presence of other species, such as homocysteine, methionine, cysteine, glutathione, N-acetyl-l-cysteine, glycine, tyrosine, tryptophan, arginine, and metal ions (Fe2+, Fe3+, Mg2+, Ca2+, and Zn2+). Fluorescence was inhibited by the presence of Cu2+, likely due to the cation's coordination with the thiol groups on SSP4. The efficacy of SSP4 towards protein-bound sulfane sulfurs was also compared to that of SSP2 and demonstrated that SSP4 was much more sensitive for detecting persulfidated proteins than SSP2 despite having two reaction sites. After determining the efficacy of SSP4 at detecting protein persulfidation on three different protein models (papain, BSA, GAPDH), SSP4 was applied to study human recombinant PDI, a protein with reactive cysteines that had not previously been evaluated for persulfidation. Results led to the discovery that persulfidation may not be restricted to PDI's active site cysteines, and that PDI reductase activity is affected by Na2S2 treatment. Overall, conditions and protocols were optimized for SSP4 to detect protein-bound sulfane sulfurs in a relatively quick and simple manner. The ability of SSP4 to detect intracellular sulfane sulfurs, both exogenously and endogenously, in a variety of cell lines (HeLa, COS-7, and HEK293T) was also evaluated and determined to be efficient and sensitive, even with low concentrations of the probe. All these results demonstrate that SSP4 has a wide application for sulfane sulfur detection.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by NSF (CHE2100870) and NIH (R01GM125968) to M.X., R35HL135775 and 5U01HL143365 to R.F., American Society of Hematology Scholar Award to M.Y., Grants-in-Aid for [Transformative Research Areas (A), Scientific Research (S) and (C) and Challenging Research (Exploratory)] from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, to T.A. (18H05277, 21H05258, 21H05263and 22K19397) and T.M. (19K07554 and 22K06893); Japan Science and Technology Agency (JST), CREST Grant Number JPMJCR2024 to T.A., Japan.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102433.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Kimura H. Role of hydrogen sulfide and polysulfide signaling regulates activity of ion channels and enzymes as well as growth of tumors in patients. Biomolecules. 2021;11:896. [Google Scholar]
- 2.Filipovic M.R., Zivanovic J., Alvarez B., Banerjee R. Chemical biology of H2S signaling through persulfidation. Chem. Rev. 2018;118:1253–1337. doi: 10.1021/acs.chemrev.7b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toohey J.I. Sulfur signaling: is the agent sulfide or sulfane? Anal. Biochem. 2011;413(1):1–7. doi: 10.1016/j.ab.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 4.Greiner R., Pálinkás Z., Bäsell K., Becher D., Antelmann H., Nagy P., Dick T.P. Polysulfides link H2S to protein thiol oxidation. Antioxidants Redox Signal. 2013;19:1749–1765. doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ide M., Ohnishi T., Toyoshima M., Balan S., Maekawa M., Shimamoto-Mitsuyama C., Iwayama Y., Ohba H., Watanabe A., Ishii T., Shibuya N., Kimura Y., Hisano Y., Murata Y., Hara T., Morikawa M., Hashimoto K., Nozaki Y., Toyota T., Wada Y., Tanaka Y., Kato T., Nishi A., Fujisawa S., Okano H., Itokawa M., Hirokawa N., Kunii Y., Kakita A., Yabe H., Iwamoto K., Meno K., Katagiri T., Dean B., Uchida K., Kimura H., Yoshikawa T. Excess hydrogen sulfide and polysulfides production underlies a schizophrenia pathophysiology. EMBO Mol. Med. 2019;11(12) doi: 10.15252/emmm.201910695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyamoto R., Koike S., Takano Y., Shibuya N., Kimura Y., Hanaoka K., Urano Y., Ogasawara Y., Kimura H. Polysulfides (H2Sn) produced from the interaction of hydrogen sulfide (H2S) and nitric oxide (NO) activate TRPA1 channels. Sci. Rep. 2017;7(1) doi: 10.1038/srep45995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin V.S., Chen W., Xian M., Chang C.J. Chemical probes for molecular imaging and detection of hydrogen sulfide and reactive sulfur species in biological systems. Chem. Soc. Rev. 2015;44:4596–4618. doi: 10.1039/c4cs00298a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takata T., Jung M., Matsunaga T., Ida T., Morita M., Motohashi H., Shen X., Kevil C.G., Fukuto J.M., Akaike T. Methods in sulfide and persulfide research. Nitric Oxide. 2021;116:47–64. doi: 10.1016/j.niox.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echizen H., Sasaki E., Hanaoka K. Recent advances in detection, isolation, and imaging techniques for sulfane sulfur-containing biomolecules. Biomolecules. 2021;11(11) doi: 10.3390/biom11111553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood J.L. Sulfane sulfur. Methods Enzymol. 1987;143:25–29. doi: 10.1016/0076-6879(87)43009-7. Academic Press. [DOI] [PubMed] [Google Scholar]
- 11.Takano Y., Shimamoto K., Hanaoka K. Chemical tools for the study of hydrogen sulfide (H2S) and sulfane sulfur and their applications to biological studies. J. Clin. Biochem. Nutr. 2016;58(1):7–15. doi: 10.3164/jcbn.15-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W., Liu C., Peng B., Zhao Y., Pacheco A., Xian M. New fluorescent probes for sulfane sulfurs and the application in bioimaging. Chem. Sci. 2013;4(7):2892–2896. doi: 10.1039/c3sc50754h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakaguchi M., Marutani E., Shin H.-s., Chen W., Hanaoka K., Xian M., Ichinose F. Sodium thiosulfate attenuates acute lung injury in mice. Anesthesiology. 2014;121(6):1248–1257. doi: 10.1097/aln.0000000000000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marutani E., Sakaguchi M., Chen W., Sasakura K., Liu J., Xian M., Hanaoka K., Nagano T., Ichinose F. Cytoprotective effects of hydrogen sulfide-releasing N-methyl-D-aspartate receptor antagonists are mediated by intracellular sulfane sulfur. MedChemComm. 2014;5(10):1577–1583. doi: 10.1039/C4MD00180J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibli S.-I., Luck B., Zukunft S., Wittig J., Chen W., Xian M., Papapetropoulos A., Hu J., Fleming I. A selective and sensitive method for quantification of endogenous polysulfide production in biological samples. Redox Biol. 2018;18:295–304. doi: 10.1016/j.redox.2018.07.016. For selected examples, see a) [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nelp M.T., Zheng V., Davis K.M., Stiefel K.J.E., Groves J.T. Potent activation of indoleamine 2,3-dioxygenase by polysulfides. J. Am. Chem. Soc. 2019;141(38):15288–15300. doi: 10.1021/jacs.9b07338. [DOI] [PubMed] [Google Scholar]; c) Marutani E., Morita M., Hirai S., Kai S., Grange R.M.H., Miyazaki Y., Nagashima F., Traeger L., Magliocca A., Ida T., Matsunaga T., Flicker D.R., Corman B., Mori N., Yamazaki Y., Batten A., Li R., Tanaka T., Ikeda T., Nakagawa A., Atochin D.N., Ihara H., Olenchock B.A., Shen X., Nishida M., Hanaoka K., Kevil C.G., Xian M., Bloch D.B., Akaike T., Hindle A.G., Motohashi H., Ichinose F. Sulfide catabolism ameliorates hypoxic brain injury. Nat. Commun. 2021;12(1):3108. doi: 10.1038/s41467-021-23363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Olson K.R., Gao Y., Arif F., Arora K., Patel S., DeLeon E.R., Sutton T.R., Feelisch M., Cortese-Krott M.M., Straub K.D. Metabolism of hydrogen sulfide (H2S) and production of reactive sulfur species (RSS) by superoxide dismutase. Redox Biol. 2018;15:74–85. doi: 10.1016/j.redox.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Olson K.R., Briggs A., Devireddy M., Iovino N.A., Skora N.C., Whelan J., Villa B.P., Yuan X., Mannam V., Howard S., Gao Y., Minnion M., Feelisch M. Green tea polyphenolic antioxidants oxidize hydrogen sulfide to thiosulfate and polysulfides: a possible new mechanism underpinning their biological action. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bekendam R.H., Iyu D., Passam F., Stopa J.D., De Ceunynck K., Muse O., Bendapudi P.K., Garnier C.L., Gopal S., Crescence L., Chiu J., Furie B., Panicot-Dubois L., Hogg P.J., Dubois C., Flaumenhaft R. Protein disulfide isomerase regulation by nitric oxide maintains vascular quiescence and controls thrombus formation. J. Thromb. Haemostasis. 2018;16(11):2322–2335. doi: 10.1111/jth.14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., Yamamoto M., Ono K., Devarie-Baez N.O., Xian M., Fukuto J.M., Akaike T. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA. 2014;111(21):7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akaike T., Ida T., Wei F.-Y., Nishida M., Kumagai Y., Alam M.M., Ihara H., Sawa T., Matsunaga T., Kasamatsu S., Nishimura A., Morita M., Tomizawa K., Nishimura A., Watanabe S., Inaba K., Shima H., Tanuma N., Jung M., Fujii S., Watanabe Y., Ohmuraya M., Nagy P., Feelisch M., Fukuto J.M., Motohashi H. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017;8(1):1177. doi: 10.1038/s41467-017-01311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng B., Chen W., Liu C., Rosser E.W., Pacheco A., Zhao Y., Aguilar H.C., Xian M. Fluorescent probes based on nucleophilic substitution-cyclization for hydrogen sulfide detection and bioimaging. Chemistry. 2014;20(4):1010–1016. doi: 10.1002/chem.201303757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C., Chen W., Shi W., Peng B., Zhao Y., Ma H., Xian M. Rational design and bioimaging applications of highly selective fluorescence probes for hydrogen polysulfides. J. Am. Chem. Soc. 2014;136(20):7257–7260. doi: 10.1021/ja502968x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Finley E.J., Chakraborty S., Fretham S.J.B., Aschner M. Cellular transport and homeostasis of essential and nonessential metals. Metallomics. 2012;4(7):593–605. doi: 10.1039/c2mt00185c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan J., Carroll K.S. Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem. Biol. 2013;8(6):1110–1116. doi: 10.1021/cb4001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vlasova I.M., Zhuravleva V.V., Saletskii A.M. Denaturation of bovine serum albumin under the action of cetyltrimethylammonium bromide, according to data from fluorescence analysis. Russ. J. Phys. Chem. A. 2013;87(6):1027–1034. doi: 10.1134/S0036024413060319. [DOI] [Google Scholar]
- 24.Yang M., Flaumenhaft R. Oxidative cysteine modification of thiol isomerases in thrombotic disease: a hypothesis. Antioxidants Redox Signal. 2021;35(13):1134–1155. doi: 10.1089/ars.2021.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furie B., Flaumenhaft R. Thiol isomerases in thrombus formation. Circ. Res. 2014;114(7):1162–1173. doi: 10.1161/CIRCRESAHA.114.301808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bekendam R.H., Iyu D., Passam F., Stopa J.D., De Ceunynck K., Muse O., Bendapudi P.K., Garnier C.L., Gopal S., Crescence L., Chiu J., Furie B., Panicot-Dubois L., Hogg P.J., Dubois C., Flaumenhaft R. Protein disulfide isomerase regulation by nitric oxide maintains vascular quiescence and controls thrombus formation. J. Thromb. Haemostasis. 2018;16(11):2322–2335. doi: 10.1111/jth.14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell L.E., Foster P.A. Protein disulphide isomerase inhibition as a potential cancer therapeutic strategy. Cancer Med. 2021;10(8):2812–2825. doi: 10.1002/cam4.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreu C.I., Woehlbier U., Torres M., Hetz C. Protein disulfide isomerases in neurodegeneration: from disease mechanisms to biomedical applications. FEBS Lett. 2012;586(18):2826–2834. doi: 10.1016/j.febslet.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 29.Ogura J., Ruddock L.W., Mano N. Cysteine 343 in the substrate binding domain is the primary S-Nitrosylated site in protein disulfide isomerase. Free Radic. Biol. Med. 2020;160:103–110. doi: 10.1016/j.freeradbiomed.2020.07.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.