Abstract
Acetyl-coenzyme A (acetyl-CoA) synthetase (ADP forming) represents a novel enzyme in archaea of acetate formation and energy conservation (acetyl-CoA + ADP + Pi → acetate + ATP + CoA). Two isoforms of the enzyme have been purified from the hyperthermophile Pyrococcus furiosus. Isoform I is a heterotetramer (α2β2) with an apparent molecular mass of 145 kDa, composed of two subunits, α and β, with apparent molecular masses of 47 and 25 kDa, respectively. By using N-terminal amino acid sequences of both subunits, the encoding genes, designated acdAI and acdBI, were identified in the genome of P. furiosus. The genes were separately overexpressed in Escherichia coli, and the recombinant subunits were reconstituted in vitro to the active heterotetrameric enzyme. The purified recombinant enzyme showed molecular and catalytical properties very similar to those shown by acetyl-CoA synthetase (ADP forming) purified from P. furiosus.
Acetyl-coenzyme A (acetyl-CoA) synthetase (ADP forming) (acetyl-CoA + ADP + Pi ⇄ acetate + ATP + CoA) has been detected in the domain Eucarya, in the protists Entamoeba histolytica and Giardia lamblia, where it is involved in acetate formation and ATP production in the course of fermentative metabolism (15, 16). In prokaryotes, this unusual synthetase was first described in detail in the hyperthermophilic archaeon Pyrococcus furiosus (17), where it represents the major energy-conserving reaction during pyruvate and sugar conversion to acetate (19). Later studies demonstrated the presence of acetyl-CoA synthetase (ADP forming) in all acetate-forming Archaea tested, including anaerobic hyperthermophiles and mesophilic aerobic halophiles (18, 19). In contrast, all acetate-forming Bacteria studied so far, including the hyperthermophile Thermotoga maritima, utilize two almost “classical” enzymes, phosphate acetyltransferase and acetate kinase, for acetate formation and ATP synthesis (3, 19). Thus, acetyl-CoA synthetase (ADP forming) represents a novel enzyme in prokaryotes, restricted to the domain of Archaea, catalyzing acetate formation and ATP synthesis via the mechanism of substrate-level phosphorylation.
Recently, Glasemacher et al. (7) purified and characterized acetyl-CoA synthetase (ADP forming) from P. furiosus. The native enzyme is a heterotetramer (α2β2) with an apparent molecular mass of 145 kDa, composed of two subunits, α and β, with apparent molecular masses of 47 and 25 kDa, respectively. The enzyme exhibits a high optimum temperature (90°C) and thermostability in accordance with its physiological function under hyperthermophilic conditions. Independently, Mai and Adams (14) purified two distinct isoforms of acetyl-CoA synthetase (ADP forming) from P. furiosus. The isoforms had similar molecular properties but showed different N-terminal amino acid sequences for the α and β subunits and different substrate specificities and kinetic properties. On the basis of substrate specificities and N-terminal amino acid sequences of the subunits, the enzyme purified by Glasemacher et al. (7) corresponded to isoform I isolated by Mai and Adams (14). In this communication, we describe the identification of the genes encoding acetyl-CoA synthetase (ADP forming) isoform I from P. furiosus via functional overexpression in Escherichia coli. The recombinant enzyme was purified and characterized.
Identification of the genes, acdAI and acdBI, encoding acetyl-CoA synthetase (ADP forming) isoform I.
The genes putatively encoding subunit α (47 kDa) and subunit β (25 kDa) of acetyl-CoA synthetase (ADP forming) were identified by using the N-terminal amino acid sequences of both subunits, as reported by Glasemacher et al. (7). Based on the sequences, two open reading frames were identified by a BLAST search in the complete sequenced genome of P. furiosus (23). The open reading frames were designated acdAI and acdBI, where “acd” indicates the genes encoding acetyl-CoA synthetase (ADP forming) to discriminate the acd genes and the “acs” genes, encoding the widely distributed AMP-forming acetyl-CoA synthetases. “A” and “B” stand for the subunits α and β, respectively, and “I” stands for isoenzyme I. acdAI comprises 1,386 bp coding for a polypeptide of 462 amino acids (aa) with a calculated molecular mass of 49,964 Da; acdBI consists of 696 bp coding for a protein of 232 aa with a calculated molecular mass of 25,878 Da. The coding sequences of acdAI and acdBI genes start with ATG and stop with either TAA (acdAI) or TAG (acdBI). Immediately upstream of the initiation codon of both genes, putative ribosome-binding sites with the sequence GAGGT were present, as reported for other Pyrococcus genes (e.g., see references 9 and 24). Archaeal promoter regions, TATA boxes, and initiator elements (21) were not found. Downstream from the acdBI gene, rather than from the acdAI gene, a pyrimidine-rich region within 16 to 19 nucleotides with the consensus sequence TTTTTYT, indicating a transcription termination site, was identified (22). The G+C contents of the acdAI and acdBI genes are 43.3 and 40.3 mol%, respectively, and thus are slightly higher than the value of 38.5 mol% reported for the total genome of P. furiosus (6).
Comparison of AcdAI and AcdBI sequences with those of other proteins.
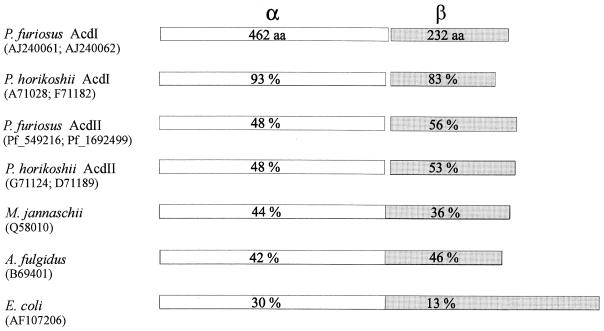
In a BLASTP search (1), the deduced amino acid sequences of the α (AcdAI) and β (AcdBI) subunits from P. furiosus were compared to those of proteins in the database derived from genome sequences (2, 5, 10, 11). Several proteins showing significant amino acid sequence identity were identified (Fig. 1). The highest degrees of identity (93 and 83%) were found with two proteins from Pyrococcus horikoshii, which indicates the presence of acdAI and acdBI homologous genes encoding an acetyl-CoA synthetase (ADP forming) isoform I in this Pyrococcus species. A lower degree of identity (about 50%) for both the α and β subunits was found within proteins from both P. furiosus and P. horikoshii. These proteins had N-terminal amino acid sequences and deduced molecular masses almost identical to those of the α and β subunits of acetyl-CoA synthetase (ADP forming) isoform II purified from P. furiosus (14). Thus, the sequence data indicate the presence of genes, acdAII and acdBII, encoding the acetyl-CoA (ADP forming) isoform II (Acd II) in both Pyrococcus species.
FIG. 1.
Comparison of deduced amino acid sequences of the α subunit (462 aa) and the β subunit (233 aa) of acetyl-CoA synthetase (ADP forming) isoform I from P. furiosus with hypothetical proteins of P. horikoshii (212 aa), M. jannaschii (704 aa), A. fulgidus (685 aa), and E. coli (886 aa) deduced from genome sequences (2, 5, 10, 11). EMBL accession numbers of the proteins are given in brackets. Amino acid sequence identities given in white boxes are for the homologous α subunits of the Pyrococcus proteins and the N-terminal parts (456 aa each) of proteins of M. jannaschii, A. fulgidus, and E. coli. Sequence identities given in shaded boxes are for the homologous β subunits of Pyrococcus proteins and the C-terminal parts of proteins of M. jannaschii (248 aa), A. fulgidus (229 aa), and E. coli (430 aa).
Homologous hypothetical proteins of the hyperthermophilic archaea Methanococcus jannaschii (704 aa) and Archaeoglobus fulgidus (685 aa) and the eubacterium Escherichia coli (886 aa) each had a size of about the sum of the amino acids of the α and β subunits of acetyl-CoA synthetase (ADP forming). The α subunit was found to align in the N-terminal part (456 aa) with a sequence identity of 30 to 44%, whereas the β subunit aligns best within the C-terminal part of the proteins (13 to 46% identity). Whether these hypothetical proteins constitute functional acetyl-CoA (or acyl-CoA) synthetases (ADP forming), in which the α and β subunits are fused, remains to be established. In the α and β subunits of the AcdI protein, several amino acid stretches which were almost completely conserved in all proteins shown aligned in Fig. 1 were identified (e.g., the α subunit contained PKXVAVIGAS [aa 9 to 18], MRILGPNXXGVV [aa 128 to 139], GXLAXISQSGA [aa 157 to 167], and IXIYMEGVXDGRRFM [aa 213 to 227]; the β subunit contained IGYPVVMKIXSPQIXHKS [aa 56 to 73] and DFQFGXXVMFGXGGI [aa 130 to 144]); however, substrate binding domains, e.g., for nucleotides, CoA, and Pi, have yet to be identified.
Expression of acdAI and acdBI genes in E. coli.
The identity of putative acdAI and acdBI genes as coding genes for the α and β subunits of acetyl-CoA synthetase (ADP forming) isoform I was proved by functional overexpression in E. coli. pET-14b and pET-17b protein expression vectors as well as E. coli JM109 and BL21(DE3) were purchased from Novagen. P. furiosus (DSM 3638) (6) was grown at 90°C with starch as a carbon and energy source, as described previously (8). The acdAI and acdBI genes were amplified by PCR with genomic DNA of P. furiosus as the template. The PCR product of acdAI was cloned in pET-17b by using the forward oligonucleotide primer 5′AATTTGACATATGAGTTTGGAGGCTCTTTTTAATC′3 extended by an NdeI restriction site and the reverse complement oligonucleotide primer 5′CCGCTCGAGTTACTTTTCTTTGTGTTTTGCTTTC′3 containing an XhoI site (restriction sites underlined). The PCR product of acdBI was cloned in pET-14b by using the restriction sites NcoI and XhoI and the primers 5′GATGCCATGGACAGGGTTGCTAAG′3 and 5′CGCCTCGAGCTAAAGAATCATCCTAGC′3. The recombinant plasmids were named pET-17b(acdAI) and pET-14b(acdBI). The inserted gene sequence was confirmed on each strand by the Sanger method. The two expression vectors pET-17b(acdAI) and pET-14b(acdBI) were transformed separately into E. coli BL21(DE3). Cells were grown in 400 ml of Luria-Bertani medium at 37°C to an optical density at 600 nm of 1.0, and expression was initiated by the addition of 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 3 h of further growth, the cells were harvested by centrifugation at 4°C. The pellet was frozen at −20°C. Cell extracts were prepared by passing cell suspensions in buffer (150 mM NaCl, 20 mM Tris HCl [pH 8.0]) through a French pressure cell at 150 MPa. After centrifugation at 40,000 × g for 1 h, the resulting supernatant (cell extract, 3 to 4 mg of protein/ml) was analyzed for overexpressed α and β subunits and used for reconstitution experiments.
After induction of the cells with IPTG, proteins with molecular masses of 47 kDa (α subunit [AcdAI]) and 25 kDa (β subunit [AcdBI]) were overexpressed as revealed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of cell extracts. Heat treatment (15 min at 80°C) of the extracts resulted in a significant enrichment (>80%) of the recombinant α or β subunit as judged by SDS-PAGE (data not shown). The oligomeric state of the recombinant subunits obtained after heat treatment was analyzed by gel filtration chromatography on a Superdex 200 HiLoad 26/60 column, equilibrated with 20 mM Tris HCl buffer, pH 8.0, containing 0.15 M NaCl. For the respective subunits more than 80% of the protein applied (1.2 mg each) was eluted at an apparent molecular mass of about 90 kDa (α subunit) or about 55 kDa (β subunit), indicating that the recombinant subunits were present predominantly as dimers.
Reconstitution and purification of recombinant acetyl-CoA synthetase (ADP forming).
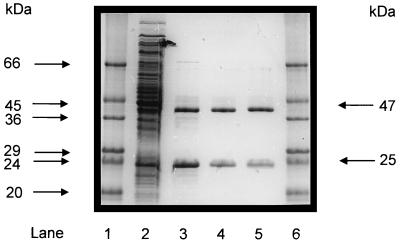
Equal amounts of E. coli extracts (3 mg/ml) containing recombinant α and β subunits (AcdAI and AcdBI) were mixed and incubated on ice (1 h). The combined extracts exhibited acetyl-CoA synthetase (ADP forming) activity of about 1 U/mg at 55°C (for direction of acetate formation, see reference 7), indicating that the subunits had been reconstituted to an active enzyme. The enzyme was purified about 10-fold by heat treatment (15 min at 80°C) and subsequent anion-exchange chromatography, as follows. Heat-precipitated host cell proteins were removed by centrifugation. The resulting supernatant was applied to a 6-ml Resource Q column equilibrated with 20 mM Tris HCl buffer, pH 8.0, containing 10 mM MgCl2. Protein was eluted with a linear gradient from 0 to 0.4 M NaCl in buffer (150 ml). Highest activity of acetyl-CoA synthetase (ADP forming) was eluted at 0.14 M NaCl. Protein purity was assessed by SDS-PAGE analysis. This two-step purification yielded a homogeneous preparation of recombinant holoenzyme, as indicated by two bands in SDS-PAGE (Fig. 2), which showed the same apparent molecular masses as the enzyme purified from P. furiosus (7).
FIG. 2.
Purification of recombinant acetyl-CoA synthetase (ADP forming) after reconstitution from its subunits (α and β) separately expressed in E. coli, as analyzed by SDS-PAGE. Protein was separated on a 12% polyacrylamide gel and subsequently stained with Coomassie brilliant blue R-250 (13). Lanes 1 and 6, molecular mass standards (Sigma); lane 2, 20 μg of protein of combined cell extract (10 μg each) of E. coli transformed with either pET-14b (acdBI) or pET-17b (acdAI) vector after induction with IPTG; lane 3, 6 μg of 13,000 × g supernatant of combined E. coli extracts (see lane 2) after heat treatment (15 min, 80°C); lane 4, 2.5 μg of purified recombinant enzyme after chromatography on Resource Q; lane 5, 2.5 μg of native acetyl-CoA synthetase (ADP forming) purified from P. furiosus (7).
Biochemical characterization of recombinant acetyl-CoA synthetase (ADP forming).
The purified recombinant acetyl-CoA synthetase (ADP forming) was biochemically analyzed with respect to molecular and catalytical properties and compared with the native enzyme purified from P. furiosus (7). Enzyme activity (acetyl-CoA + ADP + P ⇄ acetate + ATP + CoA) was measured in both directions as described previously (7). Optimum temperature and thermostability (between 80 and 110°C) of the enzyme were determined as described previously (7). The apparent molecular masses of the enzyme (determined by gel filtration) and of its subunits (determined by SDS-PAGE) and the apparent Km values of all substrates were almost identical, as reported for the enzyme isolated from P. furiosus; however, the apparent Vmax values were about 40 to 50% lower (Table 1). The recombinant enzyme showed almost identical thermostability and pattern of heat inactivation, i.e., it did not lose activity upon incubation for 3 h at 90°C, but it lost about 60% of its activity after 2 h at 100°C (see Fig. 3 in reference 7).
TABLE 1.
Comparison of the molecular and kinetic properties of recombinant acetyl-CoA synthetase (ADP forming) isoform I of P. furiosus produced in E. coli and the native enzyme isolated from P. furiosus
| Propertya | Enzyme isolated from:
|
|
|---|---|---|
| E. coli | P. furiosusb | |
| Molecular mass (kDa) of: | ||
| Subunit α | 47 | 47 |
| Subunit β | 25 | 25 |
| Enzyme (α2β2) | 145 | 145 |
| Km (μM) | ||
| Acetyl-CoA | 19 | 17 |
| ADP | 60 | 60 |
| Pi | 100 | 200 |
| Vmax (U/mg at 55°C) in the direction of acetate formation | 11 | 18 |
| Km (μM) | ||
| Acetate | 800 | 660 |
| ATP | 90 | 80 |
| CoA-SH | 21 | 30 |
| Vmax (U/mg at 55°C) in the direction of acetyl-CoA formation | 20 | 40 |
| Optimum temperature (°C) | 87 | 90 |
All values, except optimum temperatures, are apparent.
Date from reference 7.
The data indicate that in vitro reconstitution of separately expressed α and β subunits yielded a recombinant heterotetrameric acetyl-CoA synthetase (ADP forming) with properties very similar to those of the native enzyme isoform I isolated from P. furiosus (7). So far, only three heterooligomeric enzymes from hyperthermophiles have been functionally overexpressed in E. coli by processes involving reconstitution of separately expressed subunits: heterodimeric reverse gyrase from Methanopyrus kandleri (12) and heterotetrameric DNA topoisomerase VI from Sulfolobus shibatae (4) and indolepyruvate ferredoxin oxidoreductase from Pyrococcus kadokaraensis (20). In conclusion, the successful heterologous expression of acetyl-CoA synthetase (ADP forming) isoform I will allow detailed biochemical analyses of this unusual enzyme in the future, e.g., studies of structure-function relationships following crystallization and mechanistical studies involving site-directed mutagenesis.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the EMBL nucleotide sequence database with the accession no. AJ240061 (acdAI) and AJ240062 (acdBI).
Acknowledgments
We thank K. Lutter-Mohr for skillful technical assistance.
This work was supported by a grant from the European Union (“Extremophiles as cell factories”) and the Fonds der Chemischen Industrie.
Footnotes
Dedicated to Rolf Thauer on the occasion of his 60th birthday.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers W, Lipman D L. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bock A-K, Glasemacher J, Schmidt R, Schönheit P. Purification and characterization of two extremely thermostable enzymes, phosphate acetyltransferase and acetate kinase, from the hyperthermophilic eubacterium Thermotoga maritima. J Bacteriol. 1999;181:1861–1867. doi: 10.1128/jb.181.6.1861-1867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buhler C, Gadelle D, Forterre P, Wang J C, Bergerat A. Reconstitution of DNA topoisomerase VI of the thermophilic archaeon Sulfolobus shibatae from subunits separately overexpressed in Escherichia coli. Nucleic Acids Res. 1998;26:5157–5162. doi: 10.1093/nar/26.22.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bult C, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Ventner J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 6.Fiala G, Stetter K O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 7.Glasemacher J, Bock A-K, Schmid R, Schönheit P. Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur J Biochem. 1997;244:561–567. doi: 10.1111/j.1432-1033.1997.00561.x. [DOI] [PubMed] [Google Scholar]
- 8.Hethke C, Geerling A C M, Hausner W, de Vos W, Thomm M. A cell-free transcription system for the hyperthermophilic archaeon Pyrococcus furiosus. Nucleic Acids Res. 1996;24:2369–2376. doi: 10.1093/nar/24.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen S, Vorgias C E, Antranikian G. Cloning, sequencing, characterization, and expression of an extracellular α-amylase from the hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli and Bacillus subtilis. J Biol Chem. 1997;272:16335–16342. doi: 10.1074/jbc.272.26.16335. [DOI] [PubMed] [Google Scholar]
- 10.Kawarabayasi Y, et al. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;30:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 11.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Ventner J C, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 12.Krah R, O’Dea M H, Gellert M. Reverse gyrase from Methanopyrus kandleri. Reconstitution of an active extremozyme from its two recombinant subunits. J Biol Chem. 1997;272:13986–13990. doi: 10.1074/jbc.272.21.13986. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Mai X, Adams M W W. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves R E, Warren L G, Susskind B, Loh H. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. J Biol Chem. 1977;252:726–731. [PubMed] [Google Scholar]
- 16.Sanchez L B, Müller M. Purification and characterization of the acetate forming enzyme, acetyl-CoA synthetase (ADP-forming) from the amitochondriate protist, Giardia lamblia. FEBS Lett. 1996;378:240–244. doi: 10.1016/0014-5793(95)01463-2. [DOI] [PubMed] [Google Scholar]
- 17.Schäfer T, Schönheit P. Pyruvate metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus. Acetate formation from acetyl-CoA and ATP synthesis are catalyzed by an acetyl-CoA synthetase (ADP-forming) Arch Microbiol. 1991;155:366–377. [Google Scholar]
- 18.Schäfer T, Selig M, Schönheit P. Acetyl-CoA synthetase (ADP-forming) in archaea, a novel enzyme involved in acetate formation and ATP synthesis. Arch Microbiol. 1993;159:354–363. [Google Scholar]
- 19.Schönheit P, Schäfer T. Metabolism of hyperthermophiles. World J Microbiol Biotechnol. 1995;11:26–57. doi: 10.1007/BF00339135. [DOI] [PubMed] [Google Scholar]
- 20.Siddiqui M A, Fujiwara S, Takagi M, Imanaka T. In vitro heat effect on heterooligomeric subunit assembly of thermostable indolepyruvate ferredoxin oxidoreductase. FEBS Lett. 1998;434:372–376. doi: 10.1016/s0014-5793(98)00998-3. [DOI] [PubMed] [Google Scholar]
- 21.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 22.Thomm M, Hausner W, Hethke C. Transcription factors and termination of transcription in Methanococcus. Syst Appl Microbiol. 1994;16:648–655. [Google Scholar]
- 23.Utah Genome Center Website. [Online.] Sequences. Department of Human Genetics, University of Utah. http://www.genome.utah.edu. [11 May 1999, last date accessed.]
- 24.Zwickel P, Fabry S, Bogedain C, Haas A, Hensel R. Glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaebacterium Pyrococcus woesei: characterization of the enzyme, cloning and sequencing of the gene, and expression in Escherichia coli. J Bacteriol. 1990;172:4329–4338. doi: 10.1128/jb.172.8.4329-4338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]