COMPLEXITY OF NITRATE REDUCTION PATHWAYS
Nitrogen is a basic element for life because it is a component of the two preeminent biological macromolecules: proteins and nucleic acids. Nitrogen exists in the biosphere in several oxidation states, from N(V) to N(−III). Interconversions of these nitrogen species constitute the global biogeochemical nitrogen cycle, which is sustained by biological processes, with bacteria playing a predominant role (74). Briefly, inorganic nitrogen is converted to a biologically useful form by dinitrogen fixation or nitrate assimilation and the further incorporation of ammonia into C skeletons. Nitrogen is removed from the environment by both nitrification, the oxidative conversion of ammonia to nitrate, and denitrification, a respiratory process whereby nitrate is successively reduced to nitrite, N oxides (NO and N2O), and dinitrogen (N2). Nitrate reduction plays a key role in the nitrogen cycle and has important agricultural, environmental, and public health implications. Assimilatory nitrate reduction, performed by bacteria, fungi, algae, and higher plants, is one of the most fundamental biological processes, accounting for more than 104 megatons of inorganic nitrogen transformed each year (38). However, there is worldwide concern over the excessive use of fertilizers in agricultural activities, leading to nitrate accumulation in groundwater. Consumption of drinking water with high nitrate levels has been associated with methemoglobinemia and gastric cancer due to endogenous formation of genotoxic N-nitroso compounds by bacteria in the gastrointestinal tract (93). The main threat to the environment comes from eutrophication of aquatic ecosystems. Nitrogen oxides generated by denitrification are also associated with the greenhouse effect and the depletion of stratospheric ozone (100). Therefore, nitrate reduction has become an important focus for research in the last several years, generating a vast literature. The aim of this minireview is to summarize recent advances in the physiology, biochemistry, and genetics of prokaryotic nitrate reduction, emphasizing the different molecular characteristics of the bacterial nitrate reductases. Comprehensive reviews covering nitrate assimilation or denitrification have been published elsewhere (7, 17, 31, 38, 43, 49, 86, 100).
Nitrate reduction can be performed with three different purposes: the utilization of nitrate as a nitrogen source for growth (nitrate assimilation), the generation of metabolic energy by using nitrate as a terminal electron acceptor (nitrate respiration), and the dissipation of excess reducing power for redox balancing (nitrate dissimilation). Four types of nitrate reductases catalyze the two-electron reduction of nitrate to nitrite: the eukaryotic assimilatory nitrate reductases and three distinct bacterial enzymes, comprising the cytoplasmic assimilatory (Nas), membrane-bound respiratory (Nar), and periplasmic dissimilatory (Nap) nitrate reductases. All eukaryotic and bacterial nitrate reductases contain a molybdenum cofactor at their active sites. The basic structure of the eukaryotic cofactor is molybdopterin, a 6-alkyl pterin derivative with a phosphorylated C4 chain with two thiol groups binding the Mo atom. By contrast, the cofactor found in bacterial nitrate reductases and some molybdoenzymes is the bis-molybdopterin guanine dinucleotide (MGD) form (7, 24, 69, 100). Nitrite oxidase of nitrifying bacteria also shows nitrate reductase activity. This membrane-bound enzyme, which contains MGD and shows a high sequence similarity to the membrane-bound Nar, catalyzes nitrite oxidation to nitrate to allow chemoautotrophic growth, but it can also catalyze the reverse reaction (89). As nitrite oxidase is not a proper nitrate reductase, we will not consider it further.
Eukaryotic assimilatory nitrate reductases are cytosolic homodimeric enzymes that use pyridine nucleotides as electron donors. Each monomer is composed of a 100- to 120-kDa polypeptide with three prosthetic groups, flavin adenine dinucleotide (FAD), cytochrome b557, and Mo cofactor, which are located in three functional domains highly conserved among eukaryotic species. The Mo cofactor domain is located at the N-terminal end, the heme region corresponds to the middle domain, and the FAD-NAD(P)H domain is present at the C-terminal end. Structural genes coding for nitrate and nitrite reductases and for high-affinity nitrate and nitrite transporters have been cloned in several eukaryotes (Fig. 1). Biochemistry and molecular genetics of eukaryotic nitrate reduction have been investigated intensively during the last decades (17, 31, 38, 86). However, eukaryotic and prokaryotic assimilatory nitrate reductases share no sequence similarity and have little in common beyond their physiological function.
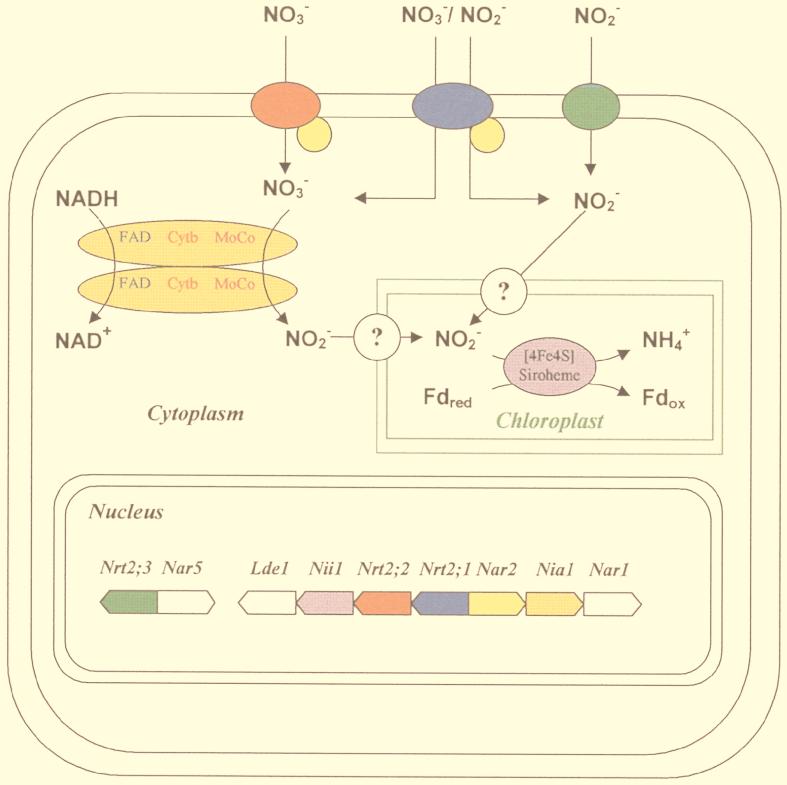
FIG. 1.
Nitrate assimilation pathway in the eukaryotic green alga Chlamydomonas reinhardtii. The components of the high-affinity nitrate and nitrite transport systems, the NAD(P)H-dependent nitrate reductase, and the ferredoxin-dependent nitrite reductase, are drawn in the same color as the corresponding nuclear genes coding for them. The functions of the products of the white genes are unknown. Cytb, cytochrome b; Fdred, reduced ferredoxin; Fdox, oxidized ferredoxin; MoCo, molybdenum cofactor.
In addition to the assimilatory enzyme, two types of dissimilatory nitrate reductases are present in bacteria: the respiratory membrane-bound Nar, which generates a transmembrane proton motive force (PMF) allowing ATP synthesis, and the periplasmic Nap of some gram-negative bacteria (Table 1). Nap seems to be a dissimilatory enzyme sensu stricto, because quinol oxidation by Nap is not directly coupled to the generation of a PMF and because it is independent of the cytochrome bc1 complex. Nap could still generate a PMF if a proton-translocating NADH dehydrogenase were involved in reducing the quinone pool, but the resulting PMF seems to be insufficient to support ATP synthesis in some bacteria [see “Dissimilatory Periplasmic Nitrate Reductases (Nap)” below] (60). However, depending on the metabolic fate of nitrite, a certain type of nitrate reductase can have different functions under various conditions. Thus, Escherichia coli assimilates nitrite generated by anaerobic nitrate respiration (88), and some denitrifiers use the nitrite formed by Nap to perform anaerobic nitrite respiration or aerobic denitrification (2–4).
TABLE 1.
Prokaryotic nitrate reduction
| Characteristic | Assimilatory, NO3− assimilation | Dissimilatory
|
|
|---|---|---|---|
| NO3− respiration | NO3− reduction | ||
| Nitrate reductase | Assimilatory Nas | Respiratory Nar | Dissimilatory Nap |
| Location | Cytoplasm | Membrane | Periplasm |
| Reaction catalyzed | NO3−⇒NO2− | NO3−⇒NO2− | NO3−⇒NO2− |
| Structural genes | nasCAa/narBb | narGHI | napAB |
| Prosthetic groups | FADc, FeSd, MGD | cytbe, FeS, MGD | cytc, FeS, MGD |
| Nitrate transport | Yes | Yes | No |
| Function | Biosynthesis of N compounds | PMF (nitrate respiration and denitrification) | 2H ⇓f and denitrification |
| Regulationg | |||
| O2 | No | Yes | No/yes |
| NH4+ | Yes | No | No |
| NO3−/NO2− | Yes | Yes | No/yes |
Following the gene designation in K. oxytoca for the NADH-nitrate reductase.
Following the gene designation in cyanobacteria for the ferredoxin-nitrate reductase.
FAD is present in the diaphorase subunit of the NADH-dependent nitrate reductases, but it is absent from the cyanobacterial ferredoxin-nitrate reductase.
FeS, iron-sulfur centers.
cytb, cytochrome b.
2H⇓, dissipation of reducing power. A PMF can be generated if a proton-translocating complex is involved in the electron transfer, but in most cases, this seems to be insufficient to support ATP synthesis coupled to nitrate reduction.
Some differences in regulation in prokaryotic organisms have been reported.
Nitrite formed by nitrate reduction can be reduced to ammonium or to nitric oxide by different types of nitrite reductases (Table 2) (15). In nitrate-assimilating bacteria, ammonium is generated in the cytoplasm by a NADH-dependent (the nasB gene product in Klebsiella oxytoca [50]) or a ferredoxin-dependent assimilatory nitrite reductase (the nirA gene product in cyanobacteria [52, 90]). In E. coli, a cytoplasmic NADH-dependent enzyme encoded by the nirB gene (42) catalyzes the reduction of nitrite to ammonium to detoxify the nitrite that accumulates in anaerobic nitrate-respiring cells and to regenerate NAD+. Although ammonium generated by this enzyme can be assimilated, the process is termed nitrite dissimilation (88). All these cytoplasmic nitrite reductases contain a single siroheme and a [4Fe-4S] center. The NADH-dependent enzymes also contain FAD (15, 17, 38). It is worth noting that the ferredoxin-nitrite reductase structure is very similar in cyanobacteria, eukaryotic algae, and vascular plants; these organisms all have conserved Cys residues for binding the Fe-S and siroheme cofactors (31). Alternatively, nitrite can be excreted to the periplasm where, depending on the bacteria, three classes of respiratory enzymes can couple its reduction to energy-conserving electron transport pathways. One of these enzymes is the E. coli multiheme cytochrome c nitrite reductase, encoded by the nrf operon, which catalyzes the formate-dependent nitrite reduction to ammonium (28, 44). This enzyme is also known as hexaheme nitrite reductase in some bacteria (15), although the E. coli enzyme binds only five heme c groups (28). Finally, two different respiratory enzymes reduce nitrite to nitric oxide in the periplasm of denitrifying bacteria: the nirS-encoded cytochrome cd1 nitrite reductase, which is found in Pseudomonas and most denitrifiers, and the nirK-encoded copper nitrite reductase, which is present in some bacteria (7, 15, 43, 100).
TABLE 2.
Prokaryotic nitrite reduction
| Characteristic | Assimilatory, NO2− assimilation | Dissimilatory
|
||
|---|---|---|---|---|
| NO2− respiration
|
NO2− reduction | |||
| Nir | Nrf | |||
| Nitrite reductase | Assimilatory Nas | Respiratory Nir | Respiratory Nrf | Dissimilatory Nir |
| Location | Cytoplasm | Periplasm | Periplasm | Cytoplasm |
| Reaction catalyzed | NO2−⇒NH4+ | NO2−⇒NO | NO2−⇒NH4+ | NO2−⇒NH4+ |
| Structural genes | nasBa/nirAb | nirS/nirK | nrfA | nirBD |
| Prosthetic groups | FADc, FeSd, siroheme | cytcd1e/Cu | cytc | FAD, FeS, siroheme |
| Nitrite transport | Yes | No | No | Yes |
| Function | Biosynthesis of N compounds | PMF (denitrification) | PMF (ammonification) | 2H ⇓f and nitrite detoxification |
| Regulationg | ||||
| O2 | No | Yes | Yes | Yes |
| NH4+ | Yes | No | No | No |
| NO3−/NO2− | Yes | Yes | Yes | Yes |
Following the gene designation in K. oxytoca for the NADH-nitrite reductase.
Following the gene designation in cyanobacteria for the ferredoxin-nitrite reductase.
FAD is present in the NADH-nitrite reductases, but it is absent from the cyanobacterial assimilatory ferredoxin-dependent nitrite reductase.
FeS, iron-sulfur centers.
cytcd1, cytochrome cd1 complex.
2H ⇓, dissipation of reducing power.
Some differences in regulation in prokaryotic organisms have been reported.
BACTERIAL ASSIMILATORY NITRATE REDUCTASES (NAS)
Structure and biochemical properties of assimilatory nitrate reductases.
Nitrate assimilation has been studied at the biochemical or genetic level in several phototrophic and heterotrophic bacteria. Two classes of assimilatory nitrate reductases are found in bacteria: the ferredoxin- or flavodoxin-dependent Nas and the NADH-dependent enzyme (Fig. 2). Both types of Nas contain MGD cofactor and one N-terminal iron-sulfur cluster but are devoid of heme groups, in contrast to eukaryotic and other bacterial nitrate reductases. The cyanobacterial ferredoxin-Nas is a single subunit of 75 to 85 kDa (59, 80), whereas the flavodoxin-Nas of Azotobacter vinelandii is a polypeptide of 105 kDa (34, 35). The purified Nas proteins of A. vinelandii and Plectonema boryanum contain one Mo, four Fe, and four acid-labile S atoms per molecule (35, 59). Amino acid sequence analysis reveals the presence of a Cys motif in the N-terminal end of the proteins, probably binding one [4Fe-4S] or [3Fe-4S] center. Ferredoxin-Nas is also present in Azotobacter chroococcum, Clostridium perfringens, and Ectothiorhodospira shaposhnikovii (38). On the other hand, the NADH-Nas proteins of Klebsiella pneumoniae (50) and Rhodobacter capsulatus (12) are heterodimers of a 45-kDa FAD-containing diaphorase and a 95-kDa catalytic subunit with MGD cofactor and a putative N-terminal [4Fe-4S] center. This NADH-Nas, as deduced by the Klebsiella nasA gene sequence, probably contains an additional [2Fe-2S] center linked to a C-terminal Cys cluster that is similar to a sequence of the NifU protein (49). This region is absent from the ferredoxin-Nas and could act as a ferredoxin-like electron transfer domain. The Bacillus subtilis NADH-Nas does not contain the NifU-like domain in the catalytic subunit but has two tandem NifU-like modules in a central region of the FAD-containing diaphorase (65).
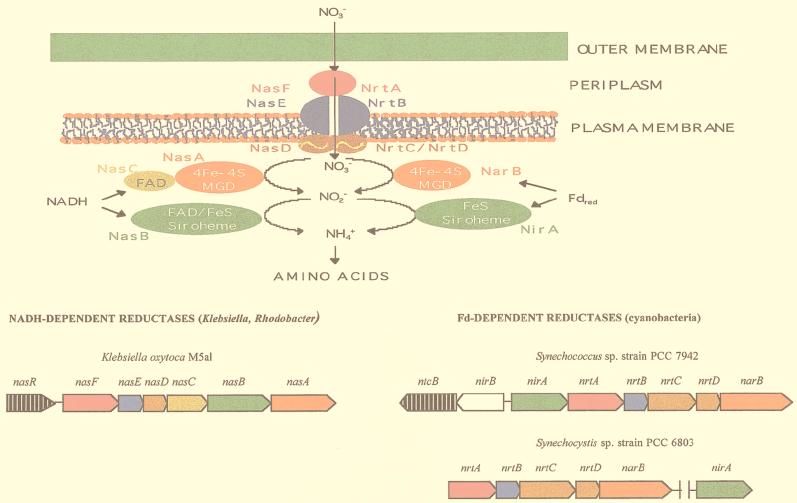
FIG. 2.
Nitrate assimilation and comparison of the organization of the nitrate assimilation gene clusters in bacteria. The scheme shows the cyanobacterial ferredoxin (Fd)-dependent assimilatory nitrate and nitrite reductases (right) and the NADH-dependent nitrate and nitrite reductases from Klebsiella and Rhodobacter (left). The organization of the Synechococcus, Synechocystis, and K. oxytoca (pneumoniae) nitrate assimilation gene clusters is shown beneath the corresponding proteins. Genes are drawn approximately to scale, and arrows show the direction of transcription. The genes and their products are shown in the same color. Regulatory genes are indicated by black arrows with white vertical lines. The product of the white gene is not shown.
Although all nitrate reductases can use reduced viologens as electron donors, the ability to use bromophenol blue as an artificial reductant is a characteristic of both eukaryotic and prokaryotic assimilatory enzymes (12, 35). In R. capsulatus, Nas is inhibited by cyanide and azide but is unaffected by cyanate and chlorate. NADH also inactivates Nas under aerobic conditions by formation of superoxide anion at the diaphorase flavin center, and this activity is protected by superoxide dismutase (12).
Organization of the genes coding for the assimilatory nitrate reductases.
The genes coding for the assimilatory nitrate-reducing system are normally clustered and have been cloned in several bacterial species. These gene clusters include regulatory and structural genes coding for proteins required for uptake and reduction of both nitrate and nitrite (Fig. 2). Nomenclature of these genes is confusing because different names have been given to homologous genes in different bacteria. In our opinion, the nas gene designation in K. pneumoniae (49, 50) is more appropriate. In this bacterium, the nasR gene encoding a transcription antiterminator is linked to the nasFEDCBA operon (36, 48–50, 98). The nasFED genes code for a multicomponent nitrate or nitrite transport system, the nasB gene encodes a siroheme-dependent assimilatory nitrite reductase, and the NADH-nitrate reductase is encoded by the nasC (diaphorase) and nasA (catalytic subunit) genes.
In B. subtilis, the nasBC genes code for the NADH-nitrate reductase: NasB is a diaphorase with NADH- and FAD-binding domains, and NasC is the catalytic subunit. The nas gene cluster also includes nasA, coding for a nitrate transporter, nasDE, coding for the subunits of a soluble NADH-nitrite reductase, and nasF, a gene involved in siroheme cofactor biosynthesis (63, 65). It is worth noting that B. subtilis contains only the NasDE nitrite reductase, but it has two different nitrate reductases, the assimilatory NasBC enzymes and the respiratory narGHJI-encoded enzymes. The Nar enzyme can function together with this NADH-nitrite reductase during anaerobic growth, although nitrite reduction does not result in a proton gradient coupled to ATP generation (64).
The structural gene encoding the flavodoxin-dependent Nas of A. vinelandii (nasB) is also cotranscribed with the nitrite reductase nasA gene (71). In cyanobacteria, the gene coding for the ferredoxin-dependent Nas is termed narB (note that the nar designation should be kept for the respiratory enzyme) and has been sequenced in several strains, including unicellular (Synechococcus and Synechocystis) and filamentous nonheterocyst (Oscillatoria) or heterocyst-forming (Anabaena) cyanobacteria (16, 45, 80, 92). In most cases, the nitrite reductase nirA gene, the nrtABCD nitrate transport genes, and the nitrate reductase narB gene constitute an operon (16, 33, 45, 51, 66, 90).
Nitrate transport in bacterial Nas systems.
Due to the cytoplasmic location of the Nas enzyme, nitrate reduction is preceded by nitrate transport into the cells (Fig. 2). In most bacterial Nas systems, nitrate seems to be transported by an ABC-type transporter requiring a periplasmic binding protein. Nitrate and nitrite transport systems (and genes coding for their components) have been thoroughly studied in cyanobacteria and Klebsiella. In Synechococcus, nitrate transport is mediated by a periplasmic binding protein (the nrtA gene product), an integral membrane protein (encoded by the nrtB gene), and two homologous ATP-binding proteins, the nrtC and nrtD gene products (53, 66). Similar nrt clusters have been reported for Synechocystis (45), Anabaena (16, 33), and Phormidium laminosarum (56). In Klebsiella, the nasFED genes encode a typical ABC transporter for both nitrate and nitrite: a 46-kDa periplasmic binding protein (NasF) homologous to NrtA, a homodimeric membrane protein (NasE) related to NrtB, and a homodimeric ATP-binding protein (NasD) similar to NrtD (49, 50, 98). A similar nitrate permease has been found in A. chroococcum (62) and a 47-kDa periplasmic protein is involved in the ATP-dependent nitrate transport system of R. capsulatus (18, 25). However, an electrogenic nitrate uptake mediated by a different transporter, the nasA gene product, is present in B. subtilis (65). NasA protein, a member of the major facilitator superfamily, shows sequence similarity to the E. coli narK gene product, a membrane potential-dependent nitrite extrusion protein (79). A narK-homologous gene encoding the putative nitrite efflux porter is also found in B. subtilis.
Regulation of bacterial assimilatory nitrate reductases.
Expression of Klebsiella nas genes is subjected to dual control: ammonia repression by the general nitrogen regulatory system (Ntr) and specific nitrate or nitrite induction (36, 49). The Ntr system regulates the synthesis of most enzymes required for utilizing alternate nitrogen sources. During nitrogen-limited growth, the NtrC protein is activated by phosphorylation mediated by NtrB and binds to upstream sequences of promoters recognized by the alternate rpoN-encoded ςN (ς54) factor, activating transcription of the Ntr-regulated genes (57). The Nac protein, a member of the LysR family, also activates expression of several nitrogen-regulated operons (49, 57). Specific nitrate or nitrite induction of nas gene expression in Klebsiella is mediated by NasR, a positive regulator that acts by a transcription attenuation mechanism. In the absence of nitrate or nitrite, a factor-independent transcription terminator present in the nasF leader region prevents nas gene transcription. When nitrate or nitrite is present, NasR promotes transcription antitermination in the leader region, increasing nasF operon expression. Thus, nitrate regulation does not act by controlling transcription initiation but by controlling transcription termination (36, 48, 88).
Ammonium-promoted repression and positive regulation of nitrate assimilation by nitrate or nitrite have also been reported for photosynthetic bacteria. In R. capsulatus, assimilatory nitrate reductase is induced by nitrate and repressed at low C/N ratios, probably through the balance of 2-oxoglutarate and glutamine. Ammonium also inhibits nitrate transport, avoiding nitrate reductase induction (18, 26). In the cyanobacterium Synechococcus sp. strain PCC 7942, ammonium represses nitrate assimilation genes (nirA-nrtABCD-narB) through its incorporation into glutamine. Positive regulation by nitrate requires nitrate reduction, and nitrite seems to be the actual activator of transcription of nitrate assimilation genes. Nitrogen control in cyanobacteria is mediated by the NtcA protein, a member of the Crp family of transcriptional activators (94). A second regulatory protein, the ntcB gene product, is a LysR family transcription factor required to activate nirA operon expression in response to nitrite (1).
In A. vinelandii, expression of the nasAB structural genes is induced by nitrate or nitrite and repressed by ammonium, through the ntr genes (71). The nasST operon is also required for expression of the nasAB genes. NasT is homologous to regulator proteins of two-component regulatory systems and is essential for nasAB operon expression, whereas NasS is similar to proteins involved in nitrate uptake but seems to play a negative regulatory role blocking NasT action in the absence of nitrate (40). In addition, the nasAB operon seems to be subjected to autogenous regulation by the nitrate reductase protein NasB, which is a negative regulatory element for nitrate and nitrite reductase synthesis (71). Molybdenum metabolism might also function as a regulatory factor, and a complex regulatory network involving the nifO gene has been proposed (41).
In B. subtilis, the nas operon is not subject to pathway-specific nitrate induction, in contrast to other bacteria (63, 64). In addition, this bacterium has no known system analogous to the Ntr system, and nitrogen control is mediated by a positive regulator, the TnrA protein, which binds DNA to activate transcription of the nas operon and several nitrogen-controlled genes (64).
RESPITATORY MEMBRANE-BOUND NITRATE REDUCTASES (NAR)
Structure and biochemical properties of membrane-bound nitrate reductases.
Membrane-bound nitrate reductases are associated with denitrification and anaerobic nitrate respiration (Fig. 3). Although the most exhaustive biochemistry and genetic studies have been performed in E. coli and Paracoccus denitrificans, Nar enzymes have been purified from several denitrifying and nitrate-respiring bacteria (100). A thermophilic Nar protein with an optimal temperature of 80°C has also been found in Thermus thermophilus (70). In E. coli, there are two different membrane-bound isoenzymes: NRA, which is expressed under anaerobiosis in the presence of nitrate and represents 90% of total activity, and NRZ, which is expressed constitutively (9, 13).
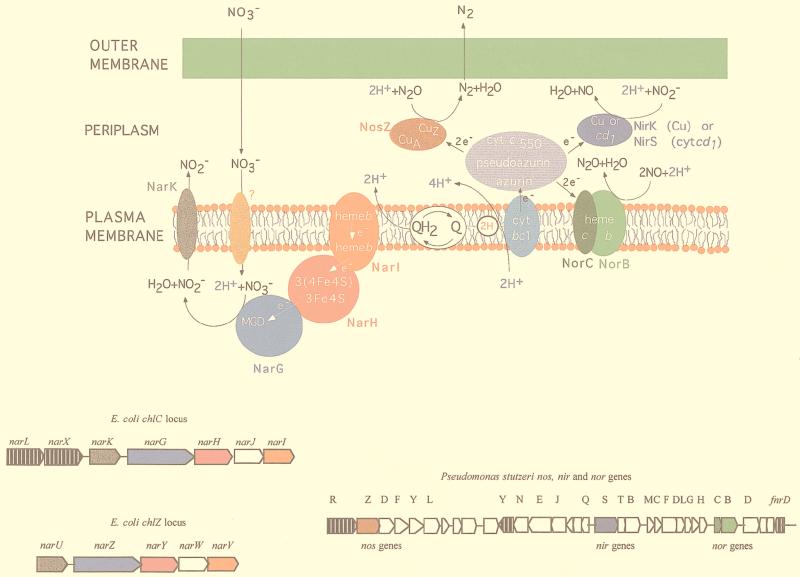
FIG. 3.
Nitrate respiration and denitrification pathways in bacteria. The organization of the anaerobic electron transport chains for nitrate respiration and denitrification is shown at the top. The E. coli respiratory nitrate reductase (nar) gene clusters and the Pseudomonas stutzeri denitrification nos, nir, and nor gene clusters are also shown. Genes are drawn approximately to scale, and arrows show the direction of transcription. The genes and their products are drawn in the same color. Regulatory genes are indicated by black arrows with white vertical lines. The products of the white genes are not shown. The quinone pool is indicated by the white oval labeled QH2⇄Q. cyt, cytochrome.
Nar enzymes are composed of three subunits: a catalytic α subunit (NarG) of 112 to 140 kDa with MGD cofactor, a soluble β subunit (NarH) of 52 to 64 kDa with one [3Fe-4S] and three [4Fe-4S] centers, and a 19- to 25-kDa membrane biheme b quinol-oxidizing γ subunit (NarI). Soluble α and β subunits are anchored to the cytoplasmic side of the membrane by the γ subunit and can be solubilized by detergents or heat. NarI is heat sensitive and can be lost during the purification procedure, leading to the isolation of a soluble αβ complex that can reduce nitrate with reduced viologens as electron donors. A δ polypeptide (NarJ), which is not part of the final enzyme, seems to participate in the assembly or stability of the αβ complex prior to its membrane attachment (11, 27). Strains with mutations at the mob locus are able to synthesize molybdopterin but do not form MGD cofactor. Inactive Nar purified from a mob mutant is activated in vitro by incubation with protein FA (the mobA gene product), GTP, and the so-called factor X. It has been recently found that NarJ is a major component of factor X and activates nitrate reductase after completing MGD cofactor synthesis (68).
In general, all membrane-bound nitrate reductases can reduce chlorate and are inhibited by azide, chlorate, cyanide, and thiocyanate (43). The Nar system of the bacteria in the intestinal tract is also involved in nitrosation of aromatic and alkyl amines by nitrite due to a weak NO-producing nitrite reductase activity associated with this enzyme (58). Formation of these N-nitroso compounds is believed to be a major cause of human gastric cancer. Curiously, although subunits of the E. coli NRA and NRZ enzymes are highly similar and can form active hybrid complex (10), only the narG operon-encoded NRA is implicated in the nitrosation activity, whereas neither the second membrane-bound enzyme NRZ nor the periplasmic nitrate reductase contributes to the nitrosation reaction (58).
Nar proteins use the quinol pool as the physiological electron donor and generate a PMF by a redox loop mechanism (7, 74). NarI oxidizes quinols at the periplasmic side of the membrane, releasing two protons into the periplasm. Electrons are passed to NarG, via the Fe-S centers of NarH, to reduce nitrate with consumption of two cytoplasmic protons. The low- and high-potential heme b groups of NarI located at opposite sides of the membrane allow an effective transmembrane electron transfer. Electron paramagnetic resonance and biochemical characterization of the wild-type and site-directed mutated NarH proteins reveal the presence of two pairs of Fe-S clusters in the β subunit (39). In addition, it has been proposed that a His-Cys3 motif in the N-terminal end of NarG could bind a [4Fe-4S] center participating in the electron transfer from the Fe-S centers of NarH to the MGD cofactor (74, 76). However, this center has not been detected by spectroscopic studies, and all the Fe-S clusters of the enzyme are ligated to the NarH subunit (54).
Organization of the genes coding for the membrane-bound nitrate reductases.
In E. coli, NRA is encoded by the narGHJI operon located in the chlC locus at 27 min on the chromosome, and NRZ is encoded by the narZYWV operon in the chlZ locus at 32.5 min (Fig. 3) (13). The presence of narGHJI-homologous genes has also been reported for other bacteria. In E. coli, NRA and NRZ show a high similarity: 76% identity for the catalytic subunits (NarG and NarZ), 75% identity for the β subunits (NarH and NarY), and 87% similarity for the NarI and NarV proteins (9). The E. coli chlC locus also includes the narK gene encoding a nitrite efflux porter (79) and the narXL genes encoding a nitrate response two-component regulatory system, in which NarX is the nitrate sensor and NarL is the DNA-binding regulator (88). The chlZ locus does not include regulatory genes, but a narK-homologous gene (narU) is located upstream of narZYWV operon. The narQP genes coding for a second nitrate sensor (NarQ) and a second nitrate response regulator (NarP) are located at 53 and 46 min on the E. coli genetic map, respectively (13, 88).
Nitrate transport in Nar systems.
As a consequence of the cytoplasmic location of the active site of NarG, nitrate has to be transported into the cells before it is reduced, and nitrite is usually excreted to the periplasm by a specific nitrite extrusion system. The respiratory nitrate uptake is poorly understood, although it is clear that the nitrate porter is highly specific for nitrate and is inhibited by oxygen (23). Oxygen inhibition of nitrate transport seems to be caused by an indirect mechanism (i.e., the diversion of electrons to oxygen), rather than causing conformational changes in the porter system (23). In contrast to assimilatory nitrate uptake, which uses an ABC-type transporter, the nitrate transport system in nitrate-respiring bacteria has not been identified, although several mechanisms for nitrate uptake have been proposed, including passive nitrate uniport, ATP-dependent uniport, PMF-dependent NO3−/H+ symport, and NO3−/NO2− antiport (7). In E. coli, the narK gene product was considered a nitrate/nitrite antiporter for several years. However, more-detailed studies have demonstrated that NarK is a nitrite exporter which mediates electrogenic nitrite excretion rather than nitrate uptake (79). A gene (narT) encoding a putative nitrate transporter involved in dissimilatory nitrate reduction has been identified in Staphylococcus carnosus (29). NarT shows homology with E. coli and B. subtilis NarK proteins and with B. subtilis NasA, suggesting a role in both nitrate import and nitrite extrusion. In addition, a putative nitrite transporter gene (nirC) has been identified in E. coli and other bacteria. However, it is unclear if NirC is a nitrite importer or exporter (42).
Regulation of respiratory membrane-bound nitrate reductases.
In E. coli, Nar proteins are synthesized during anaerobic growth, via the Fnr protein, in the presence of nitrate or nitrite, by two-component regulatory systems of sensor proteins (NarX and NarQ) and DNA-binding regulators (NarL and NarP). Synthesis of Nar enzymes is unaffected by ammonium (7, 43, 88, 100). Although both NRA and NRZ show a high identity, the narZ operon is not regulated either by O2 (Fnr) or by nitrate (9, 13). Constitutive NRZ could play a role, as proposed for the Nap system, in adaptation to anaerobic metabolism after the transition from aerobic conditions to anoxia.
The E. coli transcriptional regulator Fnr plays a central role in the expression of anaerobic metabolism genes (87). Fnr binds to a consensus sequence upstream of the Fnr-regulated promoters acting as either an activator or repressor, depending on its location. Disassembly of a labile Fe-S center has been proposed as a model for the O2-dependent Fnr inactivation. Under anoxia, dimeric Fnr binds to DNA and activates transcription of nar and other anaerobic metabolism genes. Under aerobic conditions, the [4Fe-4S]2+ clusters are converted to [3Fe-4S]2+ or [2Fe-2S]2+ centers, resulting in Fnr inactivation (47). Sequences with similarity to the E. coli Fnr box have been found upstream of anaerobic nitrate respiration and denitrification genes in many bacteria, and several Fnr-like factors have been identified in both gram-positive and -negative bacteria (87, 100).
Nitrate or nitrite regulation of nar gene expression in E. coli is mediated by a two-component signal transfer system with membrane sensor proteins (NarX and NarQ) and cytoplasmic response regulators (NarL and NarP). NarX and NarQ are homologous sensors that respond to nitrate and nitrite phosphorylating both NarL and NarP regulators (20, 88). Nitrate and nitrite bind to a periplasmic domain (the P-box element, a 17-amino-acid sequence between the two transmembrane regions of NarX and NarQ), altering the conformation of these proteins to allow autophosphorylation and subsequent phosphorylation of NarL and NarP (19). Activated NarL and NarP bind to specific DNA target sites, the so-called NarL heptamers (91). However, some operons, such as nitrate reductase narGHJI, fumarate reductase frdABCD, and nitrite export narK, are regulated by NarL alone, whereas others, such as nitrite reductase nrfABCDEFG and the periplasmic nitrate reductase aeg-46.5 locus, are controlled by both NarL and NarP (21). The NarL-binding heptamers are found as single copies, inverted repeats, or direct repeats at positions between +20 and −200 relative to the transcriptional start sites (21). NarL recognizes all heptamer arrangements, but NarP binds only to heptamers organized as an inverted repeat with 2-bp spacing (22). This complex regulatory system discriminates between nitrate and nitrite: NarL mainly serves as a nitrate regulator and becomes only weakly phosphorylated with nitrite, which induces respiratory nitrite reductase synthesis more efficiently. Sensors also provide phosphatase activity: NarQ dephosphorylates NarP, and NarX dephosphorylates NarL. In response to nitrate, NarX and NarQ protein kinase activities are practically indistinguishable and phosphorylate both NarL and NarP. In the presence of nitrite, NarX phosphorylates NarP but acts primarily as a NarL phosphatase. Thus, in response to nitrite, NarX is a positive regulator of NarP and a negative regulator of NarL. On the other hand, NarQ phosphorylates NarL and NarP in response to both nitrate and nitrite (96). Integration host factor is an additional element required for nar operon activation. Bending of DNA around the integration host factor seems to be required for contact between NarL, Fnr, and polymerase (83, 99).
DISSIMILATORY PERIPLASMIC NITRATE REDUCTASES (NAP)
Structure and biochemical properties of periplasmic nitrate reductases.
Periplasmic nitrate reductases were first reported for phototrophic and denitrifying bacteria, but they are widespread among gram-negative bacteria. Different physiological functions have been proposed for this enzyme. The Nap activity seems not to be primarily involved in nitrate assimilation or anaerobic respiration, although the nitrite generated by Nap can be used as a nitrogen source or as a substrate for anaerobic respiration depending on the organism. The Nap enzyme, as a consequence of its periplasmic location, does not directly contribute to the generation of a PMF. The Nap system is also independent of the energy-conserving cytochrome bc1 complex, but it is likely linked to the generation of a PMF when the electrons from NADH are passed through the proton-translocating NADH dehydrogenase (7, 74). However, this seems to be insufficient to support anaerobic growth on nitrate in Rhodobacter sphaeroides in the dark (46, 60). Also, the anaerobic growth rate on nitrate of a T. pantotropha NarH− mutant overexpressing Nap activity is decreased threefold on account of the reduced energy conservation by Nap relative to Nar during denitrification (4). However, in Pseudomonas sp. strain G-179, the Nap enzyme catalyzes the first step of denitrification in an energy-generating process, although the mechanism used by Nap to gain energy is unclear (2). Thus, the physiological role of the Nap system may vary in different organisms or even in the same bacterium under different metabolic conditions. There are clear evidences that Nap is a dissimilatory enzyme used for redox balancing (7, 60, 75, 77, 84). Maintenance of an appropriate redox balance can be necessary for optimal bacterial growth under some physiological conditions, particularly during fermentative processes in enteric bacteria, oxidative metabolism of highly reduced carbon substrates in aerobic heterotrophs, or anaerobic photoheterotrophic growth in photosynthetic bacteria. In addition, since oxygen primarily inhibits denitrification at the level of nitrate transport (23) and the Nap system does not require this step, some denitrifiers perform aerobic denitrification coupling the Nap enzyme to the nitrite and N-oxide reductases (3, 7). Aerobic denitrification can be a valuable feature for organisms growing under microaerobic conditions or in environments rapidly changing between aerobic conditions and anoxia. Other proposed roles for Nap are the adaptation to anaerobic metabolism after transition from aerobic conditions, the utilization of alternate reductants (85), or even a self-defense mechanism forming high nitrite levels to inhibit the growth of potential competing bacteria (46).
Nap systems have been studied at the biochemical or genetic level in Alcaligenes eutrophus (Ralstonia eutropha), T. pantotropha (P. denitrificans), E. coli, and Rhodobacter species (Fig. 4). The enzyme is a heterodimer containing a 90-kDa catalytic subunit (NapA) with MGD cofactor and an N-terminal [4Fe-4S] center and a 15-kDa biheme c cytochrome (NapB), which receives electrons from NapC, a membrane-bound tetraheme cytochrome c of 25 kDa (6, 8, 72, 73). Activity with reduced viologens as electron donors decreases when the NapB subunit is lost during the purification of the R. capsulatus enzyme (55). Evidence for a role of NapC in the electron transfer to the periplasmic enzyme complex has been presented by the results of mutational analysis in R. sphaeroides (72, 73). NapC homologues are involved in electron transfer between the membrane quinol pool and several soluble periplasmic reductases. Electron transfer by the NapC family appears to be cytochrome bc1 independent and is not coupled to proton translocation. The spectroscopic characterization of a soluble form of NapC expressed as a periplasmic protein has been recently reported (78).
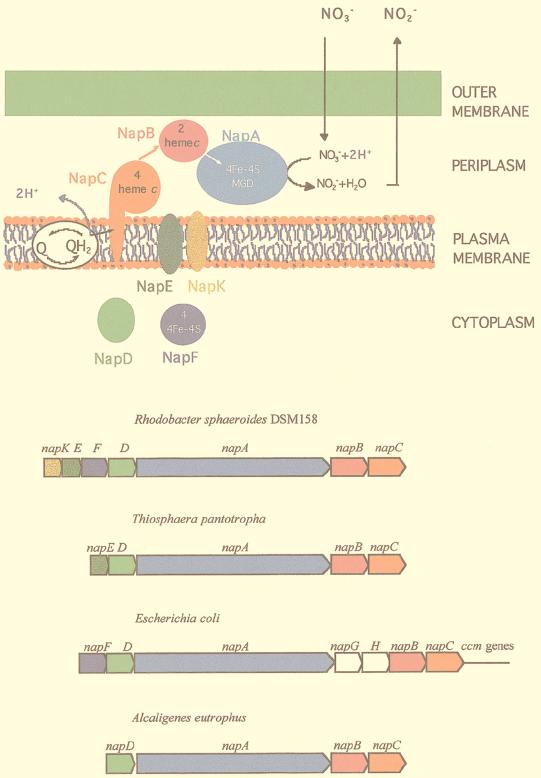
FIG. 4.
Periplasmic nitrate reduction and organization of the nap gene clusters in bacteria. The R. sphaeroides periplasmic nitrate-reducing system is shown at the top (the small black arrows indicate the electron flow). The comparative organization of the nap gene clusters of R. sphaeroides, T. pantotropha, E. coli, and A. eutrophus is also shown. Genes are drawn approximately to scale, and arrows show the direction of transcription. The genes and their products are drawn in the same color. The products of the white genes are not shown. The quinone pool is indicated by the white oval labeled Q⇄QH2.
The crystal structure of the Desulfovibrio desulfuricans Nap protein, a single-subunit form that lacks the biheme cytochrome c subunit (NapB), has been recently determined at a resolution of 1.9 Å (24). The Nap protein is folded into four domains, all of them involved in MGD cofactor binding, and its structure is more similar to that of formate dehydrogenase than to that of dimethylsulfoxide reductase. In the catalytic site, a single Mo atom is coordinated to two MGD molecules, a Cys residue, and a water/hydroxo ligand, and an electron transfer pathway through bonds connects the molybdenum and the [4Fe-4S] center (24). This structure suggests a mono-oxo/desoxo catalytic cycle, although a di-oxo/mono-oxo cycle has also been proposed for the T. pantotropha enzyme (5). The results of electron paramagnetic resonance and X-ray absorption spectroscopy analyses suggest that the Mo environments in the soluble Nas and Nap enzymes are similar to each other but distinct from that in the membrane-bound Nar (5, 14, 35). Nap and Nar are also catalytically distinct: Nap is less sensitive to inhibition by cyanide, does not use chlorate as substrate, and is slightly stimulated by thiocyanate and azide (7, 43). However, in R. sphaeroides, Nap activity is competitively inhibited by chlorate. Both nitrate and chlorate stimulate the phototrophic growth in the wild-type strain, but not in a NapA− mutant. This Nap-dependent chlorate or nitrate stimulation of bacterial growth has been explained in terms of redox balancing; the dissipation of excess photosynthetic reducing power allows optimal growth (61, 77).
Organization of genes coding for periplasmic nitrate reductases.
Since the napAB structural genes of A. eutrophus were first identified (85), several nap loci have been sequenced (Fig. 4) (2, 8, 32, 37, 72, 73). It is worth noting that nap genes are located on endogenous plasmids in R. capsulatus (97), R. sphaeroides (18), A. eutrophus (85), and P. denitrificans (76). In these bacteria, nap gene expression is unaffected by O2. However, the E. coli nap genes are clustered on the chromosome (aeg-46.5 locus) and are induced anaerobically by Fnr (21, 37). The plasmid location of most nap genes, the heterologous expression of nap genes (72), and the fact that the ability to reduce nitrate is present in only a few wild-type strains of purple bacteria suggest the possibility of horizontal transfer of nap genes within the bacterial community.
Seven genes involved in periplasmic nitrate reduction are clustered in the napKEFDABC operon in R. sphaeroides (Fig. 4) (72, 73). The structural napABC genes are essential for in vivo activity. NapE and NapK are small transmembrane proteins of unknown function. NapF, a soluble protein of 16 kDa with four Cys clusters that probably bind four [4Fe-4S] centers, could be involved in the assembly of the iron-sulfur center of NapA. Finally, NapD is a 9-kDa cytoplasmic protein that could participate in maturation or processing of NapA (73). Similar napEFDABC and napEDABC gene clusters are found in Pseudomonas sp. strain G-179 and T. pantotropha, respectively (2, 8). In the former bacterium, nap genes are linked to nir and nor genes involved in nitrite and nitric oxide reduction, respectively (2). In E. coli, seven nap genes and eight cytochrome c biogenesis genes are clustered in the anaerobically regulated aeg-46.5 locus (37). This locus lacks a napE-homologous gene but contains two additional napGH genes. NapG is a 20-kDa soluble protein with four putative [4Fe-4S] centers, and NapH is a 32-kDa membrane protein that probably binds two [4Fe-4S] centers. It has been proposed that a putative NapGH complex could act as a redox sensor controlling the electron flow to NapA (7). Sequencing of the Haemophilus influenzae genome (32) has shown the presence of a nap locus organized identically to that in E. coli but unlinked to cytochrome c biogenesis genes.
Regulation of dissimilatory periplasmic nitrate reductases.
Although there are some differences in the nap gene expression depending on the organisms, the Nap systems are normally unaffected by ammonium or oxygen. In phototrophic bacteria, the Nap activity is present under aerobic and anaerobic conditions and is unaffected by ammonium or by the intracellular C and N balance. In addition, Nap activity is stimulated by nitrate, although a basal activity is also observed in the absence of nitrate (26, 72). In P. denitrificans, the Nap activity is observed in aerobically grown cells even in the absence of nitrate. Activity is maximally expressed during growth on highly reduced carbon sources, such as butyrate, suggesting a Nap regulation in response to the redox state of the bacterium (84). Similarly, the Nap system is not induced by nitrate in A. eutrophus, and maximal expression is observed under aerobic conditions at the stationary phase (85). However, expression of the E. coli nap operon (aeg-46.5 locus) is induced during anaerobic conditions, via the Fnr regulator, and by nitrate or in a lesser extent by nitrite, via the homologous regulators NarL and NarP. Both proteins compete in vivo for a common binding site in the aeg-46.5 promoter region, but only NarP activates gene expression. Thus, NarL has a negative effect on expression of the aeg-46.5 operon because it antagonizes NarP-dependent activation (21). The nap gene cluster of Pseudomonas sp. strain G-179 could also be regulated by a Fnr-like protein under anaerobic conditions (2).
Translocation of Nap to the periplasm.
The periplasmic location of the Nap enzyme raises important questions about its export process. The cytochrome c subunit (NapB) contains the typical N-terminal signal sequence required for a Sec-dependent translocation (for a recent review, see reference 30). Therefore, heme binding to the NapB apoprotein can take place in the periplasm, as demonstrated for other cytochromes c (67). However, several observations suggest that the catalytic NapA subunit could be assembled in the cytoplasm and exported into the periplasm in a folded conformation by a Sec-independent pathway. First, NapA and other periplasmic molybdoenzymes contain N-terminal signal sequences that are unusually long and bear a twin-Arg motif. This presequence can be involved in an alternative translocation system similar to the PMF-dependent thylakoid import pathway (30). Second, MGD cofactor is synthesized in the cytoplasm by the action of five different loci (moa, mob, mod, moe, and mog), and no cofactor export system has been reported for any bacteria analyzed so far. In addition, the size of the cofactor and its almost completely buried environment within the protein (24) suggest that the MGD cofactor should be assembled in the cytoplasm prior to protein translocation. It has been demonstrated that cofactor insertion into the apoprotein is a prerequisite for the translocation of the E. coli trimethylamine N-oxide reductase by a Sec-independent pathway (81). Recently, the E. coli genes required for the Sec-independent export of cofactor-containing periplasmic proteins have been identified (82, 95).
CONCLUDING REMARKS
Recent biochemical and genetic studies of nitrate reduction have revealed an unexpected complexity for this process, which is of particular significance within the biogeochemical nitrogen cycle. Three distinct classes of bacterial nitrate-reducing systems, which are clearly different at the level of cellular location, structure, biochemical properties, regulation, and gene organization, have been described. Bacterial nitrate reductases are also different from eukaryotic nitrate reductase. The three bacterial nitrate reductases, all of which are predicted to contain a MGD cofactor, can be present in the same organism (85). Although some differences among various organisms are observed for each type of nitrate-reducing system, the properties of distinct nitrate reductases can be rationalized. Some bacteria contain a cytoplasmic assimilatory nitrate reductase that enables the utilization of nitrate as a nitrogen source. This Nas enzyme is usually induced by nitrate and repressed by ammonium but is not affected by oxygen. Depending on the organism, the enzyme uses ferredoxin, flavodoxin, or NADH as the electron donor. The ferredoxin- and flavodoxin-Nas proteins are single polypeptides with MGD and one [4Fe-4S] or [3Fe-4S] center, whereas the NADH-Nas protein is a heterodimer of a large iron-sulfur and MGD-containing catalytic subunit and a small FAD-containing diaphorase subunit. The membrane-bound respiratory nitrate reductase of some denitrifiers and nitrate-respiring bacteria allows ATP synthesis by using nitrate as an alternative electron acceptor under anaerobic conditions. This Nar system is generally induced by nitrate and repressed by oxygen, but it is insensitive to ammonium. The enzyme is a three-subunit complex of a MGD-containing catalytic subunit, an iron-sulfur subunit with one [3Fe-4S] and three [4Fe-4S] centers, and a biheme b membrane-anchoring subunit. Finally, a periplasmic dissimilatory nitrate reductase is found in many gram-negative bacteria; in most of these gram-negative bacteria, the enzyme is not repressed by either ammonium or oxygen. This Nap system seems to be involved in aerobic denitrification and/or the maintenance of an optimal redox balance. Structural differences among bacterial nitrate reductases are also revealed by comparisons of the amino acid sequences of the catalytic subunits. These comparisons show that only the enzymes of the same type are closely related (more than 60% of sequence identity), whereas Nas, Nar, and Nap are only 20 to 35% identical, which is the same degree of sequence identity found for other bacterial molybdoenzymes (72). The results of spectroscopic and sequence analyses also indicate that the soluble Nas and Nap proteins are more closely related, with essentially identical active sites, but they are distinct from the membrane-bound Nar. In addition, the crystal structure of Nap shows more resemblance to the formate dehydrogenase enzyme than to dimethylsulfoxide reductase (24). Although Nas, Nar, and Nap systems seem to perform different physiological functions, some differences are observed among the organisms. Also, the enzymes can sometimes play distinct roles under different metabolic conditions and assimilatory, respiratory, and dissimilatory pathways can be interconnected to facilitate a rapid adaptation to changing nitrogen and/or oxygen conditions, increasing the metabolic plasticity for survival in natural environments.
ACKNOWLEDGMENTS
We thank Emilio Fernández for helpful comments on the manuscript.
Work in our laboratory was supported in part by grants from DGICYT (PB95-0554-CO2-02) and Junta de Andalucía (CVI 0117), Seville, Spain, and Alexander von Humboldt Foundation, Bonn, Germany.
REFERENCES
- 1.Aichi M, Omata T. Involvement of NtcB, a LysR family transcription factor, in nitrite activation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC7942. J Bacteriol. 1997;179:4671–4675. doi: 10.1128/jb.179.15.4671-4675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedzyk L, Wang T, Ye R W. The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J Bacteriol. 1999;181:2802–2806. doi: 10.1128/jb.181.9.2802-2806.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell L C, Richardson D J, Ferguson S J. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. The periplasmic enzyme catalyzes the first step in aerobic denitrification. FEBS Lett. 1990;265:85–87. doi: 10.1016/0014-5793(90)80889-q. [DOI] [PubMed] [Google Scholar]
- 4.Bell L C, Page M D, Berks B C, Richardson D J, Ferguson S J. Insertion of transposon Tn5 into a structural gene of the membrane-bound nitrate reductase of Thiosphaera pantotropha results in anaerobic overexpression of periplasmic nitrate reductase activity. J Gen Microbiol. 1993;139:3205–3214. doi: 10.1099/00221287-139-12-3205. [DOI] [PubMed] [Google Scholar]
- 5.Bennett B, Charnock J M, Sears H J, Berks B C, Thomson A J, Ferguson S J, Garner C D, Richardson D J. Structural investigation of the molybdenum site of the periplasmic nitrate reductase from Thiosphaera pantotropha by X-ray absorption spectroscopy. Biochem J. 1996;317:557–563. doi: 10.1042/bj3170557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berks B C, Richardson D J, Robinson C, Reilly A, Aplin R T, Ferguson S J. Purification and characterization of the periplasmic nitrate reductase from Thiosphaera pantotropha. Eur J Biochem. 1994;220:117–124. doi: 10.1111/j.1432-1033.1994.tb18605.x. [DOI] [PubMed] [Google Scholar]
- 7.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 8.Berks B C, Richardson D J, Reilly A, Willis A C, Ferguson S J. The napEDABC gene cluster encoding the periplasmic nitrate reductase system of Thiosphaera pantotropha. Biochem J. 1995;309:983–992. doi: 10.1042/bj3090983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blasco F, Iobbi C, Ratouchniak J, Bonnefoy V, Chippaux M. Nitrate reductase of Escherichia coli: sequence of the second nitrate reductase and comparison with that encoded by the narGHJI operon. Mol Gen Genet. 1990;222:104–111. doi: 10.1007/BF00283030. [DOI] [PubMed] [Google Scholar]
- 10.Blasco F, Nunzi F, Pommier J, Brasseur R, Chippaux M, Giordano G. Formation of active heterologous nitrate reductase between nitrate reductases A and Z of Escherichia coli. Mol Microbiol. 1992;6:209–219. doi: 10.1111/j.1365-2958.1992.tb02002.x. [DOI] [PubMed] [Google Scholar]
- 11.Blasco F, Pommier J, Augier V, Chippaux M, Giordano G. Involvement of the narJ or narW gene product in the formation of active nitrate reductase in Escherichia coli. Mol Microbiol. 1992;6:221–230. doi: 10.1111/j.1365-2958.1992.tb02003.x. [DOI] [PubMed] [Google Scholar]
- 12.Blasco R, Castillo F, Martínez-Luque M. The assimilatory nitrate reductase from the phototrophic bacterium, Rhodobacter capsulatus E1F1, is a flavoprotein. FEBS Lett. 1997;414:45–49. doi: 10.1016/s0014-5793(97)00968-x. [DOI] [PubMed] [Google Scholar]
- 13.Bonnefoy V, DeMoss J A. Nitrate reductases in Escherichia coli. Antonie Leeuwenhoek. 1994;66:47–56. doi: 10.1007/BF00871632. [DOI] [PubMed] [Google Scholar]
- 14.Breton J, Berks B C, Reilly A, Thomson A J, Ferguson S J, Richardson D J. Characterization of the paramagnetic iron-containing redox centers of Thiosphaera pantotropha periplasmic nitrate reductase. FEBS Lett. 1994;345:76–80. doi: 10.1016/0014-5793(94)00445-5. [DOI] [PubMed] [Google Scholar]
- 15.Brittain T, Blackmore R, Greenwood C, Thomson A J. Bacterial nitrite-reducing enzymes. Eur J Biochem. 1992;209:793–802. doi: 10.1111/j.1432-1033.1992.tb17350.x. [DOI] [PubMed] [Google Scholar]
- 16.Cai Y, Wolk C P. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J Bacteriol. 1997;179:258–266. doi: 10.1128/jb.179.1.258-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell W H, Kinghorn J R. Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem Sci. 1990;15:315–319. doi: 10.1016/0968-0004(90)90021-3. [DOI] [PubMed] [Google Scholar]
- 18.Castillo F, Dobao M M, Reyes F, Blasco R, Roldán M D, Gavira M, Caballero F J, Moreno-Vivián C, Martínez-Luque M. Molecular and regulatory properties of the nitrate reducing systems of Rhodobacter. Curr Microbiol. 1996;33:341–346. doi: 10.1007/s002849900125. [DOI] [PubMed] [Google Scholar]
- 19.Chiang R C, Cavicchioli R, Gunsalus R P. ‘Locked-on’ and ‘locked-off’ signal transduction mutations in the periplasmic domain of the Escherichia coli NarQ and NarX sensors affect nitrate- and nitrite-dependent regulation by NarL and NarP. Mol Microbiol. 1997;24:1049–1060. doi: 10.1046/j.1365-2958.1997.4131779.x. [DOI] [PubMed] [Google Scholar]
- 20.Darwin A J, Stewart V. Expression of the narX, narL, narP, and narQ genes of Escherichia coli K-12: regulation of the regulators. J Bacteriol. 1995;177:3865–3869. doi: 10.1128/jb.177.13.3865-3869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darwin A J, Stewart V. Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J Mol Biol. 1995;251:15–29. doi: 10.1006/jmbi.1995.0412. [DOI] [PubMed] [Google Scholar]
- 22.Darwin A J, Tyson K L, Busby S J W, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 23.Denis K, Dias F M, Rowe J J. Oxygen regulation of nitrate transport by diversion of electron flow in Escherichia coli. J Biol Chem. 1990;265:18095–18097. [PubMed] [Google Scholar]
- 24.Dias J M, Than M E, Humm A, Huber R, Bourenkov G P, Bartunik H D, Bursakov S, Calvete J, Caldeira J, Carneiro C, Moura J J G, Moura I, Romão M J. Crystal structure of the first dissimilatory nitrate reductase at 1.9 Å solved by MAD methods. Structure. 1999;7:65–79. doi: 10.1016/s0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 25.Dobao M M, Martínez-Luque M, Castillo F. Nitrate reductase activity in spheroplasts from Rhodobacter capsulatus E1F1 requires a periplasmic protein. Arch Microbiol. 1993;160:471–476. [Google Scholar]
- 26.Dobao M M, Martínez-Luque M, Moreno-Vivián C, Castillo F. Effect of carbon and nitrogen metabolism on nitrate reductase activity of Rhodobacter capsulatus E1F1. Can J Microbiol. 1994;40:645–650. [Google Scholar]
- 27.Dubourdieu M, DeMoss J A. The narJ gene product is required for biogenesis of respiratory nitrate reductase in Escherichia coli. J Bacteriol. 1992;174:867–872. doi: 10.1128/jb.174.3.867-872.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eaves D J, Grove J, Staudenmann W, James P, Pode R K, White S A, Griffiths I, Cole J A. Involvement of products of the nrfEFG genes in the covalent attachment of haem c to a novel cysteine-lysine motif in the cytochrome c552 nitrite reductase from Escherichia coli. Mol Microbiol. 1998;28:205–216. doi: 10.1046/j.1365-2958.1998.00792.x. [DOI] [PubMed] [Google Scholar]
- 29.Fast B, Lindgren P E, Götz F. Cloning, sequencing, and characterization of a gene (narT) encoding a transport protein involved in dissimilatory nitrate reduction in Staphylococcus carnosus. Arch Microbiol. 1997;166:361–367. doi: 10.1007/BF01682980. [DOI] [PubMed] [Google Scholar]
- 30.Fekkes P, Driessen A J M. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernández E, Galván A, Quesada A. Nitrogen assimilation and its regulation. In: Rochaix J D, Goldschmidt-Clermont M, Merchant S, editors. The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 637–659. [Google Scholar]
- 32.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 33.Frias J E, Flores E, Herrero A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gangeswaran R, Eady R A. Flavodoxin 1 of Azotobacter vinelandii: characterization and role in electron donation to purified assimilatory nitrate reductase. Biochem J. 1996;317:103–108. doi: 10.1042/bj3170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangeswaran R, Lowe D J, Eady R R. Purification and characterization of the assimilatory nitrate reductase of Azotobacter vinelandii. Biochem J. 1993;289:335–342. doi: 10.1042/bj2890335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldman B S, Lin J T, Stewart V. Identification and structure of the nasR gene encoding a nitrate- and nitrite-responsive positive regulator of nasFEDCBA (nitrate assimilation) operon expression in Klebsiella pneumoniae M5a1. J Bacteriol. 1994;176:5077–5085. doi: 10.1128/jb.176.16.5077-5085.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grove J, Tanapongpipat S, Thomas G, Griffiths L, Crooke H, Cole J. Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol Microbiol. 1996;19:467–481. doi: 10.1046/j.1365-2958.1996.383914.x. [DOI] [PubMed] [Google Scholar]
- 38.Guerrero M G, Vega J M, Losada M. The assimilatory nitrate-reducing system and its regulation. Annu Rev Plant Physiol. 1981;32:169–204. [Google Scholar]
- 39.Guigliarelli B, Magalon A, Asso M, Bertrand P, Frixon C, Giordano G, Blasco F. Complete coordination of the four Fe-S centers of the β subunit from Escherichia coli nitrate reductase. Physiological, biochemical, and EPR characterization of site-directed mutants lacking the highest or lowest potential [4Fe-4S] clusters. Biochemistry. 1996;35:4828–4836. doi: 10.1021/bi952459p. [DOI] [PubMed] [Google Scholar]
- 40.Gutiérrez J C, Ramos F, Ortner L, Tortolero M. nasST, two genes involved in the induction of the assimilatory nitrate-nitrite reductase operon (nasAB) of Azotobacter vinelandii. Mol Microbiol. 1995;18:579–591. doi: 10.1111/j.1365-2958.1995.mmi_18030579.x. [DOI] [PubMed] [Google Scholar]
- 41.Gutiérrez J C, Santero E, Tortolero M. Ammonium repression of the nitrite-nitrate (nasAB) assimilatory operon of Azotobacter vinelandii is enhanced in mutants expressing the nifO gene at high levels. Mol Gen Genet. 1997;255:172–179. doi: 10.1007/s004380050486. [DOI] [PubMed] [Google Scholar]
- 42.Harborne N R, Griffiths L, Busby S J W, Cole J A. Transcriptional control, translation and function of the products of the five open reading frames of the Escherichia coli nir operon. Mol Microbiol. 1992;6:2805–2813. doi: 10.1111/j.1365-2958.1992.tb01460.x. [DOI] [PubMed] [Google Scholar]
- 43.Hochstein L I, Tomlinson G A. The enzymes associated with denitrification. Annu Rev Microbiol. 1988;42:231–261. doi: 10.1146/annurev.mi.42.100188.001311. [DOI] [PubMed] [Google Scholar]
- 44.Hussain H, Grove J, Griffiths L, Busby S, Cole J. A seven-gene operon essential for formate-dependent nitrite reduction to ammonia by enteric bacteria. Mol Microbiol. 1994;12:153–163. doi: 10.1111/j.1365-2958.1994.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 45.Kaneko T, Satoh S, Kotani H, Suzuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 46.Kerber N L, Cárdenas J. Nitrate reductase from Rhodopseudomonas sphaeroides. J Bacteriol. 1982;150:1091–1097. doi: 10.1128/jb.150.3.1091-1097.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley P J. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J T, Stewart V. Nitrate and nitrite-mediated transcription antitermination control of nasF (nitrate assimilation) operon expression in Klebsiella pneumoniae M5a1. J Mol Biol. 1996;256:423–435. doi: 10.1006/jmbi.1996.0098. [DOI] [PubMed] [Google Scholar]
- 49.Lin J T, Stewart V. Nitrate assimilation by bacteria. Adv Microb Physiol. 1998;38:1–30. doi: 10.1016/s0065-2911(08)60014-4. [DOI] [PubMed] [Google Scholar]
- 50.Lin J T, Goldman B S, Stewart V. The nasFEDCBA operon for nitrate and nitrite assimilation in Klebsiella pneumoniae M5a1. J Bacteriol. 1994;176:2551–2559. doi: 10.1128/jb.176.9.2551-2559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luque I, Herrero A, Flores E, Madueño F. Clustering of genes involved in nitrate assimilation in the cyanobacterium Synechococcus. Mol Gen Genet. 1992;232:7–11. doi: 10.1007/BF00299130. [DOI] [PubMed] [Google Scholar]
- 52.Luque I, Flores E, Herrero A. Nitrite reductase gene from Synechococcus sp. PCC 7942: homology between cyanobacterial and higher-plant nitrite reductases. Plant Mol Biol. 1993;21:1201–1205. doi: 10.1007/BF00023618. [DOI] [PubMed] [Google Scholar]
- 53.Luque I, Flores E, Herrero A. Nitrate and nitrite transport in the cyanobacterium Synechococcus sp. PCC 7942 are mediated by the same permease. Biochim Biophys Acta. 1994;1184:296–298. [Google Scholar]
- 54.Magalon A, Asso M, Guigliarelli B, Rothery R A, Bertrand P, Giordano G, Blasco F. Molybdenum cofactor properties and [Fe-S] cluster coordination in Escherichia coli nitrate reductase A: investigation by site-directed mutagenesis of the conserved His-50 residue in the NarG subunit. Biochemistry. 1998;37:7363–7370. doi: 10.1021/bi972858f. [DOI] [PubMed] [Google Scholar]
- 55.McEwan A G, Wetzstein H G, Meyer O, Jackson J B, Ferguson S J. The periplasmic nitrate reductase of Rhodobacter capsulatus: purification, characterization and distinction from a single reductase for trimethylamine-N-oxide, dimethylsulfoxide and chlorate. Arch Microbiol. 1987;147:340–345. [Google Scholar]
- 56.Merchán F, Kindle K L, Llama M J, Serra J L, Fernández E. Cloning and sequencing of the nitrate transport system from the thermophilic, filamentous cyanobacterium Phormidium laminosum: comparative analysis with the homologous system from Synechococcus sp. PCC 7942. Plant Mol Biol. 1995;28:759–766. doi: 10.1007/BF00021199. [DOI] [PubMed] [Google Scholar]
- 57.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metheringham R, Cole J A. A reassessment of the genetic determinants, the effect of growth conditions and the availability of an electron donor on the nitrosating activity of Escherichia coli K-12. Microbiology. 1997;143:2647–2656. doi: 10.1099/00221287-143-8-2647. [DOI] [PubMed] [Google Scholar]
- 59.Mikami B, Ida S. Purification and properties of ferredoxin-nitrate reductase from the cyanobacterium Plectonema boryanum. Biochim Biophys Acta. 1984;791:294–304. [Google Scholar]
- 60.Moreno-Vivián C, Ferguson S J. Definition and distinction between assimilatory, dissimilatory and respiratory pathways. Mol Microbiol. 1998;29:664–666. doi: 10.1046/j.1365-2958.1998.00946.x. [DOI] [PubMed] [Google Scholar]
- 61.Moreno-Vivián C, Roldán M D, Reyes F, Castillo F. Isolation and characterization of transposon Tn5 mutants of Rhodobacter sphaeroides deficient in both nitrate and chlorate reduction. FEMS Microbiol Lett. 1994;115:279–284. [Google Scholar]
- 62.Muñoz-Centeno M C, Cejudo F J, Ruiz M T, Paneque A. The Azotobacter chroococcum nitrate permease is a multicomponent system. Biochim Biophys Acta. 1993;1141:75–80. [Google Scholar]
- 63.Nakano M M, Yang F, Hardin P, Zuber P. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J Bacteriol. 1995;177:573–579. doi: 10.1128/jb.177.3.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakano M M, Hoffmann T, Zhu Y, Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogawa K I, Akagawa E, Yamane K, Sun Z W, LaCelle M, Zuber P, Nakano M M. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Omata T. Structure, function and regulation of the nitrate transport system of the cyanobacterium Synechococcus sp. PCC7942. Plant Cell Physiol. 1995;36:207–213. doi: 10.1093/oxfordjournals.pcp.a078751. [DOI] [PubMed] [Google Scholar]
- 67.Page M D, Sambongi Y, Ferguson S J. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem Sci. 1998;23:103–108. doi: 10.1016/s0968-0004(98)01173-6. [DOI] [PubMed] [Google Scholar]
- 68.Palmer T, Santini C L, Iobbi-Nivol C, Eaves D J, Boxer D H, Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol Microbiol. 1996;20:875–884. doi: 10.1111/j.1365-2958.1996.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 69.Rajagopalan K V, Johnson J L. The pterin molybdenum cofactors. J Biol Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- 70.Ramírez-Arcos S, Fernández-Herrero L A, Berenger J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim Biophys Acta. 1998;1396:215–227. doi: 10.1016/s0167-4781(97)00183-8. [DOI] [PubMed] [Google Scholar]
- 71.Ramos F, Blanco G, Gutiérrez J C, Luque F, Tortolero M. Identification of an operon involved in the assimilatory nitrate-reducing system of Azotobacter vinelandii. Mol Microbiol. 1993;8:1145–1153. doi: 10.1111/j.1365-2958.1993.tb01659.x. [DOI] [PubMed] [Google Scholar]
- 72.Reyes F, Roldán M D, Klipp W, Castillo F, Moreno-Vivián C. Isolation of periplasmic nitrate reductase genes from Rhodobacter sphaeroides DSM 158: structural and functional differences among prokaryotic nitrate reductases. Mol Microbiol. 1996;19:1307–1318. doi: 10.1111/j.1365-2958.1996.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 73.Reyes F, Gavira M, Castillo F, Moreno-Vivián C. Periplasmic nitrate-reducing system of the phototrophic bacterium Rhodobacter sphaeroides DSM 158: transcriptional and mutational analysis of the napKEFDABC gene cluster. Biochem J. 1998;331:897–904. doi: 10.1042/bj3310897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Richardson D J, Watmough N J. Inorganic nitrogen metabolism in bacteria. Curr Opin Chem Biol. 1999;3:207–219. doi: 10.1016/S1367-5931(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 75.Richardson D J, King G F, Kelly D K, McEwan A G, Ferguson S J, Jackson J B. The role of auxiliary oxidants in maintaining redox balance during phototrophic growth of Rhodobacter capsulatus on propionate or butyrate. Arch Microbiol. 1988;150:131–137. [Google Scholar]
- 76.Richardson D J, Sears H, Berks B, Ferguson S J, Spiro S. The bacterial nitrate reductases. In: Vega J M, Aparicio P J, Castillo F, Maldonado J M, editors. Avances en el metabolismo del nitrógeno: de la fisiología a la biología molecular. Seville, Spain: Universidad de Sevilla; 1998. pp. 565–574. [Google Scholar]
- 77.Roldán M D, Reyes F, Moreno-Vivián C, Castillo F. Chlorate and nitrate reduction in the phototrophic bacteria Rhodobacter capsulatus and Rhodobacter sphaeroides. Curr Microbiol. 1994;29:241–245. [Google Scholar]
- 78.Roldán M D, Sears H J, Cheesman M R, Ferguson S J, Thomson A J, Berks B C, Richardson D J. Spectroscopic characterization of a novel multiheme c-type cytochrome widely implicated in bacterial electron transport. J Biol Chem. 1998;273:28785–28790. doi: 10.1074/jbc.273.44.28785. [DOI] [PubMed] [Google Scholar]
- 79.Rowe J J, Ubbink-Kok T, Molenaar D, Konings W N, Driessen A J M. NarK is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol Microbiol. 1994;12:579–586. doi: 10.1111/j.1365-2958.1994.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 80.Rubio L M, Herrero A, Flores E. A cyanobacterial narB gene encodes a ferredoxin-dependent nitrate reductase. Plant Mol Biol. 1996;30:845–850. doi: 10.1007/BF00019017. [DOI] [PubMed] [Google Scholar]
- 81.Santini C L, Ize B, Chanal A, Müller M, Giordano G, Wu L F. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sargent F, Bogsch E G, Stanley N, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schröeder I, Darie S, Gunsalus R P. Activation of the Escherichia coli nitrate reductase (narGHJI) operon by NarL and Fnr requires integration host factor. J Biol Chem. 1993;268:771–774. [PubMed] [Google Scholar]
- 84.Sears H J, Spiro S, Richardson D J. Effect of carbon substrate and aeration on nitrate reduction and expression of the periplasmic and membrane-bound nitrate reductases in carbon-limited continuous cultures of Paracoccus denitrificans Pd1222. Microbiology. 1997;143:3767–3774. doi: 10.1099/00221287-143-12-3767. [DOI] [PubMed] [Google Scholar]
- 85.Siddiqui R A, Warnecke-Eberz U, Hengsberger A, Schneider B, Kostka S, Friedrich B. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J Bacteriol. 1993;175:5867–5876. doi: 10.1128/jb.175.18.5867-5876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Solomonson L P, Barber M J. Assimilatory nitrate reductase: functional properties and regulation. Annu Rev Plant Physiol Mol Biol. 1990;41:225–253. [Google Scholar]
- 87.Spiro S. The FNR family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 88.Stewart V. Regulation of nitrate and nitrite reduction in enterobacteria. Antonie Leeuwenhoek. 1994;66:37–45. doi: 10.1007/BF00871631. [DOI] [PubMed] [Google Scholar]
- 89.Sundermeyer-Klinger H, Meyer W, Warninghoff B, Bock E. Membrane-bound nitrite oxidoreductase of Nitrobacter: evidence for a nitrate reductase system. Arch Microbiol. 1984;149:153–158. [Google Scholar]
- 90.Suzuki I, Sugiyama T, Omata T. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 91.Tyson K L, Cole J A, Busby S J W. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for both NarP- and NarL-dependent regulation. Mol Microbiol. 1994;13:1045–1055. doi: 10.1111/j.1365-2958.1994.tb00495.x. [DOI] [PubMed] [Google Scholar]
- 92.Unthan M, Klipp W, Schmid G H. Nucleotide sequence of the narB gene encoding assimilatory nitrate reductase from the cyanobacterium Oscillatoria chalybea. Biochim Biophys Acta. 1996;1305:19–24. doi: 10.1016/0167-4781(95)00210-3. [DOI] [PubMed] [Google Scholar]
- 93.Van Maanen J M S, Welle I J, Hageman G, Dallinga J W, Mertens P L J M, Kleinjans J C S. Nitrate contamination of drinking water: relationship with HPTR variant frequency in lymphocyte DNA and urinary excretion of N-nitrosamines. Environ Health Perspect. 1996;104:522–528. doi: 10.1289/ehp.96104522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vega-Palas M A, Flores E, Herrero A. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 95.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 96.Williams S B, Stewart V. Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coli K-12. Mol Microbiol. 1997;26:911–925. doi: 10.1046/j.1365-2958.1997.6262002.x. [DOI] [PubMed] [Google Scholar]
- 97.Willison J C. Derivatives of Rhodobacter capsulatus strain AD2 cured of their endogenous plasmid are unable to utilize nitrate. FEMS Microbiol Lett. 1990;66:23–28. [Google Scholar]
- 98.Wu Q, Stewart V. NasFED proteins mediate assimilatory nitrate and nitrite transport in Klebsiella oxytoca (pneumoniae) M5a1. J Bacteriol. 1998;180:1311–1322. doi: 10.1128/jb.180.5.1311-1322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang X, DeMoss J A. Structure modification induced in the narG promoter by binding of integration host factor and NARL-P. J Bacteriol. 1996;178:3971–3973. doi: 10.1128/jb.178.13.3971-3973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]