Abstract
In this study, a simple and efficient miracidium hatching technique (MHT) protocol for preparing a single-genome DNA of Schistosoma japonicum was proposed. The protocol was designed with 96-well plates to collect a miracidium for single-genome DNA preparation, and the effects of lighting conditions on hatching rates were evaluated. The highest hatching rate was recorded under sunlight (92.4%), followed by fluorescent light (88.0%), and the lowest rate was recorded under the dark condition (4.7%). The results suggested for the first time, to our knowledge, that sunlight was efficient for this simple MHT protocol. Successful amplification of microsatellite marker genes using DNA isolated from a single miracidium also confirmed the quality of the single-genome DNA for subsequent applications.
Keywords: hatching, miracidia, Schistosoma japonicum
Schistosomiasis is one of the most important parasitic diseases caused by digenean trematodes of the genus Schistosoma, which are widely distributed in tropical and subtropical countries and seriously endanger the health of both humans and animals. Schistosoma japonicum, which causes Asian zoonotic schistosomiasis, infects more than 40 species of wild and domestic animals and is endemic in the Philippines and parts of China and Indonesia [2]. Information regarding the genetic diversity of S. japonicum will help in understanding the epidemiology and transmission dynamics of the parasite between humans and reservoir animal hosts in the field. To analyze the microsatellite DNA of the parasite [1], DNA samples derived from a single genome should be prepared. However, the collection of adult worms from definitive hosts directly in the field is unfeasible because the worms inhabit the inferior mesenteric and superior hemorrhoidal veins during schistosome infection of the host [8]. The adult worm developed in experimentally infected laboratory animals with cercariae obtained from naturally infected snails may pose a loss of true genetic diversity under immunological selection brought by artificial infection [10]. On the other hand, the eggs from the host feces can be singly separated, but DNA extraction from the eggs is difficult due to their thick egg-shell. Therefore, miracidium, the larval stage of the parasite formed inside the egg, may be useful for the preparation of single-genome DNA.
A previously reported miracidium hatching technique (MHT), which has been applied for diagnosis, requires specific equipment, artificial light, and is time consuming [4]. Thus, in the present study, the previous MHT protocol was modified and simplified to recover miracidia under field study for preparation of single-genome DNA that can be used for a population genetics study with microsatellite markers.
The S. japonicum Yamanashi strain was maintained using the snail intermediate host Oncomelania hupensis nosophora [6]. The snails were infected with miracidia and crushed 6 months later to collect cercariae. Eleven 5-week-old female ICR mice (Clea Inc., Tokyo, Japan) were used for S. japonicum infection. The mice were percutaneously infected with 30 cercariae each, and their fecal samples were collected after 6 weeks of infection. This study was approved by the Committee for Animal Experimentation of Obihiro University of Agriculture and Veterinary Medicine (Approval no. 20-38) and Dokkyo Medical University (Approval nos. 29-52 and 0006).
One g of fecal sample collected from the parasite-infected mice was emulsified with 10 mL of dechlorinated tap water, filtered by stainless mesh (TESTING SIEVE: wire diameter of 71 µm and aperture of 106 µm; Tokyo Screen Co., Ltd., Tokyo, Japan), and left for 20 min at room temperature (RT: 23–25°C) under room light to deposit Schistosoma eggs at the bottom of the container. After the eggs were washed with 10 mL of dechlorinated tap water, 10 eggs were aliquoted into each well of 96-well plastic plates with 350 µL of dechlorinated tap water. To evaluate the promotive effect of light from several different sources in the miracidium hatching, the eggs were exposed at RT to room light, sunlight, fluorescent light and halogen light. For the room light condition, the plastic plate was kept on a bench in the laboratory [4, 11, 14]. For the sunlight condition, the plastic plate was kept beside a window inside a room from 10 in the morning in June. For the fluorescent light condition, the plastic plate was kept under a 27-W fluorescent light at a distance of 40 cm in a room with room light. For the halogen light condition, the plastic plate was kept under a halogen light (LG-PS2, OLYMPUS Corp., Tokyo, Japan) at a distance of 30 cm in a room without room light. The light intensity on the plate position in lux of each condition was measured with a digital lux meter (AP-881D, Aoputtriver Technology Co., Ltd., Guangdong, China) and they were 410 lux, 2,800–3,700 lux, approx. 1,100 lux and approx. 2,000 lux for room light, sunlight, fluorescent light and halogen light conditions, respectively. For no light condition, the plastic plate was kept inside a dark box. Miracidia and unhatched eggs for these conditions were counted under stereomicroscopy at 2, 4, 6, and 24 hr after exposure. The experiments were conducted three times. Statistical analysis was performed using GraphPad Prism 9 (San Diego, CA, USA).
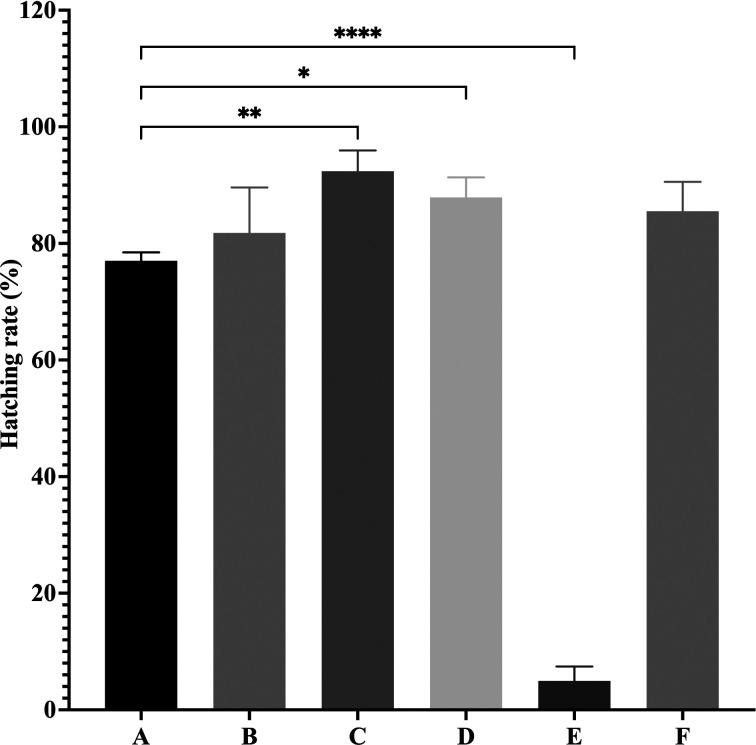
The hatching rate at 24 hr under the room lighting condition was 77.0%, whereas that under the dark condition was 4.7% (Fig. 1). This result confirmed that lighting was crucial for miracidium hatching. The sunlight and fluorescent light conditions showed higher hatching rates than the room light condition (Fig. 1, Supplementary Table 1). The sunlight condition showed the highest hatching rate, and hatching was completed within 6 hr. Interestingly, the eggs that had been kept under the dark condition for 5 days at RT could resume and complete the hatching process after being exposed to sunlight (Fig. 1, Supplementary Table 1). This condition can be used for storing eggs before the MHT can be conducted.
Fig. 1.
Comparison of Schistosoma japonicum miracidium hatching rates at 24 hr under different lighting conditions. The rates are presented as the mean ± SD. One-way ANOVA was used to analyze the data sets. A post-hoc comparison of mean values between the conditions was performed using Dunnett’s test, and a P-value of <0.05 was considered statistically significant. *P<0.05, **P<0.01 and ***P<0.001. A, room light condition (original protocol), B, halogen light condition, C, sunlight condition, D, fluorescent light condition, E, dark condition and F, eggs kept in the dark condition for 5 days before being exposed to sunlight.
Several factors are considered to affect the S. japonicum MHT in which water temperature and lighting condition are both essential in hatching of the miracidia [3, 5, 12]. However, the effects of different types of lighting on the hatching process have not yet been compared. The results in the present study confirmed that lighting condition was crucial for the miracidium hatching as only a few miracidia could hatch under the dark condition. The result also indicated for the first time, to our knowledge, that sunlight was an efficient lighting source for inducing a higher rate of hatching within a shorter time. Information regarding the specific wavelength region in sunlight that best promotes miracidium hatching may assist in the development of an automated MHT that can greatly help in the diagnosis and elimination of schistosomiasis.
Hatched miracidia were washed three times with autoclaved dechlorinated tap water, and single miracidia were picked up with a micropipette tip under stereomicroscopy and transferred to plastic tube with 1 μL of water. Genomic DNA was extracted from individual miracidia using a Stool Mini Kit and QIAamp DNA Micro kit (QIAGEN, Germantown, MD, USA). To evaluate the quality of the DNA, S. japonicum microsatellite markers including RRPS, M5A, TS2, MPA, 2AAA, J5, Sjp1, Sjp5, Sjp6, and Sjp9 were amplified by PCR [9, 13]. The amplifications were done following a previously reported protocol [7]. These 10 markers were successfully amplified for all 20 DNA samples. The amplicons showed the expected size from 220 to 550 bp (Supplementary Fig. 1), and their sequences were also confirmed by direct sequencing (data not shown). Comparative genetic studies of S. japonicum from each definitive host are important to understand the epidemiology and transmission dynamics of the parasite. The miracidium hatching protocol in the present study will help in the sampling procedure for population genetics studies of S. japonicum by avoiding bias due to experimental host-induced selection. The present protocol under the sunlight and fluorescent light conditions provides a more rapid and convenient procedure than the previously MHT and would be suitable for High-throughput DNA extraction using 96-well plates.
POTENTIAL CONFLICTS OF INTEREST
The authors declare no conflicts of interest in this study.
Supplemental Materials
Acknowledgments
The authors would like to thank Assistant Professor Keisuke Suganuma and Dr. Ehab Elnour Ahmed from the National Research Center for Protozoan Diseases, Obihiro University of Agriculture and Veterinary Medicine, for their suggestions and technical assistance. We would like to thank Assistant Professor Yuma Ohari from Rakuno Gakuen University, School of Veterinary Medicine and Associate Professor Jose Ma. M. Angeles from Department of Parasitology, College of Public Health, University of the Philippines Manila, for their suggestions. We would also like to thank the technical staff from the Department of Tropical Medicine and Parasitology, Dokkyo Medical University, for their technical assistance. We gratefully acknowledge funding from the Japan Society for the Promotion of Science to S. K. (19KK0173).
REFERENCES
- 1.Agola LE, Steinauer ML, Mburu DN, Mungai BN, Mwangi IN, Magoma GN, Loker ES, Mkoji GM. 2009. Genetic diversity and population structure of Schistosoma mansoni within human infrapopulations in Mwea, central Kenya assessed by microsatellite markers. Acta Trop 111: 219–225. doi: 10.1016/j.actatropica.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He YX, Salafsky B, Ramaswamy K. 2001. Host--parasite relationships of Schistosoma japonicum in mammalian hosts. Trends Parasitol 17: 320–324. doi: 10.1016/S1471-4922(01)01904-3 [DOI] [PubMed] [Google Scholar]
- 3.Ito J. 1955. Studies on hatchability of Schistosoma japonicum eggs in several external environmental conditions. Jpn J Med Sci Biol 8: 175–184. doi: 10.7883/yoken1952.8.175 [DOI] [PubMed] [Google Scholar]
- 4.Jurberg AD, Oliveira AA, Lenzi HL, Coelho PMZ. 2008. A new miracidia hatching device for diagnosing schistosomiasis. Mem Inst Oswaldo Cruz 103: 112–114. doi: 10.1590/S0074-02762008005000005 [DOI] [PubMed] [Google Scholar]
- 5.Kassim O, Gibertson DE. 1976. Hatching of Schistosoma mansoni eggs and observations on motility of miracidia. J Parasitol 62: 715–720. doi: 10.2307/3278948 [DOI] [PubMed] [Google Scholar]
- 6.Kirinoki M, Hu M, Yokoi H, Kawai S, Terrado R, Ilagan E, Chigusa Y, Sasaki Y, Matsuda H. 2005. Comparative studies on susceptibilities of two different Japanese isolates of Oncomelania nosophora to three strains of Schistosoma japonicum originating from Japan, China, and the Philippines. Parasitology 130: 531–537. doi: 10.1017/S0031182004006924 [DOI] [PubMed] [Google Scholar]
- 7.Moendeg KJ, Angeles JMM, Nakao R, Leonardo LR, Fontanilla IKC, Goto Y, Kirinoki M, Villacorte EA, Rivera PT, Inoue N, Chigusa Y, Kawazu SI. 2017. Geographic strain differentiation of Schistosoma japonicum in the Philippines using microsatellite markers. PLoS Negl Trop Dis 11: e0005749. doi: 10.1371/journal.pntd.0005749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross AGP, Sleigh AC, Li Y, Davis GM, Williams GM, Jiang Z, Feng Z, McManus DP. 2001. Schistosomiasis in the People’s Republic of China: prospects and challenges for the 21st century. Clin Microbiol Rev 14: 270–295. doi: 10.1128/CMR.14.2.270-295.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrivastava J, Barker GC, Johansen MV, Xiaonong Z, Aligui GD, McGarvey ST, Webster JP. 2003. Isolation and characterization of polymorphic DNA microsatellite markers from Schistosoma japonicum. Mol Ecol Notes 3: 406–408. doi: 10.1046/j.1471-8286.2003.00466.x [DOI] [Google Scholar]
- 10.Shrivastava J, Gower CM, Balolong E, Jr, Wang TP, Qian BZ, Webster JP. 2005. Population genetics of multi-host parasites--the case for molecular epidemiological studies of Schistosoma japonicum using larval stages from naturally infected hosts. Parasitology 131: 617–626. doi: 10.1017/S0031182005008413 [DOI] [PubMed] [Google Scholar]
- 11.Sugiura S, Sasaki T, Hosaka Y, Ono R. 1954. A study of several factors influencing hatching of Schistosoma japonicum eggs. J Parasitol 40: 381–386. doi: 10.2307/3273880 [DOI] [PubMed] [Google Scholar]
- 12.Ye XP, Fu YL, Wu ZX, Anderson RM, Agnew A. 1997. The effects of temperature, light and water upon the hatching of the ova of Schistosoma japonicum. Southeast Asian J Trop Med Public Health 28: 575–580. [PubMed] [Google Scholar]
- 13.Yin M, Hu W, Mo X, Wang S, Brindley PJ, McManus DP, Davis GM, Feng Z, Blair D. 2008. Multiple near-identical genotypes of Schistosoma japonicum can occur in snails and have implications for population-genetic analyses. Int J Parasitol 38: 1681–1691. doi: 10.1016/j.ijpara.2008.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu JM, de Vlas SJ, Jiang QW, Gryseels B. 2007. Comparison of the Kato-Katz technique, hatching test and indirect hemagglutination assay (IHA) for the diagnosis of Schistosoma japonicum infection in China. Parasitol Int 56: 45–49. doi: 10.1016/j.parint.2006.11.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.