Abstract
Proliferating cell nuclear antigen (PCNA) is an essential component of the DNA replication and repair machinery in the domain Eucarya. We cloned the gene encoding a PCNA homolog (PfuPCNA) from an euryarchaeote, Pyrococcus furiosus, expressed it in Escherichia coli, and characterized the biochemical properties of the gene product. The protein PfuPCNA stimulated the in vitro primer extension abilities of polymerase (Pol) I and Pol II, which are the two DNA polymerases identified in this organism to date. An immunological experiment showed that PfuPCNA interacts with both Pol I and Pol II. Pol I is a single polypeptide with a sequence similar to that of family B (α-like) DNA polymerases, while Pol II is a heterodimer. PfuPCNA interacted with DP2, the catalytic subunit of the heterodimeric complex. These results strongly support the idea that the PCNA homolog works as a sliding clamp of DNA polymerases in P. furiosus, and the basic mechanism for the processive DNA synthesis is conserved in the domains Bacteria, Eucarya, and Archaea. The stimulatory effect of PfuPCNA on the DNA synthesis was observed by using a circular DNA template without the clamp loader (replication factor C [RFC]) in both Pol I and Pol II reactions in contrast to the case of eukaryotic organisms, which are known to require the RFC to open the ring structure of PCNA prior to loading onto a circular DNA. Because RFC homologs have been found in the archaeal genomes, they may permit more efficient stimulation of DNA synthesis by archaeal DNA polymerases in the presence of PCNA. This is the first stage in elucidating the archaeal DNA replication mechanism.
In the domain Eucarya, proliferating cell nuclear antigen (PCNA) is implicated in many DNA metabolic processes, including cell cycle control, DNA replication, nucleotide excision repair, and postreplication mismatch repair (reviewed in references 22, 24, 25, and 36). Among these processes, PCNA’s role as an elongation (processivity) factor of DNA polymerase δ (Pol δ), a replicative enzyme in the eukaryotic cells, has been the most intensively investigated (reviewed in references 3 and 40).
PCNA consists of three identical subunits which form a torus-like structure with a central cavity capable of accommodating double-stranded DNA (30). In the DNA replication process in Eucarya, the multipolypeptide replication factor C (RFC) is required to load PCNA efficiently onto the DNA duplex. RFC binds to DNA at a primer-template junction and catalyzes the loading of PCNA onto DNA in an ATP-dependent manner. Pol δ then binds to PCNA to form a holoenzyme capable of processive DNA replication, either on the leading or on the lagging strand. PCNA shares an identical three-dimensional structure with the β-subunit of Escherichia coli DNA polymerase III (Pol III), the functional homolog in Eubacteria (29, 30). Eucarya and Bacteria constitute two different domains of life. However, in each of these domains, a common mechanism exists for facilitating processive DNA replication of the genome by the cooperative work of DNA polymerase and its sliding clamp (3, 26, 40).
Archaea, the third domain of life (43), resemble the Bacteria in cellular ultrastructure. However, a similarity between archaeal and eukaryotic DNA replication became evident soon after their discovery. Aphidicolin, an inhibitor of eukaryotic DNA Pol α and Pol δ, also inhibited the replication in vivo of a halophilic archaeon, Halobacterium halobium (11, 33). It was subsequently confirmed that Archaea contains a family B (α-like) DNA polymerase with an amino acid sequence similar to that of the large subunit of eukaryotic DNA replicases α, δ, and ɛ (reviewed in references 8 and 18). Interestingly, while the crenarchaeotes, a subdomain of Archaea, contain two or more homologs of this protein, their relatives in the euryarchaeotic subdomain possess only one homolog (Pol I) (8, 18). The slow rate of in vitro DNA synthesis by the archaeal family B DNA polymerases (28, 31, 37, 39) led to two hypotheses: (i) the DNA polymerases require accessory proteins to achieve their maximum processivities or (ii) an entirely different DNA polymerase other than the family B enzyme, one yet to be isolated, was responsible for replication in Archaea. Regarding the second hypothesis our search culminated in the isolation of a distinct heterodimeric DNA polymerase (Pol II) from the euryarchaeotes (5, 17, 19, 39). Pol II is composed of a small (DP1) and a large (DP2) subunit. Amino acid sequences of DP1 have some significant similarity to the small subunit of eukaryotic DNA Pol δ, even though DP2, the catalytic subunit, is not similar to any known DNA polymerases. Despite these new findings, the evidences for identifying an archaeal replicative DNA polymerase are still inconclusive.
All known replicative DNA polymerases require a sliding clamp, the β-subunit for E. coli, gp45 for bacteriophage T4, and PCNA for the eukaryotes. Therefore, a further understanding of the function of the archaeal DNA polymerases may require the identification of the archaeal sliding clamp and an investigation of its effect on the DNA polymerases. All completely sequenced and analyzed archaeal genomes contain PCNA and RFC homologs (4, 23, 27, 34). However, a direct link between these proteins and archaeal DNA polymerases has yet to be demonstrated. We describe here the cloning and characterization of a PCNA homolog from Pyrococcus furiosus (PfuPCNA) and show that this protein interacts with Pol I and Pol II in this organism. The interactions augmented DNA synthesis by both DNA polymerases. Our results, aside from showing the distinct features of the pyrococcal PCNA, also provide an initial evidence for a common role of PCNA in replication in the domains Archaea and Eucarya.
MATERIALS AND METHODS
Cloning and sequencing of P. furiosus pcna gene.
PCR was used to amplify a gene coding for a homolog of PCNA from P. furiosus genome. The PCR primers were based on a DNA sequence encoding a PCNA homolog found from the total genome sequence of Pyrococcus horikoshii (23). Two primers designed were as follows: PCNAF (5′-GCGAATTCATGCCATTCGAAATAGTCTTTGAGGGTG-3′) and PCNAR (5′-ATCGCCTCGAGTCACTCCTCAACCCTTGGGGCTAGCAGG-3′), which have an EcoRI and a XhoI site (underlined), respectively. The PCR conditions involved 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min. The PCR product was cloned into a TA-cloning vector (pT7Blue; Novagen), and the nucleotide sequences of the inserted DNA were determined from several independent clones by a capillary sequencer (ABI Prism 310 Genetic Analyzer; Applied Biosystems). The native sequences in the primer region were corrected by a genomic walking method as described previously (7). The putative gene for P. furiosus PCNA was excised by digestion with EcoRI and XhoI. It was then fused by ligation to the gene coding for maltose-binding protein in the vector pMAL (New England Biolabs) at the EcoRI and SalI sites. This construct was named pMPCNA. To insert the structural gene for the protein PfuPCNA into a pET21a (Novagen), a forward primer which was designed to contain the initiation codon (5′-ACATATGCCATTTGAAATCGTATTTGA) at an NdeI site (underlined) was used together with PCNAR to amplify the gene. The PCR product was cloned into the TA-cloning vector and nucleotide sequenced as described above. The recombinant plasmid was digested with NdeI and BamHI to yield a 123-bp fragment comprising the N-terminal region of the structural gene. In a separate reaction, the remaining 627 bp of the pcna gene was excised by digesting the recombinant plasmid with BamHI and XhoI. The two DNA fragments (123 and 627 bp) were gel purified and ligated into a pET21a vector digested with NdeI and XhoI. The correctness of the gene was confirmed by nucleotide sequencing, and the construct was designated pTPCNA.
Production of recombinant PfuPCNA.
E. coli BL21(DE3) cells harboring pTPCNA were grown in 1 liter of Luria-Bertani medium containing ampicillin (100 μg/ml) to an optical density at 600 nm of 0.3 at 37°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to the culture at a final concentration of 0.2 mM, and growth was continued for 2 h. Cells were harvested, suspended in 25 ml of buffer A (50 mM Tris-HCl, pH 8.5; 0.1 mM EDTA; 2 mM β-mercaptoethanol; 0.1 M NaCl; 10% glycerol), and lysed through sonication. Cell debris was removed by centrifugation at 48,000 × g for 15 min at 4°C. E. coli proteins were partially removed by a two-step heating program involving an initial heating at 75°C for 15 min. The supernatant after centrifugation was again heated at 80°C for 10 min, followed by recentrifugation. To the second supernatant, polyethyleneimine (Polymin P) and NaCl were added to 0.2% (wt/vol) and 0.58 M, respectively, and the mixture was stirred for 30 min at 4°C. The proteins in the supernatant (30 ml) were precipitated by adding 16.8 g of ammonium sulfate (80% saturation). The precipitate was dissolved in buffer B (50 mM Tris-HCl, pH 8.0; 0.1 mM EDTA; 10% glycerol; and 0.5 mM dithiothreitol) and dialyzed against the same buffer. The dialysate was applied to an anion-exchange column (HiTrap Q, 5 ml; Pharmacia Biotech) fitted to a high-pressure liquid chromatography apparatus (ÄKTA Explorer 10S; Pharmacia Biotech), and the chromatography was developed with a 50-ml linear gradient of 0 to 1.0 M NaCl in buffer B at a flow rate of 2 ml/min. The PfuPCNA eluted at a salt concentration of 0.5 M. The purified PfuPCNA was stored at 4°C after dialysis against buffer B. The concentration of PfuPCNA was determined by using a molar extinction coefficient ɛ of 7,250 M−1 cm−1, which was obtained by a method of Gill and von Hippel (13).
N-terminal amino acid sequencing.
A sample containing both full-length PCNA and truncated PCNA was fractionated by electrophoresis on a sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel, electroblotted onto a polyvinylidene difluoride membrane (Sequi-Blot, 0.2 μM; Bio-Rad), stained with Coomassie brilliant blue R250 (0.02% in 40% methanol), and destained with 5% methanol. The protein bands were excised and subjected to automated Edman degradation in an Applied Biosystems Procise model 492 protein sequencer.
Primer extension analysis.
The primer elongation abilities of Pol I and Pol II in the absence or presence of PfuPCNA were investigated by using three templates: poly(dA)400 (Pharmacia), linearized M13 single-stranded DNA (ssDNA), and circular M13 ssDNA. 32P-5′-end-labeled oligo(dT)30 was annealed to poly(dA)400 [poly(dA):oligo(dT), 20:1], while an end-labeled oligomer, 5′-ATTCGTAATCATGGTCATAGCTGTTTCCTG-3′, complementary to positions 6224 to 6233 of the M13mp18 (44) genome, was annealed to linearized and circular M13 ssDNA. To create a linearized M13 ssDNA, an oligonucleotide complementary to positions 6259 to 6289 (within the multicloning site) was initially annealed to circular M13 ssDNA to create a double-stranded region containing some restriction enzyme recognition sites. The DNA was then digested overnight with PstI. An aliquot of the product was digested with Exonuclease VII, a single-strand-specific exonuclease (GIBCO-BRL), for 3 h to ensure the success of the digestion. To anneal the primers to the template, 1.0 μg of M13 ssDNA and 1.0 pmol of 32P-labeled primer in DNA polymerase reaction buffer were heated at 95°C for 3 min to ensure denaturation. The mixture was then gradually cooled to room temperature, and 0.05 U of the respective DNA polymerase was added. Pol I and Pol II were prepared as described previously (38, 39). Twenty microliters from this mixture were aliquoted into the respective tubes containing either PCNA or buffer. The reaction was initiated by adding deoxynucleotide triphosphates to a concentration of 250 μM. Each assay mixture was 25 μl in volume and contained 20 mM Tris-HCl (pH 8.0) and 1.5 mM MgCl2. The reaction was carried out at 70°C. Aliquots (5 μl) were taken at 1, 2, and 3 min after initiation of the reaction and dispensed into 3 μl of stop solution (98% deionized formamide, 1 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue), and 2.5 μl of each were analyzed by polyacrylamide gel electrophoresis (PAGE) in the presence of 8 M urea. In the case of Pol II, aliquots of the reaction products were further analyzed on 1.2% alkaline agarose gel in 50 mM sodium hydroxide and 1 mM EDTA.
Production of GST fusion proteins.
The genes for Pol I (38), DP1, DP2, and DP1+DP2 (39) were amplified by PCR and inserted into the BamHI/XhoI sites of pGEX4T-2 (Pharmacia). E. coli JM109 strains carrying these plasmids were grown at 37°C to 0.5 A600, and the expression of the target genes was induced by IPTG. Glutathione S-transferase (GST)-DP1, GST-DP2, GST-DP1+DP2 (Pol II), and GST-Pol I were purified by affinity chromatography (glutathione-Sepharose 4B) according to the manufacturer’s instructions (Pharmacia).
Interaction of PfuPCNA with Pol I and Pol II in vivo.
Rabbit polyclonal antibody was raised against homogeneous PfuPCNA, as well as Pfu Pol I, DP1, and Pol II (DP1-DP2 complex). Immunoprecipitation experiments were done basically as described in our previous report (5). Thirty microliters of protein A-Sepharose (Pharmacia) was dispensed into each of five Eppendorf tubes and washed three times with phosphate-buffered saline (PBS; 10 mM sodium phosphate, pH 7.5; 150 mM NaCl). The protein A-Sepharose in each tube was then mixed with one of the above antisera and incubated at room temperature for 1 h on a rotary shaker (one did not contain antibodies as a negative control). Each mixture was washed twice with PBS and once with buffer A. The contents of each tube were mixed with 300 μl of the P. furiosus cell extract (15 ml of culture) and incubated for 30 min on a rotary shaker. Precipitates were washed twice with buffer A, and the immunoprecipitated products were eluted by boiling them in 240 μl of buffer A and 60 μl of 5× loading buffer (0.25 M Tris-HCl [pH 6.8], 5% glycerol, 5% 2-mercaptoethanol, 0.2% bromophenol blue). Three microliters of products from each tube was subjected to Western blot analysis. Protein samples were separated by SDS–10% PAGE and electroblotted onto a polyvinylidene difluoride membrane. The blots were analyzed with the enhanced chemiluminescence system (Amersham) as described earlier (5).
Interaction of PfuPCNA with Pol I and Pol II in vitro.
Glutathione-Sepharose 4B resin (100 μl/tube) was dispensed into five Eppendorf tubes and spun (500 × g for 5 min) to sediment the matrix. The supernatant was decanted, and the resin in each tube was washed with 1 ml of cold buffer C (10 mM sodium phosphate, pH 7.3; 140 mM NaCl; 2.7 mM KCl), followed by centrifugation to sediment the resin. GST (as a negative control), GST-DP1, GST-DP2, GST-DP1+DP2, and GST-Pol I produced in JM109 cells were individually immobilized onto the resin in the respective tubes. An E. coli cell extract containing recombinant PfuPCNA was reacted with each immobilized protein, followed by extensive washings with cold buffer C. The bound proteins were eluted with 50 μl of 10 mM glutathione in buffer A, and 10-μl portions of eluted fractions were used for Western blot analysis.
Sedimentation equilibrium analysis.
PfuPCNA in buffer A was analyzed for its native molecular mass by using a Beckman KL-I Optima Analytical Ultracentrifuge equipped with absorbance optics. The PCNA samples, at concentrations of 0.3 and 1.8 mg/ml, were centrifuged at three different speeds: 10,000, 14,000, and 18,000 rpm. The partial specific volume used for the analysis was 0.759 ml/g, as calculated from the weighted average of the amino acid content by using the method of Cohn and Edsall (9), and the density of the solvent was calculated to be 1.035 g/ml.
Chemical cross-linking.
For the chemical cross-linking of purified PfuPCNA, ethylene glycol-bis(succinimidyl succinate) (EGS; Sigma) was used as described earlier (46). The reaction mixtures (10 μl) for cross-linking with EGS contained 60 μg of PfuPCNA per ml and 50 μM EGS in 20 mM sodium phosphate (pH 7.5) and 150 mM NaCl. The products were analyzed by Western blotting.
Sequence analysis and model building.
The PCNA homologs retrieved from the GenBank and their accession numbers are as follows: Methanococcus jannaschii (Mja PCNA, Q57797), Methanobacterium thermoautotrophicum (Mth PCNA, sp027367), Archaeoglobus fulgidus (Afu PCNA, AE001081), Homo sapiens (Hum PCNA, P12004), Drosophila melanogaster (Dro PCNA, P17917), Caenorhabditis elegans (Cel PCNA, sp002115), and Saccharomyces cerevisiae (Sce PCNA, P15873). Amino acid sequence comparisons were carried out by using CLUSTALW (9a). A model structure of PfuPCNA was built based on the coordinates of yeast PCNA, which are available in Protein Data Bank as 1PFQ. At first, the amino acid sequence of PfuPCNA was aligned with that of 1PFQ by using CLUSTALW. The obtained alignment was modified so that gaps are not inserted into the secondary structures of 1PFQ or the corresponding regions of PfuPCNA. Based on the alignment, a model structure for a single subunit of PfuPCNA was constructed with the HOMOLOGY and the DISCOVER modules in a molecular modeling software package, Insight II (Biosym, Inc.). Then, a model for the trimer of PfuPCNA was constructed by generating other two subunits by the rotation and the translation operations described in the file of 1PFQ. The trimer was further subjected to energy minimization with the DISCOVER module.
Nucleotide sequence and accession number.
The DNA sequence reported here has been submitted to the DDBJ and has been assigned accession number AB017486.
RESULTS
Cloning, expression, and purification of recombinant PfuPCNA.
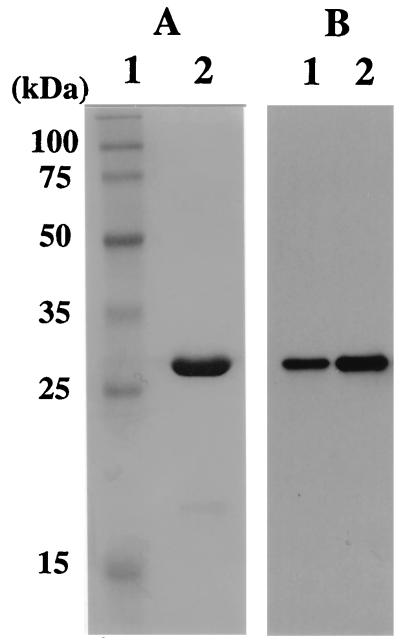
A gene encoding the homologous sequence to eukaryotic PCNA was found in the total genome sequence of Pyrococcus horikoshii (23). By use of primers based on the sequence of the gene in P. horikoshii, a gene encoding the similar sequence of PCNA was amplified by PCR from the P. furiosus genome. Figure 1 shows the nucleotide sequence of the gene and the deduced amino acid sequence. The gene coded for a protein of 249 amino acids with an estimated molecular mass of 28,004 Da. The G+C content of the gene was 40%, a finding which was in good agreement with the overall G+C content of P. furiosus genomic DNA (38%). The deduced amino acid sequence from the gene had similarity to eukaryotic PCNA and other archaeal homologs as described below (Table 1). Therefore, the gene for the PCNA homolog was cloned into the vector, pET21a and expressed in E. coli BL21(DE3). The recombinant protein of ca. 28 kDa detected by SDS-PAGE was purified to homogeneity through heat treatment to denature the majority of the host proteins, polyethyleneimine treatment to remove DNA, ammonium sulfate precipitation, and anion-exchange chromatography (Fig. 2). The N-terminal amino acid sequence was confirmed to match that of the initiation region of the PCNA-like open reading frame (ORF). About 1.5 mg of protein was purified from a 1-liter culture of E. coli cells harboring the gene (Fig. 2). Depending on the culture condition (long-time cultivation), an extensive amount of another thermostable protein of approximate molecular mass 20 kDa was produced, as determined by SDS-PAGE. The N-terminal amino acid sequence of the protein, MDHLKKILK, corresponded to positions from Met73 to Lys81 of the ORF (Fig. 1). It is not known whether the product was derived from proteolysis of the intact product or from an unexpected translational initiation. However, we had found that the genes from P. furiosus are sometimes translated in E. coli from the internal ATG or GTG in the structural genes (unpublished data). It is possible in this case that the small protein was produced by a second translation from ATG for Met73, and the -GGAG- sequence, five nucleotides upstream of this codon, could serve as a ribosome binding site. The 20-kDa band was not observed in the Western blot analysis of P. furiosus cell extract, as shown below (Fig. 2).
FIG. 1.
Nucleotide and deduced amino acid sequences of the gene for the P. furiosus PCNA homolog. The BamHI site used for cloning of the structural gene into pET21a and the determined N-terminal amino acid sequence of the truncated protein produced in E. coli are underlined. The asterisk indicates the stop codon.
TABLE 1.
Sequence comparison of eukaryotic and archaeal PCNAs
| Domain | Organism | % Amino acid identity
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Pfu | Mja | Afu | Mth | Hsa | Sce | Cel | Dro | ||
| Archaea | P. furiosus | ||||||||
| M. jannaschii | 39 | ||||||||
| A. fulgidus | 24 | 28 | |||||||
| M. thermoautotrophicum | 30 | 33 | 25 | ||||||
| Eucarya | H. sapiens | 22 | 25 | 20 | 28 | ||||
| S. cerevisiae | 25 | 28 | 20 | 22 | 35 | ||||
| C. elegans | 21 | 25 | 19 | 23 | 47 | 39 | |||
| D. melanogaster | 23 | 22 | 21 | 24 | 70 | 36 | 47 | ||
FIG. 2.
Purification of recombinant PfuPCNA from E. coli cells and identification of PfuPCNA in P. furiosus cells. (A) Recombinant PfuPCNA purified as described in the text was loaded onto an SDS–12.5% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: 1, molecular mass markers (Perfect Protein Markers; Novagen); 2, purified PfuPCNA (3 μg). The sizes of the molecular mass markers are indicated on the left. (B) Western blot analysis of PfuPCNA. P. furiosus cell extracts (35 μg of cells) and recombinant PCNA (0.01 μg) produced in E. coli were separated by SDS–12.5% PAGE and then analyzed by Western blotting with anti-PfuPCNA antiserum. Lanes: 1, P. furiosus cell extract; 2, recombinant PfuPCNA.
Detection of PCNA in P. furiosus cell extract.
To investigate whether the PCNA-like protein exists in P. furiosus cells and also whether the size matched that of the recombinant protein produced in E. coli, polyclonal antibody was prepared by using the highly purified recombinant protein. As shown in Fig. 2B, a protein of corresponding size to the recombinant PCNA-like protein was detected in the crude cell extract of P. furiosus, which indicated that the gene for the ORF is actually expressed in P. furiosus.
Effect of PfuPCNA on the processivity of DNA polymerases.
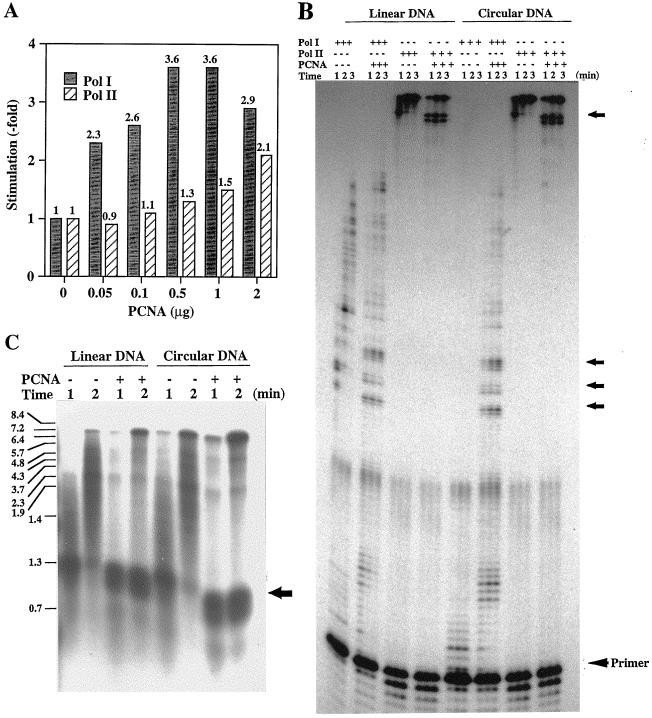
One of the biological functions of the PCNA homolog was investigated through its effect on the primer elongation abilities of each DNA polymerase found in P. furiosus as described above. When the poly(dA) sparsely primed by oligo(dT)30 was used for DNA synthesis reaction, incorporated radioactivities in 2 min was increased by the addition of PCNA homolog by 3.6-fold for Pol I and by 2.1-fold for Pol II, respectively (Fig. 3A). To confirm the effect of the PCNA homolog on the primer elongation abilities of these DNA polymerases, synthesized products were visualized by denaturing gel electrophoresis by using linear and circular ssDNA of M13 phage as substrates. As shown in Fig. 3B, longer products were observed for Pol I in the presence of the PCNA homolog. An unexpected observation was that the stimulation of Pol I was found on not only the linear substrate but also on the circular DNA and, moreover, the effect was more remarkable on circular than linear DNA. Because Pol II has a very strong extension ability by itself as described earlier (39), even though its associated 3′→5′ exonuclease activity is very strong as more primer degradation was observed in the Pol II lanes, the difference of the product sizes with or without the PCNA homolog could not be seen on PAGE, and shorter products due to pausing in the presence of the PCNA homolog looked rather remarkable (Fig. 3B). For a better resolution of the products that overcame the pausing site, the 1- and 2-min reaction products were analyzed on an alkaline agarose gel. As shown in Fig. 3C, on the linear DNA and in the absence of the PCNA homolog, Pol II synthesized products up to the approximate size of 3.7 kb in 1 min. Addition of the PCNA homolog, however, resulted in completely replicated products (7.4 kb), even though the amount was very low. The samples taken 2 min after incubation showed a higher yield of full-size products due to the addition of the PCNA homolog. A similar pattern was observed in the experiment with circular DNA as the template. When the template-primer combinations described above were used, the radioactive counts incorporated into DNA strand by Pol I increased with the PCNA homolog by 1.9- and 3.6-fold for linear and circular DNA, respectively, compared with the reactions performed without the PCNA homolog (data not shown). In the case of Pol II, the nucleotide incorporation was not so different between the reactions in the presence or absence of the PCNA (1.2- and 1.1-fold for the linear and circular forms, respectively [data not shown]). In the presence of PCNA, a strong pausing of Pol II at a specific site of the template occurred (Fig. 3C). Therefore, it was reasonable that the total incorporations of [3H]TTP into the DNA strand were not so different between the reactions with and those without the PCNA homolog, even though the longest product sizes were different. The subsequent sampling showed that Pol I and Pol II in the absence of the PCNA homolog can translocate through the pauses after 2 min. In the presence of the PCNA homolog, these pausing signals remained with the same intensity even after 3 min of incubation (Fig. 3C). Similar transient pauses were shown by eukaryotic Pol δ in the presence of PCNA (10, 32). The pauses may be due to the DNA polymerase-PCNA complex encountering secondary template structures. It has been suggested that Pol δ holoenzyme traverses the pause sites by distributive “recycling” which involves dissociation and reassociation of its components (32). Under physiological conditions, replicative helicases and ssDNA binding protein are likely to prevent or alleviate this condition.
FIG. 3.
Effect of recombinant PfuPCNA on the primer extension abilities of Pol I and Pol II. (A) The amounts of nucleotide incorporation into the DNA strand by DNA polymerases were compared in the presence or absence of PfuPCNA by using poly(dA)400-oligo(dT)30 as the substrate. (B) The primer extension abilities of Pol I and Pol II were compared with linear or circular DNA as the template in the presence (0.3 μg) or absence of PfuPCNA. Equal volumes of reaction mixtures were taken at 1, 2, and 3 min after initiation of the reaction. The products were analyzed by 6% PAGE containing 8 M urea and visualized by autoradiography. (C) The products from the Pol II reactions were separated by using 1.2% alkaline agarose gel electrophoresis. The sizes indicated on the left were from BstPI-digested λ DNA labeled by polynucleotide kinase with [γ-32P]ATP. Arrows on the right sides of panels B and C show the pausing sites.
In conclusion, even though a template without any pause site should be used to show more definite results, especially for Pol II, these observations showed that the PCNA homolog stimulated the synthesis of longer products by Pol I and Pol II both on linear and on circular DNA templates, and we therefore designated the protein PfuPCNA as a functional homolog of eukaryotic PCNA.
Interactions between PfuPCNA and DNA polymerases.
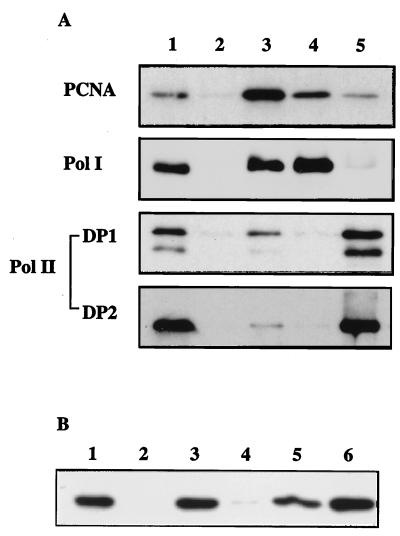
To examine whether the PCNA homolog interacts physically with Pol I or Pol II, an immunoprecipitation experiment was done with antibodies raised against the Pol I, Pol II, and PfuPCNA. As shown in Fig. 4A, both Pol I and Pol II were coprecipitated with the PfuPCNA (lane 3). Conversely, PfuPCNA was coprecipitated with Pol I (lane 4) or DP1 (lane 5). In the case of Pol II, it is known that DP1 and DP2 exist as a strong complex in vivo (5) and in vitro (39), and it was therefore not clear whether the PfuPCNA interacted with DP1 directly or whether it interacted with DP2 in its coprecipitation with DP1. To address this question, further interaction analyses were done by using GST fusion proteins of Pol I, DP1, DP2, and Pol II (DP1+DP2). Each of the fusion proteins was immobilized onto glutathione-Sepharose and then reacted with E. coli cell extract containing recombinant PfuPCNA. After the noninteracting proteins were washed out with a large volume of buffer C, the bound proteins were eluted with 10 mM glutathione and subjected to Western blot analysis with anti-PfuPCNA. As shown in Fig. 4B, PfuPCNA bound to Pol I and Pol II. Moreover, it was clear that PfuPCNA bound to DP2. A very faint band was detected in the DP1-binding fraction. Thus, DP1 may have a weak interaction with PfuPCNA.
FIG. 4.
Analysis of DNA polymerase-PCNA interaction. (A) Immunoprecipitation analysis. Total cell extracts were precipitated with anti-PfuPCNA (lane 3), anti-Pfu Pol I (lane 4), and anti-Pfu DP1 (lane 5). The total cell extract without immunoprecipitation (lane 1) or precipitated with PBS-treated protein A-Sepharose (lane 2) were loaded as controls. The immunoprecipitates were separated by SDS-PAGE and then analyzed by Western blotting by using indicated antibodies. (B) In vitro interactions. The GST fusion proteins with Pol I (lane 3), DP1 (lane 4), DP2 (lane 5), and RFC large subunit (lane 6) were first immobilized onto glutathione-Sepharose and then reacted with E. coli cell extracts containing recombinant PfuPCNA. After extensive washes, bound proteins were eluted and subjected to Western blot analysis with anti-PfuPCNA. Lane 2 shows the eluent from the GST-immobilized glutathione-Sepharose. As a positive control, P. furiosus cell extract was directly loaded onto the electrophoresis gel (lane 1).
Native molecular weight of PfuPCNA.
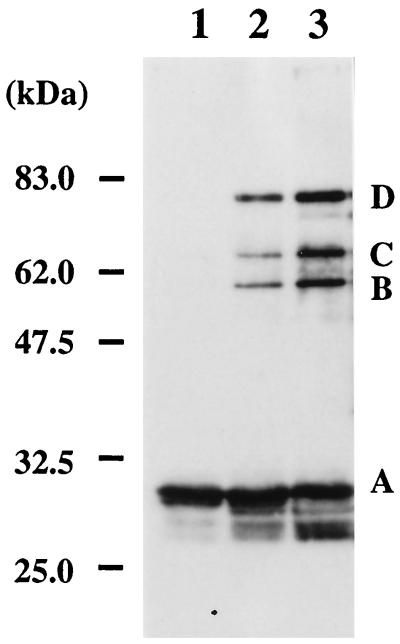
To estimate the size of native PfuPCNA, purified recombinant protein was subjected to gel filtration column chromatography. The protein eluted at ca. 75.5 kDa compared with the positions of the size marker proteins (data not shown). The sedimentation equilibrium profiles of the protein with an analytical ultracentrifuge showed that the PfuPCNA was a mixture of oligomeric forms. To visualize the oligomeric nature of PfuPCNA, a chemical cross-linking experiment was done. As shown in Fig. 5, EGS rapidly cross-linked PfuPCNA, which caused it to migrate as larger molecules in SDS-PAGE. This result supports the existence of multimeric protein species of PfuPCNA. Three major cross-linked species appeared. They can be postulated as products from a single cross-linked dimer, two cross-linked trimers (linear), and three cross-linked trimers (circle), as shown earlier of the gp45 protein from T4 phage (21). The active form of PfuPCNA may be a ring-shaped trimer as other sliding clamps.
FIG. 5.
Association state of PfuPCNA protein as revealed by chemical cross-linking. PfuPCNA was treated with EGS for 10 s (lane 2) and 1 min (lane 3) and analyzed by SDS–10% PAGE, followed by Western blot analysis. The protein without treatment with EGS is shown in lane 1. Each band is derived from PfuPCNA monomer (A), a single cross-linked dimer (B), three cross-linked circled trimers (C), and two cross-linked linear trimers, as indicated for the T4 gp45 protein (21). Indicated molecular sizes were derived from Prestained Protein Marker (New England Biolabs).
Comparison of euryarchaeotic PCNA with eukaryotic homologs.
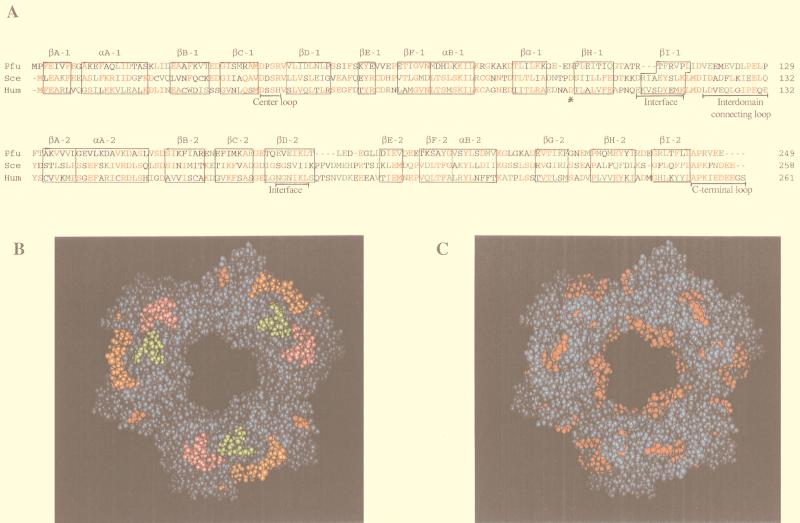
The amino acid sequence of PfuPCNA and those of the homologs which are found in the total genome sequences of the other euryarchaeotic organisms were compared with those of four eukaryotic homologs (Table 1). The average lengths of the euryarchaeotic and eukaryotic PCNA homologs were 246 and 260 amino acids, respectively. The identities within the four euryarchaeotic PCNA homologs ranged from 24 to 39%, with an average of 29.8%. The eukaryotic PCNA showed a higher conservation than those from Archaea. The predicted isoelectric points (pIs) for the archaeal PCNA ranged from 4.16 to 4.63, which shows that, like eukaryotic PCNAs, they are very acidic proteins (pH 4.07 to 4.30). From the sequence similarity of PfuPCNA to eukaryotic PCNA (Fig. 6A), a model of the ring-shaped three-dimensional structure with an inner diameter of 34 Å could be built by trimerizing the monomer protein by using the data for the crystal structure of yeast PCNA (Fig. 6B). It is quite possible that PfuPCNA has basically the same structure as eukaryotic PCNA. The functional loops to interact with Pol δ and RFC determined from yeast and human PCNA are indicated by colors. The positively charged residues are localized at the inner surface of the ring.
FIG. 6.
Structure of PfuPCNA. (A) Amino acid sequence alignment of the PfuPCNA with the human and yeast PCNA. Each PCNA has been divided into two domains labeled 1 and 2. The secondary structure elements are boxed and labeled, and the conserved and similar amino acid residues are shown in red and green, respectively. Other important regions required for the function and oligomerization of PCNA are indicated. (B and C) Model building of a ring-shaped three-dimensional structure of PfuPCNA. Pictures show the structure from the front (C-side), and the predicted functional loops indicated in panel A are colored green for the center loop (Asp42-Arg45), orange for the interdomain connecting loops (Val118-Pro129), red for Asn97, and pink for the C-terminal tail (Leu243-Glu249) in panel B. Positively charged residues (Lys and Arg) are indicated in red in panel C.
DISCUSSION
We cloned a gene coding for a homolog of PCNA, a sliding clamp of DNA polymerases from P. furiosus, and produced the protein in E. coli cells. The archaeal PCNA homolog actually stimulated the primer extension abilities of both Pol I and Pol II from P. furiosus. These effects suggested that the PCNA homolog cloned from P. furiosus works as the sliding clamp of archaeal DNA polymerases such as PCNA in Eucarya and the β-subunit in Bacteria, and the mechanism of the processive DNA synthesis necessary for DNA replication is conserved in the three domains of life.
It was necessary to analyze how PfuPCNA stimulates the DNA polymerase activity in P. furiosus to understand the mechanism of archaeal DNA replication. We demonstrated by using immunological procedures that PfuPCNA interacts directly with Pol I and Pol II in vivo and in vitro. Furthermore, an in vitro experiment suggested that the large subunit, DP2, mainly binds to PCNA in the Pol II complex. The DNA polymerase activity of the catalytic subunit of Pol δ from yeasts (expressed in E. coli) and humans (expressed by using the vaccinia virus system) were slightly stimulated by PCNA (2, 45). From these results, a consensus motif, GX4GX8GX3YFY, was proposed in the catalytic subunits of Pol δ to be important for binding to PCNA (47). However, other reports showed that the second subunit is required for the functional interaction of Pol δ from humans (48, 49), mice (14, 16), and S. pombe (1) with PCNA. It is probably now the consensus view that the second subunit is necessary for the full stimulation of Pol δ activity by PCNA (35). The stimulation of DNA synthesis by PfuPCNA observed in this study was evident but was not so salient in both cases of Pol I and Pol II compared with the case of eukaryotic Pol δ. Both DNA polymerases, especially Pol II, have much more efficient primer extension ability by themselves than Pol δ and, therefore, the effect of PCNA may be less distinct in these in vitro experiments. The other possibility is that there may be other proteins interacting with the DNA polymerases necessary for full stimulation of their activity by PCNA in Archaea.
Neither the Pol I nor the Pol II of P. furiosus has the GX4GX8GX3YFY motif in its sequences. In several eukaryotic proteins, including a cell-cycle checkpoint protein p21, endonucleases FEN1 and XPG, and cytosine-5-methyltransferase, which are known to interact with PCNA, an octapeptide sequence referred to as the PCNA-interacting protein (PIP) box is conserved (41, 42). An X-ray structure analysis of a complex between human PCNA and p21 showed that the amino acids in an octapeptide (144QTSMTDFY151) in p21 make contact with the interdomain connecting loop of PCNA. Met147, Phe150, and Tyr151 in this motif contact the inside of a hydrophobic pocket formed mainly by the interdomain connecting loop (15). The third subunit of S. cerevisiae Pol δ, which has been shown to interact with PCNA directly, also has this PCNA-binding motif (12). We examined both Pol I and Pol II by visual inspection for PIP box-like sequences, and the similar sequence motifs were found at the extreme C-terminal region of Pol I and DP2 proteins (Table 2). We then examined other archaeal forms of Pol I and euryarchaeotic DP2s. In each of the Pol I and DP2 homologs investigated, a similar octapeptide was conserved at the extreme C terminus. Interestingly, the DP2 homologs contained one more PIP box candidate very close to the first one. There is no candidate sequence of PIP box in DP1. These PIP box-like sequences in Pol I and DP2 may work for the interactions with PfuPCNA experimentally detected in vivo and in vitro in this study. Two amino acids, Leu126 and Ile128, of PCNA are universally conserved in eukaryotic PCNAs, and they are known to be involved in the formation of the hydrophobic pocket in the interdomain connecting loop. Mutation of these amino acids to alanine abolished an interaction between PCNA and the third subunit of S. cerevisiae Pol δ (10). In PfuPCNA these amino acids are conserved with two hydrophobic amino acids, Val123 and Leu125, which may not affect the nature of the hydrophobic pocket. It is, however, noteworthy that there is no candidate for the PIP box in the second subunits of eukaryotic Pol δ. Therefore, some other interaction may exist between the second subunit and PCNA in eukaryotes, except for S. cerevisiae, in which the third subunit works as described above.
TABLE 2.
Conserved sequences in Archaea resembling PCNA-binding motifs in Eucaryaa
| Domain | Organism | Protein | PIP sequence | Position (amino acids) | Size of protein (amino acids) |
|---|---|---|---|---|---|
| Archaea | P. furiosus | Pol I | QVGLTSWL | 763–770 | 775 |
| P. occultum | Pol II (B3) | QRSLFDFF | 772–779 | 803 | |
| T. litoralis | Pol I | QTGLDAWL | 765–772 | 774 | |
| A. fulgidus | Pol I | QMSLDSFF | 773–780 | 781 | |
| A. pernix | Pol II (B3) | QSTLLDFM | 761–768 | 772 | |
| P. furiosus | DP2 | ETILNSHL | 1096–1103 | 1,263 | |
| P. furiosus | DP2 | VISLDDFF | 1253–1261 | ||
| M. jannaschii | DP2 | EKVIQSHF | 1031–1038 | 1,139 | |
| M. jannaschii | DP2 | QVKLSDFF | 1129–1136 | ||
| M. thermoautotrophicum | DP2 | EGVLMSHF | 987–994 | 1,092 | |
| M. thermoautotrophicum | DP2 | QSSLDVFL | 1085–1092 | ||
| A. fulgidus | DP2 | ERVINVHF | 1037–1044 | 1,143 | |
| A. fulgidus | DP2 | QVSISDFV | 1136–1143 | ||
| Eucarya | Human | XPG | QLRIDSFF | 990–997 | 1,186 |
| Human | Fen1 | QGRLDDFF | 337–344 | 380 | |
| Human | p21 | QTSMTDFY | 144–151 | 164 | |
| Human | MCMT | QTTITSHF | 164–171 | 1,616 | |
| Consensus | QxSLdxFF |
B3, crenarchaeotic DNA polymerase B3; Pol I, euryarchaeotic family B DNA polymerase; DP2, the large subunit of euryarchaeotic heterodimeric DNA polymerase; XPG, Xeroderma pigmentosum group G-complementing protein; Fen1, 5′→3′ exo/endonuclease; p21, the cyclin-dependent kinase inhibitor protein p21; MCMT; DNA (cytosine-5) methyltransferase; x, any residue. (Sequences for Eukarya are from reference number x.)
In this study of Archaea, one more interesting finding different from the eukaryotic studies needs to be mentioned. The DNA synthesis reactions by the two P. furiosus DNA polymerases with the circular DNA as a template were stimulated by PfuPCNA. It is, however, well known that the molecule called “clamp loader” (gamma complexes in Bacteria and RFC in Eucarya) is required for the opening and loading of the ring-shaped sliding clamp onto circular DNA. There may be some different molecular mechanism in Archaea. One possible explanation is that the subunit-subunit interaction of PfuPCNA from its predicted ring structure may be less stable than other sliding clamp molecules, which causes the ring of PCNA to open spontaneously in the in vitro reactions at 72°C. In the case of yeast PCNA, an antiparallel configuration of two β-sheets of the flanking monomer produces a stable interface connection through eight clustered hydrogen bonds (15, 30). However, as shown in the sequence alignment of PCNA molecules (Fig. 6A), there are some deletions in the β-strands or in the loop constituting the interfaces. Such deletions would cause a conformational change at the interface regions and, as a result, a difference in the interaction mechanism between subunits of PfuPCNA from those of yeast and human PCNA. This may cause the less-stable connection of subunits in PfuPCNA. Further structural and biochemical analyses are necessary to understand the molecular mechanism of the clamp-loading in Archaea. The euryarchaeotic genomes sequenced so far have homologs of eukaryotic RFC components. Biochemical analyses of these proteins will expand our knowledge on the basic mechanism of this very important process in DNA replication. We observed an interaction between PfuPCNA and the large subunit of RFC in vitro (Fig. 4, lane 6).
We have cloned two family B DNA polymerase genes from a crenarchaeote, Aeropyrum pernix, and characterized the gene products (6). The A. pernix Pol I and Pol II were very similar to Pol I and Pol II from another crenarchaeote, Pyrodictium occultum, which we cloned previously (37). Interestingly, A. pernix has three PCNA homologs in its genome (20). How different are the DNA replication mechanisms between Euryarchaeota and Crenarchaeota? Studies of archaeal DNA replication are likely to yield very exciting results, including the answer to this question.
ACKNOWLEDGMENTS
We thank M. Shimizu for help in the sedimentation equilibrium analysis with analytical supercentrifugation. We are grateful to Y. Shimura, the director of BERI, for continuous encouragement.
REFERENCES
- 1.Arroyo M P, Downey K M, So A G, Wang T S-F. Schizosaccharomyces pombe proliferating cell nuclear antigen mutations affect DNA polymerase δ processivity. J Biol Chem. 1996;271:15971–15980. doi: 10.1074/jbc.271.27.15971. [DOI] [PubMed] [Google Scholar]
- 2.Brown W C, Campbell J L. Interaction of proliferating cell nuclear antigen with yeast DNA polymerase δ. J Biol Chem. 1993;268:21706–21710. [PubMed] [Google Scholar]
- 3.Brush G S, Kelly T J. Mechanisms for replicating DNA. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–43. [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Cann I K O, Komori K, Toh H, Kanai S, Ishino Y. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc Natl Acad Sci USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann I K O, Ishino S, Nomura N, Sako Y, Ishino Y. Two family B DNA polymerases from Aeropyrum pernix, an aerobic hyperthermophilic crenarchaeote. J Bacteriol. 1999;181:5984–5992. doi: 10.1128/jb.181.19.5984-5992.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cann I K O, Ishino Y. A tRNAGlu gene from the hyperthermophilic archaeon Pyrococcus furiosus contains the 3′-terminal CCA sequence of the mature tRNA. FEMS Microbiol Lett. 1998;160:199–204. doi: 10.1111/j.1574-6968.1998.tb12911.x. [DOI] [PubMed] [Google Scholar]
- 8.Cann I K O, Ishino Y. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohn E J, Edsall J T. Proteins, amino acids and peptides as ions and dipolar ions. New York, N.Y: Reinhold Publishing Corp.; 1943. pp. 370–381. [Google Scholar]
- 9a.Clustal W. Multiple Sequence Alignment. 5 February 1998, copyright date. [Online.] http://www.genome.ad.jp/SIT/CLUSTALW.html. [15 MARCH 1999, last date accessed.]
- 10.Eissenberg J C, Ayyagai R, Gomes X V, Burgers P M J. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol Cell Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forterre P, Elie C, Kohiyama M. Aphidicolin inhibits growth and DNA synthesis in halophilic archaebacteria. J Bacteriol. 1984;159:800–802. doi: 10.1128/jb.159.2.800-802.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerik K J, Li X, Pautz A, Burgers P M J. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 13.Gill S C, von Hippel P H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 14.Goulian M, Hermann S M, Sackett J W, Grimm S L. Two forms of DNA polymerase δ from mouse cells. purification and properties. J Biol Chem. 1990;265:16402–16411. [PubMed] [Google Scholar]
- 15.Gulbis J M, Kelman Z, O’Donnell J M, Kuriyan J. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 16.Hindges R, Hubscher U. Production of active mouse DNA polymerase δ in bacteria. Gene. 1995;158:241–246. doi: 10.1016/0378-1119(95)00065-e. [DOI] [PubMed] [Google Scholar]
- 17.Imamura M, Uemori T, Kato I, Ishino Y. A non-α-like DNA polymerase from the hyperthermophilic archaeon Pyrococcus furiosus. Biol Pharmacol Bull. 1995;18:1647–1652. doi: 10.1248/bpb.18.1647. [DOI] [PubMed] [Google Scholar]
- 18.Ishino Y, Cann I K O. The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: fact or farce? Genes Genet Syst. 1998;73:323–336. doi: 10.1266/ggs.73.323. [DOI] [PubMed] [Google Scholar]
- 19.Ishino Y, Komori K, Cann I K O, Koga Y. A novel DNA polymerase family found in Archaea. J Bacteriol. 1998;180:2232–2236. doi: 10.1128/jb.180.8.2232-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishino, Y., S. Ishino, I. K. O. Cann, Y. Kawarabayasi, H. Kikuchi, and Y. Sako. Unpublished data.
- 21.Jarvis T C, Paul L S, von Hippel P H. Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J Biol Chem. 1989;264:12709–12716. [PubMed] [Google Scholar]
- 22.Jonsson Z O, Hubscher U. Proliferating cell nuclear antigen: more than a clamp for DNA polymerase. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- 23.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Yoshizawa T, Nakamura Y, Robb F T, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 24.Kelman Z. PCNA: structure, functions, and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 25.Kelman Z, Hurwitz J. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biochem Sci. 1998;23:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- 26.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 27.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 28.Kong H, Kucera R B, Jack W E. Characterization of a DNA polymerase from the hyperthermophilic Archaea Thermococcus litoralis. J Biol Chem. 1993;268:1965–1975. [PubMed] [Google Scholar]
- 29.Kong X-P, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 30.Krishna T S R, Kong X-P, Gary S, Burgers P M J, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 31.Perler F B, Kumar S, Kong H. Thermostable DNA polymerases. Adv Protein Chem. 1996;48:377–435. doi: 10.1016/s0065-3233(08)60367-8. [DOI] [PubMed] [Google Scholar]
- 32.Podust V N, Podust L M, Muller F, Hubscher U. DNA polymerase δ holoenzyme: action on single-stranded DNA and on double-stranded DNA in the presence of replicative DNA helicases. Biochemistry. 1995;34:5003–5010. doi: 10.1021/bi00015a011. [DOI] [PubMed] [Google Scholar]
- 33.Schinzel R, Burger K J. Sensitivity of halobacteria to aphidicolin, an inhibitor of eukaryotic α-type DNA polymerases. FEMS Microbiol Lett. 1984;25:187–190. [Google Scholar]
- 34.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicare R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Jiang Y, Zhang P, Zhang S-J, Zhou Y, Li B Q, Toomey N L, Lee M Y W T. Expression and characterization of the small subunit of human DNA polymerase δ. J Biol Chem. 1997;272:13013–13018. doi: 10.1074/jbc.272.20.13013. [DOI] [PubMed] [Google Scholar]
- 36.Tsurimoto T. PCNA, a multifunctional ring on DNA. Biochem Biophys Acta. 1998;1443:23–39. doi: 10.1016/s0167-4781(98)00204-8. [DOI] [PubMed] [Google Scholar]
- 37.Uemori T, Ishino Y, Doi H, Kato I. The hyperthermophilic archaeon Pyrodictium occultum has two α-like DNA polymerases. J Bacteriol. 1995;177:2164–2177. doi: 10.1128/jb.177.8.2164-2177.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uemori T, Ishino Y, Toh H, Asada K, Kato I. Organization and nucleotide sequence of the DNA polymerase gene from the archaeon Pyrococcus furiosus. Nucleic Acids Res. 1993;21:259–265. doi: 10.1093/nar/21.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uemori T, Sato Y, Kato I, Doi H, Ishino Y. A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells. 1997;2:499–512. doi: 10.1046/j.1365-2443.1997.1380336.x. [DOI] [PubMed] [Google Scholar]
- 40.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 41.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Warbrick E, Lane D P, Glover D M, Cox L S. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to coordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- 43.Woese C R, Fox G E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 45.Zhang P, Frugulhetti I, Jiang Y, Holt G L, Condit R C, Lee M Y W T. Expression of the catalytic subunit of human DNA polymerase δ in mammalian cells using a vaccinia virus vector system. J Biol Chem. 1995;270:7993–7998. doi: 10.1074/jbc.270.14.7993. [DOI] [PubMed] [Google Scholar]
- 46.Zhang P, Zhang S-J, Zhang Z, Woessner J F, Jr, Lee M Y W T. Expression and physicochemical characterization of human proliferating cell nuclear antigen. Biochemistry. 1995;34:10703–10712. doi: 10.1021/bi00034a002. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S-J, Zeng X-R, Zhang P, Toomey N L, Chuag R-Y, Chang L-S, Lee M Y W T. A conserved region in the aminoterminus of DNA polymerase δ is involved in proliferating cell nuclear antigen binding. J Biol Chem. 1995;270:7988–7992. doi: 10.1074/jbc.270.14.7988. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J Q, He H, Tan C K, Doweny K M, So A G. The small subunit is required for functional interaction of DNA polymerase δ with the proliferating cell nuclear antigen. Nucleic Acids Res. 1997;25:1094–1099. doi: 10.1093/nar/25.6.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J Q, Tan C-K, So A G, Downey K M. Purification and characterization of the catalytic subunit of human DNA polymerase δ expressed in baculovirus-infected insect cells. J Biol Chem. 1996;271:29740–29745. doi: 10.1074/jbc.271.47.29740. [DOI] [PubMed] [Google Scholar]