Abstract
A teichuronopeptide (TUP) is one of major structural components of the cell wall of the facultative alkaliphilic strain Bacillus lentus C-125. A mutant defective in TUP synthesis grows slowly at alkaline pH. An upper limit of pH for growth of the mutant was 10.4, while that of the parental strain C-125 was 10.8. Gene tupA, directing synthesis of TUP, was cloned from C-125 chromosomal DNA. The primary translation product of this gene is likely a cytoplasmic protein (57.3 kDa) consisting of 489 amino acid residues. Introduction of the tupA gene into the TUP-defective mutant complemented the mutation responsible for the pleiotropic phenotypes of the mutant, leading to simultaneous disappearance of the defect in TUP synthesis, the diminished ability for cytoplasmic pH homeostasis, and the low tolerance for alkaline conditions. These results demonstrate that the acidic polymer TUP in the cell wall plays a role in pH homeostasis in this alkaliphile.
Alkaliphiles, which are capable of growth in a high alkaline milieu at pH 10 to 11, serve as a model system to understand the physiological mechanisms allowing microbial growth under extreme environmental conditions. Several studies carried out in the past decade have shown that a key feature of alkaliphiles is their ability to maintain a neutral cytoplasm pH (23). Cytoplasmic pH homeostasis is achieved as the net result of overall cell functions controlling the influx and efflux of hydrogen ions and hydroxyl ions. Among the cell components involved in these functions, the Na+/H+ antiporter seems to play a predominant role in establishing cytoplasmic pH homeostasis (23). The facultative alkaliphile Bacillus lentus C-125 grows in the pH range of 6.8 to 10.8 (4). A Δψ-dependent Na+/H+ antiport system is involved in maintaining a neutral cytoplasmic (intracellular) (pHi) pH in this bacterium (22). Its cell wall seems to play some role in the development and maintenance of alkaliphily. This consideration is supported by the observation that outgrowth of the protoplasts occurs effectively at neutral pH but poorly under alkaline conditions above pH 8 (8).
The cell wall of strain C-125 is composed mainly of peptidoglycan, teichuronic acid (TUA) and teichuronopeptide (TUP). The peptidoglycan is an A1γ type identical to that of the neutrophilic species B. lentus and Bacillus megaterium and slightly different from that of Bacillus subtilis (6). TUA is widely distributed in gram-positive bacteria, including both alkaliphiles and neutrophiles. TUP has distinctive chemical features. TUP is covalently bound to the peptidoglycan. It is a highly acidic copolymer composed exclusively of two polymers, one consisting of uronic acids and the other consisting of acidic amino acids. Several variations of TUP molecules are found in the cell walls of one-half to two-thirds of the alkaliphilic Bacillus spp. isolated from soils (5, 7). The TUP of strain C-125 is a copolymer of polyglutamic acid (PGlu) and polyglucuronic acid (PGlcU). The TUA of C-125 is composed of galacturonic acid, glucuronic acid, and N-acetylfucosamine (1–3, 14).
The amounts of TUA and TUP in the cell wall of C-125 markedly increase as the culture pH increases. The amount ratios of TUP and TUA to peptidoglycan of B. lentus C-125 grown at pH 10.5 are each three- and fivefold higher than when the alkaliphile is grown at pH 6.8 (5, 10). Mutants defective in TUP grow slowly at alkaline pH (10, 13, 19). These observations suggest that TUP in the cell wall may be important to allow the growth in an alkaline environment. We cloned a gene, tupA, complementing phenotypes of the mutant, defective TUP synthesis and low alkaliphily.
In this report, we describe the cloning and sequencing of the C-125 tupA gene, immunological detection of the translational product, and improvement of the ability to maintain both cytoplasmic pH homeostasis and alkaliphily of a TUP-defective mutant by introduction of the gene into the mutant.
MATERIALS AND METHODS
Microorganisms and plasmids.
The alkaliphilic bacterium B. lentus C-125 and its cell wall-defective derivatives were used. The following derivatives were constructed previously (13). Strain C-125-11 (Strr Thr− TUA−), a mutant lacking TUA, was isolated from a threonine-requiring, streptomycin-resistant derivative of C-125. Strain C-125-90 (Strr Thr− TUA− TUP- Glu−) was prepared from C-125-11 and lacks TUA and the PGlu moiety of TUP. Strain C-125-F19 (Strr Nalr TUP-Glu−) was constructed by cell fusion between C-125-90 and a methionine-requiring, nalidixic acid-resistant derivative of C-125 (19). This strain produces TUA but not the PGlu moiety of TUP.
Escherichia coli MV1184 {ara Δ(lac-proAB) rpsL thi φ80(lacZΔM15) Δ(srl-recA)306::Tn10(Tetr)/F′[traD36 proAB+ lacIq lacZΔM15]} was employed as the host strain for plasmids used in nucleotide sequencing. E. coli BL21(DE3) (F− ompT rB− mB−) was purchased from Takara Shuzo (Kyoto, Japan) and was used as the host strain for pCla. B. subtilis DB104 (his nprE18 nprR2 ΔaprA3) (20) was used as the host strain for plasmid pCW21-1.
Plasmid pHW1, carrying a chloramphenicol resistance gene (18), was used as a vector in B. lentus. The recombinant plasmids pCW21 and pCW21-1, containing the tupA gene, were constructed in this study. The expression vector pET-21d, purchased from Takara Shuzo, was used to construct pCla, encoding ′tupA′ (tupA with N- and C-terminal truncations).
Media and culture conditions.
Strains of alkaliphiles were grown aerobically at 37°C in an alkaline complex medium containing 13.7 g of K2HPO4, 5.9 g of KH2PO4, 10.6 g of Na2CO3, 5 g of glucose, 5 g of peptone, 1 g of yeast extract, 0.3 g of citric acid, and 0.05 g of MgSO4 · 7H2O per liter of deionized water (pH 10). A similar medium containing 11.6 g of NaCl instead of Na2CO3 was occasionally used as a neutral complex medium (pH 7.0 to 7.5) (5). E. coli and B. subtilis were grown in LB broth at 37°C. These media were solidified with 1.5% (wt/vol) agar when necessary.
When it was necessary to keep the culture pH constant during growth, bacteria were grown in 3-liter batches in a jar fermentor equipped with a pH-stat apparatus. The required pH conditions were initially established by using a medium consisting of 5 g of glucose, 5 g of peptone, 1 g of yeast extract, 0.3 g of citric acid, and 0.05 g of MgSO4 · 7H2O per liter of deionized water and one of the following buffers: pH 6.5 to 7.0, 0.1 M K2HPO4–0.2 M NaCl-HCl; pH 8.0 to 9.0, 0.1 M K2HPO4–0.14 M NaCl–0.03 M Na2CO3; pH 10.0 to 11.0, 0.2 M KCl–0.05 M Na2CO3–NaOH. The pH was maintained by automatic addition of 2 M HCl or 1 M NaOH during cultivation. Air was bubbled into the medium (4.5 liter/min). The culture was stirred at 120 rpm.
DNA manipulations.
Chromosomal DNA was prepared from C-125 cells lysed with lysozyme (0.4 mg/ml) at pH 8.0 and purified by phenol treatment. B. lentus and B. subtilis were transformed by the protoplast transformation method (16). Plasmid DNA was recovered from these Bacillus species by the method of Voskuil and Chambliss (26). DNA was digested with restriction enzymes under the conditions recommended by the manufacturers. Southern hybridization with digoxigenin-labeled DNA probes and antidigoxigenin antibody coupled to alkaline phosphatase was performed by means of a DNA labeling and detection kit (Boehringer, Mannheim, Germany). Other DNA manipulations not described in this report were performed by standard methods. The dideoxy chain termination method was used for sequencing of the gene.
Cloning of DNA fragments capable of improving the growth of B. lentus C-125-90.
C-125 chromosomal DNA was partially digested with HindIII and fractionated by agarose gel electrophoresis. Fragments with sizes in the range of 3 to 10 kb were recovered and inserted into the HindIII site of pHW1. To isolate a DNA fragment capable of complementing defective TUP synthesis, protoplasts prepared from C-125-90 by lysozyme treatment were transformed with the recombinant plasmids in the presence of 22% (wt/vol) polyethylene glycol 6000. The transformed protoplasts were plated on modified DM-3 medium (pH 6.8) containing chloramphenicol (2.5 μg/ml) to allow cell wall regeneration (8, 16). Transformants that outgrew on the medium were transferred to alkaline medium (pH 10) containing chloramphenicol.
Immunological detection of the TupA protein.
A 1.2-kb EcoRI-ClaI fragment encoding codons 11 to 425 of tupA was fused in frame with the lacZ gene under the control of the T7 promoter of pET-21d. E. coli BL21(DE3) harboring the resulting plasmid, pCla, was incubated in LB medium containing isopropyl-β-d-thiogalactoside (1 mM). The cells were broken by sonication in 20 mM Tris-HCl (pH 8.0) buffer. The unbroken cells were removed by the low-speed centrifugation, and then the lysate was centrifuged (50,000 × g, 30 min, 4°C). The precipitate was washed with 0.5% Triton X-100–6 M urea–20 mM Tris-HCl (pH 8.0) and then with 0.8% n-octyl-β-d-thioglucoside–20 mM Tris-HCl (pH 8.0). Proteins in the insoluble residue were electrophoresed on a sodium dodecyl sulfate (SDS)–10% (wt/vol) polyacrylamide gel (24). The recombinant ′TupA′ protein was recovered from the gel and used to immunize a rabbit.
Sample proteins were solubilized in SDS and electrophoresed on an SDS–10% (wt/vol) polyacrylamide gel. Proteins were detected by immunoblotting with the ′TupA′ antiserum and goat anti-rabbit horseradish peroxidase conjugate (Bio-Rad Laboratories, Hercules, Calif.). Prestained SDS-PAGE Standards (Bio-Rad Laboratories) were used as molecular mass markers for electrophoretic mobility.
Measurement of cytoplasmic pH in the alkaliphile.
B. lentus cells were loaded with a pH-sensitive fluorescent probe, 2′,7′-bis-(2-carboxyethyl)-5 (and -6)-carboxyfluorescein (BCECF). The BCECF-loaded cells were incubated in buffers [0.1 M N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid–NaOH (pH 6.0 to 8.6) or 0.1 M glycine-NaOH (pH 8.0 to 12.0)] containing 0.1 M NaCl–0.1 M KCl–0.1% glucose. The intracellular BCECF was excited at 450 or 510 nm, and emission was measured at 535 nm (9). Immediately after measurement of fluorescence, the pH of the suspension was measured by means of a glass electrode, and then the suspension was centrifuged at 6,000 × g for 5 min. Using the supernatant, the fluorescence intensity of the extracellular BCECF that had leaked from the cells was measured.
Preparation of cell walls.
Cells of each strain in the early stationary phase of growth were harvested and incubated in 2% (wt/vol) SDS–0.1 M NaCl at 80°C for 30 min. The cells were suspended in 0.1% NaN3–0.1 M NaCl and broken by sonication (20 kHz, 200 W, 10 min). After removal of unbroken cells by centrifugation (8,000 × g, 20 min, 20°C), the supernatant solution was incubated in the presence of 2% (wt/vol) SDS at 80°C for 30 min. The cell wall material was recovered by centrifugation (20,000 × g, 30 min, 20°C) and washed repeatedly with the warm NaN3-NaCl solution. The cell wall material was treated with trypsin (0.5 mg/ml) in 10 mM CaCl2–50 mM Tris–HCl (pH 7.9) at 37°C overnight (5).
Characterization of acidic polymers in the cell wall.
The cell wall material prepared from each strain grown in 2 liters of the alkaline medium was extracted with 5% (wt/vol) trichloroacetic acid. This treatment hydrolyzes linkages through which TUA and TUP bind to the peptidoglycan. The extract was held in a dialysis tube with an Mr cutoff of 3,500 (Spectropore, Los Angeles, Calif.) and dialyzed against running water (1). The dialysate was loaded onto a column of DE-52 (1.5 by 50 cm; Whatman Ltd., Maidstone, United Kingdom) equilibrated with 50 mM acetic acid-NaOH buffer (pH 5.0). The column was washed with 60 ml of buffer and with 60 ml of buffer containing 0.2 M NaCl and then eluted by a linear gradient from 0.2 to 0.6 M NaCl in the buffer (200 ml) at a flow rate of 80 ml/h. Finally, the column was eluted with the buffer containing 0.6 M NaCl. Fractions (3 ml each) was assayed for uronic acids, amino sugars, l-glutamic acid, and NaCl.
The molecular mass of each sample was estimated by gel chromatography on two columns of Shodex WS 802.5F (Showa Denko, Tokyo, Japan) connected in series (2). Degradation products of pullulan, P-5, -10, -20 and -50 (Showa Denko), were used as Mr references.
Chemical analyses.
(i) Uronic acids were assayed directly by the carbazole reagent method (17) with glucuronic acid as a reference standard. (ii) Amino sugars were liberated by hydrolysis in 4 M HCl for 16 h at 100°C and assayed by the Elson-Morgan reaction (27) with glucosamine as a reference standard. (iii) For assays of l-glutamic acid and l-alanine, samples were hydrolyzed as described above; then l-glutamic acid content was determined with l-glutamate dehydrogenase and diaphorase (15), and l-alanine content was determined with l-alanine dehydrogenase (28). (iv) The NaCl concentration was measured by determining the refractive index of the sample solution.
RESULTS
Cloning of a DNA fragment capable of improving the alkali tolerance of B. lentus C-125-90.
C-125 and C-125-11 grow rapidly at pH 10 to 10.5 (Fig. 1). C-125-90 formed tiny smooth colonies on alkaline medium after prolonged incubation, compared with the robust rough colonies formed by C-125 and C-125-11 (13). We used the growth characteristics and colony morphology as convenient indicators to clone a DNA fragment capable of restoring high alkali-tolerant growth and TUP synthesis in C-125-90.
FIG. 1.
Growth of cell wall component-defective mutants. B. lentus strain C-125, TUA-defective mutant C-125-11, TUA- and TUP-defective mutant C-125-90, and C-125-90(pCW21) were grown for 24 h (plates A and B) or 40 h (plates C and D) at pH 7.5 (plate A), 10.0 (plates B and C), or 10.5 (plate D). Compositions of the media used in plates A to C are provided in Materials and Methods; the medium in plate D contained 0.2 M Na2CO3 and no K2HPO4.
C-125 chromosomal DNA was digested partially with HindIII. The resulting fragments were inserted into the HindIII site of pHW1. Protoplasts from C-125-90 were transformed with the resulting recombinant plasmids and grown at pH 6.8 in the presence of chloramphenicol. When about 10,000 transformants were grown at pH 10, one displayed a rough colony morphology resembling that of C-125. This transformant was found to contain a recombinant plasmid with an 8-kb DNA insert composed of six HindIII segments (Fig. 2); this plasmid was designated pCW21. C-125-90(pCW21) grew as rapidly as C-125 and C-125-11 on alkaline media (Fig. 1).
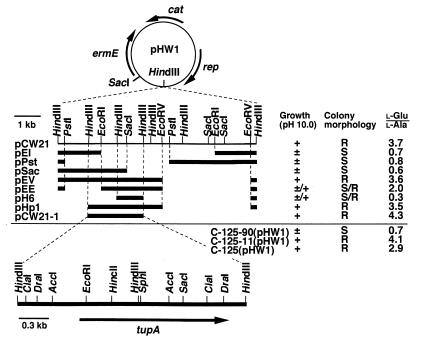
FIG. 2.
Restriction maps and functions of the inserts in pCW21 and its derivatives. pCW21 was constructed by insertion of HindIII partial digestion of C-125 chromosomal DNA into the HindIII site of pHW1. Deletion derivatives were constructed from pCW21. A black bar represents the DNA insert in each plasmid. C-125-90 harboring each plasmid was grown on agar medium (pH 10). Growth and colony morphology of the transformant were observed after 24 h. + and ±, colony formation was rapid and slow, respectively; ±/+, most colonies grew slowly and a few grew rapidly; R and S, rough and smooth colonies, respectively, were formed; S/R, the transformant formed primarily smooth colonies and a few rough colonies; l-Glu/l-Ala, molar ratio of l-glutamic acid content to l-alanine content in the cell wall of each transformant grown at initial pH 8. An arrow indicates the direction of transcription of each gene.
Restoration of TUP synthesis in B. lentus C-125-90 by transformation with pCW21.
The cell wall of each strain was incubated in warm trichloroacetic acid, and nonpeptidoglycan components released from the cell wall were fractionated by DEAE-cellulose chromatography (Fig. 3). TUA derived from C-125 was eluted from the column with 0.3 to 0.4 M NaCl. The average molecular mass of TUA was estimated to be 50 kDa by gel chromatography. TUP solubilized from the C-125 or C-125-11 cell wall was eluted with 0.4 to 0.5 M NaCl. The molecular mass of TUP was 21 kDa. The cell wall of strain C-125-90 (derived from C-125-11) was found to contain PGlcU as a major acidic component, which was eluted with 0.3 M NaCl. The PGlcU was eluted at the same concentration of NaCl as C-125 TUA but contained no galacturonic acid or fucosamine, both of which are constituents of the TUA. The molecular mass of PGlcU was 7.3 kDa. Thus, PGlcU is considered to be generated from TUP by removal of the PGlu moiety (13). A small amount of TUP was eluted with 0.4 M NaCl.
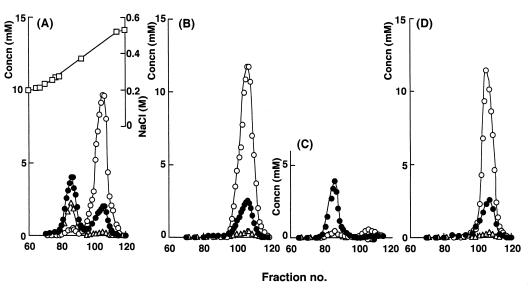
FIG. 3.
Fractionation of acidic polymers extracted from cell walls by DEAE-cellulose chromatography. Cell walls were prepared from C-125 (A), C-125-11 (B), C-125-90 (C), and C-125-90(pCW21) (D), each grown in 2 liters of alkaline medium (pH 10). Acidic polymers were extracted from the cell wall preparations and fractionated by DEAE-cellulose column chromatography. The samples were eluted in the same gradient of NaCl concentration. Each fraction was assayed for l-glutamic acid (○), uronic acids (●), amino sugars (▵), and NaCl (□).
The extract prepared from the cell wall of C-125-90(pCW21) showed a single peak of glucuronic acid residues and l-glutamic acid residues (Fig. 3D). These residues were eluted with 0.4 to 0.5 M NaCl. The extract contained no PGlcU eluted with 0.3 M NaCl. Thus, transformation of C-125-90 with pCW21 resulted in the production of a much more acidic form of PGlcU. Upon analysis of the peak fraction, the molar ratio of glutamic acid to glucuronic acid was found to be 4.3. This ratio was the same as that observed for the peak fraction in the case of C-125 or C-125-11 TUP. The molecular mass of the substance recovered from the peak fraction was 21 kDa. These results indicate that PGlcU was covalently bound to PGlu synthesized in C-125-90(pCW21) as found in TUP produced by C-125-11.
The minimum region of pCW21 needed to complement the C-125-90 mutation.
Several deletion derivatives were constructed from plasmid pCW21 DNA to determine the minimum region essential to restore alkali tolerance or TUP synthesis in C-125-90 (Fig. 2). Each derivative introduced into C-125-90 was assessed by observing the morphology of colonies formed at pH 10 and by determining the l-glutamic acid content of the cell walls of transformants grown at pH 8. Colony morphology was used as an indicator for the construct pCW21. In the cell wall preparations, the l-isomer of glutamic acid is derived only from PGlu of TUP, and l-alanine is derived only from the peptide moiety of C-125 peptidoglycan (3, 6). Thus, the molar ratio of l-glutamic acid to l-alanine is proportional to the ratio of PGlu to peptidoglycan.
The molar ratio l-glutamic acid to l-alanine was high in the cell walls prepared from all transformants that formed colonies with rough exteriors. This ratio was generally low in the case of transformants that formed tiny colonies with smooth exteriors. The exception was C-125-90(pEE), which initially formed tiny smooth colonies together with a few robust rough colonies; the exterior of the colonies became rough during prolonged incubation. Growth at alkaline pH and TUP synthesis seemed to be restored partially in C-125-90(pEE). Alternatively, the cells that formed rough colonies might have resulted from in vivo recombination between the chromosome and plasmid pEE. Such recombination was responsible for the occurrence of a few rough colonies in the case of C-125-90(pH6) because the formation of rough colonies was a heritable feature of C-125-90(pH6).
The results suggest that the hereditary determinants causing hypersensitivity to alkaline pH and defect in TUP synthesis were identical. The phenotypic characteristics of C-125-90 (defective TUP synthesis and reduction in alkaliphily) probably result from a single mutation in a certain gene that shows pleiotropic effects. Spontaneous revertants displaying rapid growth at pH 10 appeared occasionally from C-125-90 grown on the alkaline agar and formed rough colonies (10), indicating that the single mutation resulted in the pleiotropic alterations of C-125-90. This putative gene was named tupA on the basis of its involvement in TUP synthesis. It was concluded that the gene tupA was encoded within the 2.4-kb HindIII-HindIII-HindIII segment carried by pCW21-1 and mainly within the 1.6-kb EcoRI-HindIII-HindIII segment (Fig. 2). The site of the mutation in C-125-90 tupA is likely in the 0.7-kb SacI-HindIII segment common to derivatives pEE and pH6. It seems unlikely that C-125-90 has two or more mutated genes which in concert lead to decrease in alkali tolerance and to loss of PGlu synthesis. The analyses showed that pCW21-1 complemented the phenotypic characteristics of C-125-90 as did pCW21. Thus, subclone pCW21-1 was used in the following experiments.
Southern blot analysis of C-125 chromosomal DNA with the DNA insert in pCW21-1.
The DNA insert in pCW21-1 consisted of two HindIII fragments. This insert might have been artificially generated during the ligation reaction by fusion of two HindIII fragments not adjacent to each other on the chromosome. To verify that these regions are present adjacently on the C-125 chromosome, the chromosomal DNA was analyzed by Southern blot analysis using a 1.9-kb DraI fragment derived from pCW21-1 as a probe. When the chromosomal DNA was treated with HindIII, two bands with sizes of 1.1 and 1.2 kb reacted with the probe DNA (results not shown). Sizes of these bands corresponded to those of the two HindIII fragments found as the insert in pCW21-1. When the chromosomal DNA was treated with DraI, a 1.9-kb band showing a signal was found. This band was identical in size to the probe DNA prepared from pCW21-1. These results indicated that the two HindIII fragments comprising the insert are indeed neighbors on the C-125 chromosome, and the orientation of each of the two fragments is the same as that in the C-125 chromosome.
Nucleotide sequence of the tupA gene.
The insert in pCW21-1 was analyzed to determine the nucleotide sequence of tupA. Figure 4 shows the nucleotide sequence of the insert. This DNA was found to contain a long (1,485-nucleotide [nt]) open reading frame (ORF) starting at position 640 and ending at position 2125. The sizes of the other ORFs were no more than 435 nt. The long ORF occupies almost all of the region downstream of the EcoRI site, as suggested by subclone analysis. A small portion of the ORF corresponding to the N-terminal region of the encoded protein is present upstream of the EcoRI site. It seems more plausible that the long ORF is the tupA structural gene rather than that multiple genes contributing to alkali tolerance and TUP synthesis were mutated in C-125-90.
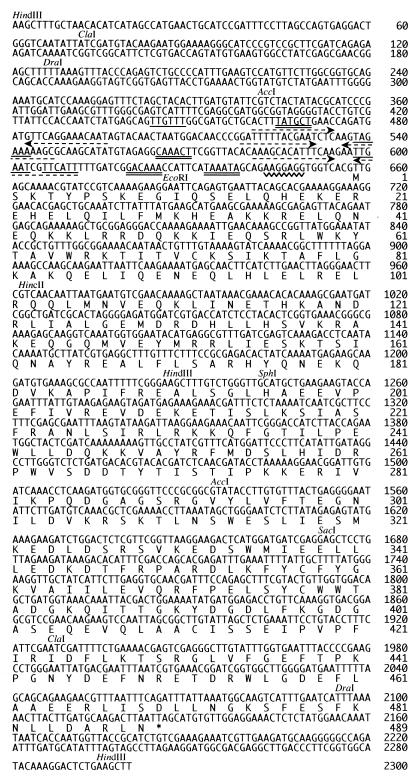
FIG. 4.
Nucleotide sequence of the DNA insert in pCW21-1 and deduced amino acid sequence of the tupA product. Nucleotides are numbered starting from the HindIII site upstream of tupA. Restriction sites shown in Fig. 2 are indicated above the sequence. Possible promoters are underlined with single (−35 sequences) or double lines (−10 sequences). Palindromic sequences in the promoter region are shown with dotted arrows. A possible Shine-Dalgarno sequence is shown with a wavy underline. The deduced amino acid sequence of TupA is shown under the nucleotide sequence of the structural gene tupA.
Many possible start codons are found in the ORF. Among them, two (GTG at position 649 and TTG at 658) are present upstream of the EcoRI site. The second possible start codon TTG is preceded at a spacing of 8 nt by a typical ribosome binding sequence (AAGGAGG at positions 644 to 650) which is complementary to the 3′ end of the 16S rRNA of E. coli (3′-AUUCCUCCACU--) or B. subtilis (3′UCUUUCCUCCA--) (25). Possible pairs of promoter elements (TTGTTT---TATGCT at positions 444 to 470, TAGAAA---CAAACT at 538 to 570, TTGAAT---GACAAA at 598 to 625, and TTCATT---TAAATA at 606 to 638) are present in the 5′-flanking region of tupA. It is interesting that palindromic sequences AXTXTXTXCTGAAC---GTTCAGXAXAXAXT (positions 461 to 496), GXTTTTTXCXAXT---AXTXGAAAAAXC (positions 519 to 547), and AAXGXACXXTTCAA---TTGAAXXGTXCXTT (positions 581 to 611) are present in the region containing the possible promoter sequences.
The primary translation product of the gene read from the start codon TTG is considered to be a protein (TupA) consisting of 489 amino acid residues with a molecular mass of 57.3 kDa. The deduced N-terminal amino acid sequence of the putative product shows no signal sequence motif for secretion, suggesting that TupA is a cytoplasmic protein. A leucine zipper motif is found from codons 80 to 101. We did not detect TupA as a major protein in B. lentus, B. subtilis, or E. coli transformed with the tupA gene when proteins were fractionated by electrophoresis and stained with dye (results not shown), suggesting that the level of expression of tupA is low probably because of the initiation codon being TTG and the putative transcriptional regulation.
Immunological detection of TupA in C-125.
We constructed the recombinant protein ′TupA′, designed to consist of 415 amino acid residues of TupA and 25 residues encoded by DNA derived from the vector. SDS-polyacrylamide gel electrophoresis showed that the molecular mass of ′TupA′ was 52 kDa, almost the same as that of the protein as designed. The N-terminal amino acid sequence (10 residues) of ′TupA′ was identical to that designed (results not shown). This recombinant protein was used to raise rabbit antiserum against TupA.
Immunoblotting with the antiserum showed that a novel protein with a molecular mass of 57 kDa was produced in B. subtilis DB104 by transformation with pCW21-1 (Fig. 5). The size of the protein expressed from the plasmid corresponded to that calculated for the tupA product, with the assumption that the initiation codon was TTG at position 658, indicating that the gene shown by nucleotide sequencing could be expressed in B. subtilis. B. lentus C-125 produced a protein of the same size, indicating that the tupA gene encoded on the chromosome is expressed in this alkaliphile. This protein was found in both the soluble and insoluble protein fractions recovered from C-125 broken by sonication (results not shown).
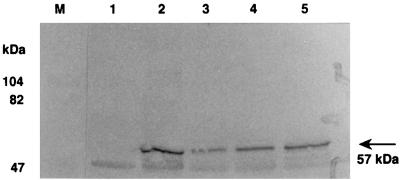
FIG. 5.
Immunological detection of TupA protein. B. subtilis and B. lentus were grown at pH 7 and 10, respectively, and the cells were broken by sonication. The cellular proteins (20 μg) were electrophoresed on an SDS–10% (wt/vol) polyacrylamide gel. Proteins with molecular masses higher than 40 kDa were transferred onto a nitrocellulose membrane and stained immunologically with antiserum against ′TupA′. Lanes: M, size markers; 1, B. subtilis DB104; 2, B. subtilis DB104(pCW21-1); 3, B. lentus C-125; 4, B. lentus C-125-90; 5, B. lentus C-125-90(pCW21-1).
C-125-90 produced a protein cross-reactive with the ′TupA′ protein, suggesting that C-125-90 is a missense mutant of tupA producing a protein of the same size as C-125. The amount of mutant TupA in C-125-90 was greater than that in C-125. This mutation is probably leaky, considering that a small amount of TUP was produced in C-125-90 (Fig. 3C). Because of the high-level production of mutant TupA and the low-level production of plasmid-encoded TupA protein, TupA production did not increase to a high level in C-125-90(pCW21-1). The amounts of TupA were estimated by immunoblot image analysis using Bio-Image Intelligent Quantifier version 2.1.2a software (B.I. Systems Corporation, Ann Arbor, Mich.). The ratio of the level of TupA produced by C-125-90 to that of C-125 and the ratio of the level of TupA produced by C-125-90(pCW21-1) to that of C-125-90 were each approximately 1.5.
Proteins obtained from C-125 grown under constant pH conditions at either pH 7.5, 9.0, or 10.0 were stained by immunoblotting (Fig. 6). TupA levels were estimated by immunoblot image analysis. The amount of TupA in the cells grown at pH 9.0 and 10.0 increased to 140 and 220% of that in cells grown at pH 7.5. The results showed that the amount of TupA produced increased as the culture pH increased.
FIG. 6.
Culture pH-dependent production of TupA protein in B. lentus. C-125 was grown at a constant pH, either 7.5 (lane 1), 9.0 (lane 2), or 10.0 (lane 3). The cellular proteins (40 μg) were electrophoresed on an SDS–7.5% (wt/vol) polyacrylamide gel. TupA was stained immunologically. Lane M, size markers.
Improvement of pHi homeostasis in C-125-90 by transformation with pCW21-1.
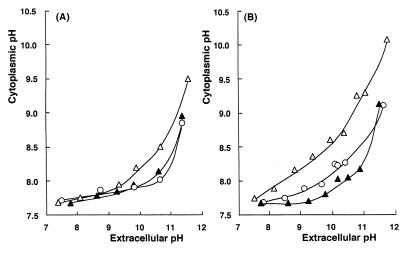
The pHi of C-125-90 was measured based on the fluorescence intensity of intracellular BCECF. The pHi of C-125(pHW1) grown at pH 10 was almost constant in a wide range of extracellular pH (pHo) under the experimental conditions used (Fig. 7A). The pHi was maintained at 7.7 to 8.0 over the range of pHo 7.3 to 10.6, which is within the physiological pH range for C-125. At pHo 11.4, C-125(pHW1) almost lost its ability to maintain moderate pHi. B. lentus C-125 does not grow above pH 10.8 (4). C-125-90(pHW1) grown at pH 10 maintained pHi below 8.0 when exposed to pHo 7.3 to 9.3. Above pHo 9.3, the pHi of C-125-90 was higher than that of C-125, indicating that the ability of C-125-90 to maintain pHi was diminished compared to that of C-125. When C-125 and C-125-90 were exposed to pHo 10.6, the pHi of C-125-90 was 0.5 units higher than that of C-125. Transformation of C-125-90 with pCW21-1 complemented the defect in pH homeostasis.
FIG. 7.
Improvement of the ability for pH homeostasis in C-125-90 by introduction of the tupA gene. Each strain was grown at pH 10.0 (A) or 8.5 (B). The cells were loaded with BCECF and incubated under various pHo conditions. The pHi was determined by the fluorescence intensity of BCECF; pH, shown was measured immediately after measurement of the fluorescence intensity. Symbols: ○, C-125(pHW1); ▵, C-125-90(pHW1); ▴, C-125-90(pCW21-1).
The ability to maintain pH homeostasis in B. lentus C-125 depends on the culture pH, being diminished after growth at pH 8.5 (9). The pHi of C-125(pHW1) grown at pH 8.5 and then exposed to pHo below 10 was almost neutral (7.5 to 8) (Fig. 7B). The cytoplasm became more alkaline when the cells were exposed to pHo above 10. The cytoplasm of C-125-90(pHW1) was more alkaline than that of C-125(pHW1) when the pHo was in the range of 7.3 to 11.6. Also, the mutation responsible for the diminished ability to maintain pH homeostasis of C-125-90 cells grown at pH 8.5 was complemented in cells transformed with pCW21-1.
Effect of transformation with tupA on the growth of C-125-90.
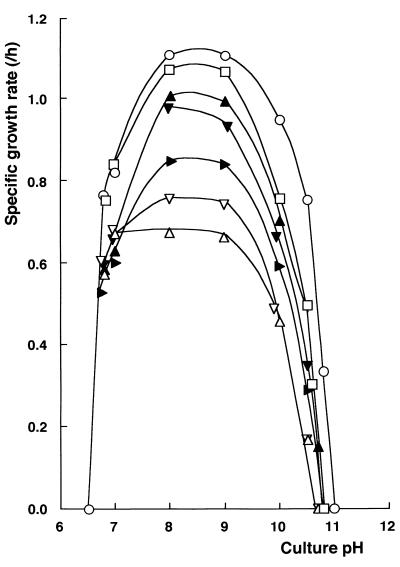
A series of growth studies was carried out at constant pH under controlled conditions. C-125(pHW1) grew at pH 6.7 to 10.8 but not at pH 11.0 (Fig. 8). Plasmid pHW1 did not alter the growth characteristics of C-125. C-125-11(pHW1) grew as rapidly as C-125(pHW1) at pH 6.7 to 9 and slower than C-125(pHW1) at pH above 9. C-125-90 grew at pH 6.7 to 10.4 but not at pH 10.5. The growth rate of C-125-90(pHW1) was significantly low above pH 7. The growth of C-125-90 above pH 7 was stimulated by introduction of pCW21-1. C-125-90(pCW21-1) grew at pH 6.7 to 10.7 at a rate similar to that of C-125-11(pHW1). At around pH 7, all of the strains examined showed similar growth rates. Thus, transformation of C-125-90 with the tupA gene increased the growth rate above pH 8.
FIG. 8.
Improvement of the growth of C-125-90 by introduction of the tupA gene. Each strain was grown under various pH conditions. Specific growth rate was calculated on the basis of increases in turbidity measured at 660 nm. Symbols: ○, C-125(pHW1); □, C-125-11(pHW1); ▵, C-125-90(pHW1); ▸, C-125-90(pCW21); ▴, C-125-90(pCW21-1); ▿, C-125-F19(pHW1); ▾, C-125-F19(pCW21-1).
A fusion strain, C-125-F19, constructed from C-125-90, produces TUA and PGlcU but not TUP (19). Its TUA and PGlcU have the same characteristics as TUA of strain C-125 and PGlcU of C-125-90, respectively. The growth characteristics of C-125-F19(pHW1) were highly similar to those of C-125-90(pHW1), suggesting that restoration of TUA synthesis does not improve the growth of C-125-90 at alkaline pH. Thus, the alkali sensitivity of C-125-90 results mainly from the loss of PGlu moiety of TUP rather than a synergistic effect of losses of TUA and PGlu. Also, the growth of C-125-F19 was stimulated by the transformation with pCW21-1. These results indicate that the slow growth of C-125-90 and C-125-F19 at alkaline pH was due to the incomplete cell wall structure, which could restored by introduction of the tupA gene.
DISCUSSION
TUP-defective mutants of alkaliphile B. lentus C-125 grow poorly at alkaline pH (13, 19). In this study, we showed that the loss of TUP synthesis and decrease in alkaliphily in the mutant C-125-90 were due to a single pleiotropic mutation in a certain gene. We cloned a DNA fragment capable of complementing the mutation responsible for the alkali hypersusceptibility and the mutation responsible for the defect in TUP synthesis in this mutant (Fig. 1 to 3). Subclone analysis showed that the complementation was associated with a single region of the insert DNA. Subclone pCW21-1, containing a 2.4-kb insert, complemented the mutation responsible for both features of the mutant phenotype of C-125-90 (Fig. 2). We obtained no subclone able to complement only one of the mutant features. Nucleotide sequencing showed that the insert in pCW21-1 encoded a novel gene, tupA, with a size of 1,470 bp, which encoded a 57-kDa protein, TupA (Fig. 4).
It was shown that pCW21-1 directed the synthesis of the 57-kDa protein in B. subtilis DB104 (Fig. 5). We observed that B. lentus C-125 produces TupA of the same size, indicating that tupA is functional but not dormant in C-125. Although we purified a candidate of the 57-kDa TupA protein reactive with the antiserum from the cytoplasm of B. lentus C-125, it seems that its N terminus was not open for analysis by automatic Edman degradation (results not shown). The N terminus of the protein might be modified in some manner. Thus, the initiation codon of tupA has not been definitively established although the molecular mass of TupA produced by B. subtilis DB104(pCW21-1) or B. lentus C-125 was same as that predicted on the basis of the deduced amino acid sequence of the product translated from the TTG codon. C-125-90 is thought to have a missense mutation downstream of the SacI site, resulting in a weakly active TupA mutant.
The N-terminal region of TupA shows low homology (18 to 21%) to moesins of human (Swiss-Prot P26038), mouse (Swiss-Prot P26041), rat (GenBank AF004811), and drosophila (GenBank DML38909) sequences, suggesting that TupA may be a peripheral membrane protein. No protein showing high homology to TupA has been deposited in databases. At the present time, tupA is an orphan gene. However, bacteria containing TUP analogues are predominant among the alkaliphiles examined (5, 7). Immunological analysis revealed that 57-kDa TupA homologues are distributed widely among strains of alkaliphilic Bacillus spp. possessing TUP (results not shown). The function of the TupA protein is not clear, although it was demonstrated that TupA is involved in TUP synthesis. TUP has been found only in certain alkaliphilic Bacillus spp., and little is known about its synthesis. At this time, various possible functions can be suggested for TupA.
The culture pH-dependent increase in the amount of TUP (10) is probably related to the increase in TupA levels depending on culture pH (Fig. 6). The mechanism of culture-pH responsive TupA production is not clear, although the palindromes found in the promoter region of tupA suggest that expression of the gene is regulated in some manner. In C-125, several biological processes are dependent on the culture pH; these include flagellin synthesis and mobility (12), Δψ-dependent Na+/H+ antiporter activity (22), oxygen uptake and NADH oxidation activity (11), pHi homeostasis (9), and sporulation (unpublished work). Elevated activity in certain of these processes seems to be advantageous in allowing the alkaliphile to adapt to an alkaline environment. Probably, elevated levels of the TupA and TUP benefit C-125 growing at high alkaline pH.
One of the biological functions of TUP, shown in this study, is its involvement in maintaining pHi homeostasis at a high level in C-125 under high pH conditions (Fig. 7). The extent of the contribution of TUP to homeostasis varied depending on the culture conditions because the levels of both TUP synthesis and Δψ-dependent Na+/H+ antiporter activity increased in response to an increase in the culture pH. TUP is essential for pH homeostasis when C-125 is grown at pH 8.5, under which conditions there is a low level of the antiporter activity. When C-125 is grown at pH 10, under which conditions there is a high level of the antiporter activity, TUP is essential for full homeostasis in cells exposed to an environmental pH above 9.5.
The results described here strongly suggest that high levels of acidic compounds in the cell wall contribute to maintaining pH homeostasis of C-125 at high alkaline pH although we cannot rule out another possibility, that the presence of PGlcU alone in the cell wall resulted in the phenotypic alterations found in C-125-90. Due to the low ability to maintain pHi homeostasis, C-125-90 shows poor growth above pH 8 compared with C-125 and C-125-11 (Fig. 8). The mutation responsible for this impairment of growth was complemented by the introduction of tupA into the cells. In this study, we examined the effect of TUP exclusively because growth of C-125-11 was similar to that of C-125 on the alkaline medium. However, TUA might contribute slightly to growth of C-125 at a high alkaline pH (Fig. 8).
It is not clear how acidic polymers in the cell walls are involved in sustaining the pH of the cytoplasm. A high density of fixed anion charges is generated by dissociation of carboxyl groups of the polymers in the cell walls of C-125 at alkaline pH. We have assumed that the fixed charges would function as an obstacle repulsing hydroxyl ions abundant in the milieu and as a reservoir capturing hydrogen ions generated intracellularly by aerobic respiration (5, 13). It is reported that the cell wall of B. subtilis has a lower pH than that of the surrounding medium (21), supposedly as a result of capturing protons by the cell wall materials. The predominant acidic polymer, the PGlu moiety of TUP (Fig. 3), likely functions to capture protons, as reported for B. subtilis. We are now trying to reveal the mechanism by which the cell wall polymers function to maintain the pH homeostasis high.
ACKNOWLEDGMENTS
This work was partially supported by a grant for the Biodesign Research Program from RIKEN to R. Aono and grant-in-aid 11876022 from the Ministry of Science, Education and Culture of Japan to R. Aono.
REFERENCES
- 1.Aono R. Isolation and partial characterization of structural components of the walls of alkalophilic Bacillus strain C-125. J Gen Microbiol. 1985;131:105–111. [Google Scholar]
- 2.Aono R. Characterization of structural component of cell walls of alkalophilic strain of Bacillus sp. C-125: preparation of poly(γ-l-glutamate) from cell wall component. Biochem J. 1987;245:467–472. doi: 10.1042/bj2450467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aono R. Characterization of cell wall components of the alkalophilic Bacillus strain C-125: identification of a polymer composed of polyglutamate and polyglucuronate. J Gen Microbiol. 1989;135:265–271. [Google Scholar]
- 4.Aono R. Assignment of facultatively alkaliphilic Bacillus sp. strain C-125 to Bacillus lentus group 3. Int J Syst Bacteriol. 1995;45:582–585. [Google Scholar]
- 5.Aono R, Horikoshi K. Chemical composition of cell walls of alkalophilic strains of Bacillus. J Gen Microbiol. 1983;129:1083–1087. [Google Scholar]
- 6.Aono R, Horikoshi K, Goto S. Composition of the peptidoglycan of alkalophilic Bacillus spp. J Bacteriol. 1984;157:688–689. doi: 10.1128/jb.157.2.688-689.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aono R, Ito M, Horikoshi K. Occurrence of teichuronopeptide in cell walls of group 2 alkaliphilic Bacillus spp. J Gen Microbiol. 1993;139:2739–2744. [Google Scholar]
- 8.Aono R, Ito M, Horikoshi K. Regeneration of protoplasts from alkaliphilic strains of Bacillus spp. Biosci Biotechnol Biochem. 1993;57:1597–1598. [Google Scholar]
- 9.Aono R, Ito M, Horikoshi K. Measurement of cytoplasmic pH of the alkaliphile Bacillus lentus C-125 with a fluorescent pH probe. Microbiology. 1997;143:2531–2536. doi: 10.1099/00221287-143-8-2531. [DOI] [PubMed] [Google Scholar]
- 10.Aono R, Ito M, Joblin K N, Horikoshi K. A high cell wall negative charge is necessary for the growth of the alkaliphile Bacillus lentus C-125 at elevated pH. Microbiology. 1995;141:2955–2964. [Google Scholar]
- 11.Aono R, Kaneko H, Horikoshi K. Alkaline growth pH-dependent increase of respiratory and NADH oxidation activities of facultatively alkaliphilic strain Bacillus lentus C-125. Biosci Biotechnol Biochem. 1996;60:1243–1247. [Google Scholar]
- 12.Aono R, Ogino H, Horikoshi K. pH-dependent flagellar formation by facultative alkaliphilic Bacillus sp. C-125. Biosci Biotechnol Biochem. 1992;56:48–53. doi: 10.1271/bbb.56.48. [DOI] [PubMed] [Google Scholar]
- 13.Aono R, Ohtani M. Loss of alkalophily in cell-wall-defective mutants derived from alkalophilic Bacillus C-125. Biochem J. 1990;266:933–936. [PMC free article] [PubMed] [Google Scholar]
- 14.Aono R, Uramoto M. Presence of fucosamine in teichuronic acid of the alkalophilic Bacillus strain C-125. Biochem J. 1986;233:291–294. doi: 10.1042/bj2330291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beutler H O, Michal G. Glutamate, determination with glutamate dehydrogenase, diaphorase and tetrazolium salts. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 2nd ed. Vol. 4. Weinheim, Germany: Verlag Chemie; 1974. pp. 1708–1713. [Google Scholar]
- 16.Chang S, Cohen S N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- 17.Davidson E A. Analysis of sugars found in mucopolysaccharides. Methods Enzymol. 1966;8:52–60. [Google Scholar]
- 18.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M, Tabata K, Aono R, Horikoshi K. Construction of a new teichuronopeptide-defective derivative from alkaliphilic Bacillus sp. C-125. Biosci Biotechnol Biochem. 1994;58:2275–2277. doi: 10.1271/bbb.58.2275. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura F, Doi R H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984;160:442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemper M A, Urrutia M M, Beveridge T J, Koch A L, Doyle R J. Proton motive force may regulate cell wall-associated enzymes of Bacillus subtilis. J Bacteriol. 1993;175:5690–5696. doi: 10.1128/jb.175.17.5690-5696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitada M, Hashimoto M, Kudo T, Horikoshi K. Properties of two different Na+/H+ antiport system in alkaliphilic Bacillus sp. strain C-125. J Bacteriol. 1994;176:6464–6469. doi: 10.1128/jb.176.21.6464-6469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krulwich T A. Alkaliphiles: ′basic′ molecular problems of pH tolerance and bioenergetics. Mol Microbiol. 1995;15:403–410. doi: 10.1111/j.1365-2958.1995.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.MacLaughlin J R, Murray C L, Rabinowitz J C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus β-lactamase. J Biol Chem. 1981;256:11283–11291. [PubMed] [Google Scholar]
- 26.Voskuil M, Chambliss G H. Rapid preparation and sequencing of purified plasmid DNA from Bacillus subtilis. Appl Environ Microbiol. 1993;59:1138–1142. doi: 10.1128/aem.59.4.1138-1142.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheat R W. Analysis of hexosamines in bacterial polysaccharides by chromatographic procedures. Methods Enzymol. 1966;8:60–78. [Google Scholar]
- 28.Williamson D H. Alanine: determination with alanine dehydrogenase. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 3rd ed. Vol. 8. Weinheim, Germany: VCH Verlagsgesellschaft; 1985. pp. 341–344. [Google Scholar]