Abstract
The extracellular chitosanase (34,000 Mr) produced by a novel gram-negative bacterium Matsuebacter chitosanotabidus 3001 was purified. The optimal pH of this chitosanase was 4.0, and the optimal temperature was between 30 and 40°C. The purified chitosanase was most active on 90% deacetylated colloidal chitosan and glycol chitosan, both of which were hydrolyzed in an endosplitting manner, but this did not hydrolyze chitin, cellulose, or their derivatives. Among potential inhibitors, the purified chitosanase was only inhibited by Ag+. Internal amino acid sequences of the purified chitosanase were obtained. A PCR fragment corresponding to one of these amino acid sequences was then used to screen a genomic library for the entire choA gene encoding chitosanase. Sequencing of the choA gene revealed an open reading frame encoding a 391-amino-acid protein. The N-terminal amino acid sequence had an excretion signal, but the sequence did not show any significant homology to other proteins, including known chitosanases. The 80-amino-acid excretion signal of ChoA fused to green fluorescent protein was functional in Escherichia coli. Taken together, these results suggest that we have identified a novel, previously unreported chitosanase.
Chitosanase (EC 3.2.1.132) catalyzes the hydrolysis of glycosidic bonds of chitosan, which is a totally or partially deacetylated derivative of chitin. Chitin is a polymer of N-acetylglucosamine. It was proposed that chitosanases are enzymes that hydrolyze GlcN-GlcN, GlcN-GlcNAc, and GlcNAc-GlcN bonds but not GlcNAc-GlcNAc bonds (5, 13). However, the difference between chitosanase and other enzymes, such as chitinases, lysozymes, and N-acetyl-β-d-glucosaminidases is sometimes obscure, especially in the case of enzymes that display activity toward multiple substrates. It is thought that the ability to hydrolyze a 100% deacetylated chitin is an important criterion for classifying an enzyme as a chitosanase. Chitosanases are produced by many organisms, including actinomycetes (2, 17), fungi (4, 27), plants (6, 18), and bacteria (7, 16, 22, 29, 30). Bacterial chitosanases have received special attention because they are important for the maintenance of the ecological balance and have been used to determine the mechanism of chitosan hydrolysis at both biochemical and molecular levels. Compared to the numerous reports on the primary structure and function of chitinases, information on chitosanases is still relatively limited.
So far, chitosanase genes have been isolated from Bacillus circulans MH-K1 (1), Nocardioides sp. strain N106 (12), Streptomyces sp. strain N174 (13), and Fusarium solani (26). Among these, the former three chitosanases show structural similarity, but the one from F. solani does not. The chitosanase from Streptomyces sp. is the only example of a chitosanase whose three-dimensional structure has been determined (11, 23). We still did not know how many different types of chitosanase exist in nature. Some chitosanases have hydrolytic activity on substrates other than chitosan, such as chitin (28) and cellulose (21). A more detailed knowledge of the structure of a number of chitosanases will be required for understanding their enzymatic differences and common structural elements.
We isolated Matsuebacter chitosanotabidus 3001 as a bacterium that produces chitosanase and classified it as a new genus and species belonging to the β-subclass of Proteobacteria (20). In this study, we report the properties of purified chitosanase from M. chitosanotabidus 3001 and describe the complete nucleotide sequence of the chitosanase (choA) gene.
MATERIALS AND METHODS
Materials.
Various degrees of deacetylated chitosan (70 to 100%) were purchased from Funakoshi Co., Ltd. (Tokyo, Japan). Chitin was obtained from San-in Kensetsu Co., Ltd. Oligomers of glucosamine were purchased from Seikagaku Kogyo Co., Ltd. (Tokyo, Japan). Glycol chitosan was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Carboxymethylchitosan and carboxymethylchitin were obtained from Nissui Co., Ltd. (Tokyo, Japan). λZapII and the packaging kit were purchased from Stratagene. Ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG) were purchased from Takara Biomedicals (Kyoto, Japan). Restriction endonuclease and bacterial alkaline phosphatase were obtained from Toyobo Co., Ltd. (Osaka, Japan), or Takara Biomedicals. The cassettes and cassette primers, 5′-d(GTACATATTGTCGTTAGAACGCGTAATACGACTCA)-3′ (C1) and 5′-d(CGTTAGAACGCGTAATACGACTCACTATAGGGAGA)-3′ (C2), were purchased from Takara Biomedicals. All other chemicals were reagent grade or molecular biological grade.
Screening and culture conditions of M. chitosanotabidus.
M. chitosanotabidus 3001 was identified as a novel bacterium on the basis of its 16S rRNA sequence, morphology, and physiological properties as described previously (20).
For growth in liquid culture, M. chitosanotabidus 3001 was grown in modified base medium containing 0.4% (wt/vol) colloidal chitosan. For chitosanase production, a single colony of M. chitosanotabidus 3001 was inoculated in 50 ml of colloidal chitosan liquid medium and grown for 4 days at 30°C with shaking (200 rpm). This medium consisted of 0.4% (wt/vol) colloidal chitosan, 0.5% MgSO4, 0.3% KH2PO4, 0.7% K2HPO4, 0.25% yeast extracts, and 0.25% polypeptone at pH 7.0.
Concentration and purification of chitosanase.
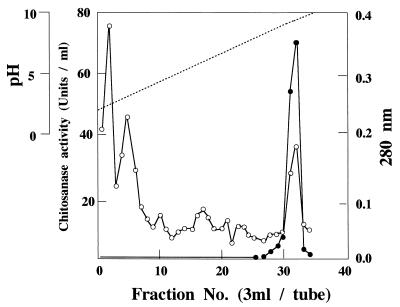
Bacterial cells were removed from culture broth by centrifugation at 12,000 rpm for 15 min in a Kubota KR-20000T rotor, and proteins in supernatant fluids were concentrated with ammonium sulfate (70% saturation). After incubation at 4°C for 1 h, the precipitates were collected by centrifugation at 12,000 rpm for 20 min. The precipitates were dissolved in an appropriate volume of 50 mM Tris-HCl buffer at pH 8.0 and dialyzed against 20 mM Tris-HCl buffer (pH 8.0) at 4°C overnight. The dialysates were centrifuged to remove the insoluble materials and were used as crude chitosanolytic enzyme fraction. Crude chitosanolytic enzyme was purified by isoelectric chromatography on a 110-ml column (LKB-Produkter) with ampholine (Sigma) as the carrier ampholite. Each fraction was collected, and the chitosanase activity was measured. Active fractions (numbers 31 and 32) were collected and used as a source of purified enzyme (Fig. 1). Proteins eluting from the column were detected by measuring the absorbance at 280 nm.
FIG. 1.
Isoelectric-focusing chromatography of chitosanase produced by M. chitosanotabidus 3001. Open circles (○) indicate the absorbance at 280 nm, and solid circles (●) indicate the activity of the enzyme. Fractions 31 and 32 were pooled and used as a source of purified enzyme.
Determination of amino acid sequence and immunoblotting.
To determine internal amino acid sequences, purified chitosanase from M. chitosanotabidus 3001 was digested with an appropriate concentration of trypsin, and the resulting peptides were separated by sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis (PAGE). Chitosanase purified as mature extracellular protein from E. coli expressing the chitosanase gene (choA) was also used for determining of the N-terminal amino acid sequence. The enzyme partially digested with trypsin was blotted to polyvinylidene difluoride (PVDF) membrane (Immobilon-PSQ; pore size, 0.45-μm; IPVH 304FO; Millipore) by using the electroblotting system according to the manufacturer’s instructions (Nippon Millipore Co.). Amino acid sequencing was performed by using a Shimadzu protein sequencer (PSQ-10). Rabbit antiserum raised against the chitosanase (anti-Cho) was prepared by Takara Biomedicals (Kyoto, Japan).
In Western blot analysis, cell extracts were subjected to SDS-PAGE (12.5%), and the protein bands were electrophoretically transferred onto a nitrocellulose membrane. For immunolabeling of ChoA, nitrocellulose membranes were incubated at room temperature with shaking in TBS-M buffer (20 mM Tris-HCl [pH 7.6], 0.137 M NaCl, 0.25% Tween 20, 5% dried milk) for at least 1 h. Afterwards, the membranes were rinsed several times with Tris-buffered saline (TBS-T,) buffer and then incubated for 1 h with rabbit antiserum. After several rinses in TBS buffer, the membranes were incubated with the horseradish peroxidase-conjugated second antibody, and the membrane-bound immunocomplexes were detected on X-ray film by the enhanced chemiluminescence method, which is based on measuring light emission from the oxidation of acridinium esters (ECL-Plus System; Amersham).
Construction and expression of ChoA-GFP fusions.
The DNA fragment corresponding to the N-terminal region of ChoA was amplified by PCR by using the primer TAGGATCCTATGCAACTTCCTCGACCTGAT (to create a BamHI site in front of the ATG codon) as the sense primer and TTGAATTCTTGGGGCCGGCCATGCC (to create an EcoRI site at the codon for the 48-amino-acid segment of ChoA) or TTGAATTCGGAATCACGCCCGCCGC (to create an EcoRI at the codon for the 88-amino-acid segment of ChoA) as the antisense primer. PCR was used to amplify the GFP (green fluorescent protein) gene from pGP110 by using the primer ACGAATTCTAGTAAAGGAGAAGAA (to create an EcoRI site before the ATG codon) and the primer ATCCCGGGAAGCTTATTTGTATAGTTCATC (to create a HindIII site at the 3′ end) (15). The two DNA fragments coding for ChoA and GFP were ligated into pBluescript SK(+) to yield pB48ChG (the N-terminal 48-amino-acid region was fused to GFP) and pB88ChG (the N-terminal 88-amino-acid region was fused to GFP).
E. coli JM109 harboring pB48ChG or pB88ChG was grown on Luria-Bertani medium to an optical density at 600 nm of 0.2 to 0.5. The cells were then further incubated for 2.5 h with 1 mM IPTG. Cells and supernatant were separated by centrifugation. Protein in the supernatant was concentrated by ultrafiltration through a Centricon 10 (Amicon) apparatus. E. coli cells were disrupted by sonication, and undisrupted cells were removed by centrifugation. Proteins (5 μg) were loaded onto each lane and electrophoresed. Western blot analysis was done by using an anti-GFP antibody (Clontech). Western blot analysis was carried out as described above.
Chitosanase assay.
Chitosanase activity was assayed by using colloidal chitosan as a substrate. The reaction mixture consisted of 0.5 ml of 0.5% colloidal chitosan, 1 ml of McIlvaine buffer (0.1 M citrate plus 0.2 M Na2HPO4) at pH 7.0, and 0.5 ml of the enzyme solution, and the mixtures were incubated at 30°C for 10 min. Reactions were stopped by boiling for 3 min, the reaction mixtures were centrifuged, and the supernatants were retained. The amount of reducing sugars produced was determined at A420 by the modified Schales method (8). One unit of chitosanase activity was taken as the amount of enzyme that produced 1 μmol of reducing sugars (expressed as glucosamine equivalents) per min.
DNA manipulations and amplification (PCR) reactions.
Oligonucleotide probes were synthesized by Takara Biomedicals. Standard procedures for restriction endonuclease digestion, agarose gel electrophoresis, purification of DNA from agarose gels, DNA ligations, and other cloning-related techniques were done as described by Sambrook et al. (24).
PCR amplification of the chitosanase gene was performed by using a DNA Thermal Cycler (Perkin-Elmer/Cetus). The two primers, designated S1 and S2 (5′-TCCGACAAGAAC(T)AAG(A)CG(T)CGCG-3′ and 5′-GTACTT(C)GTCCTGCTG(T)CGTGT-3′), were used to amplify a piece of the choA gene from M. chitosanotabidus 3001. The mixtures contained 0.1 nM concentrations of each primer, 2.5 mM concentrations of each deoxynucleoside triphosphate, 100 ng of template DNA, 0.2 U of Ex-Taq DNA polymerase (Takara Biomedicals), and a 10× concentration of reaction buffer. Amplification was allowed to proceed through 30 cycles, where each cycle consisted of denaturation (94°C, 1 min), primer annealing (45°C, 2 min), and polymerization (72°C, 3 min). For amplification of a piece of the choA gene, the cassettes and cassette primer method (9, 14) were used to amplify a neighboring DNA of the known DNA region. Chromosomal DNA of M. chitosanotabidus was digested with BamHI and ligated with double-stranded DNA cassettes possessing BamHI sites. Then, cassette-ligated DNA was amplified by PCR by using the known sequence primers (S2, R1) and cassette primers (C1, C2).
Construction of genomic libraries and hybridization conditions.
In order to construct a genome library, the total chromosomal DNA from M. chitosanotabidus 3001 was isolated by the method described by Sambrook et al. (24). Genomic DNA was completely digested with SacI, and the fractions containing 2- to 6-kbp DNA fragments were obtained by density gradient centrifugation. The fragments were cloned into the SacI site of SacI-digested λZapII. The PCR-amplified 0.3-kbp DNA fragment containing a portion of the chitosanase gene of M. chitosanotabidus 3001 was used to probe the genomic library by plaque hybridization. Southern transfer of DNA onto a nylon membrane was performed according to the method of ECL Systems (Amersham, United Kingdom).
Competent E. coli XL1-Blue MRF′ cells were transfected with the packaged phage. Approximately 1,800 plaques were obtained, and these were screened by plaque hybridization with the PCR-amplified 0.3-kbp DNA fragment as a probe. A positive clone that contained the choA gene was recovered as a plasmid by in vivo excision.
DNA sequencing and analysis.
Nested deletions of pChoU1 were generated by using the Kilo-Sequence Deletion Kit (Takara). DNA sequencing was performed on both double-stranded templates obtained after the subcloning of appropriate segments in the pBluescript II SK(−) and KS(−) vectors by using the dideoxy DNA sequencing system of the Ampli CycleR Sequence kit (Perkin-Elmer/Cetus) according to the procedure supplied by the manufacturer. DNA fragments were analyzed on an ABI Prism 377 DNA automated sequencer according to the instructions of the manufacturer. Nucleotide and amino acid sequence comparisons were made with the help of the GenBank database. The DNA sequence of the choA gene was deposited in the DDBI/EMBL/GenBank data base with the accession AB010493.
RESULTS
Purification of the chitosanase.
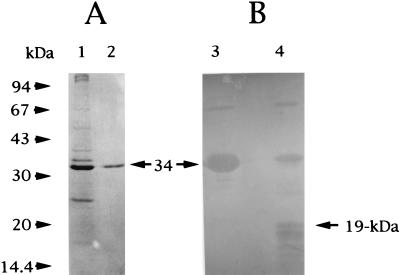
M. chitosanotabidus 3001, isolated from soil collected in Matsue, Japan, was selected for its ability to form clear zones on plates of agar medium containing 1% colloidal chitosan (20). Apparently, this bacterium was an active chitosanase producer and was chosen for further studies of this enzyme. Chitosanase was purified from the supernatant fluids of broth cultures of M. chitosanotabidus 3001 as described in Materials and Methods. The steps used to purify the chitosanase are summarized in Table 1. The specific activity of the final preparation was 250 U/mg. The homogeneity of the purified enzyme was verified by SDS-PAGE (Fig. 2A, lane 2). The molecular weight of the purified chitosanase was estimated to be 34,000. The isoelectric point (pI) of the enzyme was determined to be 9.6 by isoelectric focusing.
TABLE 1.
Purification of chitosanase from M. chitosanotabidus 3001
| Step | Protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Culture filtrate | 14.5 | 761.8 | 52.5 | 100 | 1 |
| Ammonium sulfate precipitation | 5.6 | 495.8 | 88.5 | 65.1 | 1.7 |
| Isoelectric-focusing chromatography | 3.1 | 371.0 | 250.2 | 48.7 | 4.8 |
FIG. 2.
SDS-PAGE (A) and immunoblotting (B) of the purified enzyme from M. chitosanotabidus. (A) The homogeneity of the purified enzyme was examined by using SDS-PAGE and staining with Coomassie brilliant blue R-250 (horizontal arrow). (B) Results of immunoblots of the purified enzyme and trypsin-digested proteolytic products. The positions of molecular size standards are indicated to the left of panel A. A 19-kDa protein is the digested product which was used to determine the amino acid sequence. Lanes 1, crude proteins (1.0 mg) from culture supernatant; 2, purified chitosanase (30 ng); 3, immunoblot of the purified chitosanase (50 ng) from M. chitosanotabidus 3001; 4, immunoblot of the digested products of the purified chitosanase.
To determine the pH dependence of chitosanase activity, McIlvaine buffer (pH 2.0 to 8.0), Tris-HCl buffer (pH 9.0 to 10.0), and 0.05 M Na2HPO4 plus 0.1 M NaOH buffer (pH 10.0 to 12.0) were used to create a range of pH values in the reaction mixtures. The purified chitosanase showed optimal activity at pH 4.0. The stability of the chitosanase was determined in the same buffers used in the experiments described above. After 30 min of incubation at 37°C, the residual chitosanase activity was determined after readjustment of the pH to 4.0 with 0.5 M sodium acetate buffer. Chitosanase activity was found to be stable over a wide range of pH values, ranging from 4.0 to 9.0 (data not shown).
The optimum temperature of the purified enzyme under standard assay conditions was 30 to 40°C in the presence of chitosan. To determine the temperature stability of the purified enzyme, the residual activity was measured after incubation of the enzyme at various temperatures for 1 h at pH 4.0 in the absence of substrate. Ca. 92, 95, and 95% of the initial chitosanase activity remained after incubation at 20, 30, and 40°C for 1 h, respectively. At least 35% of the initial chitosanase activity remained when reaction mixtures were kept at under 50°C (data not shown).
Effects of metal ions on chitosanase activity.
The effect of several metal ions on the activity of purified chitosanase was tested. Chitosanolytic activity was assayed after metal ions, including Ag+, Ba2+, Co2+, Hg2+, Ca2+, Cu2+, Mg2+, Fe2+, Mn2+, and Zn2+, were each added to the reaction mixtures at final concentrations of 1 mM. The enzymatic activity was assayed after preincubation at 30°C for 30 min. Chitosanolytic activity was not affected by Ba2+, Co2+, Hg2+, Ca2+, Cu2+, Mg2+, Fe2+, Mn2+, or Zn2+, but it was almost completely inhibited by Ag+ (data not shown).
Specific activities of the purified enzyme with different substrates.
Various derivatives of chitin and chitosan were used to determine the specificity of chitosanolytic activity (Table 2). Glycol chitosan and carboxymethylchitosan showed 113 and 6% susceptibilities to the enzyme, respectively. Chitin derivatives and cellulose were not degraded by the purified enzyme. The chitosanolytic enzyme, therefore, showed high specificity for chitosan and its derivatives. We also examined the ability of the enzyme to degrade chitosan that had been subjected to various degrees of deacetylation. Compared to the 90% deacetylated chitosan, 100% deacetylated preparations were poor substrates, as were preparations containing less than 70% deacetylation (Table 2).
TABLE 2.
Effect of substrate specificity on the chitosanase activitya
| Substrate | Relative activity (%) |
|---|---|
| Colloidal chitosanb | 100 |
| Glycol chitosanc | 113 |
| Flaked chitosand | 35 |
| Carboxymethylchitosan | 6 |
| Colloidal chitin | 0 |
| Ethyleneglycol chitin | 0 |
| Carboxymethyl chitin | 0 |
| Cellulose | 0 |
Relative activity determined with colloidal chitosan is indicated as the standard. Chitosan and chitin-related compounds were used as the substrates of purified enzyme.
80% deacetylated chitosan was used.
90% deacetylated chitosan was used.
100% deacetylated chitosan was used.
Reaction pattern of the chitosanase with chitosan or its oligosaccharides.
The products of hydrolysis of chitosan and GlcN oligosaccharides by the purified enzyme were analyzed by thin-layer chromatography. Table 3 shows the products of hydrolysis by chitosanase. When 80% deacetylated chitosan was employed, the amounts of tetramer, pentamer, and hexamer of glucosamine increased during the first 24 h of incubation, but their concentrations decreased after 30 h in parallel with the increase in the concentration of the dimer and trimer. The dimer and trimer of glucosamine were the final products of chitosan degradation. The monomer was never detected with the purified enzyme. The purified chitosanase could not hydrolyze the dimer and the trimer of glucosamine, but the enzyme could cleave the tetramer, pentamer, and hexamer into a dimer and a trimer. These results show that the tetramer is the minimum molecular size necessary for digestion by chitosanase. These results also suggest that chitosanase from M. chitosanotabidus 3001 is an endosplitting-type chitosanase.
TABLE 3.
Production of chitosan oligomers in the degradation of chitosan and glucosamine oligomers with chitosanase
| Substrate | Product(s)a |
|---|---|
| Chitosanb | (GlcN)∼(GlcN)6 |
| Chitosanc | (GlcN)2∼(GlcN)6 |
| (GlcN)2 | (GlcN)2 |
| (GlcN)3 | (GlcN)3 |
| (GlcN)4 | (GlcN)2 |
| (GlcN)5 | (GlcN)2, (GlcN)3 |
| (GlcN)6 | (GlcN)2, (GlcN)3 |
GlcN indicates residues of glucosamine, and the numbers outside the parentheses indicate the degree of polymerization.
Crude protein from M. chitosanotabidus 3001 was used.
Purified chitosanase from M. chitosanotabidus 3001 was used.
Immunoblotting and determination of amino acid sequence.
We next determined the internal amino acid sequence of the purified enzyme. To determine the internal amino acid sequence, purified chitosanase was digested with the appropriate concentration of trypsin and then separated by SDS–12.5% PAGE. Purified enzyme and partially degraded chitosanase were blotted onto PVDF membrane and then detected by Western blotting by using an antibody raised against chitosanase (Fig. 2B). The amino acid sequence of the amino terminus of a 19-kDa peptide (Fig. 2B, lane 4), which was obtained after treatment with trypsin, was determined to be SDKNKRAALTKIXGALQSAFDTQQDKY.
Cloning of the chitosanase gene from M. chitosanotabidus 3001.
Based on the 27-amino-acid sequence described above, a mixture of two oligonucleotide primers, 5′-TCCGACAAGAAC(T)AAG(A)CG(T)CGCG-3′ (S1) and 5′-GTACTT(C)GTCCTGCTG(T)CGTGT-3′ (S2), was prepared, and a 80-bp DNA fragment was obtained by PCR amplification and then cloned into the vector pBluescript II SK(−). The sequence of this DNA fragment matched the amino-terminal amino acid sequence of the 19-kDa peptide. Then, the cassette-cassette PCR reaction (see Materials and Methods) was done by using M. chitosanotabidus 3001 genomic DNA to extend the region of the cloned fragment. Two oligonucleotide primers, S2 and R1 (5′-AGATCTTGGTCAGCGCCG-3′), were used to amplify DNA fragments corresponding to the coding region of the choA gene sequence. The cassette-cassette primers (C1 and C2) and the S2 and R1 primers were used to amplify the flanking fragment. A single major band of 300-bp was amplified by PCR by using the two primers, C2 and R1. Then, this 300-bp was used as a hybridization probe for genomic library screening.
Southern hybridization was performed with total M. chitosanotabidus 3001 DNA digested with several restriction enzymes by using the PCR products as probes. This resulted in the appearance of 1.6-, 2.5-, 4.0-, and 10-kb bands after SalI, SacI, PstI, and EcoRI digestion, respectively (data not shown). We chose the SacI enzyme to construct the genomic library. The ∼2.5-kbp SacI DNA fragment was isolated from an agarose gel by electroelution and cloned into λZapII.
The library was screened by plaque hybridization with the 300-bp PCR-amplified fragment as a probe. A few positive plaques carried the 2.5-kb fragment in alternative orientations. The DNA in a positive plaque of λZapII was changed into a plasmid by the in vivo excision method, and the plasmid obtained was designated pChoU1. (For a restriction map of the cloned fragment in pChoU1, see Fig. 3). When E. coli cells harboring pChoU1 were plated on a medium containing colloidal chitosan as a sole carbon source for chitosanase production, a clear degradation zone was observed, confirming that pChoU1 carries the chitosanase gene.
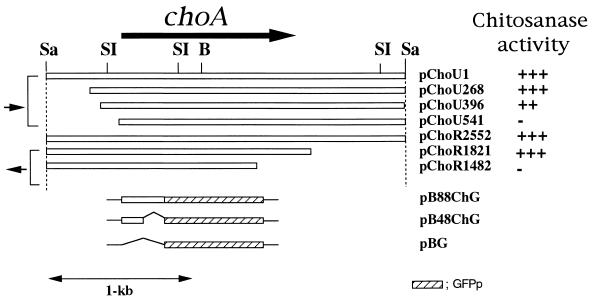
FIG. 3.
Various plasmid constructions derived from the choA gene. pChoU1 is the chitosanase-positive clone that contained a 2.5-kbp SacI fragment. The names of the clones carrying the various deletion derivatives from pChoU1 are shown on the right. The direction and coding regions of the choA gene are indicated by the large arrow. Small arrows indicate the directions of the lacZ promoter. To determine the chitosanase activity, E. coli cells carrying pChoU1 or its deletion derivatives were transferred to the colloidal chitosan plates containing 0.4% colloidal chitosan and 50 mg of ampicillin per ml. Chitosanase activities were detected by the ability to form clear halos around the colonies on colloidal chitosan plates: +, visible halo; −, no halo. pB88ChG and pB48ChG carry the fusion genes that encode the N-terminal 88-amino-acid region and the 48-amino-acid region of ChoA fused to the GFP protein, respectively. pBG carries only the GFP gene on the pBluescript SK(+). Restriction sites are designated as follows: Sa, SacI; SI, SalI; B, BamHI.
Deletion analysis and restriction mapping.
To determine the coding region of the chitosanase gene, we performed deletion analysis. Clones containing various deletion derivatives from both directions were obtained (Fig. 3). The number associated with the name of the plasmid indicates the deletion site on the nucleotide sequence shown in Fig. 4. These deletion derivatives were transformed into E. coli DH10B and were tested for their ability to form clear zones on the colloidal chitosan plate (Fig. 5).
FIG. 4.
Nucleotide sequence and deduced amino acid sequence of the choA gene from M. chitosanotabidus 3001. The putative Shine-Dalgarno sequence is indicated by a double underline. The one-letter amino acid code is aligned below the nucleotide sequence. The signal peptide sequence is underlined with a dashed line, and a signal sequence cleavage site is represented by a vertical arrow between the alanine residues at positions 80 and 81. Boxed amino acids were determined by N-terminal amino acid sequencing of the purified chitosanase from E. coli expressing the choA gene. A wavy line indicates the amino acid sequence obtained from the purified chitosanase. Two arrows indicate the nucleotide sequence of the primers that were used for PCR. The single bold arrow indicates the primer used to amplify the PCR fragment by using the cassette-cassette primer PCR amplification method.
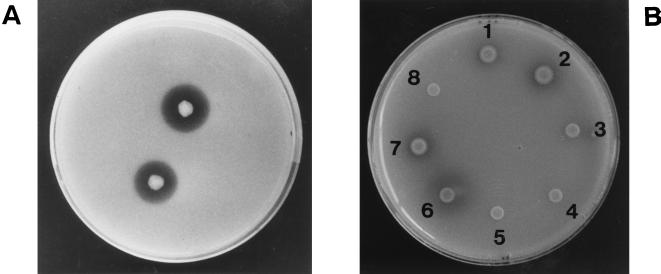
FIG. 5.
Detection of chitosanase activity on the chitosanase medium plate. M. chitosanotabidus 3001 (A) and E. coli carrying the plasmid pChoU1 and its deletion derivatives (B) were spotted onto the colloidal chitosan plate to assay for chitosanase expression. The plates were incubated at 30 and 37°C for 3 days, respectively. Chitosanase activity was identified as a clear zone around the colony on the colloidal chitosan plate. E. coli carrying the plasmid pChoU1 and its deletion derivatives are indicated as follows: 1, pChoU1; 2, pChoU268; 3, pChou396; 4, pChoU541; 5, pBluescript II SK(−); 6, pChoR2552; 7, pChoR1821; and 8, pChoR1482.
Deletion derivatives containing a 396-bp deletion from upstream to downstream (pChoU396) and a 733-bp deletion from downstream to upstream (pChoR1821) were not defective in their ability to form clear zones. However, deletion derivatives containing a 541-bp deletion (pChoU541) from upstream to downstream and a 1,072-bp deletion from downstream to upstream (pChoR1482) did not show any chitosanase activity on the colloidal chitosan plate (Fig. 5). Therefore, these results indicate that the chitosanase gene is located in the 1,425-bp region between positions 396 and 1,821.
Sequence analysis of the chitosanase gene (choA).
The nucleotide sequence of the cloned SacI fragment (2,557 bp) was determined for both strands (Fig. 4). The presumed coding sequence started with initiation codon ATG at position 529, which was preceded by a putative ribosome-binding site, GGAGAGA, and extended to the stop codon, TGA, located at position 1,702. The open reading frame of 1,173 bp would code for a 391-amino-acid polypeptide. The deduced N-terminal 80 amino acids showed the typical features of signal peptides, such as the presence of hydrophobic amino acids. A comparison of the signal sequence region of three chitosanases from M. chitosanotabidus, Nocardioides sp. strain N106, and Streptomyces sp. strain N174 is shown in Fig. 6. Although the signal sequence regions are similar among these chitosanases, other regions did not show any significant homologies.
FIG. 6.
Comparison of the N-terminal amino acid sequences of three chitosanases. N-terminal amino acid sequences of the chitosanases from M. chitosanotabidus 3001 (ChitoM), Nocardioides sp. strain N106 (12) (ChitoN), and Streptomyces sp. strain N174 (13) (ChitoS) are shown. Identical amino acids are boxed.
N-terminal cleavage site of ChoA.
We found that the signal peptide was cleaved between alanine residues 80 and 81 by determining the amino-terminal sequence of purified mature chitosanase from M. chitosanotabidus. Cleavage at amino acid position 80 would result in a protein of 311 amino acids with a calculated molecular weight of 34,000. This is consistent with the size of the 34-kDa chitosanase estimated by SDS-PAGE. The 15 N-terminal amino acids of the 34-kDa protein produced by E. coli carrying the cloned choA gene was determined to be AAAAGVIPVGDSRVY. This sequence is identical to the one predicted from the choA DNA sequence and lies between Ala-81 and Tyr-95. The 80-amino-acid sequence is considered to be the signal sequence responsible for the export of ChoA into the medium both in M. chitosanotabidus and E. coli. We determined the enzymatic activity in a culture of E. coli expressing choA. Whereas the extracellular chitosanase activity was 3.4 U/mg, the cell-associated chitosanase activity was only 0.13 U/mg, indicating that most of the enzyme had been released.
To verify that the N-terminal signal sequence of ChoA is functional in E. coli, the N-terminal sequence of ChoA was fused to GFP. GFP fusions bearing the 48-amino-acid (pB48ChG) or the 88-amino-acid (pB88ChG) region of ChoA were constructed and inserted into the expression plasmid (Fig. 3). E. coli cells harboring these plasmids were grown, and extracellular and intracellular proteins were detected by Western blot analysis with the GFP antibody. While both of these proteins were detected intracellularly (Fig. 7, lanes 2, 4, and 6), the extracellular protein contained the GFP fusion protein with the 88-amino-acid region (lane 5) but not the GFP fusion protein with the 44-amino-acid region or GFP alone (lanes 1 and 3). The cleaved ChoA protein was only seen when the 88-amino-acid–GFP fusion was employed, and a fraction of the extracellular proteins still possessed the signal sequence (lane 5). These results suggest that the N-terminal signal sequence was functional in E. coli and that this region needs to be longer than 48 amino acids to be functional.
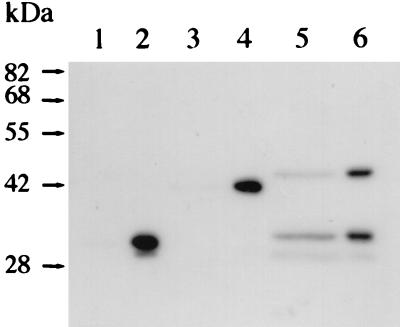
FIG. 7.
Western blot analysis of the chitosanase-GFP fusion proteins. Extracellular proteins (lanes 1, 3, and 5) and intracellular proteins (lanes 2, 4, and 6) from E. coli JM109 harboring pBG (lanes 1 and 2), pB48ChG (lanes 3 and 4), or pB88ChG (lanes 5 and 6) were loaded onto the gel. Western blot analysis was done by using an anti-GFP antibody. The extracellular ChoA-GFP protein was only seen in lane 5.
DISCUSSION
Table 4 summarizes all of the microbial chitosanases that have been identified to date. The data are still relatively limited compared to similar data for the many chitinase genes that have been analyzed thus far (19). The molecular weights of chitosanase are relatively low (26,000 to 50,000) compared to those of chitinases (31,000 to 115,000) (19). In the present study the 34-kDa chitosanase produced by M. chitosanotabidus 3001 was purified and characterized, and its corresponding gene was cloned. Although the enzymatic properties of M. chitosanotabidus 3001 chitosanase are similar to those of other chitosanases, the optimal temperature and pH of our chitosanase were the lowest among known chitosanases (Table 4). The chitosanase from M. chitosanotabidus 3001 is highly active on glycol chitosan, but it does not hydrolyze carboxymethylchitosan, colloidal chitin, or carboxymethylchitin. Most known chitosanases do not hydrolyze 100% chitin or cellulose. However, a chitosanase from Myxobacter AL-1 (7, 21) has the unique feature of being able to hydrolyze carboxymethyl cellulose, and a chitosanase from Acinetobacter sp. strain CHB101 hydrolyzes glycol chitin (28). Recently, a part of the chitosanase sequence from Myxobacter AL-1 was reported to have structural similarity to an endoglucanase from Bacillus subtilis (21). Even though there are widespread similarities in the activities of chitosanases, there seem to be many different structural types of chitosanases in nature.
TABLE 4.
Comparison of properties of purified chitosanases produced by microorganisms
| Microorganism | Mol wta | No. of amino acidsb | Optimum activity
|
||||
|---|---|---|---|---|---|---|---|
| pH | Temp | Inhibitor(s) | Substrate specificity (% deacetylation) | Source or reference | |||
| Matsuebacter chitosanotabidus 3001 | 34,000 | 311 | 4.0 | 30–40 | Ag+ | Chitosan (90), glycol chitosan | This study |
| Myxobacter AL-1 | 31,000 | 5.0, 6.8 | 70 | Hg2+, Ag+, Cd2+, Zn2+ | CMCf, chitosan | 7, 21 | |
| Acinetobacter sp. strain CHB101 | 28 | ||||||
| Chitosanase I | 37,000 | 5–9 | 65 | NDg | Chitosan (70–90) | ||
| Chitosanase II | 30,000 | 5–9 | 50 | ND | Glycol chitosan | ||
| Streptomyces sp. N174 | 29,500 | 238 | 5.5 | 65 | ND | Chitosan (40–99) | 2, 13 |
| Fusarium solani f. sp. phaseoli | 36,000 | 285 | 5.6 | 40 | No inhibitorc | Chitosan (70–100), glycol chitosan, CMC | 26 |
| Bacillus megaterium P1 chitosanase A | 43,000 | 4.5–6.5 | 50 | ND | Chitosan (81), CMC | 22 | |
| Amycolatopsis sp. strain CsO-2 | 27,000 | 5.3 | 55 | Hg2+, pCMBd, NBSe | Chitosan (100) | 16 | |
| Penicillium islandicum | 30,000 | 4.5–6.0 | 45 | Hg2+, Cu2+ | Chitosan (70) | 4 | |
| Enterobacter sp. strain G-1 | 50,000 | 7.0 | 50 | ND | Chitosan (80) | 30 | |
| Bacillus circulans MH-K1 | 30,000 | 259 | 6.5 | 50 | Cu2+, Hg2+, Ni2+, Zn2+, pCMB | Chitosan (80) | 29 |
| Nocardioides sp. | 29,500 | 237 | ND | Chitosan (100) | 12 | ||
Determined by SDS-PAGE analysis with purified enzyme.
The processed amino acid number as deduced from the gene sequence.
Tested chemicals were Ca2+, Mg2+, Co2+, Ni2+, Fe2+, Hg2+, and Cu2+.
pCMB, para-chloromercuribenzoate.
NBS, N-bromosuccinimide.
Carboxymethyl cellulose.
ND, not determined.
Purified chitosanase from M. chitosanotabidus 3001 had highest activities with 90% deacetylated chitosan and efficiently hydrolyzed 70 to 100% deacetylated chitosan. The fact that 90% deacetylated chitosan is more susceptible to hydrolysis than 100% deacetylated chitosan may suggest that N-acetylglucosamine residues in the chitosan are important in the recognition mechanism of the substrate by the enzyme. The ability to degrade 100% deacetylated chitosan is an important characteristic used to categorize enzymes as chitosanases. However, enzymes in nature have different specificities depending on the different degrees of deacetylation of chitosan, as indicated in Table 4.
The chitosanase activity of M. chitosanotabidus 3001 was not inhibited by Ba2+, Co2+, Hg2+, Ca2+, Cu2+, Mg2+, Fe2+, Mn2+, and Zn2+, but it was completely inhibited by 1 mM Ag+. Chitosanase from Myxobacter AL-1 is an example of another enzyme inhibited by Ag+ (7, 21). However, this chitosanase differs from that of M. chitosanotabidus 3001 in several ways (Table 4).
The choA gene encoding a 34-kDa chitosanase was cloned into E. coli, and the complete nucleotide sequence of the gene was determined. This is the fifth chitosanase whose primary structure has been determined (Table 4). The deduced amino acid sequence of the choA gene is identical to the amino acid sequence of the purified chitosanase (Fig. 4). The choA gene encodes a polypeptide of 391 amino acid residues which includes an 80-residue signal sequence. This 80-residue signal sequence is functional in E. coli (Fig. 7). Fusion of the signal sequence of ChoA with GFP suggested that the signal for excretion required more than 48 amino acids but fewer than 88 amino acids (Fig. 7). Surprisingly, however, the GFP fusion protein with the 88-amino-acid region, as well as the cleaved protein, was found in both the intracellular and extracellular fractions. We speculate that the signal peptidase of E. coli recognizes the cleavage site of the fusion protein but that the fusion protein may not necessarily have to be cleaved for excretion.
Computer analyses of the deduced amino acid sequence did not reveal a strong identity with any other protein sequences of glycolytic enzymes, including the chitosanases, chitinases, and N-acetylglucosaminidases included in protein databases. Three bacterial chitosanases from B. circulans MH-K1, Streptomyces sp. strain N174, and Nocardioides sp. have a common primary structure (3). However, ChoA from M. chitosanotabidus, as well as a fungal chitosanase, is different. Thus far, at least three different types of chitosanases have been identified on the basis of their primary structure.
There is an 80-amino-acid signal polypeptide at the N terminus of ChoA of M. chitosanotabidus 3001 (Fig. 6). The N-terminal 15-amino-acid sequence determined from the 34-kDa mature chitosanase produced in E. coli was identical to the N-terminal amino acid sequence of chitosanase from M. chitosanotabidus 3001, indicating that the mature chitosanase is cleaved at the same site in E. coli. This property will be useful for the mass production of chitosanase in E. coli.
Boucher et al. (3) suggested that two invariant carboxylic amino acid residues of Glu22 and Asp40 conserved in N-terminal segments of chitosanase from Streptomyces sp. strain N174 are essential for its catalytic activity. We have looked for the corresponding amino acid combination in ChoA of M. chitosanotabidus 3001. There are two candidate residues: residues Glu121 and Asp139 or residues Glu137 and Asp152. However, since the regions surrounding these amino acids are not conserved in other chitosanases, it is difficult to predict whether they may constitute the catalytic center or not. Further analysis will be required to define the active region of M. chitosanotabidus 3001 chitosanase.
ACKNOWLEDGMENTS
This work was supported by a grant-in-aid from the Ministry of Education, Science, and Culture of Japan and by San-in Kensetsu Kogyo Co., Ltd. J. K. Park was supported by the commemorative foundation of Yoneyama International Rotary and Iwatanai Naozi.
S. Imamura and F. Ishii participated in some of this work.
REFERENCES
- 1.Ando A, Noguchi K, Yanagi M, Shinoyama H, Kagawa Y, Hirata H, Yabuki M, Fujii T. Primary structure of chitosanase produced by Bacillus circulans MH-K1. J Gen Appl Microbiol. 1992;38:135–144. [Google Scholar]
- 2.Boucher I, Dupuy A, Vidal P, Neugebauer W A, Brzezinski R. Purification and characterization of a chitosanase from Streptomyces N174. Appl Microbiol Biotechnol. 1992;38:188–193. [Google Scholar]
- 3.Boucher I, Fukamizo T, Honda Y, Willick G E, Neugebauer W A, Brzezinski R. Site-direct mutagenesis of evolutionary conserved carboxylic amino acids in the chitosanase from Streptomyces sp. N174 reveals two residues essential for catalysis. J Biol Chem. 1995;270:31077–31082. doi: 10.1074/jbc.270.52.31077. [DOI] [PubMed] [Google Scholar]
- 4.Fenton D M, Eveleigh D E. Purification and mode of action of a chitosanase from Penicillium islandicum. J Gene Microbiol. 1981;126:151–165. [Google Scholar]
- 5.Fukamizo T, Ohkawa T, Ikeda Y, Goto S. Specificity of chitosanase from Bacillus pumilus. Biochim Biophys Acta. 1994;1205:183–188. doi: 10.1016/0167-4838(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 6.Grenier J, Benhamou N, Asselin A. Colloidal gold-complexed chitosanase: a new probe for ultrastructural localization of chitosan in fungi. J Gen Microbiol. 1991;137:2007–2015. [Google Scholar]
- 7.Hedges A, Wolfe R S. Extracellular enzyme from Myxobacter AL-1 that exhibits both β-1,4-glucanase and chitosanase activities. J Bacteriol. 1974;120:844–853. doi: 10.1128/jb.120.2.844-853.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imoto T, Yagishita K. A simple activity measurement of lysozyme. Agric Biol Chem. 1971;35:1154–1156. [Google Scholar]
- 9.Isegawa Y, Sheng J, Sokawa Y, Yamanishi K, Nakagomi O, Ueda S. Selective amplification of cDNA sequence from total RNA by cassette-ligation mediated polymerase chain reaction (PCR): application to sequencing 6.5-kb genome segment of hantavirus strain B-1. Mol Cell Probes. 1992;6:467–475. doi: 10.1016/0890-8508(92)90043-w. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Marcotte E M, Monzingo A F, Ernst S R, Brzezinski R, Robertus J D. X-ray structure of an anti-fungal chitosanase from Streptomyces N174. Nat Struct Biol. 1996;3:155–162. doi: 10.1038/nsb0296-155. [DOI] [PubMed] [Google Scholar]
- 12.Masson J Y, Boucher I, Neugebauer W A, Ramotar D, Brzezinski R. A new chitosanase gene from a Nocardioides sp. is a third member of glycosyl hydrolase family 46. Microbiol. 1995;141:2629–2635. doi: 10.1099/13500872-141-10-2629. [DOI] [PubMed] [Google Scholar]
- 13.Masson J Y, Denis F, Brzezinski R. Primary sequence of the chitosanase from Streptomyces sp. strain N174 and comparison with other endoglycosidases. Gene. 1994;140:103–107. doi: 10.1016/0378-1119(94)90738-2. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo Y, Kurita M, Park J K, Tanaka K, Nakagawa T, Kawamukai M, Matsuda H. Purification, characterization and gene analysis of N-acetylglucosaminidase from Enterobacter sp. G-1. Biosci Biotechnol Biochem. 1999;63:1261–1268. doi: 10.1271/bbb.63.1261. [DOI] [PubMed] [Google Scholar]
- 15.Nabeshima K, Saitoh S, Yanagida M. Use of green fluorescent protein for intracellular protein localization in living fission yeast cells. Methods Enzymol. 1997;283:459–471. doi: 10.1016/s0076-6879(97)83037-6. [DOI] [PubMed] [Google Scholar]
- 16.Okajima S, Ando A, Shinoyama H, Fujii T. Purification and characterization of an extracellular chitosanase produced by Amycolatopsis sp. CsO-2. J Ferment Bioeng. 1994;77:617–620. [Google Scholar]
- 17.Okajima S, Kinouchi T, Mikami Y, Ando A. Purification and some properties of a chitosanase of Nocardioides sp. J Gen Appl Microbiol. 1995;41:351–357. [Google Scholar]
- 18.Osswald W F, Shapiro J P, Doostdar H, McDonald R E, Niedz R P, Narin C J, Hearn C J, Mayer R T. Identification and characterization of acidic hydrolases with chitinase and chitosanase activities from sweet orange callus tissue. Plant Cell Physiol. 1994;35:811–820. doi: 10.1093/oxfordjournals.pcp.a078662. [DOI] [PubMed] [Google Scholar]
- 19.Park J K, Morita K, Fukumoto I, Yamasaki Y, Nakagawa T, Kawamukai M, Matsuda H. Purification and characterization of the chitinase (ChiA) from Enterobacter sp. G-1. Biosci Biotechnol Biochem. 1997;61:684–689. [Google Scholar]
- 20.Park, J. K., Y. Matsuo, K. Shimono, K. Tanaka, T. Nakagawa, A. Yokota, H. Matsuda, and M. Kawamukai. Matsuebacter chitosanotabidus gen. nov., sp. nov., an aerobic chitosanase producing gram-negative bacterium belonging to the beta subclass of Proteobacteria. Submitted for publication.
- 21.Pedraza-Reyes M, Gutierrez-Corona F. The bifunctional enzyme chitosanase-cellulase produced by the gram-negative microorganism Myxobacter sp. AL-1 is highly similar to Bacillus subtilis endoglucanases. Arch Microbiol. 1997;168:321–327. doi: 10.1007/s002030050505. [DOI] [PubMed] [Google Scholar]
- 22.Pelletier A, Sygusch J. Purification and characterization of three chitosanase activities from Bacillus megaterium P1. Appl Environ Microbiol. 1990;56:844–848. doi: 10.1128/aem.56.4.844-848.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertus J D, Monzingo A F, Marcotte E M, Hart P J. Structural analysis shows five glycohydrolase families diverged from a common ancestor. J Exp Zool. 1998;282:127–132. [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimosaka M, Kumehara M, Zhang X Y, Nogawa M, Okazaki M. Cloning and characterization of a chitosanase gene from the plant pathogenic fungus Fusarium solani. J Ferment Bioeng. 1996;82:426–431. [Google Scholar]
- 27.Shimosaka M, Nogawa M, Ohno Y, Okazaki M. Chitosanase from the plant pathogenic fungus, Fusarium solani f. sp. phaseoli: purification and some properties. Biosci Biotechnol Biochem. 1993;57:231–235. doi: 10.1271/bbb.57.231. [DOI] [PubMed] [Google Scholar]
- 28.Shimosaka M, Nogawa M, Wang X Y, Kumehara M, Okazaki M. Production of two chitosanases from a chitosan-assimilating bacterium, Acinetobacter sp. strain CHB101. Appl Environ Microbiol. 1995;61:438–442. doi: 10.1128/aem.61.2.438-442.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yabuki M, Uchiyama A, Suzuki K, Ando A, Fujii T. Purification and properties of chitosanase from Bacillus circulans MH-K1. J Gen Appl Microbiol. 1988;34:255–270. [Google Scholar]
- 30.Yamasaki Y, Hayashi I, Ohta Y, Nakagawa T, Kawamukai M, Matsuda H. Purification and mode of action of chitosanolytic enzyme from Enterobacter sp. G-1. Biosci Biotechnol Biochem. 1993;57:444–449. doi: 10.1271/bbb.56.1546. [DOI] [PubMed] [Google Scholar]