Abstract
Despite being the most frequently altered oncogenic protein in solid tumours, KRAS has historically been considered ‘undruggable’ owing to a lack of pharmacologically targetable pockets within the mutant isoforms. However, improvements in drug design have culminated in the development of inhibitors that are selective for mutant KRAS in its active or inactive state. Some of these inhibitors have proven efficacy in patients with KRASG12C-mutant cancers and have become practice changing. The excitement associated with these advances has been tempered by drug resistance, which limits the depth and/or duration of responses to these agents. Improvements in our understanding of RAS signalling in cancer cells and in the tumour microenvironment suggest the potential for several novel combination therapies, which are now being explored in clinical trials. Herein, we provide an overview of the RAS pathway and review the development and current status of therapeutic strategies for targeting oncogenic RAS, as well as their potential to improve outcomes in patients with RAS-mutant malignancies. We then discuss challenges presented by resistance mechanisms and strategies by which they could potentially be overcome.
Subject terms: Targeted therapies, Cancer genomics
The RAS oncogenes are among the most common drivers of tumour development and progression but have historically been considered undruggable. The development of direct KRAS inhibitors has changed this paradigm, although currently clinical use of these novel therapeutics is limited to a select subset of patients, and intrinsic or acquired resistance presents an inevitable challenge to cure. Herein, the authors provide an overview of the RAS pathway in cancer and review the ongoing efforts to develop effective therapeutic strategies for RAS-mutant cancers. They also discuss the current understanding of mechanisms of resistance to direct KRAS inhibitors and strategies by which they might be overcome.
Key points
Owing to intrinsic and extrinsic factors, KRAS and other RAS isoforms have until recently been impervious to targeting with small-molecule inhibitors.
Inhibitors of the KRASG12C variant constitute a potential breakthrough in the treatment of many cancer types, particularly non-small-cell lung cancer, for which such an agent has been approved by the FDA.
Several forms of resistance to KRAS inhibitors have been defined, including primary, adaptive and acquired resistance; these resistance mechanisms are being targeted in studies that combine KRAS inhibitors with inhibitors of horizontal or vertical signalling pathways.
Mutant KRAS has important effects on the tumour microenvironment, including the immunological milieu; these effects must be considered to fully understand resistance to KRAS inhibitors and when designing novel treatment strategies.
Introduction
RAS signalling has an essential role in driving normal physiological cellular proliferation, and dysregulation of this signalling pathway commonly occurs during tumorigenesis1,2. Indeed, alterations in components of this pathway, particularly those in the RAS proteins themselves, have far-reaching consequences in many cancers3,4. Thus, over the past few decades, substantial efforts have been made to drug RAS proteins, the central mediators of this pathway5. In 2021, the decades of research finally achieved some clinical success, with the approval of the KRASG12C inhibitor sotorasib for a specific subset of patients with non-small-cell lung cancer (NSCLC). In this Review, we provide an overview of the RAS pathway in cancer, focusing on the role of mutant KRAS. We discuss why mutant KRAS was, until recently, unamenable to pharmacological inhibition, and detail current strategies for therapeutic targeting of KRAS. We then consider mechanisms of resistance to KRAS inhibition and ways in which they might be overcome. Finally, we discuss the future of targeted therapy for RAS-mutant cancers, with a particular focus on combinations that could move beyond transient responses towards cure.
The RAS pathway
The RAS–RAF–MEK–ERK mitogen-activated kinase signalling (MAPK) pathway is activated by most, if not all, growth factor, cytokine and immunological receptors, as well as by many integrins and chemokine receptors2,5. The canonical pathway module features a receptor tyrosine kinase (RTK) coupled to the RAS–RAF–MEK–ERK cascade. The RAS family GTPases, KRAS, NRAS and HRAS, cycle between GTP-loaded ‘on’ and GDP-loaded ‘off’ states4,6, involving the action of RAS-guanine nucleotide exchange factors (RAS-GEFs) and RAS-GTPase-activating proteins (RAS-GAPs), respectively. During this activation–inactivation process, the conformations of two regions in the RAS protein, termed the ‘switch 1’ and ‘switch 2’ domains, are altered7. As detailed further below, these structural changes, particularly in switch 2, proved crucial for the eventual development of RAS inhibitors.
RAS-GEFs can promote the release of either GDP or GTP from RAS proteins in vitro; however, because the cellular GTP:GDP ratio is ~10 (refs.8,9), they catalyse GDP-to-GTP exchange in cells. SOS1 and SOS2 (SOS1/2), the major RAS-GEFs activated by RTKs and cytokine receptors, bind via C-terminal proline-rich motifs to the SRC homology 3 (SH3) domains of the adapter protein GRB2 (ref.10). GRB2 can simultaneously bind via its SH2 domain to phosphorylated tyrosine motifs (pY-X-N-X) in various other proteins, including RTKs, scaffolding adapters (such as SHC, GAB, FRS and IRS proteins) and/or the SOS1-activating non-receptor protein tyrosine phosphatase SHP2, to precisely coordinate the RAS-GEF activity of SOS1/2 and thus RAS activation (Fig. 1). Subsequently, GTP-loaded RAS can interact with RAS-binding domains (RBDs) in downstream effector proteins, including the RAF family kinases ARAF, BRAF and RAF1 (also known as CRAF). Thus, active RAS recruits RAF proteins to the cell membrane, where they interact with membrane lipids, dimerize and are activated4. The active RAF kinases phosphorylate and activate MEK1 and/or MEK2, which in turn phosphorylate and activate ERK1 and/or ERK2 (Fig. 1). ERK1/2 can then phosphorylate multiple cytosolic and nuclear proteins, including other kinases (such as RSK, MSK and MNK), transcription factors and cytoskeletal proteins2. Other important RAS effectors include the catalytic p110 subunit of phosphatidylinositol 3-kinase (PI3K), Ral guanine nucleotide dissociation stimulator (RAL-GDS) and the Rho guanine nucleotide exchange factor TIAM1 (refs.4,6). The RAS effectors mediate diverse effects on cellular phenotype, for example, activating the cell-cycle machinery in dividing cells or driving the differentiation of various other cell types.
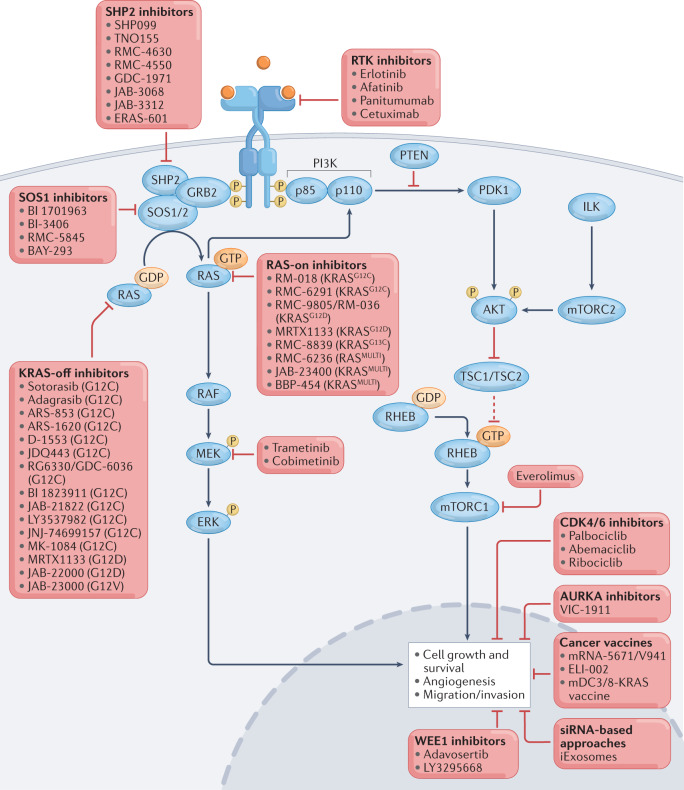
Fig. 1. The RAS signalling pathway and therapeutic approaches to target this pathway in cancer.
Numerous direct inhibitors have been developed to target mutant RAS proteins, either in their inactive, GDP-bound state (‘KRAS-off inhibitors’) or in their active, GTP-bound state (‘RAS-on inhibitors’). Many of these inhibitors are being evaluated in clinical trials. The RAS signalling pathway has many upstream and downstream mediators, which are attractive targets for combination therapies with RAS inhibitors to improve antitumour responses and to mitigate intrinsic and acquired resistance; agents that have been combined with direct KRAS inhibitors in preclinical or clinical studies are listed. Therapeutic cancer vaccines against mutant RAS epitopes and small interfering RNA (siRNA)-based approaches that target oncogenic RAS isoforms are also under ongoing development. ILK, integrin-linked kinase; mTORC2, mTOR complex 2; PI3K, phosphatidylinositol 3-kinase; RTK, receptor tyrosine kinase.
Wild-type RAS isoforms have intrinsic, albeit weak, hydrolytic (GTPase) activity and can, therefore, self-inactivate; however, RAS-GAPs stimulate this activity dramatically11–14. Indeed, the GAPs increase RAS GTPase activity fivefold relative to the intrinsic rate12. Hence, within cells, normal hydrolysis of RAS-bound GTP depends on GAPs such as neurofibromin (NF1) or Ras GTPase-activating protein 1 (RASA1; also known as p120RASGAP), which contain SH2/SH3 domains or other binding motifs and are, therefore, also recruited to activated RTKs and scaffolding adapters as a crucial feedback mechanism to switch off RAS signalling.
As alluded to previously, abnormalities in components of the RAS pathway, including activating RAS mutations, are common across a diverse range of malignancies (Fig. 2). In cells with such alterations, aberrant RAS signalling ultimately leads to activation of several key transcription factors, which in turn drive several hallmarks of cancer, including increased cell-cycle entry, metabolic reprogramming, cell growth and survival, and angiogenesis1,3,4.
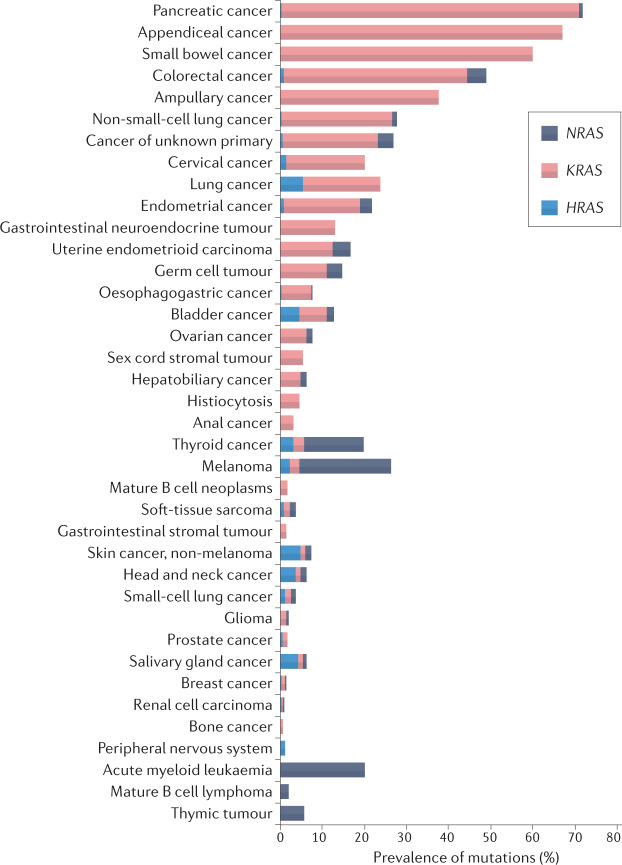
Fig. 2. The prevalence of KRAS, NRAS and HRAS mutations across cancer types.
Mutations in the RAS genes are common in gastrointestinal and lung cancers, with KRAS mutations comprising most of these mutations, but also occur more rarely in various other cancer types. The data shown in the graph are from the cBioportal TCGA and MSK-IMPACT cohorts (available via the cBioPortal for Cancer Genomics)17,18.
RAS mutations in cancer
Mutations that affect RAS–RAF–MEK–ERK pathway components, including various RTKs, SHP2, NF1, RAS proteins, RAF family members or MEK1/MEK2, can result in aberrant activation of this pathway and oncogenesis (Fig. 1). RAS mutations or amplifications are among the most frequent abnormalities in human cancers6: KRAS is most often altered, especially in solid tumours; NRAS mutations are present in melanoma and many haematological malignancies; and HRAS mutations mainly occur in bladder, thyroid, cervical, and head and neck cancers15 (Fig. 2). The MSKCC IMPACT and AACR Project GENIE clinical sequencing cohorts (data available via the cBioPortal for Cancer Genomics)16–18 provide the most accurate estimates of the prevalence of mutational events in patients with cancer; each dataset indicates that ~20% of all malignancies have a RAS abnormality. Overall, ~17% of solid tumours have KRAS mutations, including ~90% of pancreatic, ~50% of colon and ~25% of lung adenocarcinomas16,17 (Fig. 2). Indeed, KRAS mutations predominate in NSCLCs, accounting for ~78% of all RAS mutations found in such tumours17. The activating missense KRAS mutations that occur in ~25% of NSCLCs19–23 are typically mutually exclusive with other clinically actionable driver mutations, such as those in EGFR, BRAF and ALK24. Despite much effort, however, RAS gene products (HRAS, NRAS and KRAS) have been extremely challenging targets for drug development25,26.
Almost all cancer-associated RAS mutations (~95%) affect codon 12, 13 or 61, and lead to a markedly increased basal RAS-GTP:RAS-GDP ratio and constitutive activation of RAS effectors4,6. Mutations at glycine residue 12 (G12) of KRAS are most common, with glycine 13 (G13) being the next most frequently affected residue in KRAS27. Recurrent KRAS codon 12 mutations most commonly result in G12C, G12V or G12D substitutions, which account for 40%, 19% and 15% of KRAS mutations in NSCLCs, respectively. Although much attention has been focused on KRASG12C mutations in NSCLCs (prevalence of ~14%), these mutations are not unique to NSCLC and also occur less commonly in colorectal cancers (CRCs; 3%) and pancreatic ductal adenocarcinomas (PDACs; 1%)28. Unlike other clinically actionable genomic alterations found in NSCLCs (such as EGFR and ALK aberrations), KRAS mutations, particularly KRASG12C and KRASG12V, are strongly associated with smoking29–31.
Various KRAS mutations might have different prognostic and/or predictive implications in patients with NSCLC. In patients with refractory metastatic NSCLC treated with molecularly targeted therapy (either erlotinib, vandetanib, bexarotene plus erlotinib or sorafenib) in the BATTLE trial, tumours with KRASG12C or KRASG12V mutation were associated with poorer outcomes, in terms of progression-free survival (PFS), than those with wild-type KRAS or other KRAS mutations32. However, another study that involved patients from the pre-immunotherapy era found no statistically significant differences in overall survival (OS) between patients with metastatic NSCLC harbouring various KRAS codon 12 and 13 mutations33. Moreover, a study evaluating the outcomes of patients treated with immune-checkpoint inhibitors (ICIs) revealed improved PFS in those with KRASG12C mutations relative to those with non-G12C mutations34. Together, these findings highlight the need for further investigation to clarify the prognostic and predictive roles of distinct KRAS mutations.
Although mutations in KRAS and other oncogenic drivers, such as EGFR and ALK, are typically mutually exclusive, co-mutations in the tumour-suppressor genes STK11, TP53 or CDKN2A/CDKN2B are frequently found in KRAS-mutant tumours35. Patients in the KRAS–STK11 co-mutation group can be subdivided into KEAP1-wild-type or KEAP1-mutant subgroups, and KEAP1 can also be co-mutated with KRAS in the absence of STK11 mutations; each of these co-mutation patterns is associated with resistance to treatment in preclinical models and seems to confer an unfavourable prognosis and poor treatment responses in patients with NSCLC receiving ICIs with or without chemotherapy36–41. The lack of responsiveness to ICIs in patients with KRAS-mutant NSCLCs harbouring co-mutations in STK11 and/or KEAP1 has been attributed to alterations in immunophenotype in the tumour microenvironment (TME)40. Furthermore, preclinical data indicate that IGF1–IGF1R signalling functions as a bypass pathway in KRAS–STK11 co-mutated cells following molecularly targeted therapy using the MEK inhibitor trametinib42. Therefore, characterization of the entire molecular landscape of KRAS-mutant NSCLCs will be crucial to devise optimal therapeutic strategies.
Biochemical properties of mutant KRAS
Amplification of wild-type KRAS can contribute to the pathogenesis of oesophagogastric, colorectal, ovarian and endometrial tumours43. In these settings, KRAS remains under the control of its canonical regulators. By contrast, RAS mutants were once viewed as being ‘locked’ in the GTP-bound ‘on’ state, impervious to GAP-stimulated or intrinsic GTP hydrolysis, and hence undruggable. However, detailed structural and biochemical analyses of various RAS mutants associated with cancer and ‘RASopathies’, which are developmental disorders caused by germline mutations in the RAS−MAPK pathway44, have revealed subtle but important differences in their intrinsic and GAP-catalysed GTPase activity, intrinsic and SOS1/2-stimulated GDP–GTP exchange, and effector-binding profiles14,45–49. For example, KRAS mutants vary in their intrinsic GTPase activity, with KRASG12C and, to a lesser extent, KRASG12D having greater activity than other variants. KRASG12C-mediated GTP hydrolysis is, however, refractory to (and might even be inhibited by) RAS-GAPs, whereas some other KRAS variants (G12D, G12A, G12R and G12V) might remain responsive to GAPs50. Even with GAP stimulation, the GTPase activity of KRASQ61L and KRASG61H is limited51. Wild-type RAS-GTP has a >10-fold higher affinity for RAF than for p120RASGAP (at least in vitro)52. Moreover, structural studies suggest that RAS-GTP cannot undergo hydrolysis when it is bound to RAF52–55, although data from single turnover kinetic studies contest this notion45. Thus, for GAP-responsive RAS mutants, the hydrolytic rate in cells probably equates to the GAP-stimulated rate. A study published in 2021 indicates that regulator of G protein signalling 3 (RGS3), an atypical GAP that lacks the characteristic catalytic arginine finger, can enhance the GTPase activity of mutant KRAS56. This important finding implies that susceptibility to the action of RGS3, instead of intrinsic GTPase activity, is the predominant determinant of the extent to which a given KRAS mutant occupies the GDP-bound state in cells — and thus the potential sensitivity of the cells to current ‘KRAS-off’ inhibitors, which bind to the inactive conformation of mutant KRAS56.
Mutations also affect the binding affinity of RAS proteins for their effectors. For example, KRASG12D binds to the RAF1-RBD approximately fivefold less strongly than wild-type RAS does46. The biological implications of these biochemical differences were unclear until recently. In 2020, Zafra et al.57 used high-fidelity CRISPR-based engineering to create mouse models of KRAS-mutant lung, pancreas and colon cancers, and found that different KRAS mutations had distinct effects on the transformation of colonic and pancreatic epithelial cells. Moreover, data indicate that the pathogenetic effects of different KRAS mutations, and the susceptibility of KRAS mutants to direct or indirect therapeutic targeting, depend on cell lineage and the presence of specific co-mutations, as well as the precise biochemical effects of specific allelic variants57–60.
The difficulty of drugging KRAS
The several-decade struggle to drug KRAS and other RAS isoforms25,61 reflects challenges posed by three key biochemical features: first, the picomolar affinity of RAS proteins for GTP62; second, the high intracellular concentrations of GTP (~500 nM)9; and third, the absence of a deep or pharmacologically actionable small-molecule-binding pocket in the RAS proteins. The success of small-molecule drugs that compete with ATP binding (ATP-competitive kinase inhibitors) is well known. However, the first two features — the high GTP-binding affinity and concentrations — make the analogous development of GTP-competitive RAS inhibitors unfeasible, if not impossible. Although intracellular ATP levels are typically ~5 mM, the KM for ATP of typical protein kinases is in the 10–100 µM range63, enabling drugs that bind with high picomolar or low nanomolar affinity to compete effectively at pharmacologically achievable concentrations. By contrast, overcoming the picomolar affinity of RAS for GTP would require small molecules with unprecedented binding properties. Moreover, the GTP-binding site of the KRAS protein varies between specific KRAS mutants, such as the G12C, G12D, G12V, G13D and Q61H variants64, which further complicates KRAS inhibitor design. Consequently, first-generation direct KRAS inhibitors all bind to inactive, GDP-bound mutant KRAS (KRAS-off inhibitors)65–68. More recently, however, ‘KRAS-on’ inhibitors that target active KRAS-GTP, such as RM-018 (ref.69), have been developed (Fig. 1 and Table 1).
Table 1.
Selected KRAS-directed therapies
| Strategy and target | Agent | Phase of development |
|---|---|---|
| Mutant-specific direct KRAS inhibitors | ||
| KRASG12C | Sotorasib (AMG 510) | Approved for previously treated advanced-stage KRASG12C-mutant NSCLC; further clinical trials ongoing (NCT03600883, NCT04185883, NCT04303780, NCT04933695, NCT04625647, NCT05398094, NCT05074810, NCT04380753, NCT05311709, NCT05054725, NCT05400577, NCT05180422, NCT04667234, NCT05198934, NCT05374538, NCT05313009, NCT05118854, NCT05273047, NCT05251038, NCT04892017, NCT04959981, NCT04720976) |
| Adagrasib (MRTX849) | Clinical trials (NCT03785249, NCT04330664, NCT04613596, NCT04793958, NCT04685135, NCT05162443, NCT05375994, NCT05263986, NCT04975256, NCT05178888, NCT04418661) | |
| D-1553 | Clinical trials (NCT04585035, NCT05383898, NCT05379946) | |
| JDQ443 | Clinical trials (NCT04699188, NCT05132075, NCT05358249, NCT05329623) | |
| RG6330 (GDC-6036) | Clinical trials (NCT04449874, NCT03178552) | |
| LY3537982 | Clinical trials (NCT04956640) | |
| BI 1823911 | Clinical trials (NCT04973163) | |
| JAB-21822 | Clinical trials (NCT05009329, NCT05194995, NCT05002270, NCT05276726, NCT05288205) | |
| JNJ-74699157 (ARS-3248) | Clinical trials (NCT04006301) | |
| MK-1084 | Clinical trials (NCT05067283) | |
| ARS-1620 | Preclinical studies | |
| ARS-853 | Preclinical studies | |
| RM-018 | Preclinical studies | |
| RMC-6291 | Preclinical studies | |
| KRASG12D | MRTX1133 | Preclinical studies |
| JAB-22000 | Preclinical studies | |
| RMC-9805 (RM-036) | Preclinical studies | |
| KRASG13C | RMC-8839 | Preclinical studies |
| KRASG12V | JAB-23000 | Preclinical studies |
| KRASMULTI | JAB-23400 | Preclinical studies |
| Pan-RAS inhibitors | ||
| RASMULTI | RMC-6236 | Clinical trials (NCT05379985) |
| BBP-454 | Preclinical studies | |
| SOS1 inhibitors | ||
| SOS1 | BI 1701963 | Clinical trials (NCT04973163, NCT04111458, NCT04975256, NCT04835714, NCT04627142) |
| RMC-5845 | Preclinical studies | |
| BAY-293 | Preclinical studies | |
| BI-3406 | Preclinical studies | |
| SDGR5 | Preclinical studies | |
| SHP2 inhibitors | ||
| SHP2 | RMC-4630 (SAR442720) | Clinical trials (NCT03634982, NCT05054725, NCT04418661, NCT03989115, NCT04916236, NCT04185883) |
| TNO155 | Clinical trials (NCT03114319, NCT04000529, NCT04294160, NCT04956640, NCT04330664, NCT04292119, NCT04699188, NCT04185883) | |
| GDC-1971 | Clinical trials (NCT04449874) | |
| JAB-3068 | Clinical trials (NCT03518554, NCT03565003, NCT04721223) | |
| JAB-3312 | Clinical trials (NCT04045496, NCT04121286, NCT04720976, NCT05288205) | |
| SHP099 | Preclinical studies | |
| RMC-4550 | Preclinical studies | |
| RAS degraders | ||
| KRASG12C | LC-2 (PROTAC) | Preclinical studies |
| KRASG12C, KRASG12D, KRASG12V and KRASQ61H | K27-SPOP | Preclinical studies |
| RAS toxins | ||
| Pan-RAS | RRSP–DTB | Preclinical studies |
| Adoptive cell therapy | ||
| KRASG12V | Specific TCRs | Clinical trials (NCT04146298) |
| KRASG12D | Specific TCRs | Preclinical studies |
| Cancer vaccines | ||
| KRASG12C, KRASG12D, KRASG12V and KRASG13D | mRNA-5671/V941 | Clinical trials (NCT03948763) |
| KRASG12C, KRASG12V, KRASG12D, KRASG12A, KRASG13D or KRASG12R | Mutant KRAS-targeted long-peptide vaccine | Clinical trials (NCT04117087) |
| KRASG12C, KRASG12V, KRASG12D or KRASG12R | mDC3/8-KRAS vaccine | Clinical trials (NCT03592888) |
| KRASG12D or KRASG12R | ELI-002 2P | Clinical trials (NCT04853017) |
| KRAS siRNAs | ||
| Various mutant KRAS mRNAs | Various nanoparticle-based technologies | Preclinical studies |
| KRASG12D mRNA | iExosomes | Clinical trials (NCT03608631) |
NSCLC, non-small-cell lung cancer; PROTAC, proteolysis targeting chimera; siRNA, small interfering RNA; TCR, T cell receptor.
The KRASG12C breakthrough
The lack of an adequate binding pocket for small molecules posed an equally daunting challenge for RAS-targeted drug discovery. In 2013, the laboratory of K. Shokat achieved a major breakthrough in KRAS targeting. Capitalizing on a unique disulfide-fragment-based chemical library approach, they screened 480 tethering compounds coupled to a cysteine-attacking nucleophile and identified compounds that bound covalently and selectively to KRASG12C-GDP70. X-ray crystallography revealed binding of the hit compounds to an expanded switch 2 pocket created by displacement of glutamine 61 (Q61) by the mutant cysteine in KRASG12C-GDP, and coupling of the nucleophile warhead to that cysteine70. Accordingly, inhibitors of this type are KRASG12C specific; therefore, they do not affect RAS signalling in non-malignant cells65,70–72 and, at least theoretically, have a low risk of on-target, off-tumour toxicities. Extensive medicinal chemistry efforts to refine the initial lead compounds resulted in the development of ARS-853, ARS-1620 and, ultimately, all clinical KRASG12C inhibitors, including sotorasib and adagrasib65–68,70,73,74 (Fig. 1).
The mechanism of inhibition by these compounds implies that a substantial proportion of cellular KRASG12C must at least transiently reside in the GDP-bound state. As alluded to previously, data indicate that conversion of KRASG12C-GTP to KRASG12C-GDP primarily reflects the action of RGS3, although GTP hydrolysis would eventually occur owing to the preserved intrinsic GTPase activity in this mutant. Consequently, RAS-GEFs essentially compete with KRASG12C-off inhibitors for binding to KRASG12C-GDP. Direct evidence for this conclusion is provided by the finding that second-site mutations that decrease nucleotide exchange, such as Y32S, enhance ARS-853-mediated inhibition of KRASG12C-GTP formation; secondary mutations such as Y40A, N116H and A146V, which enhance intrinsic nucleotide exchange and thus result in decreased levels of KRASG12C-GDP, had the opposite effect65. Further evidence has been provided by genetic or pharmacological inhibition of SOS1 or SHP2. For example, inhibitors of SOS1 reduce the activity of this key RAS-GEF and have demonstrated preclinical efficacy in combination with trametinib in KRASG12C-mutant and KRASG13D-mutant cell lines75, suggesting the potential for synergy when combined with KRAS-off inhibitors.
KRASG12C inhibitors in the clinic
Initial first-in-human data from 22 patients with advanced-stage KRASG12C-mutant solid tumours receiving sotorasib in the phase I/II CodeBreaK 100 study indicated single-agent activity, including a partial response (PR) in one of six patients with NSCLC76. Subsequent data from 129 patients enrolled in the phase I part of this trial demonstrated an objective response rate (ORR) of 32.2%, a disease control rate (DCR) of 88.1% and a median PFS of 6.3 months in 59 patients with advanced-stage NSCLC, as well as a 7.1% ORR, a 73.8% DCR and a median PFS of 4.0 months among 42 patients with CRC60. Responses also occurred in patients with melanoma, pancreatic, endometrial or appendiceal cancer60. The phase II portion of CodeBreaK 100, which involved 124 evaluable patients with previously treated advanced-stage KRASG12C-mutant NSCLC, demonstrated an ORR of 37.1%, a median duration of response (DoR) of 11.1 months, median PFS of 6.8 months and median OS of 12.5 months67. These impressive results led to the 2021 FDA accelerated approval of sotorasib for this indication. An updated analysis encompassing 174 patients with NSCLC included in the phase I and II parts of CodeBreaK 100 revealed that 1-year OS was 50.8% and 2-year OS was 30.3%77. Furthermore, data from the PDAC cohort of CodeBreaK 100 showed an ORR of 21.1% and a DCR of 84.2% in a heavily pretreated patient population (n = 38)78. However, similar success was not seen in the CRC cohort (n = 62), in which the ORR was 9.7%79. Sotorasib is currently being compared with docetaxel in the second or later line treatment of advanced-stage KRASG12C-mutant NSCLC (after progression on both an ICI and platinum-based doublet chemotherapy, individually or combined) in the phase III CodeBreaK 200 trial, with a primary end point of PFS80. Trials in the first-line setting are also ongoing (NCT04933695), and a multitude of trials are testing sotorasib alone or in combination with other agents across various settings in NSCLC and beyond (Table 1 and Supplementary Table 1).
Adagrasib, the second KRASG12C inhibitor to enter clinical testing, received FDA breakthrough therapy designation for previously treated advanced-stage KRASG12C-mutant NSCLC on the basis of data from the phase I/II KRYSTAL-1 trial68,81. Data from this trial, presented at the European Society for Medical Oncology (ESMO) Congress 2021, demonstrated a 96% DCR in this setting: 23 (45%) of 51 evaluable patients had a PR, and an additional 26 had stable disease81. In the phase I/Ib part of KRYSTAL-1, the median PFS of 16 patients with KRASG12C-mutant NSCLC receiving adagrasib at the recommended phase II dose was 11.1 months and the median DoR was 16.4 months82. Updated data from 116 patients with previously treated NSCLC included in a registrational phase II cohort of KRYSTAL-1 showed an ORR of 42.9%, a DCR of 79.5%, median DoR of 8.5 months, median PFS of 6.5 months, median OS of 12.6 months and estimated 1-year OS of 50.8% with adagrasib83. Additionally, in the subgroup of patients with previously treated, stable brain metastases (n = 33), the intracranial ORR was 33.3%83. In another phase II cohort of KRYSTAL-1, ORRs of 50% and 35% have been reported for ten evaluable patients with KRASG12C-mutant PDAC and 17 with other KRASG12C-mutant non-CRC gastrointestinal cancers, respectively, with a DCR of 100% in both groups; median PFS was 6.6 months in the PDAC group84. In 45 evaluable patients with CRC harbouring KRASG12C mutations who were also enrolled in KRYSTAL-1, adagrasib monotherapy resulted in an ORR of 22% and a DCR of 87%, with a median DoR of 4.2 months and median PFS of 5.6 months; in an additional 28 patients who received adagrasib in combination with cetuximab, the ORR was 43%, with a 100% DCR85. Numerous trials of adagrasib in various contexts are ongoing (Table 1 and Supplementary Table 1).
Several other KRASG12C inhibitors that act through a similar mechanism have entered clinical development more recently (Table 1 and Supplementary Table 1). GDC-6036 (also known as RG6330) is being tested alone and in combination with various monoclonal antibodies and kinase inhibitors as well as the SHP2 inhibitor GDC-1971 in a phase I trial involving patients with various advanced-stage KRASG12C-mutant solid tumours (NCT04449874). JDQ443, another covalent irreversible KRASG12C-off inhibitor, has also entered clinical testing after demonstrating favourable pharmacokinetic characteristics and promising antitumour activity in preclinical models86. A phase I/II trial is evaluating JDQ443 as monotherapy and in combination with TNO155 (another SHP2 inhibitor) and/or tislelizumab (an anti-PD-1 antibody) in patients with KRASG12C-mutant NSCLC, CRC or other advanced-stage solid tumours (KontRASt-01; NCT04699188). In parallel, a confirmatory phase III trial comparing JDQ443 with docetaxel in patients with previously treated KRASG12C-mutant NSCLC is planned (KontRASt-02; NCT05132075), and a phase I/II platform study testing various other JDQ443-based combination therapies across tumour types has been initiated (KontRASt-03; NCT05358249). LY3537982, a selective KRASG12C inhibitor, is also in phase I clinical development for KRASG12C-mutant solid tumours, both alone and in combination with various other agents (NCT04956640). Other KRASG12C-targeted compounds in phase I/II trials include D-1553, JNJ-74699157, BI 1823911, JAB-21822 and MK-1084 (Supplementary Table 1) and numerous other inhibitors of KRASG12C and/or other KRAS mutants are in preclinical development (Table 1).
The dramatic conceptual advance provided by the development of KRASG12C inhibitors cannot be overstated. Nevertheless, the potential for long-term benefit from KRASG12C inhibitors, at least as single agents, is modest at best; several primary and acquired mechanisms of resistance to these mutant-specific drugs have been reported. Moreover, changes in the TME that induce an immunosuppressive state have also been associated with resistance to these drugs87–89.
Resistance to KRAS inhibition
Although KRAS is no longer undruggable, KRAS inhibitor monotherapy is far from curative. Indeed, plasticity and genetic instability enable tumours to develop resistance to all single-agent targeted therapies90–92, with KRAS-directed therapies proving no exception. The updated data from the pivotal trial that led to the approval of sotorasib for KRASG12C-mutant NSCLC, as an example, show an ORR of only ~41% and median PFS of only 6.3 months, as well as 2-year OS of ~30%77. Combining inhibitors of the same pathway (termed ‘vertical inhibition’) has the potential to improve survival outcomes, as perhaps best illustrated by BRAF and MEK inhibitor combinations for melanoma93, but resistance remains inevitable. Targeting parallel pathways (‘horizontal combination’) also invariably leads to resistance or unacceptable toxicity94,95. Here, we describe emerging data on mechanisms of resistance to single-agent KRASG12C inhibitors. Tumour cell-autonomous, as well as non-autonomous (for example, TME-mediated) resistance has been reported, and can be further categorized as being intrinsic (primary) or acquired. Acquired resistance can be further divided into mutational escape of KRAS itself or activation of upstream or downstream mediators. We also discuss potential strategies for overcoming resistance.
Primary resistance
Primary resistance to direct inhibitors of KRAS could theoretically result from mutational heterogeneity in a tumour or the presence of specific co-mutations. Understanding these mechanisms of resistance is crucial to developing informed treatment strategies. KRAS-mutation heterogeneity has been described between different sites of disease in patients with CRC and might, at least in part, explain the variable responses to EGFR-directed therapy in such patients96. Similar heterogeneity between tumour sites has been reported in patients with NSCLC97,98; however, pre-treatment intratumour heterogeneity of KRAS mutations seems to be uncommon99. Zhao et al.100 evaluated 8,750 pre-treatment KRAS-mutant tumour specimens (from 7,790 patients) and found more than one RAS mutation in only 304 specimens (3.5%); among KRASG12C-mutant tumours specifically, secondary RAS mutations were found in 3% (40 of 1,432). Cannataro et al.101 performed deep sequencing of 27 KRASG12-mutant NSCLCs and found no evidence for pre-existing resistance mutations in KRAS or downstream genes before KRASG12C-inhibitor treatment. Knowledge of the mechanisms of primary resistance to KRASG12C inhibitors remains extremely limited, and large-cohort multi-omics analyses will be required to identify pre-treatment factors associated with non-responsiveness to these agents.
Acquired resistance due to mutational escape
Mutational escape refers to resistance mutations that develop ‘de novo’ on treatment, being undetectable before therapy. The binding site for the commonly used KRAS inhibitors sotorasib and adagrasib is formed by the amino acid residues at positions 12, 68, 95 and 96 of KRASG12C; therefore, mutations that affect these residues are particularly relevant to resistance68,102,103. For example, acquired second-site Y96D mutations in KRASG12C confer clinical resistance to adagrasib by altering the switch 2 pocket such that the inhibitor can no longer bind69. In vitro experiments by Koga et al.104 identified 12 distinct secondary KRAS mutations after treatment of KRASG12C-mutant Ba/F3 cells with sotorasib or adagrasib. KRASY96D or KRASY96S mutation conferred resistance to both agents, but both mutations were associated with sensitivity to the SOS1 inhibitor BI-3406 in combination with the MEK inhibitor trametinib (although monotherapy or combination therapy with BI-3406 and/or the SHP2 inhibitor TNO155 was not efficacious)104. Interestingly, cells with secondary G13D, R68M, A59S or A59T mutations in KRAS remained sensitive to adagrasib despite showing sotorasib resistance104. Conversely, second-site Q99L mutants were sensitive to sotorasib but resistant to adagrasib104. These findings demonstrate that although sotorasib and adagrasib are similar compounds, they cannot be considered interchangeable. Perhaps unsurprisingly, given its distinct mechanism of action as a KRAS-on inhibitor, RM-018 retains activity against KRASG12C/Y96D (ref.69).
In a biomarker analysis of the CodeBreaK 100 trial, plasma next-generation sequencing at baseline and at the time of disease progression revealed heterogeneous acquired molecular alterations in 19 (28%) of 67 patients with NSCLC and in 33 (73%) of 45 patients with CRC following sotorasib monotherapy, with RTK-pathway alterations being most common105. Secondary RAS alterations were found more commonly in patients with CRC than in those with NSCLC (16% versus 3%, respectively)105. In the aforementioned study by Zhao et al.100, treatment-emergent alterations were identified in 27 (63%) of 43 patients receiving sotorasib, consisting of diverse gene amplifications (of KRAS, MET, RICTOR, MYC, MET or FGFR2), mutations (in KRAS, BRAF, NRAS, PTEN, IDH1, IDH2, TP53, EGFR, KEAP1, NF1 and CTNNB1) and deletions (loss of PTEN and CDKN2A/CDKN2B). Secondary KRAS alterations (V8L, G12D/F/V, V14I or Q61H mutation, or gene amplification) were identified in 7 (16%) of the 43 patients. Patient-derived xenograft and cell-line models were subsequently used to show that KRASG12V, as well as KRASG13D, NRASQ61K, NRASG13R, MRASQ71R or BRAFG596R mutations, conferred resistance to KRASG12C inhibition100. Similarly, Awad et al.106 studied baseline and post-treatment samples from 38 patients who developed resistance to adagrasib and found alterations suggestive of various mechanisms of resistance in 17 (45%). These alterations included secondary KRAS mutations (G12D/R/V/W, G13D, Q61H, R68S, H95D/Q/R or Y96C) in nine patients (23%); KRASG12C and MET amplifications; activating mutations in NRAS, BRAF, MAP2K1 and RET; oncogenic fusions involving ALK, RET, BRAF, RAAF1 and FGFR3; and loss-of-function mutations in NF1 and PTEN106. Notably, in vitro validation studies confirmed that R68S, H95D/Q/R and Y96C second-site mutations affecting the switch 2 pocket of KRASG12C result in resistance to adagrasib106. By contrast, the G12C/H95D, G12C/H95Q and G12C/H95R double mutants remained sensitive to sotorasib, suggesting the potential for benefit from this agent in some patients with resistance to adagrasib (although co-existing cross-resistance mechanism might be common). Feng et al.107 performed a mutagenesis screen of >3,500 missense mutations in KRASG12C using the H358 cell line (containing KRASG12C in one allele and KRASWT in the other) and found that secondary C12F, R68S, H95D/R and Y96C mutations were strongly associated with resistance to KRAS inhibitors107, consistent with the clinical findings of Awad and colleagues106. As clinical use of KRAS inhibitors expands and more patients undergo molecular testing at the time of disease progression, these and other similar alterations should be more easily studied and their relative frequency better defined.
Adaptive resistance
Attempts have also been made to target KRAS-mutant cancers by inhibiting upstream and downstream pathway components; however, alterations in these mediators, as well as a type of intrinsic resistance termed ‘adaptive resistance’, inevitably result in a lack of efficacy, and recurrence and progression of these tumours5,90,91,108. Adaptive resistance refers to the rapid reactivation of the RAS–MAPK pathway at some level, typically as a result of derepression of MYC target genes, such as those encoding RTKs and their ligands, upon suppression of ERK activity109. Initially described with BRAF and MEK inhibitors108,109, several studies have shown that similar pathway reactivation occurs upon KRASG12C-inhibitor treatment50,110–113. The precise mechanisms that underlie adaptive resistance to KRASG12C inhibitors remain a subject of controversy and seem to differ in at least some respects from those that occur with BRAF and MEK inhibition. For example, the group led by Lito110 hypothesizes that induction of new KRASG12C proteins at the transcriptional level and a subsequent KRAS-GTP rebound have an important role in adaptive resistance to KRASG12C inhibitors, whereas others groups did not find such upregulation50,111,112. Indeed, whether adaptive resistance is mediated by reactivation of mutant KRAS or activation of the remaining wild-type KRAS, HRAS and/or NRAS remains unclear110–112. In this regard, Ryan et al.112 described adaptive resistance to sotorasib and ARS-1620 resulting from activation of wild-type NRAS and HRAS in KRASG12C-mutant lung, colon and pancreatic cancer cell lines. Also uncertain is whether adaptive resistance alone can eventually result in regrowth of KRASG12C-mutant tumours, without secondary genetic and/or epigenetic alterations, or if this phenomenon instead enables tumour cells to survive until such alterations transpire.
EMT and adeno–squamous transformation
Epithelial-to-mesenchymal transition (EMT) is another potential mechanism of both intrinsic and acquired resistance to KRASG12C inhibitors. During EMT, cells downregulate expression of epithelial genes and upregulate expression of mesenchymal ones, gaining increased mobility and invasiveness114. EMT is not unique to KRAS-mutated cancers. For example, the process has been described in patients with EGFR-mutant NSCLC after treatment with EGFR inhibitors115 and results in resistance to these agents, which seems to be mediated by TGFβ116. Seminal work by Singh et al.117 identified a gene-expression signature of KRAS dependency in KRAS-mutant cell lines and demonstrated that loss of expression of certain signature genes was associated with EMT. Moreover, induction of EMT seemed to decrease KRAS dependency117, supporting the idea that EMT can confer resistance to KRAS inhibition. Adachi et al.118 used gene set-enrichment analyses to demonstrate EMT as a mechanism of both intrinsic and acquired resistance to sotorasib. The PI3K pathway was found to be activated in EMT-induced KRASG12C-mutant cell lines via bypass IGFR–IRS1 pathway signalling, and combining sotorasib with the PI3K inhibitor GDC-0941 blocked AKT activation and suppressed cellular proliferation118. Adachi and colleagues118 also found that combining sotorasib, GDC-0941 and the SHP2 inhibitor tool compound SHP099 potently inhibited both AKT and ERK activation, resulting in regression of tumours with acquired resistance to sotorasib in mouse models. These preclinical observations and combination strategies warrant further clinical investigation.
Adeno–squamous transformation, a change in histology from adenocarcinoma to squamous cell carcinoma, has been observed in two (22%) of nine patients with KRASG12C-mutant NSCLC following acquired resistance to adagrasib, in the absence of genomic resistance mechanisms described previously106. Similar histological transformation has been associated with resistance to EGFR inhibitors119,120.
Overcoming KRASG12C inhibitor resistance
Vertical combinations
Several strategies to limit adaptive resistance and prolong responses to KRASG12C inhibitors are being explored. Although adaptive resistance typically involves the upregulation of several RTKs and RTK ligands, one or more RTK–ligand combinations can predominate in individual tumours. In NSCLCs and CRCs, KRASG12C inhibition leads to an accumulation of activated upstream EGFR and/or other ERBB family members, which facilitates escape from KRASG12C-inhibitor monotherapy110,121–124. Accordingly, combined KRASG12C and EGFR inhibition is currently under investigation in several clinical trials (Fig. 1 and Supplementary Table 1). In CodeBreaK 101 (NCT04185883), sotorasib is being combined with the EGFR/HER2 tyrosine kinase inhibitor afatinib, or with the anti-EGFR monoclonal antibody panitumumab (with or without FOLFIRI chemotherapy)125. Data on the sotorasib–afatinib combination from CodeBreaK 101 were presented in late 2021 and showed a manageable safety profile, with the most common treatment-related adverse effects (TRAEs) being diarrhoea and nausea (in 69.7% and 21.2% of patients, respectively, grade ≥3 in 21.2% and 0%)126. An efficacy signal was observed with ORRs of 20.0% and 34.8% and DCRs of 70.0% and 73.9% across two dose cohorts, including stable disease in three of five patients who had previously received sotorasib monotherapy126. Adagrasib is being combined with cetuximab, another anti-EGFR monoclonal antibody, in KRYSTAL-1 and KRYSTAL-10 (NCT03785249 and NCT04793958, respectively); KRYSTAL-1 is also investigating adagrasib in combination with afatinib for the treatment of NSCLC specifically. GDC-6036 is also being tested in combination with cetuximab as well as with the EGFR tyrosine kinase inhibitor erlotinib (NCT04449874).
Since its discovery in 1992 (ref.127), SHP2 has emerged as a key ‘positive’ upstream regulator of the RAS–MAPK pathway and thus an essential component of signalling by multiple oncogenic driver kinases (Fig. 1), as well as a complex positive and negative regulator of immune cell signalling128–132. SHP2 is required for tumorigenesis in several models of KRAS-mutant NSCLC133,134, suggesting that SHP2 inhibition could have a role in the treatment of KRAS-mutant cancers. SHP2 also has KRAS-independent effects on JAK–STAT signalling, which might mediate aspects of immune-checkpoint receptor action50. Highly selective, potent, bioavailable inhibitors of SHP2 are now in clinical testing (Table 1 and Supplementary Table 1), after many years of preclinical development128. These agents inhibit RAS GDP–GTP exchange (Fig. 1) and, accordingly, have single-agent activity against cycling KRAS mutants, especially KRASG12C, in preclinical models50. Furthermore, studies using the SHP2 inhibitor RMC-4550 have shown that class III BRAF-mutant, NF1-mutant and select RAS-mutant cancers remain dependent on SHP2 function135. The fact that SHP2 acts downstream of RTKs and upstream of SOS1/2 in the RAS signalling pathway makes this GEF an attractive target for preventing adaptive resistance to KRASG12C inhibitors. Misale et al.113 demonstrated that the mechanism by which KRASG12C-mutant NSCLC cells develop adaptive resistance to ARS-1620 involves reactivation of the MAPK pathway facilitated by incomplete inhibition of PI3K–AKT signalling. As well as discovering the role of wild-type RAS in this adaptive resistance, Ryan et al.112 demonstrated that the activation of wild-type RAS could be suppressed through co-inhibition of KRAS with ARS-1620 and SHP2 with SHP099. Fedele et al.50 reported similar findings in cell lines, extending their observations to immunocompetent mouse models of pancreatic and lung cancer.
SHP2 and KRAS co-inhibition not only alters cancer cell signalling, but also has important effects on the TME. Specifically, the studies by Fedele et al.50 in syngeneic orthotopic and subcutaneous KRASG12C-mutant tumours revealed depletion of myeloid-derived suppressor cells (MDSCs) and enrichment of cytotoxic T cells in the TME, which was associated with increased sensitivity to PD-1 blockade. Differences were noted in the effects of SHP099 alone and in combination with ARS-1620 in models of PDAC and NSCLC, including variations in the relative contribution of SHP2 inhibition in cancer cells and cells of the TME between the models50. The findings of this study emphasize the importance of considering not only cell of origin, but also the injection site (subcutaneous or orthotopic) and the precise tumour genetics.
On the basis of the improved efficacy of combined SHP2 and KRAS inhibition in preclinical studies, TNO155 is currently under investigation in combination with adagrasib in the KRYSTAL-2 trial (NCT04330664)136 and with sotorasib in CodeBreak 101 (NCT04185883), as well as with JDQ443 in KontRASt-01 (NCT04699188). CodeBreaK 101 is also testing the combination of sotorasib with RMC-4360 (a SHP2 inhibitor similar to RMC-4550)125. Several other clinical trials are evaluating combinations of various SHP2 and KRASG12C inhibitors (Supplementary Table 1).
Indirect targeting of RAS via SOS1 constitutes another form of vertical inhibition of this signalling pathway. SOS1 and its paralogue, SOS2, are crucial RAS-GEFs; thus, SOS1/2 inhibition presents a different strategy for suppressing the activity of RAS mutants that retain at least some capacity to undergo GTP hydrolysis, either through intrinsic or GAP-stimulated GTPase activity (Fig. 1). BI-3406 is an orally available small-molecule inhibitor that binds selectively within the active site of SOS1 (but not SOS2) and disrupts its interaction with RAS-GDP. SOS1 activation has been shown to be a key mechanism of adaptive resistance to MEK inhibitors; therefore, BI-3406 has been evaluated in combination with trametinib in KRASG12C-mutant and KRASG13D-mutant cell lines and found to synergistically inhibit their growth75. Indeed, given that SOS1 inhibitors affect KRAS interactions, they might be active across many KRAS mutations, including G12C, G12V, G12S, G12A and G12D, but notably not G12R as this variant does not bind to the catalytic domain of SOS1 (ref.137). In preclinical studies, BI-3406 decreased cellular proliferation in a range of KRAS-mutant NSCLC, PDAC and colon cancer cell lines137. Interestingly, BI-3406 was more effective in decreasing RAS-GTP levels and in arresting cellular proliferation in SOS2-knockout models, suggesting SOS2 induction as a potential resistance mechanism; however, no (immediate) upregulation of SOS2 mRNA levels was observed137. BI 1701963, a similar small molecule to BI-3406, also impairs KRAS–SOS1 binding138 and has preclinical activity in inhibiting KRAS with activating mutations139. This agent has entered early phase clinical trials as monotherapy (NCT04111458) as well as in combination with the KRASG12C inhibitors adagrasib (KRYSTAL-14; NCT04975256) and BI 1823911 (NCT04973163) (Table 1 and Supplementary Table 1). Another SOS1 inhibitor, BAY-293, has been shown to have activity against KRASG12C-mutant cell lines when combined with the KRASG12C inhibitor ARS-853 (ref.140). BAY-293 also has demonstrated cytotoxicity against other KRAS-mutant NSCLC and PDAC cell lines, and synergy with MEK, CDK4/6, topoisomerase I or EGFR inhibitors141.
The merits of targeting SOS1 should be considered in the context of strategies that target SHP2 (ref.142). SHP2 is required for full activation of SOS1/2, whereas SOS1 inhibitors are inactive against SOS2. Therefore, on the one hand, SHP2 inhibition could be viewed as superior to SOS1 inhibition. On the other hand, SOS1 inhibitors target RAS GDP–GTP exchange directly, whereas SHP2 is indirectly required for this process and thus might be susceptible to bypass mechanisms. Notably, however, SHP2 inhibition has the potential to induce favourable effects within the TME, such as depletion of MDSCs and enhancement of cytotoxic lymphocytes via actions on the IL-6–JAK–STAT3, TNF and IFNγ signalling pathways50, which are not directly regulated by SOS1. Preclinical studies that directly compare SHP2 and SOS1 inhibitors in immunocompetent models are warranted, although ultimately, this theoretical argument will be best resolved in clinical trials. Furthermore, on-target, off-tumour toxicity is another important consideration with both SOS1 and SHP2 inhibitors that will be evaluated in clinical trials.
The CodeBreaK 101 trial is also evaluating trametinib in combination with sotorasib, with or without panitumumab, in patients with various KRASG12C-mutant solid tumours (NCT04185883; Supplementary Table 1). The rationale for this combination is predicated on evidence that KRAS-mutant tumour cells are inherently resistant to MEK inhibitors owing to RAF-mediated MEK activation (Fig. 1), which might be abrogated by simultaneous inhibition of mutant KRAS143,144. Preliminary results from 41 patients receiving sotorasib plus trametinib in CodeBreak 101, including 18 with NSCLC and 18 with CRC, were presented in late 2021 (ref.145). The combination was found to be safe and tolerable, with predominantly grade ≤2 diarrhoea (in 43.9% of patients), rash (34.1%), nausea (29.3%) and vomiting (22.0%) being the most commonly reported TRAEs145. This combination was associated with clinical benefit in 15 patients with CRC and 15 with NSCLC, including two and three PRs, respectively145. Importantly, even patients previously treated with a KRASG12C inhibitor derived clinical benefit (including a PR in the CRC cohort)145. Additionally, the aforementioned SOS1 inhibitor, BI 1701963, is also being evaluated with trametinib in a phase I trial (NCT04111458; Supplementary Table 1), based on a similar mechanistic rationale.
Horizontal combinations
Inhibition of mTOR has been proposed as an alternative strategy to overcome adaptive resistance to KRAS (or MEK) inhibitors (Fig. 1). In several preclinical models of PDAC, resistance to KRAS or MEK inhibition purportedly occurs through bypass signalling via increased integrin-linked kinase (ILK)-mediated phosphorylation of Rictor, a component of the mTOR complex 2 (mTORC2), and subsequent mTORC2-mediated phosphorylation of AKT. Accordingly, co-inhibition of KRASG12C or MEK and mTORC1/2 synergistically impaired ERK and AKT activation, resulting in durable inhibition of PDAC growth and metastasis in mouse models146. In CodeBreak 101, the mTOR inhibitor everolimus is being evaluated in combination with sotorasib (NCT04185883; Supplementary Table 1).
Cell-cycle inhibition provides another potential horizontal combination strategy for synergy with KRAS inhibitors (Fig. 1). CDK4/6 inhibitors, such as palbociclib, ribociclib and abemaciclib, are effective in the treatment of some cancers, particularly oestrogen receptor-positive breast cancers147. These inhibitors impair cell-cycle progression driven by D-type cyclins148, which are a convergent node of the RAS–MAPK and PI3K–AKT pathways. Moreover, in vitro and in vivo preclinical studies suggest additive and synergistic benefit of CDK4/6 and KRASG12C co-inhibition in models of NSCLC and PDAC149. CodeBreak 101 is testing this concept in patients by combining sotorasib with palbociclib (NCT04185883; Supplementary Table 1).
Aurora kinases (AURKA, AURKB and AURKC) have crucial cell-cycle regulatory functions that support mitotic progression, and dysregulation of these kinases seems to promote tumorigenesis by both enhancing cell proliferation when activated and contributing to aneuploidy when dysfunctional150. Furthermore, AURKA expression is a poor prognostic indicator in patients with NSCLC151. Aurora kinase signalling has also been implicated in KRAS-mutant malignancies, and these kinases are upregulated by mutant KRAS in NSCLC cell lines152. Knockdown of AURKA or dual inhibition of AURKA and AURKB has antitumour activity in preclinical models involving such cell lines152. In keeping with the intertwined roles of AURKA and KRAS, AURKA activity seems to be a mechanism of not only adaptive but also intrinsic and acquired resistance to KRAS inhibition, with increased expression of AURKA seen in sotorasib-resistant cell lines153. In preclinical models, combined inhibition of AURKA and KRASG12C showed synergy110. Moreover, the selective AURKA inhibitor VIC-1911 and sotorasib had synergistic activity in KRAS-mutant NSCLC cell lines intrinsically resistant to sotorasib, presenting a viable strategy to negate resistance to KRASG12C inhibitors153.
The mitotic checkpoint kinase WEE1 is another important cell-cycle regulator and target for cancer therapy154. The multi-kinase inhibitor sorafenib has poor activity in KRASG12V-mutant cell lines, and high-throughput screening identified WEE1 as a potential modulator of sorafenib response that could be targeted pharmacologically (with the WEE1 inhibitor adavosertib)155. Given these interactions, the role of WEE1 has been evaluated in models of acquired resistance to KRASG12C inhibitors, with synergistic activity of adavosertib and VIC-1911 against sotorasib-resistant NSCLC cells in vitro and in vivo, suggesting the potential therapeutic utility of this approach153.
Immune-mediated escape and immunotherapy combination strategies
Mutant KRAS, like other driver oncogenes, has effects on cells of the TME, in addition to altering the behaviour of cancer cells themselves87,156 (Fig. 3). Accordingly, mutant-selective KRAS inhibitors can have indirect effects on immune cells, fibroblasts and endothelial cells in the TME (including the lymphatic epithelium) by reversing the actions of mutant KRAS87,156 — albeit mitigated by adaptive and acquired resistance. Other agents that target RAS signalling, presumably including pan-RAS inhibitors, are likely to have additional direct effects on cells of the TME, which might vary by tumour type and location (for example, primary versus metastatic). A detailed understanding of these pleiotropic actions could facilitate the rational design of curative combinations.
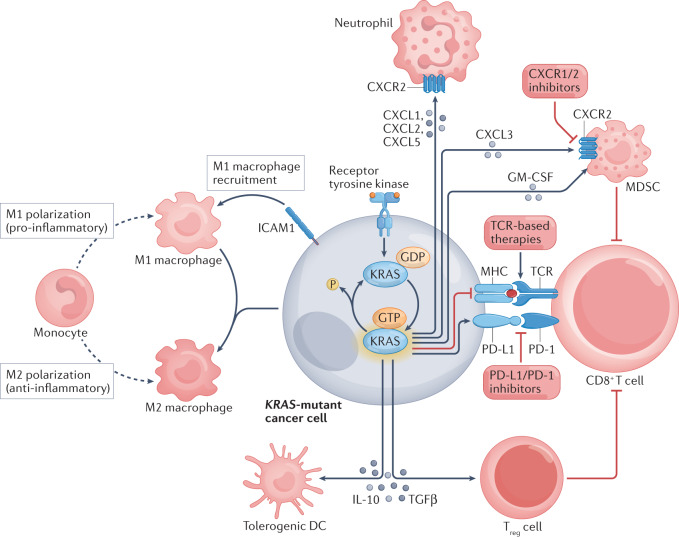
Fig. 3. The influence of mutant KRAS on the tumour immune microenvironment.
Activating KRAS mutations have numerous implications for the tumour immune microenvironment87,156, including activation and recruitment of macrophages, polarization of M1 to M2 macrophages, and suppression of CD8+ T cells via effects on MHC–T cell receptor (TCR) and PD-L1–PD-1 signalling as well as via activation of myeloid-derived suppressor cells (MDSCs) and regulatory T (Treg) cells. Together, these alterations in the tumour microenvironment present opportunities for intervention in the treatment of KRAS-mutated malignancies, and pertinent examples are provided. DC, dendritic cell; ICAM1, intercellular adhesion molecule 1.
KRAS has multiple immunomodulatory effects, mediated via several mechanisms (Fig. 3). Activation of KRAS increases production of the neutrophil chemoattractants CXCL1, CXCL2 and CXCL5; promotes recruitment of pro-inflammatory M1 macrophages via upregulation of intercellular adhesion molecule 1 (ICAM1) expression (conversely, co-activation of KRAS and MYC increases anti-inflammatory M2 macrophage recruitment via release of CCL9 and IL-23); induces immunosuppressive regulatory T (Treg) cell differentiation via secretion of TGFβ and IL-10; and enhances tumour infiltration of MDSCs via GM-CSF-dependent and IRF2/CXCL3-dependent mechanisms87,156. Additionally, Ischenko et al.157 used CRISPR–Cas9-mediated gene editing in PDAC models to demonstrate a key role of mutant KRAS in tumour immune evasion, which was attributed to alterations in both the cancer cells themselves and immune cells, and also implicated downstream BRAF and MYC signalling in this process. Accordingly, loss of oncogenic KRAS or BRAF abrogated tumour growth in immunocompetent syngeneic mice157.
The aforementioned study by Fedele et al.50 demonstrated that SHP2 inhibition is synergistic with KRASG12C inhibition through cancer cell-autonomous mechanisms as well as distinct effects on the TME in models of PDAC and NSCLC. Using PDAC cells expressing drug-resistant SHP2, these researchers demonstrated that all of the effects on immune components of the TME were dependent on inhibition of SHP2 in tumour cells; however, SHP2 inhibitors had direct anti-angiogenic actions in this model50. A subsequent study confirmed and extended these findings, implicating SHP2 inhibitors in suppression of VEGFR signalling158. By contrast, in the NSCLC model, substantial effects on the TME were observed even when SHP2 could not be inhibited in tumour cells; remarkably, SHP2 inhibition enhanced lung tumour vascularity, instead of reducing it50.
Building on these findings, Tang et al.159 found that in genetically engineered mouse models, SHP2 inhibition in KRAS-mutated or EGFR-mutated NSCLCs depleted alveolar and M2-like macrophage populations, increased T cell and B cell infiltration, but also enhanced accumulation of potently immunosuppressive granulocytic MDSCs (gMDSCs). The latter effect was attributable to NF-κB-induced secretion of the neutrophil lineage chemoattractants CXCL1 and CXCL5 (among other chemokines) by tumour cells159. Remarkably, production of these chemokines and subsequent CXCR2-dependent gMDSC infiltration required SHP2 inhibition in tumour cells, whereas the effects on lymphocytes, macrophages and monocytic MDSCs occurred even when tumour cells could not respond to the inhibitor159. These findings suggest that oncogenic KRAS promotes immune escape by fostering an immunosuppressive TME that can be mitigated through SHP2 inhibition — albeit subject to resistance mediated by CXCR2 signalling, which could potentially be a target of combinatorial strategies159 (Fig. 3).
ICIs, including those that target PD-1/PD-L1 and CTLA4, have shown efficacy in treating various cancers, most prominently NSCLC and melanoma160. Indeed, ICIs are the standard-of-care first-line treatment for advanced-stage NSCLCs that lack targetable oncogenic mutations, with or without chemotherapy depending on tumoural PD-L1 expression level161. As a result of the first-line approvals, many patients with KRAS-mutant NSCLCs will receive ICIs, given that sotorasib is currently approved only as a second-line treatment. Considering the various mechanisms by which mutant KRAS has direct and indirect immunomodulatory effects, particularly as they pertain to cytotoxic T cell activity, combining KRAS inhibitors with ICIs is a rational strategy. In mouse models of KRASG12C-mutant tumours treated with sotorasib, pro-inflammatory changes were seen within the TME, and combination therapy with ICIs resulted in synergistic tumour cell killing102. Furthermore, mice cured with a combination of sotorasib and an anti-PD-1 antibody rejected isogenic KRASG12C and KRASG12D cells upon re-challenge, suggesting adequate memory immune cell responses against common antigens102. Briere et al.162 demonstrated that adagrasib induces MHC class I (MHC I) expression and promotes a pro-inflammatory state in human KRASG12C-mutant tumour cell xenografts. Moreover, adagrasib depleted intratumoural MDSCs while increasing cytotoxic immune cell infiltration in syngeneic mouse models; tumour responses were observed in these immunocompetent BALB/c mice, but were attenuated in T cell-deficient mice162. Furthermore, combination treatment with adagrasib and an anti-PD-1 antibody resulted in durable antitumour responses, again, with evidence of immunological memory responses162. Of note, however, both of these studies used subcutaneous syngeneic models, the clinical relevance of which is uncertain. Nevertheless, this approach of combining a KRASG12C inhibitor with an ICI is being evaluated in several clinical trials across KRASG12C-mutant solid tumours (Supplementary Table 1), including with sotorasib in CodeBreaK 100 (NCT03600883) and CodeBreaK 101 (NCT04185883), adagrasib in KRYSTAL-1 (NCT03785249) and KRYSTAL-7 (NCT04613596), GDC-6036 in GO42144 (NCT04449874) and JDQ443 in KontRASt-01 (NCT04699188).
RAS-driven cancers rely on autophagy for survival163, and this complex cellular process also seems to have immunomodulatory effects that promote tumour immune evasion. In models of KRASG12D-mutant PDAC, MHC I is degraded in an autophagy-dependent manner, with inhibition of autophagy enhancing MHC I expression, antigen presentation, cytotoxic T cell activation and subsequent antitumour immunity164. Moreover, ICIs were found to have encouraging efficacy in these models when combined with autophagy inhibition164. Additionally, MEK and autophagy inhibitors have synergistic activity against PDAC cell lines165. These data highlight the complex interplay of mediators in KRAS-driven cancers but also point towards vulnerabilities that can be exploited.
As noted previously, Tang et al.159 discovered that RAS–MAPK pathway inhibition can result in CXCL1/CXCL5-dependent infiltration of KRAS-mutant NSCLCs by gMDSCs, depletion of which markedly increases accumulation of cytotoxic CD8+ T cells in the TME. Using their mouse models, these researchers also demonstrated that tumour control was markedly improved by combining SHP2 and/or EGFR inhibitors with SX-682, a novel oral small-molecule inhibitor of CXCR1 and CXCR2. The mice were not cured, however, potentially owing to T cell exhaustion mediated by PD-1 and/or by two other inhibitory immune-checkpoint receptors, NKG2A and/or KLRG1, which were found to be expressed on the CD8+ T cells. Notably, data from various mouse models indicate that disrupting the CXCR1/2–MDSC axis in tumours is therapeutically efficacious across a wide variety of solid cancers166–172. By reversing MDSC-mediated immunosuppression, SX-682 is also predicted to unmask greater efficacy of chemotherapy and immunotherapies (such as anti-CTLA4 and anti-PD-1 antibodies) that act by mechanisms that are non-redundant and indeed complementary to those of CXCR1/2 antagonists173. Tang et al.159 also showed that adagrasib increased CXCL1 and CXCL6 mRNA levels (both encoding ligands of CXCR1/2) in KRASG12C-mutant cell lines, as well as tumour infiltration by gMDSCs in preclinical models. Furthermore, analyses of matched pre-treatment and post-treatment biopsy samples provided evidence of these effects of adagrasib in patients with NSCLC159, highlighting the promise of combining CXCR1/2 inhibitors with KRAS inhibition.
Extending the breadth of RAS inhibitors
KRAS-on inhibitors
Given that KRASG12D has lower intrinsic GTPase activity than KRASG12C, most KRASG12D proteins will be GTP bound. Thus, agents that target KRASG12D-GTP, as well as other RAS isoforms (RAS-on inhibitors; Fig.1), have been a focus of drug development. In 2020, Zhang et al.174 identified three cyclic peptide ligands that are unique in that they bind preferentially within the switch 2 groove of KRASG12D-GTP and inhibit its interaction with RAF proteins. Notably, these compounds have no discernable effect on wild-type KRAS, exemplifying the distinct features of the GTP-bound state of KRAS mutants174. By contrast, the crystal structure of KRASG12D-GDP in a complex with KRpep-2d revealed that this KRASG12D-selective inhibitory peptide binds to a region near switch 2 and competes with GEF binding175. In 2021, Wang et al.176 reported the discovery and characterization of MRTX1133 as a noncovalent, selective KRASG12D inhibitor. MRTX1133 binds to the switch 2 pocket of GDP-bound KRASG12D as well as GTP-bound KRASG12D and was shown to successfully arrest KRAS-mediated signalling, resulting in substantial tumour regression in mouse xenograft models176. Vasta et al.177 have used NMR spectroscopy and competitive bioluminescence resonance energy transfer (BRET)-based engagement assays to demonstrate that the switch 2 pocket of several other non-G12C mutants of KRAS can be targeted in a noncovalent manner that is not reliant on the protein being GDP bound.
Thus, most small-molecule inhibitors of mutant KRAS currently in clinical development target the expanded pocket near the switch 2 region. However, an alternative and unique approach of targeting a switch 1/2 pocket is currently being evaluated. This pocket was initially described by Kessler et al.178,179 during the development of BI-2852, a small molecule that induces nonfunctional dimerization of KRAS. This approach offers the advantage, shared by MRTX1133, of targeting both the GTP-bound and GDP-bound states of KRAS178,179. Notwithstanding, successful clinical application of this approach remains to be reported.
Another novel strategy for targeting mutant KRAS and/or other mutant RAS isoforms, originally conceived by the group of G. Verdine180, uses a mechanism analogous to that of the immunosuppressants cyclosporin, FK506 and rapamycin. Cyclosporin binds to the chaperone protein cyclophilin A (also known as peptidyl-prolyl cis-trans isomerase A), and the cyclosporin–cyclophilin complex in turn binds to and inhibits the serine/threonine phosphatase calcineurin. FK506 binds to a different chaperone, FKBP12, although the resultant complex also inhibits calcineurin. Rapamycin also interacts with FKBP12, but the rapamycin–FKBP12 complex binds to and inhibits mTORC1 (ref.181). Various compounds that bind to cyclophilin A and subsequently form inhibitory trimeric complexes with various RAS proteins have been developed. For example, RM-018 is a covalent KRASG12C inhibitor, but through its interaction with cyclophilin A, targets KRASG12C in the GTP-bound state; hence, RM-018 is a KRASG12C-on inhibitor69. Preclinical data on the later-generation ‘tri-complex’ KRASG12C-on inhibitor, RMC-6291, were presented in April 2022 and suggest superiority over the KRAS-off inhibitor adagrasib182, supporting the viability of this targeting approach. Other tri-complex agents that target specific KRAS mutants in their GTP-bound state include RMC-9805 (KRASG12D) and RMC-8839 (KRASG13C). Moreover, pan-RAS-mutant targeting might also be possible using this tri-complex strategy, as exemplified by RMC-6236. This agent is a RASMULTI-on inhibitor that forms tri-complexes with cyclophilin A and several KRAS mutants, with activity against KRASG12D, KRASG12V and KRASG12R mutations demonstrated in preclinical models183,184. Of note, this compound is also active against ‘RAS oncogene switch’ mutations (within the switch 2 pocket), which confer resistance to adagrasib106, and it shows synergy with anti-PD-1 antibodies185.
Tri-complex RAS inhibitors are now entering clinical trials (NCT05379985; Table 1 and Supplementary Table 1); it will be interesting to see whether their impressive preclinical efficacy translates into improved patient outcomes. Importantly, cyclophilin A is expressed ubiquitously186, and thus diverse RAS-mutant tumour types could be amenable to these agents. Toxicity might be an issue, particularly with RASMULTI-on inhibitors, given that the clinical effect of wild-type RAS inhibition at this level is not known. Moreover, mutations in PPIA (which encodes cyclophilin A) could conceivably result in loss of drug binding and/or activity, although such resistance mutations would have to be homozygous, given that tri-complex inhibitors only need to bind to a small fraction of cellular cyclophilin A to fully inhibit their RAS targets. Whether homozygous PPIA mutations are consistent with cell viability remains unclear. It will also be interesting to see whether similar tri-complex strategies can be developed using compounds that bind to FKBP12 or other cellular chaperones, or that target cellular oncoproteins other than KRAS.
RAS degraders and toxins
RAS degraders are another novel approach to targeting RAS-mutant malignancies. Proteolysis-targeted chimeras (PROTACs) promote the proteasomal degradation of disease-associated proteins via bivalent small molecules that bind to and bring together the protein of interest and an E3 ubiquitin ligase. LC-2 is a PROTAC designed to target KRASG12C (Table 1). This compound combines adagrasib and a ligand for the E3 ligase VHL, enabling depletion of KRASG12C protein and thus attenuation of cellular proliferation in KRASG12C-mutant cell lines187. Similarly designed pan-KRAS188 and other mutant-specific KRAS degraders189 are in preclinical development. Given the size and molecular structure of these compounds, oral bioavailability might be limited, constituting a potential challenge of this approach190.
The use of chimeric toxins is an alternative approach to RAS targeting. RRSP–DTB is one such agent (Table 1), consisting of a RAS/RAP1-specific endopeptidase (RRSP) derived from the bacillus Vibrio vulnificus combined with translocation B fragment of diphtheria toxin (DTB)191. This toxin enters cells via heparin-binding EGF-like growth factor (HB-EGF)-mediated endocytosis and subsequently cleaves RAS within the switch 1 region191. Uniquely, RRSP–DTB is able to cleave HRAS, NRAS and KRAS irrespective of GTP or GDP binding191. Vidimar et al.191 have shown that this agent inhibits ERK signalling via RAS inactivation in KRAS-mutant PDAC models, resulting in ≥95% tumour regression without evidence of resistance191. Notably, when studied in immunocompetent mice, the toxin was not detectable in serum at 16 hours after administration, suggesting the need for frequent dosing191. Furthermore, efforts to increase the selectivity of RRSP uptake into malignant cells are currently under way191 and might improve the toxicity profile of this agent.
Immunotherapies targeting KRAS
Adoptive cell therapy (ACT), pioneered by the Rosenberg laboratory, involves isolating immune cells, typically tumour-infiltrating lymphocytes (TILs), expanding them ex vivo and then reintroducing them into the patient. This approach, initially used in patients with melanoma192, has more recently been extended to several other cancer types, including KRAS-mutant tumours. Endogenous immune responses against KRAS-mutant cancer cells were first described in 2016 by Tran et al.193, who identified CD8+ TILs with T cell receptors (TCRs) that recognize MHC I (HLA-C*08:02)-presented peptide neoepitopes derived from KRASG12D. ACT with these specific TILs resulted in regression of all seven pulmonary metastases in a patient with KRASG12D-mutant CRC from whom they had been isolated193. Notably, loss of MHC was identified as an immune-evasion mechanism in a tumour that progressed 9 months after TIL infusion193.
Other MHC I-restricted mutant KRAS epitopes have since been characterized, presenting opportunities for ACT against various KRAS-mutant malignancies. For example, Bear et al.194 identified TCRs that recognize G12V–HLA-A*03:01, G12V–HLA-A*11:01 and G12R–HLA-B*07:02 complexes, and further validated this approach by showing that exogenous expression of these TCRs in primary TCRαβnull cytotoxic T cells redirects their activity towards KRAS-mutant cell lines of various histologies. Furthermore, adoptive transfer of TCR-engineered CD8+ T cells specific for G12V–HLA-A*03:01 or G12V–HLA-A*11:01 showed efficacy in xenograft models of metastatic KRASG12V-mutant NSCLC194. ACT with TCR-engineered T cells that target KRASG12V-mutant cells has entered clinical trials (Table 1 and Supplementary Table 1).
Therapeutic cancer vaccines constitute another immune-based approach to target KRAS-mutant tumours. mRNA-based vaccine technology has the potential to introduce strong immune responses, as exemplified by the recent success of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines195. mRNA-5671/V941, a liquid nanoparticle-formulated mRNA neoantigen vaccine that targets the G12D, G12V, G13D and G12C variants of KRAS, has been developed and has strong mechanistic rationale196. On this basis, mRNA-5671/V941 was moved into phase I clinical testing, both as a monotherapy and in combination with PD-1 blockade (NCT03948763; Table 1), although the trial was subsequently closed to enrolment. Additionally, a long-peptide vaccine with activity against the G12C, G12V, G12D, G12A, G13D and G12R variants of KRAS is currently being evaluated in combination with ICIs in a phase I trial (NCT04117087). A dendritic cell-based vaccine that targets G12C, G12D, G12R and G12V neoepitopes of mutant KRAS is also in a phase I trial (NCT03592888; Supplementary Table 1). ELI-002 2P (two peptide) is a novel vaccine consisting of lipid amphiphile (Amph)-modified G12D-mutant and G12R-mutant KRAS peptides with an immunostimulatory Amph-modified CpG oligonucleotide adjuvant (Amph-CpG-7909) that has been shown to result in increased mutant KRAS-specific cytotoxic T cell activity197. ELI-002 2P is currently being evaluated in the adjuvant treatment of patients with minimal residual disease after standard treatment for KRASG12D/G12R-mutant PDACs or other solid tumours (AMPLIFY-201, NCT04853017; Table 1 and Supplementary Table 1). A future iteration of this trial will test ELI-002 7P (seven peptide), a vaccine that contains G12D, G12R, G12V, G12A, G12C, G12S and G13D neoepitopes of KRAS.
Antibody-based therapeutics that target mutant KRAS peptide–MHC complexes have also been proposed. As noted above, peptides derived from KRASG12 variants193,194, and also from KRASQ61H/L/R (ref.198), have been demonstrated to be presented on the surface of malignant cells by various MHC I (HLA-A) molecules. Bispecific dimeric single-chain antibody fragment constructs, so-called ‘diabodies’, have been engineered to recognize such MHC I–peptide complexes on KRAS-mutant cells and recruit T cells by simultaneously binding to CD3; these agents induced T cell-mediated cytotoxicity in vitro, with some anticancer activity observed in mouse models (although the effect did not reach statistical significance)198. Whether this approach will be intrinsically limited by the low level of neoepitope presentation remains to be seen. Soluble TCR therapies present an alternative, although related, strategy to bispecific antibodies. Notably, tebentafusp-tebn, a bispecific fusion protein consisting of a soluble TCR specific to gp100 (expressed by melanoma cells) linked to an anti-CD3 single-chain antibody fragment that targets T cells, has been approved for the treatment of metastatic uveal melanoma based on an OS benefit observed in a phase III trial199. A similar approach might be possible for KRAS-mutant tumours. Of note, with all these immunotherapy approaches, the immunosuppressive TME, to which mutant KRAS directly contributes, must be overcome if their efficacy is to be realized.
siRNA-based approaches
Preclinical efforts have been made to develop nanoparticle-based platforms to deliver KRAS-specific small interfering RNAs (siRNAs). This technology has been shown to be adequately delivered to cancer cells and to effectively decrease their KRAS levels, resulting in anticancer activity200–202 (Table 1). Exosomes are natural nanoparticles and have been engineered to deliver specific siRNAs to downregulate production of mutant KRAS (known as iExosomes) and are under clinical investigation for the treatment of KRASG12D-mutant PDAC203,204 (NCT03608631; Supplementary Table 1). These siRNA-based approaches might circumvent some of the difficulties in drug delivery, with endocytosis resulting in increased concentrations of the active agent being brought to tumour cells, in which this uptake mechanism is typically upregulated. Exosomes are associated with their own delivery challenges, mostly owing to phagocytosis by immune cells, although certain surface markers can be leveraged to mitigate these challenges. Thus, siRNA-based approaches against KRAS-driven cancers might present viable treatment strategies.
Conclusions
Since the identification of KRAS mutations in lung cancer more than three decades ago, KRAS-targeted drug discovery has come a long way, with a plethora of inhibitors, combination approaches and novel alternative targeting methods now under clinical investigation. However, the data on KRASG12C inhibitors, which have advanced the furthest in development and are entering clinical practice, emphasize that these agents are far from curative67. The modest benefits are attributable, at least in part, to resistance that almost invariably develops with single-agent therapy90–92. Herein, we have taken a reductionist approach to understanding resistance to KRAS-targeted therapies. However, recent work by Tsai et al.88 has revealed the true scope of the challenge ahead. In a rapid autopsy study using RNA and whole-exosome sequencing in a patient with KRASG12C-mutant NSCLC that developed resistance to sotorasib, they found that some areas of tumour growth had a decreased allele frequency of KRASG12C, others had reactivation of MAPK signalling in the absence of mutations in components of this pathway, while different areas had evidence of EMT and remodelling of the TME, including immune escape88. This study provides what is perhaps a much-needed reality check, highlighting the complexity and plasticity of the RAS pathway as well as the likelihood that substantial work lies ahead to further advance KRAS-targeted therapy.
In this Review, we describe strategies for overcoming resistance mechanisms through vertical inhibition of multiple nodes of the RAS pathway, with specific learnings from the saga of combined BRAF and MEK targeting in melanoma, which increases efficacy but is still not curative93. In addition, we describe combination strategies for horizontal inhibition of parallel pathways, an approach also marred by resistance in addition to toxicity94,95. Most likely, curative strategies will require the induction of a durable antitumour memory immune response via rational and dynamic combination strategies. Lastly, we discuss novel approaches for combating resistance as well as novel treatment strategies that can inject further promise into a field that has ballooned with optimism for the first time in decades, now that KRAS has officially lost its title as ‘undruggable’. We look forward to new approaches capable of achieving this long-sought goal of cure in populations of patients with KRAS-mutant malignancies.
Supplementary information
Author contributions
All authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks F. Skoulidis, T. Yoshino and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
V.V. has received fees for consulting or serving on advisory boards from Amgen, AstraZeneca, Bristol Myers Squibb (BMS), EMD Serono, Foundation Medicine, GlaxoSmithKline, Iteos Therapeutics, Merck, Novartis and Novocure. B.G.N. is a co-founder, holds equity in and receives consulting fees from Lighthorse Therapeutics and Navire Pharmaceuticals; is a co-founder and holds equity in Northern Biologics; is a scientific advisory board (SAB) member for, holds equity in and receives consulting fees from Arvinas; and is an SAB member for and holds equity in Recursion Pharma. K.-K.W. is a co-founder of and holds equity in G1 Therapeutics; has received grants from Alkermes, Ansun, AstraZeneca, BMS, Dracen, Merus, Mirati, Takeda and Tvardi; grants and personal fees from Delfi, Janssen, Pfizer and Zentalis; personal fees from Navire, Prelude and Recursion; and grants from Ono outside the submitted work. S.R.P. declares no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
cBioPortal for Cancer Genomics: http://www.cbioportal.org/
Supplementary information
The online version contains supplementary material available at 10.1038/s41571-022-00671-9.
References
- 1.Barbacid M. ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gimple RC, Wang X. RAS: striking at the core of the oncogenic circuitry. Front. Oncol. 2019;9:965. doi: 10.3389/fonc.2019.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hymowitz SG, Malek S. Targeting the MAPK pathway in RAS mutant cancers. Cold Spring Harb. Perspect. Med. 2018;8:a031492. doi: 10.1101/cshperspect.a031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantsar T. The current understanding of KRAS protein structure and dynamics. Comput. Struct. Biotechnol. J. 2020;18:189–198. doi: 10.1016/j.csbj.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zala D, et al. The advantage of channeling nucleotides for very processive functions. F1000Res. 2017;6:724. doi: 10.12688/f1000research.11561.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 10.Nimnual A, Bar-Sagi D. The two hats of SOS. Sci. STKE. 2002;2002:PE36. doi: 10.1126/stke.2002.145.pe36. [DOI] [PubMed] [Google Scholar]
- 11.Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–4541. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- 12.Gideon P, et al. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol. Cell Biol. 1992;12:2050–2056. doi: 10.1128/mcb.12.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheffzek K, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 14.Smith MJ, Ikura M. Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat. Chem. Biol. 2014;10:223–230. doi: 10.1038/nchembio.1435. [DOI] [PubMed] [Google Scholar]
- 15.Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium APG. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan EJ, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell JD, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox AD, Der CJ. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRASG12C somatic mutations across race, sex, and cancer type. N. Engl. J. Med. 2021;384:185–187. doi: 10.1056/NEJMc2030638. [DOI] [PubMed] [Google Scholar]
- 29.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo KY, Jelinsky SA, Loechler EL. Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat. Res. 2000;463:215–246. doi: 10.1016/s1383-5742(00)00047-8. [DOI] [PubMed] [Google Scholar]
- 31.Le Calvez F, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 32.Ihle NT, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J. Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu HA, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J. Thorac. Oncol. 2015;10:431–437. doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang S, et al. Impact of KRAS mutational variant on response to immunotherapy in metastatic NSCLC. J. Clin. Oncol. 2021;39:e21127. [Google Scholar]
- 35.Yang H, Liang SQ, Schmid RA, Peng RW. New horizons in KRAS-mutant lung cancer: dawn after darkness. Front. Oncol. 2019;9:953. doi: 10.3389/fonc.2019.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skoulidis F, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bange E, et al. Impact of KRAS and TP53 co-mutations on outcomes after first-line systemic therapy among patients with STK11-mutated advanced non-small-cell lung cancer. JCO Precis. Oncol. 2019;3:PO.18.00326. doi: 10.1200/PO.18.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero R, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krall EB, et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. eLife. 2017;6:e18970. doi: 10.7554/eLife.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricciuti B, et al. Diminished efficacy of programmed death-(ligand)1 inhibition in STK11- and KEAP1-mutant lung adenocarcinoma is affected by KRAS mutation status. J. Thorac. Oncol. 2022;17:399–410. doi: 10.1016/j.jtho.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arbour KC, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin. Cancer Res. 2018;24:334–340. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitajima S, et al. Overcoming resistance to dual innate immune and MEK inhibition downstream of KRAS. Cancer Cell. 2018;34:439–452.e436. doi: 10.1016/j.ccell.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong GS, et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat. Med. 2018;24:968–977. doi: 10.1038/s41591-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tajan M, Paccoud R, Branka S, Edouard T, Yart A. The RASopathy family: consequences of germline activation of the RAS/MAPK pathway. Endocr. Rev. 2018;39:676–700. doi: 10.1210/er.2017-00232. [DOI] [PubMed] [Google Scholar]
- 45.Johnson CW, et al. The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects. J. Biol. Chem. 2017;292:12981–12993. doi: 10.1074/jbc.M117.778886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter JC, et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 47.Mazhab-Jafari MT, et al. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl Acad. Sci. USA. 2015;112:6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandaru P, et al. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife. 2017;6:e27810. doi: 10.7554/eLife.27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gremer L, et al. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011;32:33–43. doi: 10.1002/humu.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fedele C, et al. SHP2 inhibition diminishes KRASG12C cycling and promotes tumor microenvironment remodeling. J. Exp. Med. 2021;218:e20201414. doi: 10.1084/jem.20201414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gebregiworgis T, et al. The Q61H mutation decouples KRAS from upstream regulation and renders cancer cells resistant to SHP2 inhibitors. Nat. Commun. 2021;12:6274. doi: 10.1038/s41467-021-26526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herrmann C, Martin GA, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J. Biol. Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 53.Block C, Janknecht R, Herrmann C, Nassar N, Wittinghofer A. Quantitative structure–activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat. Struct. Biol. 1996;3:244–251. doi: 10.1038/nsb0396-244. [DOI] [PubMed] [Google Scholar]
- 54.Vetter IR, et al. Structural and biochemical analysis of Ras-effector signaling via RalGDS. FEBS Lett. 1999;451:175–180. doi: 10.1016/s0014-5793(99)00555-4. [DOI] [PubMed] [Google Scholar]
- 55.Wohlgemuth S, et al. Recognizing and defining true ras binding domains I: biochemical analysis. J. Mol. Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 56.Li C, et al. The G protein signaling regulator RGS3 enhances the GTPase activity of KRAS. Science. 2021;374:197–201. doi: 10.1126/science.abf1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zafra MP, et al. An in vivo kras allelic series reveals distinct phenotypes of common oncogenic variants. Cancer Discov. 2020;10:1654–1671. doi: 10.1158/2159-8290.CD-20-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cook JH, Melloni GEM, Gulhan DC, Park PJ, Haigis KM. The origins and genetic interactions of KRAS mutations are allele- and tissue-specific. Nat. Commun. 2021;12:1808. doi: 10.1038/s41467-021-22125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Roock W, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 60.Hong DS, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westcott PM, To MD. The genetics and biology of KRAS in lung cancer. Chin. J. Cancer. 2013;32:63–70. doi: 10.5732/cjc.012.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.John J, et al. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29:6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 63.Smyth LA, Collins I. Measuring and interpreting the selectivity of protein kinase inhibitors. J. Chem. Biol. 2009;2:131–151. doi: 10.1007/s12154-009-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu S, Jang H, Nussinov R, Zhang J. The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci. Rep. 2016;6:21949. doi: 10.1038/srep21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janes MR, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172:578–589 e517. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 67.Skoulidis F, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hallin J, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka N, et al. Clinical acquired resistance to KRAS(G12C) inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021;11:1913–1922. doi: 10.1158/2159-8290.CD-21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Bryan JP. Pharmacological targeting of RAS: recent success with direct inhibitors. Pharmacol. Res. 2019;139:503–511. doi: 10.1016/j.phrs.2018.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim SM, et al. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew. Chem. Int. Ed. Engl. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patricelli MP, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 74.Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat. Rev. Drug Discov. 2016;15:771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 75.Lake D, Correa SA, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol. Life Sci. 2016;73:4397–4413. doi: 10.1007/s00018-016-2297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fakih M, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J. Clin. Oncol. 2019;37:3003. [Google Scholar]
- 77.Dy G, et al. Abstract CT008: Long-term outcomes with sotorasib in pretreated KRASp.G12C-mutated NSCLC: 2-year analysis of CodeBreaK100. Proc. 113th Annu. Meet. Am. Assoc. Cancer Res. 2022 doi: 10.1158/1538-7445.AM2022-CT008. [DOI] [Google Scholar]
- 78.Strickler JH, et al. First data for sotorasib in patients with pancreatic cancer with KRAS p.G12C mutation: a phase I/II study evaluating efficacy and safety. J. Clin. Oncol. 2022;40:360490. [Google Scholar]
- 79.Fakih MG, et al. Sotorasib for previously treated colorectal cancers with KRASG12C mutation (CodeBreaK100): a prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol. 2022;23:115–124. doi: 10.1016/S1470-2045(21)00605-7. [DOI] [PubMed] [Google Scholar]
- 80.Reck M, et al. MO01.32 CodeBreaK 200: a phase 3 multicenter study of sotorasib, a KRAS(G12C) inhibitor, versus docetaxel in patients with previously treated advanced non-small cell lung cancer (NSCLC) harboring KRAS p.G12C mutation. J. Thorac. Oncol. 2021;16:S29. [Google Scholar]
- 81.Riely GJ, et al. 99O_PR KRYSTAL-1: activity and preliminary pharmacodynamic (PD) analysis of adagrasib (MRTX849) in patients (Pts) with advanced non–small cell lung cancer (NSCLC) harboring KRASG12C mutation. J. Thorac. Oncol. 2021;16:S751–S752. [Google Scholar]
- 82.Ou S-HI, et al. First-in-human phase I/IB dose-finding study of adagrasib (MRTX849) in patients with advanced KRASG12C solid tumors (KRYSTAL-1) J. Clin. Oncol. 2022 doi: 10.1200/JCO.21.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jänne PA, et al. Adagrasib in non–small-cell lung cancer harboring a KRASG12C mutation. N. Engl. J. Med. 2022;387:120–131. doi: 10.1056/NEJMoa2204619. [DOI] [PubMed] [Google Scholar]
- 84.Bekaii-Saab TS, et al. KRYSTAL-1: updated activity and safety of adagrasib (MRTX849) in patients (Pts) with unresectable or metastatic pancreatic cancer (PDAC) and other gastrointestinal (GI) tumors harboring a KRASG12C mutation. J. Clin. Oncol. 2022;40(4 Suppl.):Abstr. 519. [Google Scholar]
- 85.Weiss J, et al. LBA6 KRYSTAL-1: adagrasib (MRTX849) as monotherapy or combined with cetuximab (Cetux) in patients (Pts) with colorectal cancer (CRC) harboring a KRASG12C mutation. Ann. Oncol. 2021;32(suppl. 5):S1294–S1346. [Google Scholar]
- 86.Brachmann SM, et al. JDQ443, a covalent irreversible inhibitor of KRAS G12C, exhibits a novel binding mode and demonstrates potent anti-tumor activity and favorable pharmacokinetic properties in preclinical models. Mol. Cancer Ther. 2021;20(12 Suppl.):Abstr. P124. [Google Scholar]
- 87.Hamarsheh S, Gross O, Brummer T, Zeiser R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun. 2020;11:5439. doi: 10.1038/s41467-020-19288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai YS, et al. Rapid idiosyncratic mechanisms of clinical resistance to KRAS G12C inhibition. J. Clin. Invest. 2022;132:e155523. doi: 10.1172/JCI155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Maldegem F, et al. Characterisation of tumour microenvironment remodelling following oncogene inhibition in preclinical studies with imaging mass cytometry. Nat. Commun. 2021;12:5906. doi: 10.1038/s41467-021-26214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ahronian LG, Corcoran RB. Strategies for monitoring and combating resistance to combination kinase inhibitors for cancer therapy. Genome Med. 2017;9:37. doi: 10.1186/s13073-017-0431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Konieczkowski DJ, Johannessen CM, Garraway LA. A convergence-based framework for cancer drug resistance. Cancer Cell. 2018;33:801–815. doi: 10.1016/j.ccell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 2018;15:709–720. doi: 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 93.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther. Adv. Med. Oncol. 2016;8:48–56. doi: 10.1177/1758834015616934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wainberg ZA, et al. A multi-arm phase I study of the PI3K/mTOR inhibitors PF-04691502 and gedatolisib (PF-05212384) plus irinotecan or the MEK inhibitor PD-0325901 in advanced cancer. Target. Oncol. 2017;12:775–785. doi: 10.1007/s11523-017-0530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grilley-Olson JE, et al. A phase Ib dose-escalation study of the MEK inhibitor trametinib in combination with the PI3K/mTOR inhibitor GSK2126458 in patients with advanced solid tumors. Invest. N. Drugs. 2016;34:740–749. doi: 10.1007/s10637-016-0377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watanabe T, et al. Heterogeneity of KRAS status may explain the subset of discordant KRAS status between primary and metastatic colorectal cancer. Dis. Colon Rectum. 2011;54:1170–1178. doi: 10.1097/DCR.0b013e31821d37a3. [DOI] [PubMed] [Google Scholar]
- 97.Sun L, et al. Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J. Exp. Clin. Cancer Res. 2011;30:30. doi: 10.1186/1756-9966-30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmid K, et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin. Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 99.Alsdorf WH, et al. Intratumoral heterogeneity of KRAS mutation is rare in non-small-cell lung cancer. Exp. Mol. Pathol. 2013;94:155–159. doi: 10.1016/j.yexmp.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 100.Zhao Y, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature. 2021;599:679–683. doi: 10.1038/s41586-021-04065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cannataro VL, et al. Heterogeneity and mutation in KRAS and associated oncogenes: evaluating the potential for the evolution of resistance to targeting of KRAS G12C. Oncogene. 2018;37:2444–2455. doi: 10.1038/s41388-017-0105-z. [DOI] [PubMed] [Google Scholar]
- 102.Canon J, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 103.Pantsar T. KRAS(G12C)-AMG 510 interaction dynamics revealed by all-atom molecular dynamics simulations. Sci. Rep. 2020;10:11992. doi: 10.1038/s41598-020-68950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koga T, et al. KRAS secondary mutations that confer acquired resistance to KRAS G12C inhibitors, sotorasib and adagrasib, and overcoming strategies: insights from in vitro experiments. J. Thorac. Oncol. 2021;16:1321–1332. doi: 10.1016/j.jtho.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 105.Li BT, et al. Largest evaluation of acquired resistance to sotorasib in KRAS p.G12C-mutated non–small cell lung cancer (NSCLC) and colorectal cancer (CRC): plasma biomarker analysis of CodeBreaK100. J. Clin. Oncol. 2022;40(16 Suppl.):Abstr. 102. [Google Scholar]
- 106.Awad MM, et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feng S, et al. A saturation mutagenesis screen uncovers resistant and sensitizing secondary KRAS mutations to clinical KRASG12C inhibitors. Proc. Natl Acad. Sci. USA. 2022;119:e2120512119. doi: 10.1073/pnas.2120512119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat. Rev. Cancer. 2015;15:577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 109.Tripathi R, et al. Combating acquired resistance to MAPK inhibitors in melanoma by targeting Abl1/2-mediated reactivation of MEK/ERK/MYC signaling. Nat. Commun. 2020;11:5463. doi: 10.1038/s41467-020-19075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xue JY, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 2020;577:421–425. doi: 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fedele C, et al. SHP2 inhibition prevents adaptive resistance to MEK inhibitors in multiple cancer models. Cancer Discov. 2018;8:1237–1249. doi: 10.1158/2159-8290.CD-18-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ryan MB, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRASG12C inhibition. Clin. Cancer Res. 2020;26:1633–1643. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Misale S, et al. KRAS G12C NSCLC models are sensitive to direct targeting of KRAS in combination with PI3K inhibition. Clin. Cancer Res. 2019;25:796–807. doi: 10.1158/1078-0432.CCR-18-0368. [DOI] [PubMed] [Google Scholar]
- 114.Yang J, et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020;21:341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weng C-H, et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38:455–468. doi: 10.1038/s41388-018-0454-2. [DOI] [PubMed] [Google Scholar]
- 116.Soucheray M, et al. Intratumoral heterogeneity in EGFR-mutant NSCLC results in divergent resistance mechanisms in response to EGFR tyrosine kinase inhibition. Cancer Res. 2015;75:4372–4383. doi: 10.1158/0008-5472.CAN-15-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adachi Y, et al. Epithelial-to-mesenchymal transition is a cause of both intrinsic and acquired resistance to KRAS G12C inhibitor in KRAS G12C-mutant non-small cell lung cancer. Clin. Cancer Res. 2020;26:5962–5973. doi: 10.1158/1078-0432.CCR-20-2077. [DOI] [PubMed] [Google Scholar]
- 119.Levin PA, et al. Histologic transformation from adenocarcinoma to squamous cell carcinoma as a mechanism of resistance to EGFR inhibition. J. Thorac. Oncol. 2015;10:e86–e88. doi: 10.1097/JTO.0000000000000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Q, Wu L, Zhang S. Transformation of advanced lung adenocarcinoma to acquired T790M resistance mutation adenosquamous carcinoma following tyrosine kinase inhibitor: a case report. Tumori. 2021;107:NP5–NP10. doi: 10.1177/0300891620973262. [DOI] [PubMed] [Google Scholar]
- 121.Amodio V, et al. EGFR blockade reverts resistance to KRAS(G12C) inhibition in colorectal cancer. Cancer Discov. 2020;10:1129–1139. doi: 10.1158/2159-8290.CD-20-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun C, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 123.Turke AB, et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res. 2012;72:3228–3237. doi: 10.1158/0008-5472.CAN-11-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moll HP, et al. Afatinib restrains K-RAS-driven lung tumorigenesis. Sci. Transl. Med. 2018;10:eaao2301. doi: 10.1126/scitranslmed.aao2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fakih M, et al. Trial in progress: a phase Ib study of AMG 510, a specific and irreversible KRASG12C inhibitor, in combination with other anticancer therapies in patients with advanced solid tumors harboring KRAS p.G12C mutation (CodeBreak 101) J. Clin. Oncol. 2020;38:TPS3661. [Google Scholar]
- 126.Gandara D, et al. Abstract P05-02: A phase 1b study evaluating the combination of sotorasib, a KRASG12C inhibitor, and afatinib, a pan-ErbB tyrosine kinase inhibitor, in advanced KRAS p.G12C mutated non-small cell lung cancer (NSCLC) Mol. Cancer Ther. 2021;20(12 Suppl.):Abstr. P05-02. [Google Scholar]
- 127.Freeman RM, Plutzky J, Neel BG. Identification of a human src homology 2-containing protein tyrosine-phosphatase: a putative homolog of Drosophila corkscrew. Proc. Natl Acad. Sci. USA. 1992;89:11239–11243. doi: 10.1073/pnas.89.23.11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ran H, Tsutsumi R, Araki T, Neel BG. Sticking it to cancer with molecular glue for SHP2. Cancer Cell. 2016;30:194–196. doi: 10.1016/j.ccell.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan, G. & Neel, B. G. in Protein Tyrosine Phosphatases in Cancer (eds Neel, B. G. & Tonks, N. K.) 115–143 (Springer, 2016).
- 130.Pandey R, Saxena M, Kapur R. Role of SHP2 in hematopoiesis and leukemogenesis. Curr. Opin. Hematol. 2017;24:307–313. doi: 10.1097/MOH.0000000000000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang J, Zhang F, Niu R. Functions of Shp2 in cancer. J. Cell. Mol. Med. 2015;19:2075–2083. doi: 10.1111/jcmm.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Neel BG, Gu H, Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 133.Mainardi S, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat. Med. 2018;24:961–967. doi: 10.1038/s41591-018-0023-9. [DOI] [PubMed] [Google Scholar]
- 134.Ruess DA, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med. 2018;24:954–960. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 135.Nichols RJ, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 2018;20:1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sabari JK, et al. KRYSTAL-2: a phase I/II trial of adagrasib (MRTX849) in combination with TNO155 in patients with advanced solid tumors with KRAS G12C mutation. J. Clin. Oncol. 2021;39:TPS146. [Google Scholar]
- 137.Hofmann MH, et al. BI-3406, a potent and selective SOS1-KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 2021;11:142–157. doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Salgia R, Pharaon R, Mambetsariev I, Nam A, Sattler M. The improbable targeted therapy: KRAS as an emerging target in non-small cell lung cancer (NSCLC) Cell Rep. Med. 2021;2:100186. doi: 10.1016/j.xcrm.2020.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gerlach D, et al. Abstract 1091: BI-3406 and BI 1701963: potent and selective SOS1::KRAS inhibitors induce regressions in combination with MEK inhibitors or irinotecan. Cancer Res. 2020;80(16 Suppl.):Abstr. 1091. [Google Scholar]
- 140.Hillig RC, et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS–SOS1 interaction. Proc. Natl Acad. Sci. USA. 2019;116:2551–2560. doi: 10.1073/pnas.1812963116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Plangger A, Rath B, Hochmair M, Funovics M, Hamilton G. Cytotoxicity of combinations of the pan-KRAS inhibitor BAY-293 against primary non-small lung cancer cells. Transl. Oncol. 2021;14:101230. doi: 10.1016/j.tranon.2021.101230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc. Natl Acad. Sci. USA. 1994;91:7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Friday BB, et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 2008;68:6145–6153. doi: 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- 144.Lito P, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ramalingam S, et al. Abstract P05-01: A phase 1b study evaluating the safety and efficacy of sotorasib, a KRASG12C inhibitor, in combination with trametinib, a MEK inhibitor, in KRAS p.G12C-mutated solid tumors. Mol. Cancer Ther. 2021;20(12 Suppl.):Abstr. P05-01. [Google Scholar]
- 146.Brown WS, et al. Overcoming adaptive resistance to KRAS and MEK inhibitors by co-targeting mTORC1/2 complexes in pancreatic cancer. Cell Rep. Med. 2020;1:100131. doi: 10.1016/j.xcrm.2020.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Burstein HJ. Systemic therapy for estrogen receptor-positive, HER2-negative breast cancer. N. Engl. J. Med. 2020;383:2557–2570. doi: 10.1056/NEJMra1307118. [DOI] [PubMed] [Google Scholar]
- 148.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016;13:417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 149.Lou K, et al. KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Sci. Signal. 2019;12:eaaw9450. doi: 10.1126/scisignal.aaw9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Willems E, et al. The functional diversity of Aurora kinases: a comprehensive review. Cell Div. 2018;13:7. doi: 10.1186/s13008-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Al-Khafaji ASK, et al. AURKA mRNA expression is an independent predictor of poor prognosis in patients with non-small cell lung cancer. Oncol. Lett. 2017;13:4463–4468. doi: 10.3892/ol.2017.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dos Santos EO, Carneiro-Lobo TC, Aoki MN, Levantini E, Basseres DS. Aurora kinase targeting in lung cancer reduces KRAS-induced transformation. Mol. Cancer. 2016;15:12. doi: 10.1186/s12943-016-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee JW, Kim S, Yang C, Burtness B. Abstract P078: Aurora A kinase inhibition with VIC-1911 overcomes intrinsic and acquired resistance to KRASG12C inhibition in KRAS(G12C)-mutated lung cancer. Mol. Cancer Ther. 2021;20(12 Suppl.):Abstr. P078. [Google Scholar]
- 154.De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJF, Würdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin. Cancer Res. 2011;17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 155.Caiola E, et al. Wee1 inhibitor MK1775 sensitizes KRAS mutated NSCLC cells to sorafenib. Sci. Rep. 2018;8:948. doi: 10.1038/s41598-017-18900-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dias Carvalho P, et al. KRAS oncogenic signaling extends beyond cancer cells to orchestrate the microenvironment. Cancer Res. 2018;78:7–14. doi: 10.1158/0008-5472.CAN-17-2084. [DOI] [PubMed] [Google Scholar]
- 157.Ischenko I, et al. KRAS drives immune evasion in a genetic model of pancreatic cancer. Nat. Commun. 2021;12:1482. doi: 10.1038/s41467-021-21736-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Y, et al. Targeting the SHP2 phosphatase promotes vascular damage and inhibition of tumor growth. EMBO Mol. Med. 2021;13:e14089. doi: 10.15252/emmm.202114089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tang KH, et al. Combined Inhibition of SHP2 and CXCR1/2 promotes antitumor T-cell response in NSCLC. Cancer Discov. 2022;12:47–61. doi: 10.1158/2159-8290.CD-21-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shi Y. Regulatory mechanisms of PD-L1 expression in cancer cells. Cancer Immunol. Immunother. 2018;67:1481–1489. doi: 10.1007/s00262-018-2226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ettinger DS, et al. NCCN guidelines insights: non-small cell lung cancer, version 2.2021. J. Natl Compr. Canc Netw. 2021;19:254–266. doi: 10.6004/jnccn.2021.0013. [DOI] [PubMed] [Google Scholar]
- 162.Briere DM, et al. The KRAS(G12C) inhibitor MRTX849 reconditions the tumor immune microenvironment and sensitizes tumors to checkpoint inhibitor therapy. Mol. Cancer Ther. 2021;20:975–985. doi: 10.1158/1535-7163.MCT-20-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yamamoto K, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature. 2020;581:100–105. doi: 10.1038/s41586-020-2229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kinsey CG, et al. Protective autophagy elicited by RAF–>MEK–>ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019;25:620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Di Mitri D, et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 2014;515:134–137. doi: 10.1038/nature13638. [DOI] [PubMed] [Google Scholar]
- 168.Highfill SL, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 2014;6:237ra267. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ijichi H, et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J. Clin. Invest. 2011;121:4106–4117. doi: 10.1172/JCI42754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jamieson T, et al. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J. Clin. Invest. 2012;122:3127–3144. doi: 10.1172/JCI61067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Katoh H, et al. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell. 2013;24:631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Greene S, et al. Inhibition of MDSC trafficking with SX-682, a CXCR1/2 inhibitor, enhances NK-cell immunotherapy in head and neck cancer models. Clin. Cancer Res. 2020;26:1420–1431. doi: 10.1158/1078-0432.CCR-19-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang Z, et al. GTP-state-selective cyclic peptide ligands of K-Ras(G12D) block its interaction with Raf. ACS Cent. Sci. 2020;6:1753–1761. doi: 10.1021/acscentsci.0c00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sogabe S, et al. Crystal structure of a human K-Ras G12D mutant in complex with GDP and the cyclic inhibitory peptide KRpep-2d. ACS Med. Chem. Lett. 2017;8:732–736. doi: 10.1021/acsmedchemlett.7b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang X, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRAS(G12D) inhibitor. J. Med. Chem. 2022;65:3123–3133. doi: 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- 177.Vasta JD, et al. KRAS is vulnerable to reversible switch-II pocket engagement in cells. Nat. Chem. Biol. 2022;18:596–604. doi: 10.1038/s41589-022-00985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kessler D, et al. Drugging an undruggable pocket on KRAS. Proc. Natl Acad. Sci. USA. 2019;116:15823–15829. doi: 10.1073/pnas.1904529116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tran TH, et al. The small molecule BI-2852 induces a nonfunctional dimer of KRAS. Proc. Natl Acad. Sci. USA. 2020;117:3363–3364. doi: 10.1073/pnas.1918164117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.McGee JH, et al. Exceptionally high-affinity Ras binders that remodel its effector domain. J. Biol. Chem. 2018;293:3265–3280. doi: 10.1074/jbc.M117.816348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schreiber SL. The rise of molecular glues. Cell. 2021;184:3–9. doi: 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- 182.Nichols RJ, et al. Abstract 3595: RMC-6291, a next-generation tri-complex KRASG12C (ON) inhibitor, outperforms KRASG12C (OFF) inhibitors in preclinical models of KRASG12C cancers. Cancer Res. 2022;82(Suppl. 12):Abstr. 3595. [Google Scholar]
- 183.Koltun E, et al. Abstract 1260: First-in-class, orally bioavailable KRASG12V(ON) tri-complex inhibitors, as single agents and in combinations, drive profound anti-tumor activity in preclinical models of KRASG12V mutant cancers. Cancer Res. 2021;81(13 Suppl.):Abstr. 1260. [Google Scholar]
- 184.Gustafson WC, et al. Direct targeting of RAS in pancreatic ductal adenocarcinoma with RMC-6236, a first-in-class, RAS-selective, orally bioavailable, tri-complex RASMULTI(ON) inhibitor. J. Clin. Oncol. 2022;40(4 Suppl.):Abstr. 591. [Google Scholar]
- 185.Koltun ES, et al. Abstract 3597: Direct targeting of KRASG12X mutant cancers with RMC-6236, a first-in-class, RAS-selective, orally bioavailable, tri-complex RASMULTI(ON) inhibitor. Cancer Res. 2022;82(12 Suppl.):Abstr. 3597. [Google Scholar]
- 186.Zhang Z, Shokat KM. Bifunctional small-molecule ligands of K-Ras induce its association with immunophilin proteins. Angew. Chem. Int. Ed. Engl. 2019;58:16314–16319. doi: 10.1002/anie.201910124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bond MJ, Chu L, Nalawansha DA, Li K, Crews CM. Targeted degradation of oncogenic KRAS(G12C) by VHL-recruiting PROTACs. ACS Cent. Sci. 2020;6:1367–1375. doi: 10.1021/acscentsci.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bery N, Miller A, Rabbitts T. A potent KRAS macromolecule degrader specifically targeting tumours with mutant KRAS. Nat. Commun. 2020;11:3233. doi: 10.1038/s41467-020-17022-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lim S, et al. Exquisitely specific anti-KRAS biodegraders inform on the cellular prevalence of nucleotide-loaded states. ACS Cent. Sci. 2021;7:274–291. doi: 10.1021/acscentsci.0c01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Poongavanam V, Kihlberg J. PROTAC cell permeability and oral bioavailability: a journey into uncharted territory. Future Med. Chem. 2022;14:123–126. doi: 10.4155/fmc-2021-0208. [DOI] [PubMed] [Google Scholar]
- 191.Vidimar V, et al. Proteolytic pan-RAS cleavage leads to tumor regression in patient-derived pancreatic cancer xenografts. Mol. Cancer Ther. 2022;21:810–820. doi: 10.1158/1535-7163.MCT-21-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bear AS, et al. Biochemical and functional characterization of mutant KRAS epitopes validates this oncoprotein for immunological targeting. Nat. Commun. 2021;12:4365. doi: 10.1038/s41467-021-24562-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hall V, et al. Protection against SARS-CoV-2 after Covid-19 vaccination and previous infection. N. Engl. J. Med. 2022;386:1207–1220. doi: 10.1056/NEJMoa2118691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nagasaka M. ES28.04 emerging mechanisms to target KRAS directly. J. Thorac. Oncol. 2021;16:S96–S97. [Google Scholar]
- 197.Pant S, et al. First-in-human phase 1 trial of ELI-002 immunotherapy as treatment for subjects with Kirsten rat sarcoma (KRAS)-mutated pancreatic ductal adenocarcinoma and other solid tumors. J. Clin. Oncol. 2022;40(16 Suppl.):Abstr. TPS2701. [Google Scholar]
- 198.Douglass J, et al. Bispecific antibodies targeting mutant RAS neoantigens. Sci. Immunol. 2021;6:eabd5515. doi: 10.1126/sciimmunol.abd5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nathan P, et al. Overall survival benefit with Tebentafusp in metastatic uveal melanoma. N. Engl. J. Med. 2021;385:1196–1206. doi: 10.1056/NEJMoa2103485. [DOI] [PubMed] [Google Scholar]
- 200.Mehta A, Dalle Vedove E, Isert L, Merkel OM. Targeting KRAS mutant lung cancer cells with siRNA-loaded bovine serum albumin nanoparticles. Pharm. Res. 2019;36:133. doi: 10.1007/s11095-019-2665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Strand MS, et al. Precision delivery of RAS-inhibiting siRNA to KRAS driven cancer via peptide-based nanoparticles. Oncotarget. 2019;10:4761–4775. doi: 10.18632/oncotarget.27109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tirella A, et al. CD44 targeted delivery of siRNA by using HA-decorated nanotechnologies for KRAS silencing in cancer treatment. Int. J. Pharm. 2019;561:114–123. doi: 10.1016/j.ijpharm.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 203.Bradley CA. Pancreatic cancer: iExosomes target the ‘undruggable’. Nat. Rev. Cancer. 2017;17:452–453. doi: 10.1038/nrc.2017.54. [DOI] [PubMed] [Google Scholar]
- 204.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.