Abstract
Salmonella enterica subsp. enterica serotype Enteritidis BM4361 and BM4362 were isolated from the same patient. BM4361 was susceptible to aminoglycosides, whereas BM4362 was resistant to tobramycin owing to synthesis of a 6′-N-acetyltransferase type I [AAC(6′)-I]. Comparative analysis of nucleotide sequences, pulsed-field gel electrophoresis patterns, and Southern hybridizations indicated that the chromosomal aac(6′)-Iy genes for the enzyme in both strains were identical and that BM4362 derived from BM4361 following a ca. 60-kb deletion that occurred 1.5 kb upstream from the resistance gene. Northern hybridizations showed that aac(6′)-Iy was silent in BM4361 and highly expressed in BM4362 due to a transcriptional fusion. Primer extension mapping identified the transcriptional start site for aac(6′)-Iy in BM4362: 5 bp downstream from the promoter of the nmpC gene. Study of the distribution of aac(6′)-Iy by PCR and Southern hybridization with a specific probe indicated that the gene, although not found in S. enterica subsp. arizonae, was specific for Salmonella. In this bacterial genus, aac(6′)-Iy was located downstream from a cluster of seven open reading frames analogous to an Escherichia coli locus that encodes enzymes putatively involved in carbohydrate transport or metabolism. This genomic environment suggests a role in the catabolism of a specific sugar for AAC(6′)-Iy in Salmonella.
Bacterial resistance to aminoglycosides is mainly due to enzymatic detoxification of the drugs. The corresponding genes are often part of plasmids (11) or transposons (17), a structural feature which accounts for the dissemination of resistance. However, in recent years, a number of aminoglycoside resistance genes, in particular those encoding acetyltransferases, were found to be chromosomal and species specific both in gram-negative (33) and gram-positive bacteria (10), including mycobacteria (1). The presence of these genes does not correlate with resistance since they are often weakly expressed or not expressed (24, 33). Aminoglycoside resistance in these strains is usually secondary to increased gene expression following regulatory mutations (24).
Acetyltransferases are involved in a variety of cellular processes including acetylation of ribosomal proteins (38), of peptidoglycan (13), and of numerous intermediates in sugar metabolic pathways (18). To account for the diversity and ubiquity of aminoglycoside acetyltransferases, it has been proposed that certain of them were derived from enzymes involved in the primary or intermediary metabolism of bacteria (21, 23). The first evidence for this notion came from the contribution of aminoglycoside acetyltransferase AAC(2′)-Ia to the O acetylation of the peptidoglycan in Providencia stuartii (20).
Until now, aminoglycoside resistance by inactivation in Salmonella spp. was attributed to the acquisition of exogenous DNA (4, 16). In this report, we have analyzed Salmonella enterica subsp. enterica serotype Enteritidis BM4361 and BM4362, which were isolated from the same patient and which differed in their susceptibilities to aminoglycosides. We have characterized the chromosomal aac(6′)-Iy gene in these strains and the molecular rearrangement responsible for its expression in the aminoglycoside-resistant strain BM4362. The distribution of this gene in the Salmonella genus and its genomic environment suggest that its product may play a physiological role in sugar metabolism.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Table 1. S. enterica subsp. enterica serotype Enteritidis BM4361 and BM4362 were isolated in 1996 from stool cultures of a patient at the Saint-Michel Hospital in Paris, France. The strains were grown in brain heart infusion broth and agar (Difco Laboratories, Detroit, Mich.) at 37°C. Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marnes-la-coquette, France). The MICs of aminoglycosides were determined by the method of Steers et al. (34).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| JM83 | F−ara Δ(lac-proAB) rpsL [F80dlacΔ(lacZ)M15] | 37 |
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X79 rpsL hsdR2(rK−mK+) mcrA mcrB1 | 36 |
| C600 | F− e14− (mcrA) thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21 | 3 |
| Cla | F− restriction defective | 31 |
| C. freundii | ||
| ATCC 8090 | Reference strain | 8 |
| Salmonella | ||
| BM4361 | S. enterica subsp. I serotype Enteridis; clinical isolate | This study |
| BM4362 | S. enterica subsp. I serotype Enteridis; clinical isolate; Tm | This study |
| LT2 | S. enterica subsp. I serotype Typhimurium | WHOCCSb |
| BM4410 | S. enterica subsp. II; 6.8:m.t:1.5 | WHOCCS |
| BM4411 | S. enterica subsp. IIIa; 21:f.z51 | WHOCCS |
| BM4412 | S. enterica subsp. IIIb; 11:1.u:z | WHOCCS |
| BM4413 | S. enterica subsp. IV; 6.7:z4.z24 | WHOCCS |
| BM4414 | S. enterica subsp. VI; 1.6.14.25:Q:e.u.x | WHOCCS |
| BM4415 | S. bongori; 44:r | WHOCCS |
| Plasmids | ||
| pAT713 | 2.2-kb SspI fragment from BM4361 cloned into pUC18 | This study |
| pAT714 | 1.6-kb SalI fragment from BM4361 cloned into pUC18 | This study |
| pAT715 | 1.3-kb PCR fragment from BM4361 cloned into pCR-Blunt vector; Tm | This study |
| pAT716 | 438-bp PCR fragment of aac(6′)-Iy from BM4361 cloned into pCR-Blunt vector | This study |
| pAT703 | 2.8-kb Sau3AI fragment from BM4362 cloned into pUC18; Tm | This study |
| pAT711 | 438-bp PCR fragment of aac(6′)-Iy from BM4362 cloned into pUC19; Tm | This study |
| pAT712 | 6-kb SalI-BamHI fragment from BM4362 cloned into pUC18 | This study |
| pAT718 | 1.17-kb HindIII PCR fragment of nmpC from BM4361 cloned into pSU19 | This study |
Tm, tobramycin resistance. For Salmonella, the antigenic formula is indicated.
WHOCCS, World Health Organization Collaborating Center for Salmonella.
Assay for aminoglycoside-acetylating enzymes.
Crude bacterial extracts were obtained by ultrasonic disruption and ultracentrifugation, and aminoglycoside-acetylating activity was searched for by the phosphocellulose paper-binding technique (14).
DNA manipulations.
Total DNA and the plasmid content of transformants were prepared as described previously (5). Purification of plasmid DNA was performed by using the Wizard Minipreps DNA kit (Promega, Madison, Wis.). Restriction by endonucleases was according to the supplier’s recommendations (Life Technologies Inc., Gaithersburg, Md.). Extraction of DNA fragments separated by agarose gel electrophoresis was carried out by using the Sephaglas BandPrep kit (Pharmacia Biotech, Saint-Quentin-en-Yvelines, France).
PCR was performed in a GeneAmp PCR system 2400 (Perkin-Elmer Cetus, Norwalk, Conn.) with Pfu DNA polymerase (Stratagene, La Jolla, Calif.) according to the manufacturer’s recommendations. Annealing steps were performed at 55°C with specific primers (Unité de Chimie Organique, Institut Pasteur, Paris, France).
For Southern hybridization, DNA fragments were transferred from agarose gel to Hybond N+ membrane (Amersham International, Little Chalfont, Buckinghamshire, England) by vacuum with a Trans Vac TE80 apparatus (Hoefer Scientific Instruments, San Francisco, Calif.). The amplification products used to generate the probes (Table 2) were labeled with [α-32P]dCTP (3,000 Ci/mmol; Amersham Radiochemical Center, Amersham, England) by using a nick translation kit (Amersham). Prehybridization and hybridization were performed under high- (65°C) or low- stringency (45°C) conditions (28).
TABLE 2.
Probes
| Probe | Positiona | Size (bp) | Location |
|---|---|---|---|
| A | 3034–3471 | 438 | aac(6′)-Iy |
| B1 | 1674–2294 | 621 | sgcEs 3′ end |
| B2 | 1552–2176 | 625 | Upstream from sgcAs to sgcEs 5′ end |
| C | 840–1534 | 695 | nmpC 3′ end |
| D | 112–688 | 577 | 140 bp upstream from the nmpC start codon |
| E | 3664–4190 | 527 | Downstream from aac(6′)-Iy |
| F | 731–1154 | 424 | Internal to sgcQs |
Preparation and digestion of embedded DNA in a 1% agarose block were performed as described previously (19). Large restriction fragments were separated by pulsed-field gel electrophoresis (PFGE) according to the recommendations of the supplier of the Autobase system for zero-integrated-field gel electrophoresis (TechGen, les Ulis, France).
Cloning and sequencing.
PCR products were cloned into pCR-Blunt vector (Zero Blunt cloning kit; Invitrogen Corp., San Diego, Calif.), pUC19, or pSU19 (7), and chromosomal DNA fragments were cloned into pUC18 (Table 1). Ligation reactions were performed with T4 DNA ligase (Pharmacia). Transformation of Escherichia coli was performed as described previously (28). Antibiotic concentrations for selection were as follows: ampicillin, 100 μg/ml; tobramycin, 8 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 12 μg/ml. When required, transformants were screened by dot blot hybridization. DNA was immobilized on Biotrans nylon membranes (ICN Pharmaceuticals Inc., Costa Mesa, Calif.) and hybridized with probes at 65°C as described previously (28). Sequencing reactions were performed by the dideoxynucleotide chain termination method (30) with T7 DNA polymerase (Pharmacia) and α-35S-dATP (400 Ci/mmol; Amersham Radiochemical Center). DNA fragments were resolved by electrophoresis on 6% vertical polyacrylamide gel with the Genomyx system (Beckman Instruments, Inc., Palo Alto, Calif.).
RNA techniques.
Total RNA was extracted from BM4361 and BM4362 grown to an optical density at 600 nm of 0.7, separated by electrophoresis on a formaldehyde agarose gel, transferred to a Hybond N+ membrane, and hybridized as described previously (2). Washes were performed twice with 1× SSC (0.15 M NaCl plus 15 mM sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at room temperature and twice with 0.1× SSC–0.1% SDS at 37°C, each for 20 min.
For primer extension, oligodeoxynucleotide O4 was 5′-end labeled with [γ-32P]ATP (6,000 Ci/mmol; Amersham Radiochemical Center) and T4 polynucleotide kinase (Amersham), 105 cpm was incubated with 50 μg of RNA overnight at 30°C, and extension was performed with 40 U of avian myeloblastosis virus reverse transcriptase (Boehringer, Mannheim, Germany) for 90 min at 42°C as described previously (2). Primer elongation products were analyzed by electrophoresis on 6% denaturating polyacrylamide gels.
Computer analysis of sequence data.
Nucleotide and amino acid sequence data were analyzed with the GCG sequence analysis software package, version 7 (Genetics Computer Group, Madison, Wis.). The GenBank and SwissProt databases were screened for sequence similarities.
Nucleotide sequence accession numbers.
The 5,327-bp sequence of BM4361 and the 4,819-bp sequence of BM4362 have been deposited in the GenBank data library (Los Alamos, N. Mex.) under accession no. AF144880 and AF144881, respectively.
RESULTS AND DISCUSSION
Phenotypes of S. enterica subsp. enterica serotype Enteritidis BM4361 and BM4362 towards aminoglycosides.
S. enterica subsp. enterica serotype Enteritidis BM4361 and BM4362 were isolated from the same patient. Strain BM4361 was susceptible to aminoglycosides, whereas BM4362 was resistant to tobramycin and dibekacin and had reduced susceptibility to netilmicin and amikacin (Table 3). Extracts from BM4361 were devoid of aminoglycoside acetyltransferase activity, whereas gentamicin C1a, dibekacin, amikacin, and 2′-N-ethylnetilmicin were acetylated by BM4362 extracts (data not shown). Since gentamicin C1a and 2′-N-ethylnetilmicin were modified and gentamicin C1 and 6′-N-ethylnetilmicin, each of which has a substitution at the 6′ position, were not, the 6′ amino group appears to be the site of acetylation. Thus, the resistance phenotype and the enzymatic substrate profile for BM4362 were consistent with production of a 6′-N-aminoglycoside acetyltransferase of type I [AAC(6′)-I].
TABLE 3.
Susceptibilities of strains to selected aminoglycosides
| Strain | MIC (μg/ml) of:
|
|||
|---|---|---|---|---|
| Amikacin | Gentamicin | Netilmicin | Tobramycin | |
| S. enterica BM4361 | 1.5 | 0.5 | 0.5 | 1 |
| S. enterica BM4362 | 8 | 1 | 2 | 16 |
| S. enterica BM4362/pAT718 | 8 | 1 | 2 | 16 |
| E. coli JM83 | 1 | ≤0.25 | ≤0.25 | ≤0.25 |
| E. coli JM83/pAT711 | 32 | ≤0.25 | 4 | 16 |
Characterization of the aac(6′)-Iy gene in BM4362.
Total DNA from BM4362 partially digested with Sau3AI and pUC18 DNA linearized by BamHI were mixed, ligated, and introduced into E. coli MC1061 by transformation. The smallest recombinant plasmid conferring resistance to tobramycin, pAT703, was found to contain a 2.8-kb Sau3AI insert (Fig. 1). MICs and the acetyltransferase substrate profile indicated that pAT703 conferred aminoglycoside resistance on the new host by synthesis of an AAC(6′)-I enzyme (data not shown). Nucleotide sequence analysis of the insert revealed three adjacent open reading frames (ORFs) (Fig. 1). A search of GenBank indicated that the main part of the central ORF was homologous to aac(6′)-I genes. A fragment delimited by the ATG and TGA codons at positions 3034 and 3469 (numbering in accordance with that for the sequence with GenBank accession no. AF144881) within this ORF was amplified by PCR from BM4362 DNA with oligodeoxynucleotides O1 and O2 and cloned into the SmaI site of pUC19, downstream from a ribosome binding site (RBS) and under the control of the plac promoter. The resulting recombinant plasmid, pAT711 (Fig. 1; Table 1), conferred to E. coli JM83 resistance to the expected set of aminoglycosides (Table 3) by production of a 6′-acetylating activity, which confirms that expression of this coding sequence, designated aac(6′)-Iy, was responsible for the aminoglycoside resistance of BM4362. Comparison of the deduced AAC(6′)-Iy sequence with those of proteins in the databases indicated that the closest enzyme, with 60% identity, was AAC(6′)-If, encoded on a plasmid in Enterobacter cloacae (35). The percentages of identity to the other AAC(6′)-I sequences found in gram-negative bacteria ranged from 40 to 50%.
FIG. 1.
Schematic representation of the environments of aac(6′)-Iy in BM4361 and BM4362 and of the 97.6-min chromosomal region of E. coli K12. Arrows indicate the direction of transcription. The ORFs upstream from aac(6′)-Iy in BM4361 (open arrows) had ca. 75% identity with the 97.6-min chromosomal region of E. coli K12. The nucleotides adjacent to the deletion are indicated. The inserts of recombinant plasmids are represented by lines between vertical lines, and the sequenced portions are indicated by thick lines. The oligodeoxynucleotides used for PCR amplification are indicated (O1 from 3034 to 3053, O2 from 3451 to 3470, and O3 from 2177 to 2196; the numbering is in accordance with that for the sequence with GenBank accession no. AF144881). Probes B1 and B2 used for screening recombinant plasmids and probes A to F used for Southern and Northern analyses are indicated. The ca. 60-kb deletion is indicated by a double-headed arrow.
Characterization of aac(6′)-Iy in BM4361.
The aac(6′)-Iy gene was detected in aminoglycoside-susceptible strain BM4361 by Southern hybridization with aac(6′)-Iy-specific probe A (Table 2 and data not shown). Nucleotide sequence determination of the PCR product obtained from BM4361 total DNA with primers O1 and O2 indicated perfect identity with the resistance gene in BM4362. These data suggest that the phenotypic change between BM4361 and BM4362 involved expression of the aac(6′)-Iy gene.
Comparison of the genomic environments of aac(6′)-Iy in BM4361 and BM4362.
The regions flanking the aac(6′)-Iy gene in BM4361 and BM4362 were studied by cloning overlapping purified PCR products and total DNA fragments, screened with probes B1 and B2 (Fig. 1; Table 2), which were then partially or entirely sequenced. Recombinant plasmids pAT713, pAT714, and pAT715 were used to determine the sequence of 3,534 contiguous base pairs upstream from aac(6′)-Iy in BM4361 (Fig. 1). A search for stop codons in the three reading frames of each DNA strand identified five ORFs with the same orientation as that of aac(6′-Iy, the upstream one being truncated at its 5′ end by the SspI cloning site. They had, from 5′ to 3′, 78, 78, 74, 65, and 78% identity to the 3′ portion of sgcC and the sgcQ, sgcA, sgcE, and sgcR genes of E. coli K12, respectively (9). These ORFs were named sgcCs, sgcQs, sgcAs, sgcEs, and sgcRs, respectively.
A BM4362 chromosomal region of 4,819 bp, including aac(6′)-Iy, was sequenced with plasmids pAT703 and pAT712 (Fig. 1), and four contiguous ORFs were identified. Two were located upstream from aac(6′)-Iy and were transcribed in the same direction. The ORF immediately upstream from aac(6′)-Iy corresponded to the sgcRs gene. Comparative analysis of the second ORF revealed a hybrid sequence with 721 bp at the 5′ end corresponding to the 5′ end of the nmpC gene from Salmonella (15) and 625 bp at the 3′ end composed of 32 bp from the 3′ end of sgcAs, 11 bp intergenic to sgcAs and sgcEs, and the entire sgcEs gene. The partially characterized nmpC gene (15) is located at 38.7 centisomes on the genetic map of S. enterica subsp. I serotype Typhimurium (29). The E. coli nmpC homologue was characterized as a cryptic porin gene (22). The presence of the hybrid nmpC-sgcEs ORF indicated that a genetic rearrangement occurred 1,482 bp upstream from the aac(6′)-Iy start codon. Expression of this hybrid ORF would result in synthesis of a fusion protein consisting of the N-terminal part of NmpC fused with the entire SgcEs by a junction containing 14 amino acids encoded by the sgcA end and the intergenic region. Downstream from aac(6′)-Iy, a perfect inverted-repeat (IR) sequence of 10 bp could constitute a rho-independent transcriptional termination signal. A fourth ORF (ORF D; Fig. 1) in opposite orientation relative to aac(6′)-Iy and encoding a protein with a C-terminal part 40% identical to that of E. coli l-lactate dehydrogenase was identified downstream from the IR sequence. In BM4361, the region downstream from aac(6′)-Iy was found by PCR to be similar to that in BM4362 (data not shown).
In summary, a genomic alteration between BM4361 and BM4362 upstream from aac(6′)-Iy was characterized. In BM4361, aac(6′)-Iy was distal to the sgcs cluster, which is homologous to the sgc cluster of E. coli K12 located at min 97.6. This cluster was truncated in BM4362 by a recombination event which generated a fused ORF between the nmpC and the sgcEs genes.
Characterization of a chromosomal deletion in BM4362.
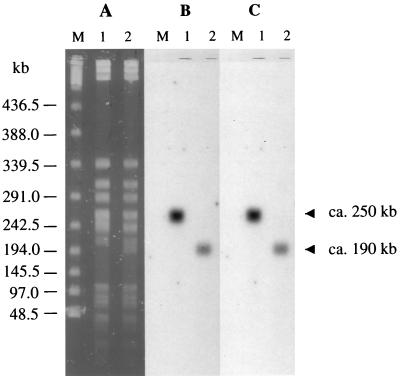
Total DNA from BM4361 and BM4362 restricted with XbaI was separated by PFGE and analyzed by Southern hybridization with aac(6′)-Iy probe A and nmpC 5′ end probe C (Fig. 2, Table 2). The two probes hybridized with single bands of 250 kb in BM4361 and of 190 kb in BM4362, indicating that BM4362 suffered a ca. 60-kb deletion internal to the 250-kb chromosomal fragment carrying both nmpC and aac(6′)-Iy. The sgcQs-specific probe F (Table 2) hybridized with the 250-kb fragment from BM4361 but not with the 190-kb band from BM4362, showing that sgcQs was included in the deletion (data not shown). The recombination event in BM4362 therefore appears to be the result of a deletion.
FIG. 2.
PFGE (A) and Southern hybridization (B and C) of total DNA from BM4361 (lanes 1) and BM4362 (lanes 2) restricted with XbaI and bacteriophage lambda concatamers (lanes M). (B) Hybridization with a probe for aac(6′)-Iy. (C) Hybridization with a probe for the nmpC 5′ end.
Transcriptional analysis of the aac(6′)-Iy gene.
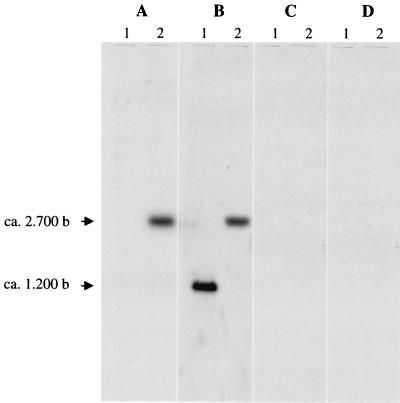
Total RNA from BM4361 and BM4362 was extracted from exponentially growing cells and analyzed by Northern hybridization with aac(6′)-Iy probe A (Fig. 3A). The lack of a detectable transcript in aminoglycoside-susceptible BM4361 indicated that aac(6′)-Iy was weakly expressed or not expressed in this strain. By contrast, a major transcript of approximately 2,700 nucleotides which cohybridized with nmpC 5′ end probe C (Table 2) was detected in BM4362, indicating that aac(6′)-Iy and the nmpC-sgcEs hybrid ORF were cotranscribed. This result also suggests that, in BM4361, expression of aac(6′)-Iy probably relied on the sgc cluster since there was no transcriptional termination signal upstream from aac(6′)-Iy. Lack of a transcript hybridizing with probes D and E (Fig. 3C and D; Table 2) indicated that the transcriptional start site of aac(6′)-Iy-specific mRNA was located between probes C and D and did not extend beyond that gene.
FIG. 3.
Analysis of aac(6′)-Iy transcription by Northern hybridization. Total RNA from BM4361 (lanes 1) and BM4362 (lanes 2) was hybridized with aac(6′)-Iy probe A (A), nmpC 5′ end probe C (B), probe D (C), and probe E (D) (Table 2). The sizes of the transcripts relative to the RNA molecular weight marker I were determined (Boehringer).
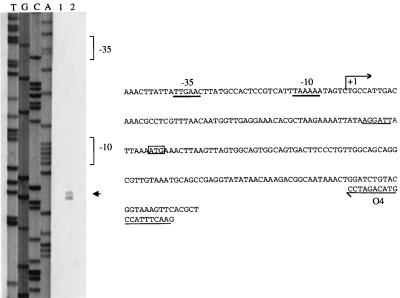
Based on these results, oligodeoxynucleotide O4 (Fig. 4B), complementary to the 5′ end of nmpC, was used as a primer for extension mapping. The 184-bp DNA fragment generated allowed exact positioning of the transcriptional start site (Fig. 4A). The −35 (TTGAAC) and −10 (TAAAAA) ς70 recognition sequences, separated by 17 bp, which formed the nmpC promoter (Fig. 4B) were then located by a computer search.
FIG. 4.
Identification of the transcriptional start site for aac(6′)-Iy in BM4362 by primer extension analysis. (Left panel) Lane 1, control without RNA; lane 2, primer elongation product obtained with oligodeoxynucleotide O4 and 50 μg of total RNA from BM4362 (arrowhead); lanes T, G, C, and A, results of sequencing reactions performed with pAT711 DNA as the template and O4 as the primer. (Right panel) Sequence from nucleotide positions 721 to 960 (numbering in accordance with that for sequence with GenBank accession no. AF144881). +1, transcriptional start site for aac(6′)-Iy mRNA in BM4362. The −35 and −10 promoter sequences upstream from the transcriptional start site are underlined with thick lines. The ATG start codon of nmpC is boxed, and the RBS is underlined with a thin line.
In addition, a ca. 1,200-nucleotide fragment corresponding in length to nmpC was detected in BM4361 by Northern hybridization with probe C (Fig. 3B), indicating that the gene was expressed in this strain, as opposed to being expressed in E. coli (6, 22). In order to test if aminoglycoside sensitivity in BM4362 was unaffected by loss of the NmpC porin, the corresponding gene and its RBS were amplified from BM4361 DNA and cloned into the HindIII site of pSU19 under the control of the plac promoter (Table 1). Strain BM4362 was then transformed with DNA of the resulting recombinant plasmid, pAT718. MICs of selected aminoglycosides for BM4362 and BM4362/pAT718 grown on medium containing IPTG (isopropyl-β-d-thiogalactopyranoside) (Table 3) indicated that NmpC had no effect on the aminoglycoside susceptibility of the host.
It thus appears that aac(6′)-Iy was cryptic in BM4361 and that its expression in BM4362 leading to aminoglycoside resistance was due to a transcriptional fusion secondary to a chromosomal deletion in which the downstream aac(6′)-Iy gene was placed under the control of the upstream nmpC promoter.
Distribution of the aac(6′)-Iy gene.
Total DNA from seven strains representative of the various species and subspecies of Salmonella was digested with PstI and studied by Southern hybridization with aac(6′)-Iy probe A. The aac(6′)-Iy gene was detected in all studied strains (Table 1), except for strains of S. enterica subsp. arizonae (data not shown). In addition, PCR with various primers specific for aac(6′)-Iy and the sgcs cluster indicated that, in every strain containing aac(6′)-Iy, the gene had the same genomic environment and that S. enterica subsp. arizonae did not harbor aac(6′)-Iy or the sgcs cluster (data not shown). No hybridization with aac(6′)-Iy, even under low-stringency conditions, was observed with total DNA from E. coli C1a, E. coli C600, and Citrobacter freundii ATCC 8090 (data not shown), the two bacterial genera phylogenetically most closely related to Salmonella (12). The aac(6′)-Iy gene appears, therefore, endogenous to and specific for Salmonella.
Salmonella bongori was formerly subspecies V of S. enterica, the only species of the genus. More recently, S. bongori strains were found to be the most divergent forms of Salmonella and thus were elevated to the species level (32). Since aac(6′)-Iy was present in both S. enterica and S. bongori, the gene must have appeared early in the evolution of the genus. In addition, the fact that aac(6′)-Iy was retained during evolution suggests a cellular function apart from aminoglycoside resistance for AAC(6′)-Iy. Nevertheless, aac(6′)-Iy does not appear to be essential since it is absent from S. enterica subsp. arizonae.
In conclusion, we have detected and characterized the cryptic aac(6′)-Iy gene endogenous to the Salmonella genus. In the deletion derivative BM4362, this gene was activated by a transcriptional fusion which led to aminoglycoside resistance. The genetic organization upstream from the aac(6′)-Iy gene (Fig. 1) and the transcriptional study of this gene in BM4362 (Fig. 3A) suggest that it is part of the sgcs cluster. The sequence of the homologous sgc locus in E. coli, which consists of seven ORFs, has been recently determined and analyzed (25, 26). Although the function of the deduced products remains unknown, they are related to enzymes involved in carbohydrate transport or metabolism. The gene organization, from 5′ to 3′, is sgcX encoding a homologue of FrvX, a protein of E. coli with an unknown function; sgcB and sgcC encoding homologues of galactitol-specific enzymes IIB and IIC of the phosphotransferase system (PTS), respectively; sgcQ encoding a protein with no homology to any sequence in the databases; sgcA encoding a protein homologous to the mannitol- and fructose-specific enzyme IIA of the PTS; sgcE encoding a pentulose-5-phosphate-3-epimerase homologue; and sgcR encoding a putative transcriptional regulatory protein which possesses the helix-turn-helix binding DNA motif of the DeoR family proteins. Interestingly, no aac(6′) gene is present at the 3′ extremity of this gene cluster in E. coli. The chromosomal environment of aac(6′)-Iy in Salmonella strongly suggests that the gene may play a physiological role in specific environmental conditions. Based on homology, the sgc cluster appears to deal with reception, transport, and degradation of a specific carbohydrate, presumably a pentose or a pentitol, and it is conceivable that the AAC(6′)-Iy activity may be part of this catabolic pathway. However, secondary functions attributed more recently to the PTS, including various ramifications of metabolic and transcriptional regulation (27), could also be envisaged for sgc. Whatever its true role, aac(6′)-Iy may turn out to be an interesting tool, as a reporter, to study the expression of the sgcs cluster in Salmonella.
ACKNOWLEDGMENTS
We thank M. Popoff for the gift of Salmonella strains, M.-C. Ploy for help with PCR, and M. Arthur for helpful discussions.
This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Diseases. S.M. was a recipient of a doctoral fellowship from the Ministère de l’Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Ainsa J A, Pérez E, Pelicic V, Berthet F X, Gicquel B, Martin C. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of aac(2′)-Ic from Mycobacterium tuberculosis and aac(2′)-Id gene from Mycobacterium smegmatis. Mol Microbiol. 1997;24:431–441. doi: 10.1046/j.1365-2958.1997.3471717.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhartdt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium, cellular and molecular microbiology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 4.Baquero F, Saldana M A, Blasquez J, Palacios R G, Aguiar J M, Martinez J L, Vicente M F, Rubio C, Gomez-Lus R. Bleomycin-kanamycin resistance as a marker of the presence of transposon Tn5 in clinical strains of Escherichia coli. Eur J Clin Microbiol Infect Dis. 1989;8:995–998. doi: 10.1007/BF01967573. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasband A J, Marcotte W R J, Schnaitman C A. Structure of the lc and nmpC outer membrane porin protein genes of Lambdoid bacteriophage. J Biol Chem. 1986;261:12723–12732. [PubMed] [Google Scholar]
- 7.Borja B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 8.Brenner D J, Grimont P A D, Steigerwalt A G, Fanning G R, Ageron E, Riddle C F. Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmeri sp. nov., Citrobacter braakii sp. nov., Citrobacter werkmanii sp. nov., Citrobacter sedlakii sp. nov., and three Citrobacter genomospecies. Int J Syst Bacteriol. 1993;43:645–658. doi: 10.1099/00207713-43-4-645. [DOI] [PubMed] [Google Scholar]
- 9.Burland V, Plunkett III G, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome VI: DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother. 1993;37:1896–1903. doi: 10.1128/aac.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courvalin P, Carlier C, Collatz E. Plasmid-mediated resistance to aminocyclitol antibiotics in group D streptococci. J Bacteriol. 1980;143:541–551. doi: 10.1128/jb.143.2.541-551.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosa J H, Brenner D, Ewing W H J, Falkow S. Molecular relationships among the Salmonellaceae. J Bacteriol. 1973;115:307–315. doi: 10.1128/jb.115.1.307-315.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupont C, Clarke A J. Evidence for N→O acetyl migration as the mechanism for O acetylation of peptidoglycan in Proteus mirabilis. J Bacteriol. 1991;173:4318–4324. doi: 10.1128/jb.173.14.4318-4324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas M J, Dowding J E. Aminoglycoside-modifying enzymes. Methods Enzymol. 1975;43:611–628. doi: 10.1016/0076-6879(75)43124-x. [DOI] [PubMed] [Google Scholar]
- 15.Hongo E, Morimyo M, Mita K, Machida I, Hama-Inaba H, Tsuji H, Ichimura S, Noda Y. The methyl viologen-resistance gene smvA of Salmonella typhimurium. Gene. 1994;148:173–174. doi: 10.1016/0378-1119(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 16.Labigne-Roussel A, Briaux-Gerbaud S, Courvalin P. Tn1525, a kanamycin R determinant flanked by two direct copies of IS15. Mol Gen Genet. 1983;189:90–101. doi: 10.1007/BF00326060. [DOI] [PubMed] [Google Scholar]
- 17.Lambert T, Gerbaud G, Courvalin P. Characterization of transposon Tn1528, which confers amikacin resistance by synthesis of aminoglycoside 3′-O-phosphotransferase type VI. Antimicrob Agents Chemother. 1994;38:702–706. doi: 10.1128/aac.38.4.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 307–342. [Google Scholar]
- 19.Liu S, Hessel A, Sanderson K E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4104–4120. doi: 10.1128/jb.175.13.4104-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payie K G, Clarke A J. Characterization of gentamicin 2′-N-acetyltransferase from Providencia stuartii: its use of peptidoglycan metabolites for acetylation of both aminoglycosides and peptidoglycan. J Bacteriol. 1997;179:4106–4114. doi: 10.1128/jb.179.13.4106-4114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piepersberg W, Distler J, Heinzel P, Perez-Gonzalez J A. Antibiotic resistance by modification: many resistance genes could be derived from cellular control genes in Actinomycetes. A hypothesis. Actinomycetologica. 1988;2:83–98. [Google Scholar]
- 22.Prilipov A, Phale P S, Koebnik R, Widmer C, Rosenbusch J P. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli. J Bacteriol. 1998;180:3388–3392. doi: 10.1128/jb.180.13.3388-3392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rather P N. Origins of the aminoglycoside modifying enzymes. Drug Resistance Updates. 1998;1:285–291. doi: 10.1016/s1368-7646(98)80044-7. [DOI] [PubMed] [Google Scholar]
- 24.Rather P N, Orosz E, Hare R S, Miller G, Shaw K J. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene of Providencia stuartii. J Bacteriol. 1993;175:6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reizer J, Charbit A, Reizer A, Saier M H., Jr Novel phosphotransferase system genes revealed by bacterial genome analysis: operons encoding homologues of sugar-specific permease domains of the phosphotransferase system and pentose catabolic enzymes. Genome Sci Technol. 1996;1:53–75. [Google Scholar]
- 26.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saier M H, Jr, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sanderson K E, Hessel A, Liu S, Rudd K E. The genomic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 1903–1999. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki I, Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965;40:365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- 32.Selander R K, Li J, Nelson K. Evolutionary genetics of Salmonella enterica. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 2691–2707. [Google Scholar]
- 33.Shaw K J, Rather P N, Sabatelli F, Mann P, Munayyer H, Mierzwa R, Petrikkos G, Hare R S, Miller G H, Bennett P, Downey P. Characterization of the chromosomal aac(6′)-Ic gene from Serratia marcescens. Antimicrob Agents Chemother. 1992;36:1447–1455. doi: 10.1128/aac.36.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steers E, Foltz E L, Graves B S, Riden J. An inocula replicating apparatus for routine testing of bacterial susceptibility to antibiotics. Antibiot Chemother (Basel) 1959;9:307–311. [PubMed] [Google Scholar]
- 35.Teran F G, Suarez J E, Mendoza M C. Cloning, sequencing, and use as a molecular probe of a gene encoding an aminoglycoside 6-N-acetyltransferase of broad substrate profile. Antimicrob Agents Chemother. 1991;35:714–719. doi: 10.1128/aac.35.4.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wertman K F, Wyman A R, Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]
- 37.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 38.Yoshikawa A, Isono S, Sheback A, Isono K. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol Gen Genet. 1987;209:481–488. doi: 10.1007/BF00331153. [DOI] [PubMed] [Google Scholar]